Abstract
Francisella tularensis is a highly infectious pathogen that infects animals and humans, causing tularemia. The ability to replicate within macrophages is central for virulence and relies on expression of genes located in the Francisella pathogenicity island (FPI), as well as expression of other genes. Regulation of FPI-encoded virulence gene expression in F. tularensis involves at least four regulatory proteins and is not fully understood. Here we studied the RNA-binding protein Hfq in F. tularensis and particularly the role that it plays as a global regulator of gene expression in stress tolerance and pathogenesis. We demonstrate that Hfq promotes resistance to several cellular stresses (including osmotic and membrane stresses). Furthermore, we show that Hfq is important for the ability of the F. tularensis vaccine strain LVS to induce disease and persist in organs of infected mice. We also demonstrate that Hfq is important for stress tolerance and full virulence in a virulent clinical isolate of F. tularensis, FSC200. Finally, microarray analyses revealed that Hfq regulates expression of numerous genes, including genes located in the FPI. Strikingly, Hfq negatively regulates only one of two divergently expressed putative operons in the FPI, in contrast to the other known regulators, which regulate the entire FPI. Hfq thus appears to be a new pleiotropic regulator of virulence in F. tularensis, acting mostly as a repressor, in contrast to the other regulators identified so far. Moreover, the results obtained suggest a novel regulatory mechanism for a subset of FPI genes.
Regulation of gene expression by noncoding or small RNAs (sRNAs) is prevalent in both prokaryotes and eukaryotes, and recent studies have identified numerous sRNAs in different organisms. Most sRNAs encoded by bacterial chromosomes act by base pairing with mRNA targets that are encoded in trans, and these sRNAs commonly require Hfq in order to function (47). Hfq is a bacterial RNA-binding protein that was initially recognized as a host factor for replication of the Qβ RNA phage in Escherichia coli (17). The Hfq protein is very abundant, and it belongs to the eukaryotic and archaeal families of Sm and Sm-like proteins, respectively, that form homohexameric structures (for reviews, see references 5 and 53). The importance of Hfq became clear when an E. coli hfq null mutant was created. This mutant had pleiotropic phenotypes, such as a decreased growth rate, increased sensitivity to cellular stresses, and increased cell length (51). Hfq is a posttranscriptional regulator that binds sRNAs and mRNA and facilitates RNA-RNA interaction (1, 18, 23, 32, 35, 55). For the most part, sRNA-mRNA interactions result in mRNA degradation and/or inhibition of translation, but they can also increase translation (for reviews, see references 1, 20, and 48). As might be expected from the pleiotropic phenotypes of an hfq mutant, deletion of hfq has been correlated with considerable changes in gene expression observed at the mRNA or protein level (14, 15, 21, 42, 46).
In the last few years, the role of Hfq in the pathogenesis of several bacterial species has been examined (11, 14, 30, 37, 39, 41, 42, 45). A Salmonella enterica serovar Typhimurium hfq mutant was found to be severely attenuated for intracellular replication in vitro and for virulence in mice (42). Similarly, decreased virulence was observed for hfq mutants of Brucella abortus, Listeria monocytogenes, Pseudomonas aeruginosa, Vibrio cholerae, Shigella flexneri, and Legionella pneumophila (11, 14, 30, 39, 41, 45). In many cases, the hfq mutation also rendered the bacteria more sensitive to cellular stress. In striking contrast, deletion of hfq in three different Staphylococcus aureus strain backgrounds did not result in any detectable phenotype (4).
Bacteria have developed numerous regulatory mechanisms so that they are capable of a rapid molecular response with coordinated gene expression in order to survive in a variety of hostile environments. A macrophage is probably one of the most stressful environments that facultative intracellular bacterial pathogens encounter. The aim of the present work was to understand how Hfq contributes to the stress response and virulence of Francisella tularensis, a gram-negative facultative intracellular pathogen that causes tularemia in humans and a large number of animal species. Three F. tularensis subspecies are capable of inducing disease in humans: F. tularensis subsp. tularensis (type A), F. tularensis subsp. holarctica (type B), and F. tularensis subsp. mediasiatica. The fourth subspecies of F. tularensis (“F. tularensis subsp. novicida”) causes disease in mice, but it is rarely pathogenic in humans. In humans, an F. tularensis infection can be acquired by ingestion of contaminated water or food, through arthropod bites, from contaminated animal carcasses, or by inhalation of bacteria. Due to its high infectivity and the severity of the disease, F. tularensis has been classified by the Centers for Disease Control and Prevention as a potential biological weapon (for reviews, see references 29 and 43).
The pathogenicity of F. tularensis is believed to be dependent largely on its ability to replicate inside host cells, particularly macrophages. Several factors important for virulence have been identified, but the molecular mechanisms underlying disease are unclear. Several of the genes located in the Francisella pathogenicity island (FPI) (38), such as iglC, iglD, iglA, pdpA, pdpB, and pdpD, have been demonstrated to be required for virulence in at least one subspecies (12, 19, 24, 27, 38, 40, 49, 54).
There are only three known regulators of virulence in F. tularensis, MglA, SspA, and PmrA, all of which control expression of FPI genes (3, 7, 10, 25, 34). Each of these proteins also regulates expression of genes outside the FPI; however, whereas MglA and SspA regulate the same set of genes outside the FPI, PmrA controls a separate group of genes (7, 10, 34). Recently, a new protein regulator of FPI gene expression was identified (6). This protein, FevR, controls the genes in the MglA-SspA regulon and seems to act in parallel with MglA (6).
The genome of F. tularensis does not encode any complete regulatory two-component systems (34) and encodes only one putative alternative sigma factor, σ32 (31), suggesting that this organism has an exceptionally limited repertoire of pleiotropic regulatory mechanisms. Expression of the alternative sigma factors σS and/or σE is controlled by the regulatory protein Hfq in several gram-negative bacteria, such as E. coli, S. Typhimurium, P. aeruginosa, and V. cholerae, and the sigma factors play an integral role in the Hfq regulatory network in these species (8, 14, 21, 36, 45). As the F. tularensis genome does not encode either σS or σE, the Hfq regulatory network could be expected to be different from the regulatory networks in other bacterial species.
This prompted us to address the role of Hfq in stress resistance and virulence in F. tularensis subsp. holarctica. For this, we created an hfq deletion mutant of the live vaccine strain, strain LVS, and demonstrated that it has pleiotropic phenotypes, such as slower growth and increased sensitivity to cellular stress, and is severely attenuated for virulence in mice. A similar mutant of the virulent clinical isolate F. tularensis subsp. holarctica FSC200 was also investigated. Using this mutant, we could verify that Hfq is also important for growth, stress tolerance, and full virulence in a human-pathogenic Francisella strain. Finally, we show that Hfq is a global regulator in F. tularensis LVS and that it controls expression of a subset of genes located in the FPI. Thus, Hfq is a novel pleiotropic regulator of virulence in F. tularensis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. F. tularensis LVS and a Δhfq derivative were routinely grown in liquid Schaedler medium containing vitamin K3 (Schaedler K3 medium) (Biomerieux), in Chamberlain medium (9), on premade chocolate agar plates (Biomerieux), or on chocolate agar plates prepared from Difco GC medium supplemented with hemoglobin and IsoVitaleX enrichment (BD Diagnostics). F. tularensis FSC200 and an Δhfq derivative (FSC768) were routinely grown in Chamberlain medium or on McLeod agar plates. E. coli strains were grown in Luria-Bertani medium or on Luria-Bertani agar plates. When required, the medium was supplemented with kanamycin (10 to 20 μg/ml for F. tularensis and 50 μg/ml for E. coli), chloramphenicol (1.75 μg/ml for LVS and 10 μg/ml for E. coli), or polymyxin B (100 μg/ml).
TABLE 1.
Plasmids, strains, and primers used in this study
| Plasmid, strain, or primer | Genotype, description, or sequence | Reference, source, or comment |
|---|---|---|
| Plasmids | ||
| pFNLTP6 gro-gfp | Shuttle plasmid with LVS groE operon promoter upstream of gfp, Kmr Apr | 28 |
| p6-gro | Shuttle plasmid derivative with groE promoter, Kmr Apr | This study |
| p6-gro-hfq | Shuttle plasmid derivative with groE promoter upstream of hfq, Kmr Apr | This study |
| pPV | oriT sacB, Apr Cmr, Francisella suicide plasmid | 19 |
| pPV-hfq | pPV carrying hfq deletion fragment for mutagenesis of LVS | This study |
| pDMK2 | mobRP4oriR6KsacB, Kmr, Francisella suicide vector | K. Kadzhaev, unpublished |
| pDMK-hfq | pDMK2 carrying hfq deletion fragment for mutagenesis of FCS200 | This study |
| pCR4TOPO | PCR cloning vector, Ampr Kmr | Invitrogen |
| pCR2.1TOPO | PCR cloning vector, Ampr Kmr | Invitrogen |
| Strains | ||
| F. tularensis LVS | F. tularensis subsp. holarctica live vaccine strain, Pmr | A. Sjöstedt |
| F. tularensis Δhfq mutant | LVS strain with deletion of codons 5 to 71 of hfq | This study |
| F. tularensis FSC200 | F. tularensis subsp. holarctica, clinical isolate | Human ulcer, Sweden |
| F. tularensis FSC768 | FSC200 with deletion of codons 9 to 113 of hfq | This study |
| E. coli DH5α | F− φ80lacZΔM15 endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 (lacZYA-argF)U169 | Invitrogen |
| E. coli TOP10 | (F−) mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 (Δara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| E. coli S17-1 | (F−) RP4-2-Tc::Mu aph::Tn7 recA, Smr | Lab collection |
| E. coli S17-1λpir | (F−) RP4-2-Tc::Mu aph::Tn7 recA λpir lysogen, Smr | Lab collection |
| Primers | ||
| Hfq-AF | 5′- TGGGTCGACTACAGCCAAGCCAACTTTACAAGa | Mutant construction |
| Hfq2 | 5′-TATTCTTGACATTATCTCACTTC | Mutant construction |
| Hfq-BR | 5′-CGGCTAGCAAGATCAGACTCTATAGCCTCTTCb | Mutant construction |
| Hfq3 | 5′-GAAGTGAGATAATGTCAAGAATAGTTTATAGCTCTTTTAATCCATAc | Mutant construction |
| Hfq_FNdeI | 5′-GGGAATTCCATATGTTGCTAGAATATACGCGAAGd | Complementing plasmid |
| Hfq_RBamHI | 5′-CGCGGATCCTTACTCGTGAATATTACCTTCe | Complementing plasmid |
| 5up-HfqA | 5′-AATTAATGCAGAAATTATTAGTG | Mutant verification |
| 3down-HfqB | 5′-CTTTTTCAACCTGCTCAATGTAGC | Mutant verification |
| Hfq-F22 | 5′-CAAGACCCGTTCTTAAATGCT | RT-PCR |
| Hfq-R79 | 5′-CTAGATATACAGATACGCTAAC | RT-PCR |
| Hfq-R133 | 5′-CGATACAGAATTGGTCAAAAGC | RT-PCR/5′ RACE |
| Hfq-R194 | 5′-GCTGGAACTATAGTAGAAATAG | RT-PCR, 5′ RACE |
| miaA-F870 | 5′-GCTATGGAGAACGAAACTAAAGA | RT-PCR |
| HflX-R170 | 5′-GATATCTGGCTCAGGATGG | RT-PCR |
| Hfq200-A_for | 5′-AACAGTCCAGGGTGACTAAAATCGAGCCAAT | Mutant construction |
| Hfq200-B_rev | 5′-ATATTACTCGTGTGACATTATCTCACTTCCTTTTA | Mutant construction |
| Hfq200-C_for | 5′-GAGATAATGTCACACGAGTAATATATCTTTTCTTTf | Mutant construction |
| Hfq200-D_rev | 5′-TAACGTCGACTAACACCAAGCTCATACATCT | Mutant construction |
| Hfq200-E_For | 5′-GCGGAAAGAAGATAGATAAGA | Mutant verification |
| Hfq200-F_Rev | 5′-TATCTTCGACTCCTGTTATTTC | Mutant verification |
| TUL4-435 | 5′-GCTGTATCATCATTTAATAAACTGCTG | 44 |
| TUL4-863 | 5′-TTGGGAAGCTTGTATCATGGCACT | 44 |
| hel_F | 5′-GGGATGTCGCCTTTTGATTTTC | qRT-PCRg |
| hel_R | 5′-CTCTTTTGTCCCTTGTGCTTGC | qRT-PCRg |
| iglC_F | 5′-TGGGTGTTTCAAAAGATGGAATG | qRT-PCR |
| iglC_R | 5′-TGCGCAACATACTGGCAAGC | qRT-PCR |
| FTL_0994_F | 5′-CGCTAATGCAATCGTTGATGG | qRT-PCR |
| FTL_0994_R | 5′-CCGGTGACATGATCAACCTCAT | qRT-PCR |
| FTL_0120_F | 5′-CAAGCACCTCATTTGCTTCGAT | qRT-PCR |
| FTL_0120_R | 5′-TTCTATTGCTTGAGCATCATCTTTTTG | qRT-PCR |
| pdpB_F | 5′-TGAATCCGAAACCAATTCATCT | qRT-PCR |
| pdpB_R | 5′-GGCGCCCACAATGACTTATC | qRT-PCR |
| FTL_0317_F | 5′-GTCGTGCGGACCAAACAAAA | qRT-PCR |
| FTL_0317_R | 5′-CGCAGCATCAGTTGTGAGATTG | qRT-PCR |
| katG_F | 5′-ATACGCCGCAACACAAATGG | qRT-PCR |
| katG_R | 5′-CATCCCCATAGCTCGGAAGG | qRT-PCR |
| iglA_F | 5′-TCAAAAGGAAGATCTGTGGATGC | qRT-PCR |
| iglA_R | 5′-CAAAGTTTGGTGCCTCAAAATCA | qRT-PCR |
| iglB_F | 5′-GGAATGGGTATGGTGGCAAA | qRT-PCR |
| iglB_R | 5′-GGATGCTCAAGCAAAGCTTCAA | qRT-PCR |
The SalI site is underlined.
The NheI site is underlined.
The sequence overlapping Hfq2 is underlined.
The NdeI site is underlined.
The BamHI site is underlined.
The sequence that overlaps part of Hfq200-B_rev is underlined.
See reference 7.
Construction of hfq mutant strains.
The counterselectable plasmid pPV-hfq used to generate a nonpolar hfq deletion mutant of strain LVS was constructed by overlap PCR. Primers Hfq-AF and Hfq2 amplify the 912-bp region upstream of position 13, and primers Hfq3 and Hfq-BR amplify the 1,021-bp region downstream of position 210 (relative to the hfq translation start site). Primers Hfq3 and Hfq2 have 23 nucleotides of overlapping sequence, resulting in fusion of hfq codon 4 to hfq codon 71 after crossover PCR. PCR with primers Hfq-AF and Hfq2 and with primers Hfq3 and Hfq-BR were performed with Mango Taq polymerase (Bioline, Massachusetts), and the products were purified using a QIAquick PCR purification kit (Qiagen, California). Two microliters of each purified product was included in a PCR mixture without primers and subjected to 20 cycles of PCR (94°C for 40 s, 48°C for 40 s, and 72°C for 90 s) to anneal and extend the crossover product. Next, 2 μl of the extended crossover product was used as a template for PCR using primers Hfq-AF and Hfq-BR. The gel-purified fragment was digested with NheI and SalI and cloned into XbaI-SalI-digested pPV (19). The plasmid was introduced into F. tularensis LVS by conjugation from E. coli strain S17-1. S17-1/pPV-hfq and LVS were grown to exponential phase, mixed (ratio, approximately 1:20) in 0.9% NaCl, and spotted onto Schaedler medium agar plates (Bio-Rad) supplemented with MEM vitamins (Sigma-Aldrich). The plates were incubated at 25°C for 16 h, and bacterial material was recovered in Schaedler K3 medium (Biomerieux) supplemented with 100 μg/ml polymyxin B (Sigma). After 3 h of incubation at 37°C (to eliminate the majority of the E. coli cells), bacteria were spread on chocolate agar plates containing 100 μg/ml polymyxin B and 1.75 μg/ml chloramphenicol. Colonies appeared after 5 to 6 days of incubation at 37°C and were subsequently passaged once on plates with selection, followed by one passage in liquid medium without selection (to allow recombination to occur). Next, bacteria were passaged once on agar plates containing 5% sucrose. Isolated colonies were checked for loss of the wild-type hfq gene by analyzing the size of the fragment obtained after PCR using primers 5up-hfqA and 3down-hfqB (which annealed to the regions outside the fragment cloned in pPV-hfq). Furthermore, colonies were verified to be F. tularensis colonies using primers TUL4-435 and TUL4-863 (44). One colony harboring an hfq gene that was smaller, as determined by PCR analysis, was used for further study. Genomic DNA was isolated and used as a template in a PCR with the 5up-hfqA/3down-hfqB primer pair. The PCR product was directly sequenced using primers Hfq-AF, Hfq-BR, and miaA-F870 to verify the presence of an hfq gene with an in-frame deletion of codons 5 to 70. As a control, genomic DNA from the LVS wild-type strain was used in the same PCR, and the product was sequenced. Sequencing confirmed the deletion of Hfq amino acids 5 to 70 and revealed that a single nucleotide substitution resulted in an amino acid substitution (Gly→Cys) in the remaining peptide in the mutant clone.
The hfq mutant of the FSC200 strain was constructed essentially as previously described (19). Briefly, a deletion construct was amplified by PCR using primer pairs Hfq200-A/Hfq200-B and Hfq200-C/Hfq200-D with strain FSC200 genomic DNA as the template. The resulting overlap PCR fragment, the Hfq200-A/Hfq200-D fragment, was cloned into the pCR4TOPO cloning vector (Invitrogen) and subsequently cloned as a 2.4-kb PmeI-NotI fragment into SmaI-NotI-digested suicide mutagenesis vector pDMK2 (K. Kadzhaev, unpublished). pDMK2-hfq, carrying the deletion fragment, was introduced into recipient strain FSC200 by conjugal mating with E. coli S17-1λpir as the donor strain. Single-crossover allelic exchange of the suicide plasmid was selected by plating bacteria on McLeod agar plates containing kanamycin and polymyxin B. To complete the allelic exchange, a second recombination event was selected for utilizing sacB-dependent sucrose sensitivity as described previously (33). The resulting mutant strain (FSC768) was verified by PCR using primers Hfq200-E_For and Hfq200-F_Rev and by loss of kanamycin resistance.
Determination of the transcription start site of hfq.
The 5′ end of the hfq mRNA was determined by 5′ rapid amplification of cDNA ends (5′-RACE) using a 5′/3′ RACE kit from Roche Applied Science. One microgram of RNA isolated from LVS grown in Schaedler K3 medium at exponential phase was used as the template for cDNA synthesis using the gene-specific primer hfq_R194. The cDNA was purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA), a poly(A) tail was added at the 3′ end, and the cDNA was PCR amplified using the gene-specific primer hfq_R133 and the poly(A) tail-specific primer oligo dT-anchor. An aliquot of the PCR product was used as the template in a second PCR with primers hfq_R133 and PCR-anchor, and the fragment obtained was directly sequenced. Additionally, the PCR product was ligated into pCR2.1-TOPO and several clone sequences. This confirmed that in five clones the transcription start site was at position −65 relative to the ATG start codon, as determined by direct sequencing. Three clones had 5′ ends at positions −29, +22, and +110, indicating that these clones were degraded transcripts.
Hfq antiserum and immunoblotting.
The hfq gene of LVS was cloned into the pET-28a expression vector (Novagen) to create a protein with a histidine tag at the N terminus. This protein was expressed in E. coli strain BL21(DE3), and a gel piece containing the overexpressed protein was used for production of polyclonal antisera in rabbits (Centre de Production Animale, Olivet, France). Antiserum was purified by incubation with an F. tularensis Δhfq bacterial lysate before it was used for immunoblotting. Aliquots of F. tularensis LVS (and the Δhfq strain as a control) were harvested at different cell densities, and equal amounts of cell lysate (corresponding to 2 × 108 bacteria per well) were deposited on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gel. After electrophoresis, proteins were transferred to a nitrocellulose membrane using a semidry Transblot apparatus (Bio-Rad) and incubated with the Hfq antiserum. After washing, horseradish peroxidase-linked secondary antibody was added, and binding was detected using an ECL Plus kit (Amersham).
Construction of complementing plasmid p6gro-hfq.
Plasmid pFNLTP6-gro-gfp (28) was digested with BamHI, which excised the gfp gene, and was subsequently self-ligated, yielding plasmid p6-gro. The hfq gene was PCR amplified using primers Hfq_FNdeI and Hfq_RBamHI and the Expand high-fidelity PCR system (Roche Applied Science). The product was digested with NdeI and BamHI and ligated into NdeI-BamHI-digested p6-gro, producing p6-gro-hfq. This construct was verified by sequencing and introduced into F. tularensis LVS by electroporation (28).
Stress resistance.
F. tularensis LVS and its Δhfq derivative were grown overnight in Schaedler K3 medium. For experiments with liquid media, the cultures were diluted 10-fold in fresh medium containing either 1% or 2% NaCl and incubated at 37°C with agitation. Growth was assessed by measuring the optical density at 600 nm (OD600). For heat resistance tests, overnight cultures were diluted in fivefold steps, and 3 μl of each dilution was spotted onto chocolate agar plates and incubated at 37°C or 40°C. For experiments with the complemented mutant strain, wild-type and mutant bacteria containing p6gro or p6gro-hfq were grown overnight in Schaedler K3 medium supplemented with kanamycin. The resulting cultures were diluted in 10-fold steps, and 3 μl of each dilution was spotted onto chocolate agar containing kanamycin and either 2% NaCl, 0.2% SDS, or water (as a control). F. tularensis FSC200 and FSC768 were grown on McLeod agar plates and suspended in Chamberlain medium. Each suspension was used to inoculate Chamberlain medium containing 1 or 2% NaCl and to prepare fivefold serial dilutions and was incubated at 37°C without agitation. NaCl resistance and heat resistance were assessed as described above. All the experiments described above were performed at least twice, and results of a single representative experiment are presented.
Multiplication in macrophages.
J774 cells were propagated in RPMI medium or Dulbecco's modified Eagle's medium containing 10% fetal calf serum. Cells were seeded at a concentration of ∼2 × 105 cells per well in 12-well (for experiments with LVS strains) or 24-well (for experiments with FSC200 strains) tissue plates (Falcon), and monolayers were used 24 h after seeding. Bone marrow-derived macrophages (BMM) from BALB/c mice were obtained and cultured as described previously (13). RAW macrophages were propagated in RPMI medium. J774 macrophage monolayers were incubated for 90 min (LVS) or 120 min (FSC200), RAW macrophages were incubated for 90 min, and BMM were incubated for 60 min at 37°C with the bacterial suspensions (multiplicities of infection, approximately 100) to allow the bacteria to enter. After washing (time zero of the kinetic analysis), the cells were incubated in fresh culture medium containing gentamicin (10 μg/ml) to kill extracellular bacteria. At several time points, cells were washed three times in RPMI medium or phosphate-buffered saline (PBS) and processed for counting of surviving intracellular bacteria. For this, bacteria were recovered by lysis of macrophages with distilled water (LVS) or 0.1% sodium deoxycholate in PBS (FSC200). The titer of viable bacteria released from the cells was determined by spreading preparations on agar plates. For each strain and time in an experiment, the assay was performed in triplicate. Each experiment was independently repeated at least two times, and the data presented below are the averages of two experiments (six wells).
Infection of mice and virulence assays.
Bacterial stocks of LVS and the LVS Δhfq mutant (in 16 to 20% glycerol) stored at −80°C were thawed and diluted to obtain the appropriate concentration (CFU/ml) in 0.15 M NaCl. Six- to 8-week-old female BALB/c mice (Janvier, Le Genest St. Isle, France) were inoculated intraperitoneally (i.p.) with 200 μl of a bacterial suspension. Groups of five mice were inoculated with various doses of bacteria, and the mortality was followed for 9 days. The actual number of viable bacteria was determined by plating dilutions of a bacterial suspension on chocolate plates. Animal experiments were performed according to the INSERM guidelines for laboratory animal husbandry.
For experiments examining the kinetics of infection, BALB/c mice were infected with ∼3 × 104 CFU of LVS or hfq mutant bacteria. At 2, 3, 4, and 7 days after infection, groups of five mice were sacrificed, and the numbers of bacteria in the liver and spleen were determined by plating homogenized samples on chocolate agar plates. Four mice infected with LVS died before day 7.
Infection with F. tularensis strain FSC200 and the FSC200 Δhfq mutant was performed approximately as described above, except that the mice were infected i.p. or subcutaneously with 100 μl of the appropriate bacterial suspension. For the competitive index (CI) infection studies, mice in groups of five animals were infected subcutaneously with approximately 100 bacteria using a 1:1 mixture of the wild-type and mutant strains (FSC200 and FSC768). Five days after infection mice were sacrificed, and spleens were isolated and homogenized in PBS. The homogenates were serial diluted and spread on McLeod agar plates, and colonies were analyzed by using PCR to distinguish the mutant from the wild type. The output value after infection was determined by dividing the number of mutant CFU by the number of wild-type CFU. This ratio was divided by the initial ratio of the inoculum (verified by viable counting) to obtain a final CI. C57Black/6 female mice (Scanbur BK AB, Sollentuna, Sweden) were housed under conventional conditions, given food and water ad libitum, and allowed to acclimatize for at least 7 days before infection. The study was approved by the Local Ethics Committee on Laboratory Animals in Umeå, Sweden.
Transcriptional profiling using DNA microarrays.
RNA was isolated from LVS and the LVS Δhfq mutant grown to exponential phase in Schaedler K3 medium (OD600, ∼0.25) or Chamberlain medium (OD600, ∼0.35) and from colonies grown on chocolate agar using Trizol reagents (Invitrogen), followed by purification of the aqueous phase on RNeasy columns (Qiagen). RNA samples were treated with DNase I (Ambion), and 3 μg of each sample was used in a reverse transcription (RT) reaction. Cy3-labeled cDNA (LVS) and Cy5-labeled cDNA (LVS Δhfq mutant) were prepared using the protocol for aminoallyl labeling of microbial RNA (The Institute for Genomic Research [TIGR]; http://pfgrc.jcvi.org/index.php/microarray/protocols.html). The labeled samples were combined and hybridized as described at the TIGR website (http://pfgrc.jcvi.org/index.php/microarray/protocols.html) with an F. tularensis microarray provided by The Pathogen Funtional Genomic Resource Center (Rockville, MD). This microarray contains 2,331 oligonucleotides (70 nucleotides) representing open reading frames from strains LVS and Schu S4, as well as plasmids pFNL10 and pOM1, each spotted four times. The experiments were each performed two or three times, and duplicate labeling and hybridization experiments were performed for each set of RNAs. The microarray slides were scanned using a Genepix 4000B microarray scanner (Molecular Devices), and the images were analyzed using the Spotfinder and MIDAS software (TIGR). Statistically significant changes in gene expression under each condition were identified by performing a one-class analysis using the significance analysis of microarray (SAM) program (52) with a 0% false discovery rate. Genes with an average change of at least twofold (calculated using all microarray slides and the four values for each slide) in at least two experiments are shown in Table 2.
TABLE 2.
Hfq-regulated genesa
| Locus | Gene product | Gene | KEGG classification | Change (fold)b
|
||
|---|---|---|---|---|---|---|
| Defined medium | Agar plates | Broth | ||||
| Genes significantly downregulated in the hfq mutant (n = 16) | ||||||
| FTL_0194 | Cytochrome o ubiquinol oxidase subunit IV | cyoD | Metabolism | 0.21 | 0.42 | |
| FTL_0195 | Protoheme IX farnesyltransferase | cyoE | Metabolism | 0.28 | 0.32 | |
| FTL_0221 | Amino acid permease | 0.50 | 0.41 | |||
| FTL_0317 | Hypothetical protein | 0.41 | 0.41 | 0.36 | ||
| FTL_0392 | Type IV pilus fiber building block protein | 0.42 | 0.47 | 0.33 | ||
| FTL_0421 | Conserved hypothetical lipoprotein | lpnA | 0.46 | 0.31 | 0.47 | |
| FTL_0568 | Transposase | isftu2 | 0.84 | 0.48 | 0.47 | |
| FTL_0570 | Hypothetical protein | 0.34 | 0.42 | 0.39 | ||
| FTL_0898 | Host factor I for bacteriophage Qβ replication | hfq | 0.12 | 0.04 | 0.08 | |
| FTL_0899 | Protease, GTP-binding subunit | hflX | 0.37 | 0.47 | 0.75 | |
| FTL_1016 | Short-chain dehydrogenase | 0.31 | 0.50 | 0.48 | ||
| FTL_1394 | Galactose-proton symporter, major facilitator superfamily transport protein | galP2 | 0.47 | 0.49 | ||
| FTL_1504 | Peroxidase/catalase | katG | Metabolism | 0.37 | 0.49 | 0.64 |
| FTL_1555 | Hypothetical membrane protein | 0.53 | 0.49 | 0.36 | ||
| FTL_1678 | Conserved membrane hypothetical protein | 0.53 | 0.47 | 0.36 | ||
| FTL_1764 | Pseudo | 0.13 | 0.37 | |||
| Genes significantly upregulated in the hfq mutant (n = 88) | ||||||
| FTL_0045 | Orotidine 5′-phosphate decarboxylase | pyrF | Metabolism | 2.60 | 3.30 | |
| FTL_0046 | Dihyroorotate dehydrogenase | pyrD | Metabolism | 2.58 | 3.44 | |
| FTL_0117/ FTL_1163 | Hypothetical protein | 4.83 | 3.92 | 3.72 | ||
| FTL_0118/ FTL_1164 | Hypothetical protein | 3.24 | 1.91 | 2.28 | ||
| FTL_0119/ FTL_1165 | Hypothetical protein | 4.21 | 3.19 | 2.68 | ||
| FTL_0120/ FTL_1166 | Hypothetical protein | 6.02 | 5.82 | 2.46 | ||
| FTL_0121/ FTL_1167 | Hypothetical protein | 7.72 | 4.08 | 4.55 | ||
| FTL_0122/ FTL_1168 | Conserved hypothetical protein | 9.35 | 6.49 | 4.94 | ||
| FTL_0123/ FTL_1169 | Hypothetical protein | 3.49 | 4.03 | 3.53 | ||
| FTL_0124/ FTL_1170 | Hypothetical protein | 5.21 | 5.35 | 3.64 | ||
| FTL_0125/ FTL_1171 | Hypothetical protein | pdpB | 10.7 | 6.00 | 5.15 | |
| FTL_0126/ FTL_1172 | Hypothetical protein | pdpA | 8.19 | 3.63 | 3.69 | |
| FTL_0237 | 50S ribosomal protein L4 | Genetic information processing | 1.23 | 2.61 | 2.51 | |
| FTL_0239 | 50S ribosomal protein L2 | rplB | Genetic information processing | 2.24 | 2.77 | |
| FTL_0240 | 30S ribosomal protein S19 | Genetic information processing | 1.23 | 2.48 | 3.26 | |
| FTL_0242 | 30S ribosomal protein S3 | rpsC | Genetic information processing | 2.93 | 2.64 | |
| FTL_0261 | DNA-directed RNA polymerase, alpha subunit | Metabolism and genetic information processing | 1.25 | 2.66 | 3.30 | |
| FTL_0396 | Fusion protein PurC/PurD | Metabolism | 1.46 | 2.45 | 2.16 | |
| FTL_0414 | GTP-binding protein | engA | 2.75 | 3.46 | 1.53 | |
| FTL_0437 | Riboflavin biosynthesis protein RibF | ribF | Metabolism | 3.27 | 5.02 | 2.06 |
| FTL_0460 | Hypothetical protein | 3.31 | 2.74 | 2.71 | ||
| FTL_0461 | Hypothetical protein | 1.88 | 2.60 | 2.22 | ||
| FTL_0534 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | dxr | Metabolism | 2.97 | 3.69 | 1.62 |
| FTL_0535 | Outer membrane protein | 2.14 | 5.31 | |||
| FTL_0603 | O-antigen flippase | wzx | 2.02 | 3.43 | ||
| FTL_0604 | Glycosyltransferase | wbtK | 1.67 | 2.04 | 2.01 | |
| FTL_0605 | Glucose-1-phosphate thymidylyltransferase | wbtL | Metabolism | 2.95 | 2.24 | |
| FTL_0669 | Exodeoxyribonuclease V, beta chain | recB | Genetic information processing | 1.75 | 2.17 | 2.23 |
| FTL_0670 | Exodeoxyribonuclease V, alpha subunit | recD | Genetic information | 4.16 | 4.04 | 1.77 |
| FTL_0737 | Hypothetical membrane protein | processing | 5.81 | 4.63 | 1.79 | |
| FTL_0739 | Glucose-inhibited division protein A | gidA | Cellular processes | 3.18 | 2.03 | |
| FTL_0788 | Glutamine amidotransferase class II family protein | 4.09 | 3.28 | 1.94 | ||
| FTL_0797 | Type IV pili associated protein | Genetic information processing | 2.99 | 2.35 | 1.32 | |
| FTL_0798 | Type IV pilus glycosylation protein | Genetic information processing | 4.06 | 3.38 | 1.81 | |
| FTL_0827 | Type IV pilus polytopic inner membrane protein | Genetic information processing | 2.32 | 4.31 | 2.32 | |
| FTL_0882 | Apolipoprotein N-acyltransferase | 6.09 | 7.01 | |||
| FTL_0883 | Metal ion transporter protein | Environmental | 3.33 | 3.15 | 1.74 | |
| FTL_0884 | Hypothetical protein | information processing | 1.49 | 3.09 | 2.55 | |
| FTL_0885 | PhoH-like protein | phoH | 2.31 | 4.40 | 1.77 | |
| FTL_0893 | ATP-dependent protease, ATP-binding subunit | clpX | 2.22 | 3.42 | 1.81 | |
| FTL_0894 | DNA-binding, ATP-dependent protease La | lon | 2.16 | 2.76 | 1.92 | |
| FTL_0921 | Endonuclease III | Genetic information | 2.78 | 2.11 | 1.75 | |
| FTL_0993 | HesA/MoeB/ThiF family protein | processing | 3.61 | 3.38 | 2.75 | |
| FTL_0994 | Hypothetical protein | 5.66 | 4.91 | 2.55 | ||
| FTL_1004 | d-Alanyl-d-alanine carboxypeptidase | vanY | 2.44 | 2.08 | 1.79 | |
| FTL_1005 | Hypothetical protein | 10.6 | 4.06 | 1.86 | ||
| FTL_1043 | Hypothetical protein | 7.12 | 5.79 | 3.75 | ||
| FTL_1044 | Hypothetical protein | 6.70 | 3.46 | 4.03 | ||
| FTL_1045 | Conserved hypothetical lipoprotein | 3.27 | 3.01 | 2.27 | ||
| FTL_1046 | d-Alanyl-d-alanine carboxypeptidase (penicillin-binding protein) family protein | dacB | 11.6 | 3.70 | 2.78 | |
| FTL_1049 | DNA primase | dnaG | Genetic information processing | 4.16 | 4.04 | |
| FTL_1050 | RNA polymerase sigma-70 factor | rpoD | Genetic information | 2.47 | 3.04 | 1.75 |
| FTL_1064 | Hypothetical protein | processing | 4.10 | 4.47 | 2.10 | |
| FTL_1065 | ABC transporter, ATP-binding protein | yhbG | 2.64 | 7.30 | 3.39 | |
| FTL_1066 | Fumarylacetoacetate hydrolase family protein | 2.37 | 2.59 | |||
| FTL_1067 | Hypothetical protein | 2.79 | 5.48 | 2.32 | ||
| FTL_1068 | tRNA pseudouridine synthase A | truA | Genetic information | 1.46 | 3.27 | 3.56 |
| FTL_1074 | GDSL-like lipase/acylhydrolase family protein | processing | 1.72 | 2.54 | 2.51 | |
| FTL_1106 | Alanyl-tRNA synthetase | alaS | Metabolism and genetic | 2.41 | 2.80 | 2.06 |
| FTL_1108 | Cytosol aminopeptidase family protein | information processing | 2.49 | 2.65 | 1.65 | |
| FTL_1174 | Cystathionine beta-synthase (cysteine synthase) | cbs | Metabolism | 6.30 | 3.86 | 3.20 |
| FTL_1177 | tRNA modification GTPase | trmE | 2.71 | 2.37 | ||
| FTL_1190 | Chaperone protein GrpE (heat shock protein family 70 cofactor) | grpE | 2.65 | 2.03 | 3.27 | |
| FTL_1228 | Hypothetical protein | 3.15 | 1.43 | 2.41 | ||
| FTL_1229 | ABC transporter, ATP-binding protein | 4.57 | 1.35 | 3.29 | ||
| FTL_1230 | Cysteine desulfurase activator complex subunit SufB | 2.49 | 2.63 | 1.44 | ||
| FTL_1261 | Anthranilate synthase component II | trpG | Metabolism | 6.60 | 3.05 | 3.42 |
| FTL_1262 | Chorismate binding family protein | 10.2 | 4.00 | 2.90 | ||
| FTL_1266 | Lipase/esterase | 5.90 | 2.91 | |||
| FTL_1267 | Hypothetical protein | 3.19 | 1.88 | 2.81 | ||
| FTL_1271 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | bioA | Metabolism | 2.71 | 2.03 | |
| FTL_1273 | 8-Amino-7-oxononanoate synthase | bioF | Metabolism | 2.57 | 2.00 | 1.49 |
| FTL_1274 | Biotin synthesis protein BioC | bioC | 5.02 | 3.25 | 3.12 | |
| FTL_1275 | Dethiobiotin synthetase | bioD | Metabolism | 3.23 | 2.66 | 3.89 |
| FTL_1276 | Bifunctional protein BirA | Metabolism | 5.74 | 2.66 | ||
| FTL_1283 | Glutamate-1-semialdehyde-2,1-aminomutase | hemL | Metabolism | 2.02 | 2.90 | 1.76 |
| FTL_1284 | Glutathione synthetase | Metabolism | 2.56 | 4.62 | 2.20 | |
| FTL_1285 | Methionyl-tRNA formyltransferase | Metabolism and genetic information processing | 5.04 | 4.87 | 2.52 | |
| FTL_1372 | Hypothetical lipoprotein | 2.65 | 3.61 | 1.14 | ||
| FTL_1396 | Galactose-1-phosphate uridylyltransferase | galT | Metabolism | 2.54 | 2.03 | 1.28 |
| FTL_1397 | Galactokinase | galK | Metabolism | 3.37 | 3.93 | 1.33 |
| FTL_1484 | Methylase | Metabolism | 4.56 | 4.53 | 1.36 | |
| FTL_1530 | Pseudo | 1.61 | 2.88 | 2.36 | ||
| FTL_1539 | Penicillin-binding protein (peptidoglycan synthetase) | ftsI | Metabolism and cellular processes | 2.39 | 2.65 | 1.79 |
| FTL_1616 | Phosphoenolpyruvate carboxykinase | pckA | Metabolism | 2.33 | 3.78 | |
| FTL_1639 | Hypothetical protein | 2.59 | 2.99 | 1.59 | ||
| FTL_1806 | Major facilitator superfamily (MFS) transport protein | 3.37 | 5.45 | 1.22 | ||
| FTL_1826 | NADH dehydrogenase I, E subunit | Metabolism | 2.13 | 2.18 | 1.72 | |
Genes whose expression is significantly changed (at least twofold, as determined using the SAM program) in LVS and the hfq mutant under at least two conditions are included.
The values are average changes between the hfq mutant and LVS in all microarrays. Bold type indicates a gene whose expression is significantly changed (as determined using the SAM program), but the change is less than twofold.
Quantitative RT-PCR (qRT-PCR).
One microgram of RNA was reverse transcribed using random hexamers and Superscript II reverse transcriptase (Invitrogen) according to the protocol provided by the manufacturer. Real-time PCR was performed with gene-specific primers using an ABI PRISM 7700 and SYBR green PCR master mixture (Applied Biosystems, Foster City, CA). To calculate the amount of transcript, a standard curve was created for each gene-specific primer set using a series of diluted genomic DNA from LVS. To compare the transcript levels of LVS and the hfq mutant, the amounts of transcript were normalized using the DNA helicase gene (FTL_1656), as the expression of this gene has been shown to change little during growth (7). The differences between LVS and the hfq mutant were calculated using triplicate samples, and the results were expressed as averages ± standard deviations. Student's t test was used to determine if the difference between the normalized transcript level of LVS and the normalized transcript level of the hfq mutant is significant.
Microarray data accession numbers.
The raw and processed microarray data have been submitted to the ArrayExpress database under accession numbers E-MEXP-1884, E-MEXP-1891, and E-MEXP-1892.
RESULTS
Promoter analysis and construction of hfq deletion mutant of F. tularensis LVS.
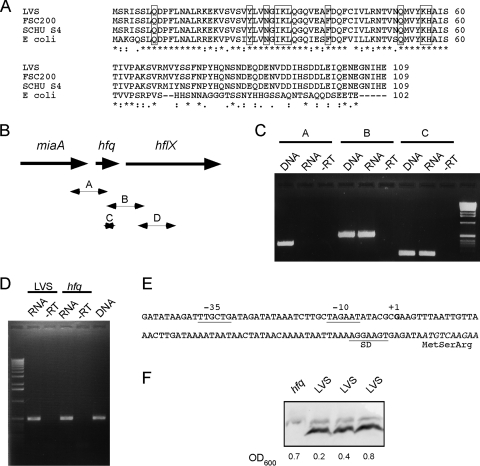
The F. tularensis LVS Hfq protein (encoded by FTL_0898) was identified by in silico analysis of the LVS genome. It consists of 109 amino acids and is 100% identical to Hfq of another F. tularensis subsp. holarctica strain (FSC200) and 98% identical to the protein from F. tularensis subsp. tularensis strain Schu S4. Furthermore, it is 53% identical (78% similar) to the protein from E. coli, and all residues proposed to contribute to binding of RNA are conserved (Fig. 1A) (5). The hfq gene is located downstream of the miaA gene [encoding tRNA delta(2)-isopentenylpyrophosphate transferase] and upstream of hflX (encoding GTP-binding protein HflX) (Fig. 1B), a gene organization that is conserved in many bacterial species. RT-PCR experiments demonstrated that there is cotranscription of hfq and hflX, but not of miaA and hfq (Fig. 1B and C). 5′ RACE experiments showed that the start site of the hfq transcript is located at position −65 relative to the ATG codon in the 110-bp miaA-hfq intergenic region (Fig. 1E). Putative −10 and −35 boxes are similar to the motifs recognized by the σ70-containing RNA polymerase and are separated by 17 nucleotides, indicating that hfq is not dependent on σ32.
FIG. 1.
Francisella Hfq protein and hfq locus. (A) Alignment of the Hfq proteins of F. tularensis strains LVS, FSC200, and Schu S4 and Hfq of E. coli performed using the ClustalW program. Residues that are identical in all strains are indicated by asterisks, conserved substitutions are indicated by colons, and semiconserved substitutions are indicated by periods. Amino acids which have been implicated in binding of RNA (5) are enclosed in boxes. (B) Genetic organization of the hfq locus in F. tularensis. miaA encodes tRNA delta(2)-isopentenylpyrophosphate transferase, and hflX encodes a GTP-binding protein. Arrows indicate fragments A, B, C, and D amplified in PCR as shown in panels C and D. (C) RT-PCR of the hfq locus. PCR to amplify fragments A, B, and C (see panel B) was performed with genomic DNA from LVS (DNA), with RNA from LVS after RT (RNA), or with RNA from LVS without RT (−RT) as a control. (D) Deletion in hfq has no polar effect on the downstream gene hflX. RT-PCR to amplify fragment D (see panel B) was performed with RNA isolated from either LVS or the hfq mutant after RT (RNA) or with RNA without RT (−RT). A control reaction was performed with DNA isolated from LVS (DNA). (E) Sequence of the promoter region of hfq. The transcription start site (+1) as determined by 5′ RACE is indicated by bold type, and putative −10 and −35 sequences are underlined. SD, Shine-Delgarno sequence. (F) Immunoblotting of bacterial lysates from strain LVS and the hfq mutant using anti-Hfq antiserum. The same amount of bacterial material, based on the optical density of the culture, was deposited in each lane for bacteria at different growth phases, including early exponential phase (OD600, 0.2), exponential phase (OD600, 0.4), and early stationary phase (OD600, 0.8).
To study the role of Hfq in F. tularensis, we constructed an hfq mutant of F. tularensis LVS in which there was an in-frame deletion of amino acids 5 to 70. Using RT-PCR, we verified that the deletion in hfq had no polar effect on transcription of the downstream gene hflX (Fig. 1D). In some bacteria, expression of Hfq has been reported to depend on the growth phase or growth rate. In P. aeruginosa the level of Hfq is higher in stationary phase (46), whereas in E. coli the level of this protein has been reported both to decrease in stationary phase (2, 22) and to increase in stationary phase (50). We assessed, using immunoblotting, the level of the Hfq protein in F. tularensis LVS in different growth phases. For this, we constructed a recombinant Hfq protein with an N-terminal histidine tag that was used for production of polyclonal anti-Hfq antibodies in rabbits (see Materials and Methods for details). As shown in Fig. 1F, we did not observe any major differences between the protein levels at the beginning of exponential phase (OD600, 0.2), in mid-exponential phase (OD600, 0.4), and in early stationary phase (OD600, 0.8).
Growth characteristics of the hfq mutant.
On chocolate agar plates, the colonies formed by the hfq mutant are smaller than the colonies formed by the wild-type strain (Fig. 2A). When grown in broth, the mutant exhibited a decreased growth rate and grew to a slightly lower density (Fig. 2B). Growth phenotypes on plates and/or in broth have also been observed for hfq mutants of E. coli, S. Typhimurium, V. cholerae, B. abortus, P. aeruginosa, and L. pneumophila (14, 30, 39, 42, 45, 51). We also checked by using transmission electron microscopy whether hfq inactivation had an impact on cell division, as previously reported for E. coli hfq mutants (51). Under the conditions tested (bacteria scratched from chocolate agar plates), the morphology of the hfq mutant was undistinguishable from that of parental strain LVS, indicating that the lack of Hfq had no effect on bacterial septation (Fig. 2C).
FIG. 2.
Growth characteristics of the LVS hfq mutant. (A) Colonies of wild-type F. tularensis LVS, the hfq mutant, and the hfq mutant containing a plasmid containing hfq on chocolate plates after 72 h of growth at 37°C. (B) Growth of F. tularensis LVS and the hfq mutant in Schaedler K3 broth at 37°C. (C) Transmission electron micrographs of LVS and the hfq mutant after growth on solid medium.
To ensure that the observed growth phenotype resulted from inactivation of hfq and not from polar effects on downstream genes, we complemented the mutant strain with a plasmid copy of hfq. Introduction of plasmid p6gro-hfq restored growth on plates to a level that was the same as the level for the wild-type parent (Fig. 2A), confirming the specific involvement of hfq.
Hfq contributes to stress resistance in F. tularensis.
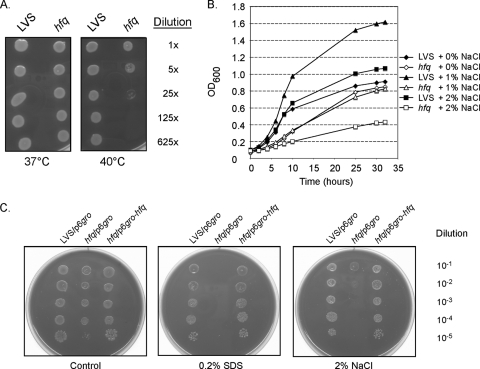
We next addressed the role of Hfq in stress resistance in F. tularensis LVS. Serial dilutions of wild-type and mutant bacterial stationary-phase cultures were spotted onto chocolate agar plates and incubated at 37°C or 40°C to compare the abilities of the organisms to grow at an elevated temperature. At 37°C, we observed a slight difference in growth between the two strains at the highest dilution (78,125-fold) (data not shown) but not at lower dilutions (Fig. 3A). At 40°C, the hfq strain grew poorly, and growth ceased completely at dilutions higher than 25-fold (Fig. 3A). In contrast, the growth of the wild-type strain at 40°C was comparable to that at 37°C (Fig. 3A and data not shown). The growth of the two strains was also compared in medium supplemented with NaCl (Fig. 3B). The wild-type strain grew to a higher density in medium with 1% NaCl than in medium without this supplement (Fig. 3B), whereas addition of 1% NaCl had no effect on the growth of the hfq strain. In medium containing 2% NaCl, the wild-type strain exhibited slightly enhanced growth compared to the growth in medium without this supplement, but the enhancement was less than that seen with the 1% NaCl supplement. In contrast, the hfq strain grew poorly in medium containing 2% NaCl, and the final density was about 50% of the density in regular medium. When the mutant strain was spotted on solid medium containing 2% NaCl, the growth was again severely affected, but complementation with a plasmid containing the hfq gene (p6gro-hfq) restored growth (Fig. 3C). We further assessed the effects of the membrane-perturbing agent SDS by spotting dilutions of stationary-phase cultures onto solid media containing 0.2% SDS. As observed in the presence of NaCl, growth of the mutant strain was almost not noticeable, whereas only slightly fewer bacteria were observed for the highest dilution on control plates (Fig. 3C). The increased sensitivity of the hfq mutant to SDS was alleviated by introduction of plasmid p6gro-hfq, further demonstrating that the observed phenotypes were due to a lack of hfq.
FIG. 3.
The hfq mutant is more sensitive to stress than the LVS parent strain. (A) Serial fivefold dilutions of stationary-phase cultures were spotted onto chocolate agar plates and incubated at 37°C or 40°C. (B) Growth of LVS and the hfq mutant in complex broth supplemented with 0%, 1%, or 2% NaCl. (C) Serial 10-fold dilutions of stationary-phase cultures of LVS, the hfq mutant, and the hfq mutant containing a plasmid with hfq were spotted onto chocolate agar plates containing 0.2% SDS, 2% NaCl, or H2O (control) and incubated at 37°C.
Hfq does not play a major role in intracellular multiplication in vitro.
To examine if Hfq contributes to Francisella pathogenesis, we initially compared the abilities of F. tularensis LVS and the hfq mutant to multiply inside macrophages in vitro (Fig. 4). For this, we infected murine macrophage-like J774 cells, RAW murine macrophages, and BMM. In all three types of macrophages, the wild-type strain grew to significantly higher levels after 24 h (P < 1 × 10−5, Student's t test), but at 48 h the difference was insignificant in J774 and BMM and only slightly a higher level was seen in RAW macrophages (P = 0.005). The slight growth defect of the hfq mutant during the initial stages of intracellular multiplication might be related to the slight growth defect seen in broth.
FIG. 4.
Intracellular multiplication of LVS and the hfq mutant in murine macrophage cell lines J774 and RAW and in murine BMM. Macrophages were infected by LVS or hfq mutant bacteria at a multiplicity of infection of ∼100, and the number of intracellular bacteria was determined after 0, 4, 24, and 48 h of infection. The results are expressed as average log10 (CFU/well) ± standard deviation for two experiments, each performed in triplicate.
The hfq mutant is attenuated for virulence in mice.
To determine if Hfq is important for the ability of F. tularensis to cause disease, we infected 6- to 8-week-old BALB/c mice with strain LVS and the hfq mutant. Groups of five mice were inoculated by the i.p. route with different numbers of bacteria, and the survival of the mice was followed for 9 days (Fig. 5A). One of the mice infected with LVS at the highest dose (∼3 × 104 bacteria) survived this infection, whereas all of the mice infected with the lowest dose (∼3 × 101 bacteria) survived. In contrast, all of the mice survived infection with the lower doses of the hfq mutant (∼3 × 102 to ∼3 × 105 bacteria), and only infection with a high dose of the hfq mutant (∼3 × 106 bacteria) resulted in death. These results demonstrate that virulence is attenuated in the hfq mutant.
FIG. 5.
The LVS hfq mutant is attenuated for virulence in mice. (A) Survival of mice after infection with LVS or the hfq mutant for 9 days after infection. Groups of five mice were infected by the i.p. route with different numbers of bacteria. The numbers of bacteria (in log10 CFU) used for infection are indicated on the right. (B) Multiplication of LVS and the hfq mutant in target organs of mice after i.p. infection with ∼3 × 104 CFU. Five mice were sacrificed after 2, 3, 4, and 7 days of infection, and the bacterial burdens in the spleen and liver were determined. Only one mouse survived after infection by LVS to day 7, and therefore no data for LVS at day 7 are shown.
To investigate the fate of the bacteria inside host tissues, we infected mice with ∼3 × 104 LVS or hfq mutant bacteria and followed the kinetics of infection by assessing the number of viable bacteria in the spleen and liver. Groups of five mice were sacrificed 2, 3, 4, and 7 days after infection, and numbers of viable bacteria were determined by plating diluted tissue homogenates (Fig. 5B). The number of LVS bacteria increased from day 2 to day 4, reaching about 108 bacteria per organ, and only one of the mice infected with LVS survived until day 7. The hfq strain was initially detected in both the spleen and liver at levels similar to the wild-type strain levels, but the numbers of bacteria declined after 2 days and all mice survived. These data indicate that the hfq mutant is able to infect mice and to persist for some period of time in the infected organs. However, it is unable to multiply efficiently inside host tissues and cause disease.
Hfq plays a role in the virulent F. tularensis clinical isolate FSC200 similar to the role that it plays in LVS.
To further study the role of Hfq in F. tularensis, we created an hfq mutant of F. tularensis subsp. holarctica clinical isolate FSC200. As observed for the LVS genetic background, the FSC200 hfq mutant formed colonies on nutrient agar that were smaller than the wild-type parent colonies and grew to a lower density in broth (Fig. 6A and data not shown). When organisms were grown in broth supplemented with 1% NaCl, the growth of FSC200 and the growth of the FSC200 hfq mutant, FSC768, were unaffected (Fig. 6A). However, in broth supplemented with 2% NaCl, the growth of FSC768 decreased compared to the growth in nonsupplemented broth, whereas the growth of FSC200 was unchanged. These results are similar to the results obtained for the hfq mutant of the LVS strain (Fig. 3B); likewise, we observed increased sensitivity of the hfq mutant to SDS (data not shown). In contrast to our observations for the LVS strain, we did not observe increased sensitivity to growth at an elevated temperature for FSC768 compared to FSC200 (Fig. 6B).
FIG. 6.
Growth characteristics and stress resistance of the FSC200 hfq mutant. (A) Growth of FSC200 and FSC768 in defined medium with additional 0%, 1%, or 2% NaCl. (B) Serial fivefold dilutions of stationary-phase cultures were spotted onto McLeod agar plates and incubated at 37°C or 40°C.
The involvement of Hfq of FSC200 in pathogenesis was addressed by assessing multiplication in macrophages in vitro and by performing virulence experiments with mice. The FSC768 mutant strain multiplied in J774 murine macrophage-like cells at levels that resemble the wild-type strain levels, although a slightly lower level was seen after 48 h (P < 3 × 10−5, Student's t test) (Fig. 7A). Mice that were infected via the i.p. route by FSC200 and FSC768 died within 9 days, even when they were infected with the lowest dose (∼2 CFU). However, the mice infected with FSC768 survived longer than the mice infected with FSC200 (Fig. 7B). The average time to death was 1 to 2 days longer for the mice infected with the hfq mutant than for the mice infected with the wild-type strain at the same dose. Similar results were obtained when mice were infected subcutaneously. The lowest dose of FSC768 resulted in a survival rate of 40%, whereas the same dose of FSC200 killed all mice. At a higher dose (∼2 × 101 CFU), all mice died, but the time to death was shorter for mice infected with the wild-type strain than for mice infected with the mutant strain. To further establish the role of Hfq in the pathogenesis of the human-pathogenic strain, we performed competition infection experiments with mice. For these experiments, a mixture of FSC200 and FSC768 bacteria (1:1 ratio; 102 CFU) was used to infect groups of five mice by the subcutaneous route. After 5 days of infection, the number of viable bacteria was determined by plating dilutions of spleen homogenates, and this was followed by PCR to distinguish wild-type bacteria from mutant bacteria. The average CI (Fig. 7C), defined as the ratio of mutant bacteria to wild-type bacteria, is 0.09, clearly demonstrating that the hfq mutant is attenuated for virulence.
FIG. 7.
Virulence of FSC200 is affected by the hfq mutation. (A) Intracellular replication in the J774 murine macrophage cell line. (B) Survival of mice after infection with FSC200 or FSC768 for 11 days after infection. Groups of five mice were infected by the i.p. or subcutaneous route with different numbers of bacteria. The numbers of bacteria (in log10 CFU) used for infection are indicated in parentheses after the strain designation. (C) Five mice were inoculated by the subcutaneous route with approximately 100 bacteria from a 1:1 mixture of FSC200 and FSC768. Spleens were homogenized after 5 days of infection, and the numbers of wild-type and mutant bacteria were determined. The results are expressed as the CI, which was calculated using the ratio of mutant bacteria to wild-type bacteria. Each symbol represents data for a single mouse, and the average CI is indicated by a solid line.
Taken together, these results show that Hfq in the clinical isolate of F. tularensis subsp. holarctica, like Hfq in LVS, contributes to virulence in mice, as well as to growth in vitro and resistance to stress.
Transcriptome analyses demonstrate global changes in gene expression in the hfq mutant.
Hfq is a global regulatory protein that influences gene expression by affecting translation and/or mRNA stability. To identify genes that are regulated by Hfq, we compared the transcriptome of LVS to that of the hfq mutant using DNA microarrrays. The microarrays (supplied by the Pathogen Functional Genomics Resource Center) contain oligonucleotides (four copies of each oligonucleotide) representing the open reading frames of F. tularensis strains LVS and Schu S4. To minimize any specific effect of a certain medium or growth condition on the results, we isolated RNA from bacteria grown under three different conditions: after growth on chocolate agar plates, after growth to exponential phase in defined medium (9), and after growth to exponential phase in complex broth (Schaedler K3 broth). All growth experiments were repeated at least twice, and each sample was labeled and hybridized to the microarrays in duplicate. The data obtained were analyzed using the SAM program (52) to identify genes for which there were statistically significant changes in expression under specific growth conditions. Genes with significantly changed expression (≥2-fold) under at least two conditions are shown in Table 2. We found that 16 genes were expressed at a lower level in the hfq mutant and 88 genes were expressed at a higher level in the mutant strain, suggesting that Hfq more often acts as a repressor than as an activator of expression in F. tularensis. However, we cannot conclude from this analysis which genes Hfq directly regulates and which genes have altered expression as a result of other changes. Among the downregulated genes were, in addition to hfq itself, hflX located downstream of hfq, as well as genes encoding putative membrane proteins, lipoproteins, and transport proteins. A number of genes that were upregulated in the hfq mutant are involved in metabolic functions (e.g., four genes encoding enzymes of the biotin metabolic pathway). Also, several genes in the “genetic information and processing” category according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg-bin/show_organism?org=ftl) were upregulated. These genes include genes encoding type IV pilus proteins, ribosomal proteins, and nucleases. Most of the genes (48 genes) are not assigned to a KEGG category, and 25 of these genes encode hypothetical proteins and conserved hypothetical proteins. Strikingly, in the group of hypothetical genes are 10 genes that are part of the FPI. These 10 genes are located in the same orientation on the genome in the proposed “pdp” operon, while the expression of none of the genes in the “igl” operon changes significantly (Fig. 8).
FIG. 8.
Schematic diagram of the FPI. Genes are numbered according to the LVS strain nomenclature for one of the two copies of the FPI. The two proposed divergent operons are indicated at the bottom.
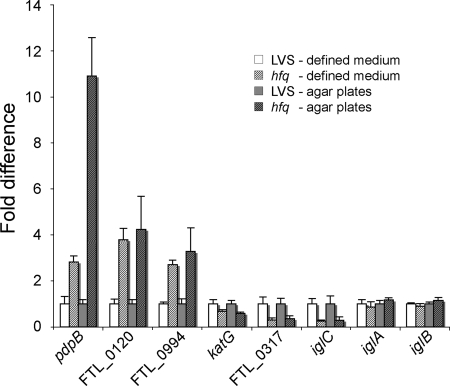
To verify the microarray results, we selected eight genes for qRT-PCR experiments. Two of these genes were downregulated in the hfq mutant (FTL_0317 and katG [FTL_1504]), three were upregulated in the hfq mutant (FTL_0120, pdpB [FTL_0125], and FTL_0994), and the expression of three genes did not change (iglC [FTL_0113], iglA [FTL_0111], and iglB [FTL_0112]). Five of the selected genes are part of the FPI (pdpB, FTL_0120, iglA, iglB, and iglC). The results obtained with RNA samples prepared from bacteria grown on agar plates or in defined medium are shown in Fig. 9 and, in general, confirm the data obtained in the microarray analysis. We similarly observed increased expression of pdpB, FTL_0120, and FTL_0994 and lower expression of katG and FTL_0317 in the hfq mutant. However, the expression of iglC was found to be lower in the hfq mutant than in LVS by qRT-PCR, whereas as determined by microarray analysis the expression of this gene was not significantly changed. However, when we performed qRT-PCR with the iglA and iglB genes, which are part of the “igl” operon of the FPI and are adjacent to the iglC gene, we did not observe any difference in expression between the wild-type strain and the hfq mutant.
FIG. 9.
Quantification of transcription of selected genes by real-time RT-PCR. Transcript levels were normalized to the level of DNA helicase (FTL_1656), and the differences (relative to LVS) and standard deviations are indicated for eight genes. RNAs from strains grown under two different growth conditions (defined medium and solid medium) were used for the analysis. The data are data for triplicate samples analyzed at the same time. The difference between the transcript levels of the LVS and hfq strains was significant (P < 0.05) for both growth conditions for pdpB, FTL_0120, FTL_0994, katG, FTL_0317, and iglC, as determined using Student's t test.
DISCUSSION
The Hfq protein contributes to the stress resistance and virulence of several pathogenic bacteria, including extracellular as well as intracellular organisms. The impact of hfq inactivation in these bacteria appears to be quite variable in terms of both the extent and the severity. A paradigm for the action of Hfq cannot be established from the data because the amount of data obtained in each case is quite variable and because our knowledge concerning the regulatory mechanisms and pathogenicities of these organisms is also quite heterogeneous.
Here we describe for the first time characterization of hfq mutants of two different strains of F. tularensis subsp. holarctica, the vaccine strain LVS and the human-pathogenic clinical isolate FSC200. We demonstrate that Hfq has a role in stress resistance, as well as in the ability to cause disease in mice. We further show that Hfq regulates, directly or indirectly, the expression of several genes in LVS, acting mainly as a repressor. Strikingly, we find that Hfq regulates expression of a subset of genes located in the FPI, thereby demonstrating that there is differential regulation of the two putative divergent operons in the FPI.
Hfq is involved in virulence.
In F. tularensis LVS, hfq gene expression is likely driven by a sigma 70 promoter, and Hfq production is constant in all growth phases in broth. Similar to what has been observed for several other bacteria, hfq inactivation results in a slight growth defect in LVS, both on solid media and in liquid media. The hfq mutant appeared to be highly sensitive to heat treatment (40°C) and was also more sensitive than the parental LVS strain to other stresses, including SDS and a high NaCl concentration (2%). These experiments demonstrate that Hfq contributes to resistance to various stress conditions in F. tularensis LVS.
We examined the contribution of Hfq to Francisella intracellular survival by comparing the abilities of the vaccine strain LVS and the isogenic hfq mutant to multiply inside J774 and RAW macrophage cell lines and in BMM. In all three types of macrophages, we found that inactivation of hfq led to an intracellular growth defect, particularly after 24 h. However, at 48 h the growth differences between the two strains were far less pronounced, suggesting that Hfq has a dominant role during the initial stages of intracellular multiplication.
We also evaluated the importance of Hfq in a mouse infection model of tularemia. Vaccine strain LVS, which is considered avirulent for humans, has significant virulence for mice, and four of five mice infected by the i.p. route with a dose of ∼3 × 104 bacteria succumbed to the infection. In contrast, an infecting dose of >106 bacteria was required to cause death when mice were infected with the hfq mutant by the i.p. route, demonstrating the severe attenuation of virulence of the hfq mutant with the LVS background. We also studied the infection kinetics of bacterial multiplication in the liver and spleen after i.p. infection with ∼3 × 104 LVS or hfq mutant bacteria. Strikingly, while the number of LVS bacteria was up to 108 bacteria per organ after 4 days (ultimately leading to death in most cases), the number of hfq mutant bacteria started to decrease after 2 days in both the spleen and liver, and all mice survived. These data indicate that the hfq mutant was unable to multiply efficiently inside host tissues and cause disease.
We further evaluated the role of Hfq in F. tularensis by creating an hfq mutant of the human-pathogenic clinical isolate FSC200. Like the mutant with the LVS genetic background, the FSC200 hfq mutant showed a slight growth defect in both solid and liquid media. The FSC768 mutant strain was also more sensitive than its isogenic parent to a high NaCl concentration (2%) or SDS, but it did not show increased sensitivity to an elevated temperature. In J774 macrophages, growth of the mutant was altered very moderately (after 48 h), reflecting the milder impact of the hfq deletion on the intracellular survival of FSC200. In mice, the hfq mutant also showed some attenuation, but the virulence defect was clearly less pronounced than that in the mutant with the LVS background. However, by performing a more sensitive CI analysis for spleens of animals infected with a 1:1 mixture of the wild-type and mutant strains, we could conclusively demonstrate that the hfq mutant was attenuated for virulence and more than 10 times less competitive for establishing an infection than the wild-type strain.
Strain FSC200 is extremely virulent in mice, and less than five bacteria (in this study the typical low doses were two to four bacteria) are required for a lethal infection by either the i.p. or subcutaneous route. Therefore, it is difficult to compare the levels of attenuation for this highly virulent strain and LVS. Importantly, the hfq mutant with the virulent FSC200 background was also attenuated compared to parental strain FSC200, both in single-strain infections and in competition infection experiments with the wild-type strain. This strongly supports the conclusion that Hfq is also important for virulence in human-pathogenic F. tularensis strains.
The fact that Hfq appeared to be less important for virulence in virulent strain FSC200 than in LVS is likely related to the fact that the mouse is far from the ideal model for tularemia. We have seen similar effects in other studies where we examined the role of a pilin gene, pilA, in virulence. In one study, where we used a moderately virulent strain isolated from a hare, we found that PilA mutants were severely attenuated compared to a strain expressing PilA (16). In contrast, when pilA mutants of virulent strains like FSC200 and the type A strain Schu S4 were used to infect mice, the level of attenuation was lower (unpublished results). Thus, it appears that the extremely low infection doses for these human-pathogenic strains in mice make it more difficult to establish the role in virulence of genes that do not have a major effect on intracellular multiplication and survival. A major strength of this work is that we have established that Hfq contributes to virulence both in the widely studied model strain LVS and in a clinical isolate.
Hfq, a novel regulator of the FPI.
Several studies, including proteomic and microarray analyses, with various bacteria have shown that Hfq controls the expression of numerous genes, including genes encoding virulence factors (14, 15, 26, 42, 46). Here, we compared the transcriptome of LVS to that of an hfq mutant using DNA microarrrays. Total RNA was extracted from bacteria grown on solid medium or in liquid culture (complex broth or chemically defined medium). Of the 104 genes with significantly changed expression, only 16 were expressed at a lower level in the hfq mutant (and are thus positively regulated by Hfq in a wild-type context), suggesting that Hfq acts more often as a repressor than as a positive regulator of gene expression in F. tularensis. However, we expect that some of these genes are not directly regulated by Hfq but the expression is changed indirectly. The 88 genes upregulated in the hfq mutant belong to a variety of functional categories, ranging from metabolic functions to type IV pilus biogenesis or ribosomal proteins, but the majority of them (55%) have not been assigned to any putative function. Strikingly, within the group of hypothetical genes, a cluster of 10 contiguous genes that are in the same orientation is part of the FPI. These genes belong to the proposed “pdp” operon. Remarkably, none of the genes in the “igl” operon of the FPI, which are in the opposite orientation, were expressed at higher levels in the hfq mutant (Fig. 9).
Of note, the iglA-iglD operon has a G+C content of 30.6% (compared to an average G+C content of 33% for the Francisella chromosome), while the pdpA-FTL_0115 region (“pdp” operon) has a G+C content of 26.6% and is the region with the lowest G+C content in the chromosome. For example, the G+C content of the pdpA gene (26.99%) is strikingly different from that of the adjacent rRNA genes (52.07% for FTL_R0003, the first gene of the rrn operon in the opposite orientation). The 374-bp intergenic region between FTL_R0003 and pdpA (FTL_0126) comprises two distinct halves, one immediately upstream of pdpA with a very low G+C content (ca. 20%) and the other with a significantly higher G+C content (>35%), suggesting that there is a putative promoter region immediately upstream of pdpA.
The pdpA and pdpB genes have been identified in at least two different screens of transposon insertion mutant libraries. Inactivation of pdpA in F. tularensis subsp. novicida led to a severe intramacrophage growth defect, and, in vivo, the pdpA mutant was highly attenuated (38, 54). Inactivation of pdpB in F. tularensis subsp. novicida also led to a severe in vitro growth defect in J774 and RAW cells, as well as in primary murine BMM, and to strong attenuation of virulence in the mouse (49, 54).
Four different regulatory proteins have been shown to control expression of the FPI. MglA interacts with the SspA protein, forming a heterodimer that binds RNA polymerase to control virulence gene expression positively (10). MglA and SspA seem to act in parallel with the FevR protein, which controls the same set of genes as these proteins (6). The fourth regulatory protein, PmrA, activates virulence gene expression as well, but it controls another set of genes in addition to the FPI (34). All four proteins affect virulence gene expression positively, and they all seem to regulate the entire FPI (although not every gene was found in each study, each study identified several genes in both of the putative divergent operons).
Our study is the first study in which it was shown that expression of only one part of the FPI is affected by mutation of a regulatory protein. Furthermore, the expression of the virulence genes is increased in the mutant strains, implying that Hfq is a negative regulator of virulence gene expression, in contrast to the other regulators of the FPI.
Even though the virulence genes are expressed at higher levels in the hfq mutant, it is possible that the deregulation is the background for the virulence attenuation that we observe in mice, as strict control of the FPI may be a requirement for bacterial survival in host organs. However, the virulence attenuation could be an effect of Hfq regulation of other target genes that impair intracellular multiplication in vivo.
Altogether, the capacity of Hfq to affect various cellular functions, such as metabolism, type IV pilus production, and virulence-related phenotypes, in F. tularensis probably reflects the ability of this chaperone to interact with a wide range of different regulatory RNAs, which remain to be identified. Of note, no homologues of sRNAs previously identified in other bacteria could be identified by in silico analysis of F. tularensis genomes, suggesting that the sRNAs produced by F. tularensis possess unique regulatory properties.
Acknowledgments
This study was funded by INSERM, CNRS, and University René Descartes Paris V. Parts of this study were funded by the Swedish Research Council (Å.F.). K.A. was supported by a fellowship from the Syrian government.
We thank Solveig Linder for excellent assistance with the animal infection studies. We thank T. C. Zahrt, Medical College of Wisconsin, for providing plasmid pFNLTP6-gro-gfp. DNA microarrays were obtained through NIAID's Pathogen Functional Resource Center, managed and funded by Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by TIGR.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10134-139. [DOI] [PubMed] [Google Scholar]
- 2.Ali Azam, T., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1816361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29247-259. [DOI] [PubMed] [Google Scholar]
- 4.Bohn, C., C. Rigoulay, and P. Bouloc. 2007. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10125-133. [DOI] [PubMed] [Google Scholar]
- 6.Brotcke, A., and D. M. Monack. 2008. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 763473-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brotcke, A., D. S. Weiss, C. C. Kim, P. Chain, S. Malfatti, E. Garcia, and D. M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 746642-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 1783763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charity, J. C., M. M. Costante-Hamm, E. L. Balon, D. H. Boyd, E. J. Rubin, and S. L. Dove. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christiansen, J. K., M. H. Larsen, H. Ingmer, L. Sogaard-Andersen, and B. H. Kallipolitis. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 1863355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 15.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the σE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62838-852. [DOI] [PubMed] [Google Scholar]
- 16.Forslund, A. L., K. Kuoppa, K. Svensson, E. Salomonsson, A. Johansson, M. Bystrom, P. C. Oyston, S. L. Michell, R. W. Titball, L. Noppa, E. Frithz-Lindsten, M. Forsman, and A. Forsberg. 2006. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 591818-1830. [DOI] [PubMed] [Google Scholar]
- 17.Franze de Fernandez, M. T., L. Eoyang, and J. T. August. 1968. Factor fraction required for the synthesis of bacteriophage Qβ RNA. Nature 219588-590. [DOI] [PubMed] [Google Scholar]
- 18.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to mRNA determines access for small RNA regulator. EMBO J. 23396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222273-280. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 21.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli J. Bacteriol. 1891963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajitani, M., A. Kato, A. Wada, Y. Inokuchi, and A. Ishihama. 1994. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Qβ. J. Bacteriol. 176531-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamoto, H., Y. Koide, T. Morita, and H. Aiba. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 611013-1022. [DOI] [PubMed] [Google Scholar]
- 24.Lai, X. H., I. Golovliov, and A. Sjostedt. 2004. Expression of IglC is necessary for intracellular growth and induction of apoptosis in murine macrophages by Francisella tularensis. Microb. Pathog. 37225-230. [DOI] [PubMed] [Google Scholar]
- 25.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoeba and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 11869-82. [DOI] [PubMed] [Google Scholar]
- 27.Ludu, J. S., O. M. de Bruin, B. N. Duplantis, C. L. Schmerk, A. Y. Chou, K. L. Elkins, and F. E. Nano. 2008. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 1904584-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 707511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLendon, M. K., M. A. Apicella, and L. A. Allen. 2006. Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 1871527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meibom, K. L., I. Dubail, M. Dupuis, M. Barel, J. Lenco, J. Stulik, I. Golovliov, A. Sjostedt, and A. Charbit. 2008. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 671384-1401. [DOI] [PubMed] [Google Scholar]
- 32.Mikulecky, P. J., M. K. Kaw, C. C. Brescia, J. C. Takach, D. D. Sledjeski, and A. L. Feig. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 111206-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1781310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohapatra, N. P., S. Soni, B. L. Bell, R. Warren, R. K. Ernst, A. Muszynski, R. W. Carlson, and J. S. Gunn. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes Infect. Immun. 753305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Møller, T., T. Franch, P. Højrup, D. R. Keene, H. P. Bächinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 923-30. [DOI] [PubMed] [Google Scholar]
- 36.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 101143-1151. [DOI] [PubMed] [Google Scholar]
- 37.Nakao, H., H. Watanabe, S. Nakayama, and T. Takeda. 1995. yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq). Mol. Microbiol. 18859-865. [DOI] [PubMed] [Google Scholar]
- 38.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson, G. T., and R. M. Roop, Jr. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34690-700. [DOI] [PubMed] [Google Scholar]
- 40.Santic, M., M. Molmeret, J. R. Barker, K. E. Klose, A. Dekanic, M. Doric, and Y. A. Kwaik. 2007. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell. Microbiol. 92391-2403. [DOI] [PubMed] [Google Scholar]
- 41.Sharma, A. K., and S. M. Payne. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62469-479. [DOI] [PubMed] [Google Scholar]
- 42.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjostedt, A. 2006. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8561-567. [DOI] [PubMed] [Google Scholar]
- 44.Sjostedt, A., U. Eriksson, L. Berglund, and A. Tarnvik. 1997. Detection of Francisella tularensis in ulcers of patients with tularemia by PCR. J. Clin. Microbiol. 351045-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35217-228. [DOI] [PubMed] [Google Scholar]
- 46.Sonnleitner, E., M. Schuster, T. Sorger-Domenigg, E. P. Greenberg, and U. Blasi. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 591542-1558. [DOI] [PubMed] [Google Scholar]
- 47.Storz, G., and D. Haas. 2007. A guide to small RNAs in microorganisms. Curr. Opin. Microbiol. 1093-95. [Google Scholar]
- 48.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7140-144. [DOI] [PubMed] [Google Scholar]
- 49.Tempel, R., X. H. Lai, L. Crosa, B. Kozlowicz, and F. Heffron. 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 745095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsui, H. C., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1797476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1335-49. [DOI] [PubMed] [Google Scholar]
- 52.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 54.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 1046037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 911-22. [DOI] [PubMed] [Google Scholar]