Abstract
Peroxisomes are single-membraned organelles ubiquitous to eukaryotic cells that house metabolic reactions that generate and destroy harmful oxidative intermediates. They are dynamic structures whose morphology, abundance, composition and function depend on the cell type and environment. Perhaps due to the potentially damaging and protective metabolic roles of peroxisomes and their dynamic presence in the cell, peroxisome biogenesis is emerging as a process that involves complex underlying mechanisms of regulated formation and maintenance. There are roughly 30 known peroxins, proteins involved in peroxisome biogenesis, many of which have been conserved from yeast to mammals (Table I). This review focuses on the biogenesis of peroxisomes with an emphasis on the regulation of peroxisome formation and the import of peroxisomal matrix proteins in the model organism Saccharomyces cerevisiae.
Introduction
Cells are made up dynamic assemblies that continually sense their environment and adjust their functioning in response to countless stimuli. To do this, cells rely on regulated expression of stored information in their genomes from their ancestors and on epigenetic memory of their own history. This information is so powerful that cells can not only replicate existing cellular components, but in some cases, they can regenerate complex subcellular structures that have been lost. Studies of the biogenesis of peroxisomes suggest that this process is a paradigm for general organellar biogenesis, but may also involve mechanisms borrowed from other biological processes. Over the past couple of decades, several aspects of peroxisome biogenesis have been elucidated using a variety of model organisms. Two areas have been of particular interest; the origin of peroxisomes and the mechanism of import of proteins into the peroxisomal matrix. The question of peroxisome origin has been the most fundamental: whether or not peroxisomes can be generated in the absence of pre-existing peroxisomes. It now appears that there are two pathways of peroxisome biogenesis - de novo generation and fission of pre-existing peroxisomes. In addition, it appears that the endoplasmic reticulum (ER) is both the source of new peroxisomes and the source of membrane material for growth of peroxisomes that have been formed by fission. These findings raise fundamental questions about the regulated molecular mechanisms underlying each pathway.
The other basic and unresolved question is the mechanism of protein translocation into peroxisomes. Early work suggested that cytoplasmic proteins destined for the peroxisome matrix are targeted by a classic signal peptide in the protein sequence and thus seemed typical of other subcellular organelles; however, the translocation of these proteins into peroxisomes appears unique. Unlike that of virtually all other organelles with a sealed membrane, peroxisomes can import larger particles (up to 9 nm) [1–3]. They do so without unfolding protein complexes, or a large stable membrane spanning pore assemblage like the nuclear pore complex. Recent data suggest this ‘slight of hand’ is accomplished by a dynamic pore structure constituted of predominantly soluble import receptors that transiently become transmembrane proteins [4]. Ironically, while the translocon has been sought for many years, it may be that it has been studied throughout this time, but disguised as the targeting signal receptor Pex5p, first identified in 1993 [5,6].
Generation of peroxisomes
A long standing question in cell biology is whether peroxisomes replicate autonomously like mitochondria and chloroplasts, or derive from a preexisting lipid bilayer such as that of the ER, like endosomes and lysosomes. The key difference between these two mechanisms is that autonomously replicating organelles cannot be regenerated by the cell in the event that they are lost, whereas other organelles can be reformed. Historically, peroxisomes were considered to be autonomously replicating organelles, but more recently evidence has accumulated to support a model in which peroxisomes arise de novo from the ER as well as by fission of pre-existing peroxisomes (reviewed in [7,8]).
Generation of peroxisomes de novo
The source of peroxisomes has been debated for many years. While peroxisomes can evidently arise from fission of preexisting peroxisomes, mutant mammalian or yeast cells lacking recognizable peroxisomes (i.e. devoid of vesicles containing any known PMPs) can generate new mature organelles upon genetic complementation [9,10], suggesting that peroxisomes can arise from an internal membrane system, and that they are not strict endosymbionts like mitochondria or chloroplasts [11]. This idea is also supported by evidence demonstrating that mutants defective in peroxisomal inheritance [12,13] or peroxisomal fission [14] contain peroxisomes, and phylogenetic analyses of peroxisomal proteins, which suggests that peroxins are of eukaryotic origin and that some even have an evolutionary link with the ER [15].
The most compelling evidence for the ER derivation of peroxisomes stems from advances in live cell imaging. Controlled expression and monitoring of Pex19p, and Pex3p, which are required for the very early steps of peroxisome biogenesis in yeast, revealed a process of peroxisome formation whereby Pex3p first localizes to perinuclear and cortical ER, and then colocalizes with Pex19p to a few dot-like subcompartments of the ER before trafficking, in a Pex19p dependent manner, to separate vesicles which mature into import competent peroxisomes [16]. The data suggest that Pex3p establishes a subcompartment of the ER as pre-peroxisomal, and acts as a docking receptor for complexes of Pex19p. Pex19p thus functions as a primarily cytosolic chaperone involved in targeting of PMPs from the cytosol to maturing peroxisomes [17,18]. Pex3p appears to play a critical role in forming the nascent organelle; without Pex3p, peroxisomes do not form and Pex19p does not associate with the ER.
The mechanisms of de novo peroxisome formation and the involvement of other pathway components remain to be established, however some sec pathway components have been implicated: in S. cerevisiae, Emp24p, a component of coat protein (COP) II-coated vesicles involved in ER to Golgi transport, is also associated with small, apparently immature, peroxisomes [19], and is required for budding of peroxisomes from the ER in Hansenula polymorpha [20]. In addition, it has been found that COP I components Arf and coatomer are involved in the biogenesis of rat peroxisomes (reviewed in [21]). But other reports suggest that peroxisome emergence from the ER may not involve COP I or COP II coated vesicles [22]. There is similar conflicting evidence for Sec61p, an essential component of the ER translocon, which has been shown to not be involved in peroxisome biogenesis in S. cerevisiae [23]. However, a recent study implicates Sec61p in the ER to peroxisome trafficking of Pex30p, a peroxin regulating the size and number of peroxisomes, suggesting a second Pex3p- and Pex19p-independent targeting mechanism for PMPs [24].
The precise mechanisms of ER involvement in peroxisome biogenesis in mammalian cells appears to be different than that of yeast cells. PEX16, which is not present in most yeasts, is cotranslationally inserted into the ER and recruits other PMPs, including PEX3 to the ER [25]. As in yeast, PEX19 enables this process by forming complexes with newly synthesized PMPs in the cytoplasm, but it docks to the PEX16 membrane receptor instead of PEX3 [26].
Generation of peroxisomes by growth and division
In addition to de novo generation, peroxisomes can also arise through the growth and division of preexisting peroxisomes. This process is positively controlled by Pex11p, and a pair of partially functionally redundant proteins, Pex25p and Pex27p. Pex11p mediates peroxisome elongation [27], which is followed by fission facilitated by dynamin related proteins (DRPs) Vps1, in yeast and Dlp1, in mammalian cells (reviewed in [28]). But how do these newly formed ‘daughter peroxisomes’ increase their membrane component during their subsequent growth? Again, developments in direct live cell imaging have illuminated this process [29]. In this assay, different colors of fluorescently labeled peroxisomes from two yeast cells are combined into the same cytoplasm by mating. The authors show that pulse labeled Pex3p-GFP is released from ER in a PEX19Δ cell after mating with a WT cell and associates with preexisting peroxisomes derived by fission from the other cell labeled with HcRed-PTS1. These data demonstrate that the fission of preexisting peroxisomes, is coupled to the transfer of membrane material from the ER to peroxisomes through budding of Pex3p-containing vesicles from the ER and subsequent heterotypic fusion to mature peroxisomes.
Regulation of the origin of peroxisomes
These live cell imaging assays promise to open new areas of research in peroxisome biogenesis. One area of investigation is the conditional regulation of peroxisome origin. How are the de novo and fission biogenesis pathways each regulated? To date, there have been a few studies that provide some insight into this process. A pulse-chase assay, using peroxisomal version of photo-activatable GFP to study the origin of peroxisomes in human fibroblasts suggests that new peroxisomes are primarily derived by de novo generation rather than fission [25]. By contrast, a similar assay in yeast suggests that peroxisomes are generated by division of preexisting peroxisomes, and that de novo peroxisome generation occurs only in mutant cells without any detectable preexisting peroxisomes [29] (Fig. 1). While the cell types and specific experimental conditions may contribute to this discrepancy, perhaps the state of the existing peroxisomes is also important. Through their metabolic functions, peroxisomes are exposed to significant potentially damaging reactive oxygen species (ROS), necessitating turnover of the organelle. Thus, under conditions of low peroxisomal oxidation, fission may be favored because de novo generation is relatively slow [16,29] and may have a higher biosynthetic cost to the cell. But, under conditions of high peroxisomal oxidative activity, de novo generation may be favored because it results in a complete turn-over of peroxisomal constituents that may have been damaged by oxidation. This is consistent with an earlier hypothesis that Pex11p senses oxidation and responds by limiting peroxisomal fission through conditional homodimerization [30]. This hypothesis is based the finding that Pex11p is predominantly a monomer early after oleate induction and in peroxisomes of low density, but is a homodimer after longer induction times (16 h) and in end-stage peroxisomes of higher density.
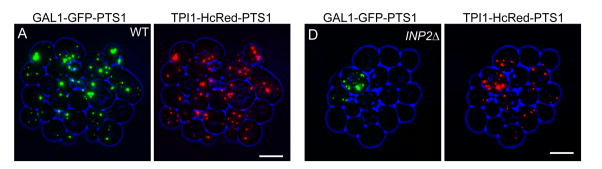
Figure 1.
Peroxisomes arise de novo only in the absence of preexisting peroxisomes. WT (left panels) and INP2Δ cells (right panels) constitutively expressing HcRed-PTS1 (under control of the TPI1 promoter) were pulse labeled with GFP-PTS1 (under control of the GAL1 promoter) and then allowed to form a colony over 6–8 h on glucose containing medium. All peroxisomes in the colony of wild-type cells were labeled with both markers indicating that all peroxisome in the colony had derived by fission of the peroxisomes present in the parent cells. However, in the INP2Δ colony of cells defective in peroxisome inheritance, many peroxisomes contained only HcRed-PTS1, indicating de novo generation of peroxisomes in the absence of preexisting peroxisomes [29] (reproduced with permission).
Regulation of peroxisome abundance and turnover
Unlike most other organelles, peroxisomes differ dramatically in size, number and content depending on the cell types and various environmental stimuli. Control of this regulation appears complex, involving signaling networks, transcriptional and post-translational control. Signaling network control is poorly understood. While certainly these networks feed into transcriptional control, it also appears that they influence peroxisomal morphology and abundance as well [31]. Transcriptional regulators are also conditionally controlled by exogenously added fatty acids and thus lead to coordinated regulation of peroxins and peroxisomal metabolic enzymes, which affects the size and number of peroxisomes (reviewed in [32]). An additional level of control involves the conditional use of proteins in peroxisomal fission. In mammalian cells, Pex11p isoforms are conditionally involved in fission; Pex11-α controls constitutive peroxisome fission, while Pex11-β and Pex11-γ control fission in response to peroxisome proliferation. In yeast, there appears to be conditional involvement of DRPs involved in fission [33]; Dnm1p, a DRP required for mitochondrial fission, is a major contributor to peroxisomal fission in response to mitochondrial dysfunction, whereas involvement of Vps1p is more pronounced in fission under non-proliferative conditions. It is thus suggested that Dnm1p might help to coordinate mitochondrial and peroxisome biogenesis [34].
The dynamics of peroxisome abundance are also controlled by vacuolar degradation by an autophagic process called pexophagy (reviewed in [35,36]). It has been established by studies in methylotropic yeasts, H. polymorpha and Pichia pastoris, that peroxisomes are degraded by one of two mechanisms, micropexophagy or macropexophagy. These processes occur in a peroxisome selective manner and are regulated by various environmental stimuli. It is likely that the generation of peroxisomes is coordinated with these processes of degradation to maintain or change peroxisome abundance. Such a link has yet to be characterized, but two peroxins involved in the formation of peroxisomes, are also involved in degradation; Pex3p removal from a peroxisomal membrane is an early step in the autophagic process [37] and Pex14p is required for macroautophagy [38], suggesting that the processes are coordinated.
Matrix protein import
Import of proteins into peroxisomes is a remarkable process unlike import into other organelles and involves accommodation of very large protein assemblies and a dynamic translocation mechanism that has thus far remained elusive to detection. Matrix proteins are imported post-translationally into peroxisomes in an ATP-dependent manner. They are recognized by predominantly soluble receptors in the cytoplasm that bind to one of two types of targeting signals, peroxisomal targeting signal 1 (PTS1) or PTS2. Most peroxisomal matrix proteins have a PTS1 and are recognized and imported by Pex5p, whereas less common PTS2-containing proteins are recognized and imported by Pex7p. Pex7p-dependent import requires a co-receptor, Pex20p, present in most fungi studied. S. cerevisiae is lacking this protein and instead has a pair of functional homologs, Pex18p and Pex21p. The currently proposed import model is based on extensive analysis of Pex5p, the major receptor protein. According to this model, import occurs by a mechanism that involves passing of the receptor through a cascade of three different membrane associated complexes; the docking complex, which binds to cargo-loaded receptors before translocation, and the RING finger and receptor recycling complexes, which are involved in receptor recycling after translocation of cargo.
The import process involves three distinct steps (reviewed in [39]): 1. Receptor-cargo complexes dock to the outside surface of peroxisomal membranes to a docking complex made up of Pex13p, Pex14p and Pex17p. 2. Cargoes are translocated into the peroxisomal matrix in a manner that exposes the receptor to the matrix of the peroxisome. This was first suggested by data showing that Pex5p interacts with Pex8p, an intraperoxisomal component of the import apparatus [40], and recently demonstrated in human cells by appending a motif on the Pex5p receptor that is recognized and cleaved by an enzyme found inside peroxisomes [41]. There is now evidence that other PTS2 shuttling receptors, Pex7p in S. cerevisiae [42] and Pex20p of P. pastoris [43], also reach the peroxisomal interior. 3. Cargoes are released on the matrix side of the membrane (perhaps by competitive binding to the receptor by intraperoxisomal Pex8p, which has a PTS1 and PTS2 [4,44]) and receptors are recycled to the cytoplasm. Receptor recycling involves the RING finger complex comprised of Pex2p, Pex10p and Pex12p transiently anchored to the docking complex by Pex8p [45], the receptor recycling complex comprised of the ubiquitin-conjugating enzyme Pex4p anchored by Pex22p, and a complex comprised of AAA ATPases, Pex1p and Pex6p, anchored to the membrane by Pex15p. Ubiquitination of the import receptor is emerging as an important aspect of import. Monoubiquitination of Pex5p by Pex4p is required for release of Pex5p from peroxisomal membranes [46,47], whereas polyubiquitation of Pex5p by Ubc4p [48–50] and E3 ligase, Pex10p [51] is required for a mechanism to target receptors for degradation when receptor recycling is impaired.
While this model continues to gain detail the nature of the translocon is still unknown. While 12 membrane-associated proteins of the import complex have been identified and characterized, not one component of a stably-associated translocation complex is evident. The absence of an obvious protein conducting channel and the shuttling of import receptors have led to the proposal that an import receptor, itself becomes a transient pore [4]. This model posits that a complex of Pex5p molecules forms a transient pore in the membrane that creates a passage for cargo-loaded receptors to pass through, perhaps similar to the action of pore-forming toxin proteins (Box1; [52]). The pore is then disassembled by ubiquitination of Pex5p followed by its ATP-dependent dislocation from the membrane to the cytoplasm by AAA ATPases, Pex1p and Pex6p. This step is modeled after other AAA proteins that are involved in membrane protein dislocation from the inner mitochondrial membrane and the ER for subsequent proteolytic degradation [53–55].
A major piece of evidence supporting a dual role for Pex5p as a soluble receptor and as a pore component is that it has two conformations; it is predominantly soluble, but a study of rat liver Pex5p shows that upon interaction with Pex14p, it behaves like an integral membrane protein [56]. In addition, S. cerevisiae Pex5p can spontaneously insert into lipid monolayers and liposomes in vitro [57]. Structural studies of mammalian and yeast Pex5p also support two conformations; electron microscopic analysis of recombinant Pex5p reveals a tetramer conformation [58,59] whereas other studies indicate that soluble Pex5p is monomeric [60]. Interestingly, the H. polymorpha Pex5p tetramer has a closed conformation that opens in the presence of Pex20p (Fig. 2) [59], perhaps hinting at a pore-forming mechanism during protein translocation in vivo.
Figure 2.
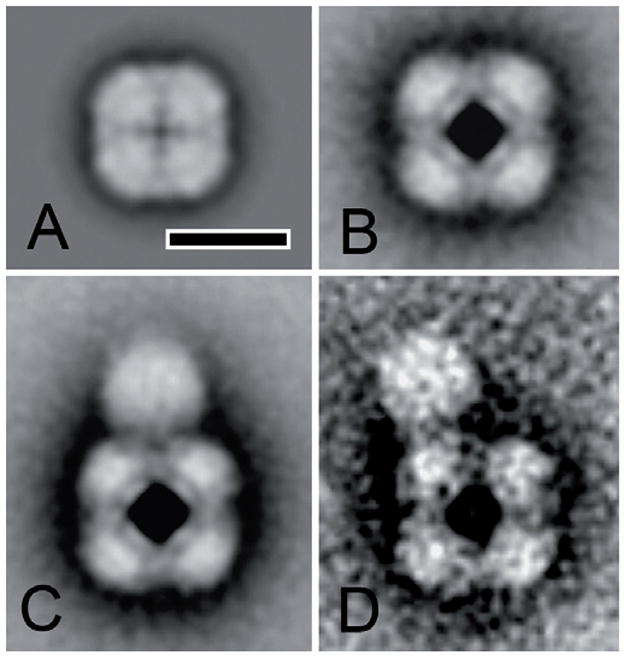
Single particle analysis of purified recombinant H. polymorpha Pex5p. (A) Average projection map of Pex5p in the closed conformation. (B–D) Pex5p in the open conformation in the presence of Pex20p. Some complexes appear to have no Pex20p association as in panel B; but 75% of the Pex5p tetramers are associated with between 1–3 globular mass of about 80 Ǻ shown in panels C and D. The bar equals 10 nm. [59] (reproduced with permission)
If Pex5p forms a transient pore, what about Pex7p-mediated PTS2 import? In most organisms PTS2-mediated import depends on a physical interaction of Pex7p with either Pex5p or a Pex5p-like protein: In most yeasts PTS2-mediated import is dependent on interaction of Pex7p with a co-receptor Pex20p. Pex20p has sequence homology to Pex5p at the N-terminus and has recently been shown to physically interact with Pex5p in H. polymorpha [59]. Plants, mammals and some yeasts lack Pex20p, but have a version of Pex5p that has a Pex7p binding site (reviewed in [61]). S. cerevisiae is the only organism for which no Pex5p- or Pex20p-dependence has been shown to be necessary for PTS2-mediated targeting. Perhaps the co-receptor Pex18p also has pore forming ability; it is able to functionally replace Pex5p when appended to PTS1 recognition site of Pex5p [62]. Alternatively Ymr018p, which is an uncharacterized protein with sequence similarity to Pex5p [61], might function in this regard.
Conclusions and future directions
Extensive progress has been made in the field of peroxisome biogenesis since the first peroxin involved in controlling this process was reported in 1991. Recent advances in the characterization of the origin of peroxisomes and import of matrix proteins illuminate these previously elusive processes and open new avenues of research. Data demonstrating that new peroxisomes are derived de novo and by fission of pre-existing peroxisomes raise the question of how each process contributes to peroxisome abundance under different conditions and how these processes are coordinated with other processes, such as peroxisome degradation, to control dynamics of peroxisome proliferation and peroxisome turnover.
Insight into the regulatory mechanisms controlling global peroxisome dynamics raises the subject of the regulation and heterogeneity of individual peroxisomes within cells. For example, peroxisome degradation might coordinate with a mechanism to discriminate peroxisomes based on a quality such as oxidative damage. Peroxisome discrimination could also be an aspect of regulation of mitotic inheritance of peroxisomes in yeast (reviewed in [63]) or peroxisome movement on microtubules in mammalian cells [64]. Such a regulatory mechanism controlling the selective inheritance of specific peroxisomes could also impact the process of replicative aging of cells. It has been proposed that normally functioning mitochondria are specifically segregated to daughter cells, but in old cells, this process is disrupted due to loss of asymmetrical division, leading to the formation of daughters with shorter life spans (reviewed in [65]). Perhaps a similar process exists for peroxisomes. Developments in cellular imaging, combined with ‘omics’ and systems approaches hold promise for new insights into the complex regulation and biogenesis of this ubiquitous, medically important, yet still somewhat obscure organelle.
Box 1.
The transient pore model suggests that Pex5p forms pores in peroxisomal membranes similar to the mechanism used by pore forming toxins (PFTs) (reviewed in [52] [71]). PFTs are a class of proteins that can switch between a soluble form and a pore-forming transmembrane form. They are used by pathogenic bacteria to invade host cells, and by higher eukaryotes for immunity, invasion and development. There are over 80 different families that, despite sharing little sequence homology with each other, have similar mechanisms of action: First, soluble proteins interact with the target membrane by binding to membrane bound receptors on the surface. After interaction, many PFTs form oligomers that are inserted into the membrane to form a channel. The structure that crosses the membrane is either alpha helical or beta-barrel. Beta-barrels are often assembled via oligomerization of proteins that each contains pairs of amphipathic beta-strands, which are independently not sufficiently hydrophobic to span a membrane bilayer. Below is a model of a member of the mammalian PFT super family MACPF involved in the lysis of bacteria and protozoan pathogens. It is shown in the pore forming conformation in a lipid bilayer, [71] (reproduced with permission).
Table I.
Proteins involved in peroxisome biogenesis
| Proteins involved in formation of peroxisomes and pre-peroxisomal vesicles from the ER | |
| Pex19p | PMP chaperone and transporter; farnesylated and mostly cytosolic [17] |
| Pex16p | PMP that traffics through the ER; membrane receptor for PMP recruitment in mammalian cells [25,26]. |
| Pex3p | Traffics through the ER; membrane receptor for PMP recruitment in yeast [16] |
| Proteins involved in controlling peroxisome size and number [28] | |
| Pex11p | Intraperoxisomal PMP involved peroxisomal fission |
| Pex25p | Peripheral PMP; interacts with paralog Pex27p [66]; recruits Rho1p to peroxisomes [19] |
| Pex27p | interacts with paralog Pex25p [66] |
| DRPs | Dynamin-related proteins includeVps1p and Dnm1p in yeast and Dlp1p in mammalian cells |
| Fis1p/Mdv1p/Caf4p | DRP recruitment to peroxisomes [33] |
| Rho1p | Guanosine triphosphatase involved in actin assembly on peroxisomes; interacts with Pex25p and Pex30p[19] |
| Pex30p/Pex23p | PMP; known as Pex30p in S. cerevisiae and Pex23p in Y. lipolytica [61]; contains dysferlin domain; partially functionally redundant with Pex31p and Pex32p [67] |
| Pex31 p | PMP; contains dysferlin domain [67] |
| Pex32p | PMP; contains dysferlin domain[67] |
| Pex24p/Pex28p | PMP known as Pex28p in S. cerevisiae and Pex24p in Y. lipolytica. [61] |
| Pex29p | PMP |
| Proteins involved in import of matrix proteins [39] | |
| Pex5p | PTS1 receptor; predominantly cytoplasmic |
| Pex7p | PTS2 receptor; predominantly cytoplasmic |
| Pex20p | Co-receptor required for Pex7p binding cargo; S. cerevisiae has functional homologs Pex21p/Pex18p instead [61] |
| Cytosolic chaperones | Chaperones including members of Hsp70 family and DnaJ-like proteins may be involved in import [39] |
| Pex13p | PMP; docking complex component |
| Pex14p | PMP; docking complex component |
| Pex17p | PMP; docking complex component |
| Pex8p | Intraperoxisomal PMP; links docking and RING finger complex [45] |
| Pex2p | PMP; RING finger protein |
| Pex12p | PMP; RING finger protein |
| Pex10p | PMP; RING finger protein; E3 ligase for Ubc4p dependent ubiquitination of Pex5p[51] |
| Pex4p | PMP anchored by Pex22p; ubiquitin-conjugating enzyme involved in receptor recycling [46,47]; interacts with Pex10p [68]) |
| Pex22p | PMP that anchors Pex4p to the membrane |
| Pex15p/Pex26p | functional homologs in yeast and mammals, respectively; PMP involved in membrane anchor of Pex6p [69,70] |
| Pex6p | AAA ATPase; PMP |
| Pex1p | AAA ATPase; PMP |
| Proteins involved in peroxisome inheritance [63] | |
| Myo2p | Motor that propels peroxisomes along actin cables [12] |
| Inp1 | PMP involved in retention of peroxisomes in mother and daughter cells by attaching peroxisomes to cell cortex [13] |
| Inp2 | PMP involved in peroxisome movement through interaction with Myo2 [12] |
PMP, peroxisomal membrane protein; peroxins in bold are known to have similar roles in yeast and in higher eukaryotes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc Natl Acad Sci U S A. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walton PA, Hill PE, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6:675–683. doi: 10.1091/mbc.6.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•••.Erdmann R, Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol. 2005;6:738–742. doi: 10.1038/nrm1710. This manuscript suggests a mechanism of import into peroxisomes that is consistent with the absence of an identifiable stable translocon in the peroxisomal membrane. By this model, the predominantly soluble import receptor Pex5p transiently inserts into peroxisomal membranes and forms a pore complex. Pore disassembly occurs by ubiquitination of transmembrane receptors followed by extraction from the membrane by ATPases Pex1p and Pex6p and recycling of the receptors to the cytoplasm. [DOI] [PubMed] [Google Scholar]
- 5.Van der Leij I, Franse MM, Elgersma Y, Distel B, Tabak HF. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993;90:11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells--the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoms S, Erdmann R. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. Febs J. 2005;272:5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 8.Titorenko VI, Mullen RT. Peroxisome biogenesis. the peroxisomal endomembrane system and the role of the ER. J Cell Biol. 2006;174:11–17. doi: 10.1083/jcb.200604036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. Embo J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 12.Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell. 2006;10:587–600. doi: 10.1016/j.devcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Fagarasanu M, Fagarasanu A, Tam YY, Aitchison JD, Rachubinski RA. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J Cell Biol. 2005;169:765–775. doi: 10.1083/jcb.200503083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 15.Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA. Origin and evolution of the peroxisomal proteome. Biol Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•••.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. This is the first demonstration that full length Pex3p expressed at endogenous levels traffics through the ER. It is also the first example of pulse-chase tracking of a newly made PMP by fluorescence microscopy of live cells. Further analyses of PEX3Δ and PEX19Δ strains show that Pex3p traffics through the ER, then colocalizes with Pex19p in foci that bud off in a Pex19p dependent manner to form peroxisomes. [DOI] [PubMed] [Google Scholar]
- 17.Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girzalsky W, Hoffmann LS, Schemenewitz A, Nolte A, Kunau WH, Erdmann R. Pex19p-dependent targeting of Pex17p, a peripheral component of the peroxisomal protein import machinery. J Biol Chem. 2006;281:19417–19425. doi: 10.1074/jbc.M603344200. [DOI] [PubMed] [Google Scholar]
- 19.Marelli M, Smith JJ, Jung S, Yi E, Nesvizhskii AI, Christmas RH, Saleem RA, Tam YY, Fagarasanu A, Goodlett DR, et al. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J Cell Biol. 2004;167:1099–1112. doi: 10.1083/jcb.200404119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otzen M, Krikken AM, Ozimek PZ, Kurbatova E, Nagotu S, Veenhuis M, van der Klei IJ. In the yeast Hansenula polymorpha, peroxisome formation from the ER is independent of Pex19p, but involves the function of p24 proteins. FEMS Yeast Res. 2006;6:1157–1166. doi: 10.1111/j.1567-1364.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 21.Lay D, Gorgas K, Just WW. Peroxisome biogenesis: where Arf and coatomer might be involved. Biochim Biophys Acta. 2006;1763:1678–1687. doi: 10.1016/j.bbamcr.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 22.South ST, Sacksteder KA, Li X, Liu Y, Gould SJ. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol. 2000;149:1345–1360. doi: 10.1083/jcb.149.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.South ST, Baumgart E, Gould SJ. Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc Natl Acad Sci U S A. 2001;98:12027–12031. doi: 10.1073/pnas.221289498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mast FD, Perry RJ, Rachubinski RA. A Novel Import and Trafficking Pathway for the Peroxisomal Membrane Protein, Pex30p, in Saccharomyces cerevisiae. Mol Biol Cell. 2008 [Google Scholar]
- 25••.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. This is the first characterization of peroxisome generation from the ER in mammalian cells. They show using a live cell pulse-chase assay that de novo peroxisome generation is different in mammalian cells than in yeast, and requires Pex16p, which is cotranslationally inserted into the ER and recruits other PMPs (including Pex3p) to membranes. Importantly, they find this de novo generation is the primary source of new peroxiosmes in wild type cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiki Y, Matsuzaki T. Peroxisome Membrane Biogenesis: The Peroxisomal Membrane Transport Receptor Pex3p is Directly Imported Via Pex16p in a Pex19p-dependent Manner. Mol Biol Cell. 2008 doi: 10.1083/jcb.200806062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch A, Schneider G, Luers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117:3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- 28.Yan M, Rayapuram N, Subramani S. The control of peroxisome number and size during division and proliferation. Curr Opin Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 29•••.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. The describe the use of a live cell pulse-chase assay and the development of a mating assay in S. cerevisiae to track de novo- and fission-derived peroxisome and preperoxisomal structures that are derived from the ER. They show that in wild type cells, peroxisomes are derived by DRP-dependent fission, but in mutants without preexisting peroxisomes, they arise de novo independently of DRPs. Importantly, peroxisomes derived by fission fuse with vesicles containing Pex3p from the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall PA, Dyer JM, Quick ME, Goodman JM. Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J Cell Biol. 1996;135:123–137. doi: 10.1083/jcb.135.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saleem RA, Knoblach B, Mast FD, Smith JJ, Boyle J, Dobson CM, Long-O’Donnell R, Rachubinski RA, Aitchison JD. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J Cell Biol. 2008;181:281–292. doi: 10.1083/jcb.200710009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurvitz A, Rottensteiner H. The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochim Biophys Acta. 2006;1763:1392–1402. doi: 10.1016/j.bbamcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epstein CB, Waddle JA, Hale Wt, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 37.Bellu AR, Salomons FA, Kiel JA, Veenhuis M, Van Der Klei IJ. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J Biol Chem. 2002;277:42875–42880. doi: 10.1074/jbc.M205437200. [DOI] [PubMed] [Google Scholar]
- 38.de Vries B, Todde V, Stevens P, Salomons F, van der Klei IJ, Veenhuis M. Pex14p is not required for N-starvation induced microautophagy and in catalytic amounts for macropexophagy in Hansenula polymorpha. Autophagy. 2006;2:183–188. doi: 10.4161/auto.2549. [DOI] [PubMed] [Google Scholar]
- 39.Brown LA, Baker A. Shuttles and cycles: transport of proteins into the peroxisome matrix (review) Mol Membr Biol. 2008;25:363–375. doi: 10.1080/09687680802130583. [DOI] [PubMed] [Google Scholar]
- 40.Rehling P, Skaletz-Rorowski A, Girzalsky W, Voorn-Brouwer T, Franse MM, Distel B, Veenhuis M, Kunau WH, Erdmann R. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor pex5p. J Biol Chem. 2000;275:3593–3602. doi: 10.1074/jbc.275.5.3593. [DOI] [PubMed] [Google Scholar]
- 41.Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 42.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leon S, Zhang L, McDonald WH, Yates J, 3rd, Cregg JM, Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D, Visser NV, Veenhuis M, van der Klei IJ. Physical interactions of the peroxisomal targeting signal 1 receptor pex5p, studied by fluorescence correlation spectroscopy. J Biol Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- 45.Agne B, Meindl NM, Niederhoff K, Einwachter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell. 2003;11:635–646. doi: 10.1016/s1097-2765(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 46.Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 48.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 49.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 50.Platta HW, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 52.Iacovache I, van der Goot FG, Pernot L. Pore formation: an ancient yet complex form of attack. Biochim Biophys Acta. 2008;1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 53.Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- 54.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 56.Gouveia AM, Reguenga C, Oliveira ME, Sa-Miranda C, Azevedo JE. Characterization of peroxisomal Pex5p from rat liver. Pex5p in the Pex5p-Pex14p membrane complex is a transmembrane protein. J Biol Chem. 2000;275:32444–32451. doi: 10.1074/jbc.M004366200. [DOI] [PubMed] [Google Scholar]
- 57•••.Kerssen D, Hambruch E, Klaas W, Platta HW, de Kruijff B, Erdmann R, Kunau WH, Schliebs W. Membrane association of the cycling peroxisome import receptor Pex5p. J Biol Chem. 2006;281:27003–27015. doi: 10.1074/jbc.M509257200. This manuscript demonstrates that yeast and human PTS1 receptors can insert spontaneously into artificial phospholipid membranes. They also show that Pex5p can insert into peroxisomal membrane in an import competent state without stable association with constituents of docking and RING finger complexes. These data support the transient pore model by suggesting that a translocation-competent form of Pex5p enters the peroxisomal membrane via protein-lipid interactions independently of interactions with other peroxins. [DOI] [PubMed] [Google Scholar]
- 58.Schliebs W, Saidowsky J, Agianian B, Dodt G, Herberg FW, Kunau WH. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J Biol Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- 59.Moscicka KB, Klompmaker SH, Wang D, van der Klei IJ, Boekema EJ. The Hansenula polymorpha peroxisomal targeting signal 1 receptor, Pex5p, functions as a tetramer. FEBS Lett. 2007;581:1758–1762. doi: 10.1016/j.febslet.2007.03.061. [DOI] [PubMed] [Google Scholar]
- 60.Costa-Rodrigues J, Carvalho AF, Fransen M, Hambruch E, Schliebs W, Sa-Miranda C, Azevedo JE. Pex5p, the peroxisomal cycling receptor, is a monomeric non-globular protein. J Biol Chem. 2005;280:24404–24411. doi: 10.1074/jbc.M501985200. [DOI] [PubMed] [Google Scholar]
- 61.Kiel JA, Veenhuis M, van der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 62.Schafer A, Kerssen D, Veenhuis M, Kunau WH, Schliebs W. Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol Cell Biol. 2004;24:8895–8906. doi: 10.1128/MCB.24.20.8895-8906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 64.Rapp S, Saffrich R, Anton M, Jakle U, Ansorge W, Gorgas K, Just WW. Microtubule-based peroxisome movement. J Cell Sci. 1996;109 (Pt 4):837–849. doi: 10.1242/jcs.109.4.837. [DOI] [PubMed] [Google Scholar]
- 65.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tam YY, Torres-Guzman JC, Vizeacoumar FJ, Smith JJ, Marelli M, Aitchison JD, Rachubinski RA. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4089–4102. doi: 10.1091/mbc.E03-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vizeacoumar FJ, Torres-Guzman JC, Bouard D, Aitchison JD, Rachubinski RA. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:665–677. doi: 10.1091/mbc.E03-09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eckert JH, Johnsson N. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci. 2003;116:3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- 69.Birschmann I, Stroobants AK, van den Berg M, Schafer A, Rosenkranz K, Kunau WH, Tabak HF. Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol Biol Cell. 2003;14:2226–2236. doi: 10.1091/mbc.E02-11-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsumoto N, Tamura S, Fujiki Y. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol. 2003;5:454–460. doi: 10.1038/ncb982. [DOI] [PubMed] [Google Scholar]
- 71.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, et al. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]