Abstract
Many microbial pathogens alter expression and/or posttranslational modifications of their surface proteins in response to dynamics within their host microenvironments to retain optimal interactions with their host cells and/or to evade the humoral immune response. Anaplasma phagocytophilum is an intragranulocytic bacterium that utilizes sialyl Lewis x (sLex)-modified P-selectin glycoprotein ligand 1 as a receptor for infecting myeloid cells. Bacterial populations that do not rely on this receptor can be obtained through cultivation in sLex-defective cell lines. A. phagocytophilum major surface protein 2 [Msp2(P44)] is encoded by members of a paralogous gene family and is speculated to play roles in host adaptation. We assessed the complement of Msp2(P44) paralogs expressed by A. phagocytophilum during infection of sLex-competent HL-60 cells and two HL-60 cell lines defective for sLex expression. Multiple Msp2(P44) and N-terminally truncated 25- to 27-kDa isoforms having various isoelectric points and electrophoretic mobilities were expressed in each cell line. The complement of expressed msp2(p44) paralogs and the glycosyl residues modifying Msp2(P44) varied considerably among bacterial populations recovered from sLex-competent and -deficient host cells. Thus, loss of host cell sLex expression coincided with both differential expression and glycosylation of A. phagocytophilum Msp2(P44). This reinforces the hypothesis that this bacterium is able to generate a large variety of surface-exposed molecules that could provide great antigenic diversity and result in multiple binding properties.
Anaplasma phagocytophilum is a tick-transmitted obligate intracellular bacterium belonging to the family Anaplasmataceae that infects neutrophils and myeloid precursor cells in the bone marrow, causing the emerging and potentially fatal infectious disease human granulocytic anaplasmosis (9, 12). The A. phagocytophilum major surface protein 2 [MSP2(P44)] gene family consists of more than 100 genes and functional pseudogenes that encode 42- to 49-kDa surface-exposed transmembrane proteins (9, 12, 18, 19, 31, 47). The Msp2(P44) proteins, which have been linked to antigenic variation, consist of conserved N- and C-terminal domains and a central hypervariable region (HVR) that is predicted to be surface exposed (9, 12, 18, 24). Msp2(P44) diversity results from RecF-mediated gene conversion of a single genomic msp2(p44) expression site by partially homologous sequences (1-3, 25, 26). Therefore, only one Msp2(P44) paralog is expressed per bacterium at any given time. Certain Msp2(P44) paralogs (3, 20, 21, 27, 45, 46), as well as paralogs of other Anaplasmataceae pathogens' outer membrane protein families (41, 42), exhibit exclusive expression patterns in tick or mammalian environments, which suggests that they may have host cell-specific roles. Indeed, msp2(p44)-18 transcription is specific to and dominates the msp2(p44) expression profile during A. phagocytophilum HZ, HGE1, and HGE2 infection of HL-60 cells (3, 21, 38, 46), and Msp2(P44)-18 is the predominant, if not the only, paralog expressed by HGE1 during growth in HL-60 cells (38). HGE1 also expresses 18- to 27-kDa Msp2 proteins [Msp2(P25)], and both Msp2(P44) and Msp2(P25) are posttranslationally modified into multiple isoforms.
P-selectin glycoprotein ligand 1 (PSGL-1) and sialyl Lewis x (sLex), a tetrasaccharide that modifies the N terminus of PSGL-1 and other selectin ligands (30), are confirmed human myeloid cell receptors for A. phagocytophilum (6, 13, 16, 34, 44). Two sugars that comprise sLex, α2,3-sialic acid and α1,3-fucose, are critical for A. phagocytophilum recognition of sLex. This bacterium is capable of using other receptors. By cultivating different A. phagocytophilum strains in HL-60 cells unable to construct sLex, we enriched for organisms capable of sLex-modified PSGL-1-independent infection (34, 35, 37). Two enriched populations are NCH-1A, which was obtained by growing the NCH-1 strain in sialylation-defective HL-60 sLex(−/low) cells (34), and NCH-1A2, which was selected for by cultivating NCH-1A in sialylation- and α1,3-fucosyltransferase-defective HL-60 A2 cells (13, 35).
The specific Msp2(P44) paralogs expressed by A. phagocytophilum NCH-1 during HL-60 cell infection are unknown, as is whether NCH-1 Msp2(P44) proteins are modified into multiple and/or truncated isoforms. Moreover, NCH-1A and NCH-1A2 provide unique means for assessing the impact of infection of host cells lacking sLex and other sialylated and/or α1,3-fucosylated glycans on Msp2(P44) expression. We therefore determined the complement of Msp2(P44) paralogs expressed by NCH-1 during cultivation in HL-60 cells, by NCH-1A during cultivation in HL-60 sLex(−/low) and HL-60 cells, and by NCH-1A2 during cultivation in HL-60 A2 cells and assessed their carbohydrate compositions. The resulting data are the first data that provide direct evidence for glycosylation of any native Anaplasmataceae protein and demonstrate that the A. phagocytophilum Msp2(P44) expression profile changes upon introduction into human myeloid cells unable to express sLex or other sialylated glycans.
MATERIALS AND METHODS
In vitro cultivation of A. phagocytophilum.
A. phagocytophilum NCH-1, NCH-1A, and NCH-1A2 were cultivated in HL-60, HL-60 sLex(−/low), and HL-60 A2 cells as described previously (34).
Preparation of A. phagocytophilum outer membrane fractions, 2DE, and gel staining.
Host cell-free bacteria were prepared as described previously (7). Bacterial outer membrane fractions were enriched for by temperature-dependent Triton X-114 phase partitioning and were resolved by two-dimensional electrophoresis 2DE and visualized exactly as described previously (38).
Cloning and expression of recombinant Msp2(P44) and portions of recombinant Msp2(P44).
Generation of recombinant full-length Msp2(P44)-18 has been described previously (38). Regions of msp2(p44)-18 corresponding to the segments encoding the N-terminal region, C-terminal region, HVR, N-terminal region through HVR, and HVR through C-terminal region were PCR amplified and cloned, and the products were expressed as recombinant proteins in Escherichia coli as described previously (38). The primers used were p44-5′-F (5′-GACGACGACAAGATGAA TGATGTCAGGGCTCATGATGACG-3′), p44-5′-R (5′-GAGGAGAAGCCCGGTATCTCCACGGCCTTAGCAAACTG-3′), p44-HVR-F (5′-GACGACGACAAGATAGCTAAGGAGTTAGCTTATGATGTTGTT ACTGG-3′), p44-HVR-R (5′-GAGGAGAAGCCCGGTGTCTTAGCTAGTAACCCTGCTACTATGG-3′), p44-3′-F (5′-GACGACGACAAGATCATAGTAGCAGGGTTACTAGCTAAGACTATTGAAG G-3′), and p44-3′-R (5′GAGGAGAAGCCCGGTAAAGCAAACCTAACACCAAATTCCCC-3′); the underlined nucleotides correspond to enterokinase/ligation-independent cloning vector pET-51b(+) (Novagen, Madison, WI)-compatible sequences.
Western blot analysis.
A. phagocytophilum whole-cell lysates, uninduced and induced E. coli cell lysates, or Triton X-114-fractionated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or 2DE and transferred to nitrocellulose at 15 V for 60 min using a Trans-Blot semidry transfer cell (Bio-Rad). Blots were screened using either mouse monoclonal antibody (MAb) 20B4 (32, 40) or MAb 8E8 (both gifts from J. Stephen Dumler of Johns Hopkins University), followed by goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Cell Signaling, Boston, MA) or horseradish peroxidase-conjugated Strep-Tag II MAb (Novagen). The blots were incubated with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) and exposed to film.
LC-MS/MS and database searching with Msp2(P44) HVR peptide.
Spot excision, liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analyses, and protein assignment were performed exactly as described previously (38). Mass spectrometric analysis was performed at the University of Kentucky Center for Structural Biology Protein Core Facility. To ascertain which Msp2(P44) paralogs are expressed in a given host cell type, the HVR peptides were searched against the GenBank database (www.ncbi.nlm.nih.gov/blast/Blast.cgi) and the annotated A. phagocytophilum HZ strain genome (www.tigrblast.tigr.org/cmr-blast) (17) using the BLASTP algorithm. We recorded only paralogs with which the peptide exhibited 100% amino acid identity. Because of the vast number of nonannotated GenBank entries that are designated simply “P44” or “Msp2,” we used only the paralogs that have been annotated and assigned a specific paralog number.
Cloning and sequencing of msp2(p44) RT-PCR products and statistical analyses.
Extraction of total RNA from A. phagocytophilum-infected host cells, reverse transcription PCR (RT-PCR) of msp2(p44) HVR transcripts, TA cloning, and sequence analyses were performed as described previously (38). Thirty-eight, 32, 50, and 59 TA clones were sequenced for NCH-1 in HL-60 cells, NCH-1A in HL-60 sLex(−/low) cells, NCH-1A in HL-60 cells, and NCH-1A2 in HL-60 A2 cells, respectively. Fifty-nine and 60 TA clones were sequenced for NCH-1 and NCH-1A2 following 100 additional passages in HL-60 and HL-60 A2 cells, respectively. The amplicon sequences were translated into predicted amino acid sequences using Editseq (part of the Lasergene 7.1 software package [DNASTAR, Madison, WI]), and the amino acid sequences were subsequently used to search the GenBank database and the annotated A. phagocytophilum genome and aligned using Megalign (Lasergene 7.1). A binomial test was conducted using the SPSS 15.0 software package (SPSS, Inc., Chicago, IL) to determine the statistical significance of the dominant msp2(p44) paralog. We assumed that there was a binomial distribution with a probability of success of 0.5 for each test. Statistical significance was defined as a P value of <0.05 (two tailed and based on Z approximation).
In silico glycosylation analyses.
In silico analyses for N-linked glycosylation motifs were performed with the amino acid sequences predicted for the HVRs of all msp2(p44) paralogs identified in this study using NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc/), while assessments for potential O-linked glycosylation sites were performed using NetOGlyc 3.1 (www.cbs.dtu.dk/services/NetOGlyc/) (22) and Support Vector Machines models (www.biosino.org/Oglyc) (23). Because of the high degree of conservation among the N and C termini of all Msp2(P44) paralogs, analyses were performed for these representative regions from APH_1221.
Monosaccharide analysis.
Three hundred fifty micrograms of an outer membrane-enriched hydrophobic pellet fraction was resolved by 2DE on five 4 to 20% polyacrylamide gradient gels (70 μg per gel; Bio-Rad). The spots corresponding to Msp2(P44) were excised from each gel and were processed for and subjected to gas chromatography-mass spectroscopy analyses exactly as described previously (38). Monosaccharide analysis was performed at the University of Georgia Complex Carbohydrate Research Center, Athens.
RESULTS
A. phagocytophilum NCH-1 and NCH-1A express full-length and N-terminally truncated Msp2 proteins.
We previously determined that A. phagocytophilum HGE1 expresses full-length 42- to 44-kDa Msp2 proteins, as well as 18- to 27-kDa isoforms that we proposed to be N-terminally truncated (38). Screening whole-cell lysates with MAb 20B4 (33), which recognizes both Msp2 isoforms, demonstrated that NCH-1 and NCH-1A also express both full-length and truncated isoforms (Fig. 1A). We next used MAb 8E8, which recognizes full-length Msp2 but not truncated Msp2 (Fig. 1A), to screen whole-cell lysates of E. coli expressing full-length Msp2(P44) or portions of Msp2(P44). Antibody targeting the Strep-Tag II epitope confirmed that there was recombinant Msp2(P44) expression (Fig. 1B). MAb 8E8 recognized only recombinant Msp2(P44) fragments corresponding to the N-terminal conserved region or the N terminus through the HVR, confirming that the smaller A. phagocytophilum Msp2 isoforms are N terminally truncated (Fig. 1C).
FIG. 1.
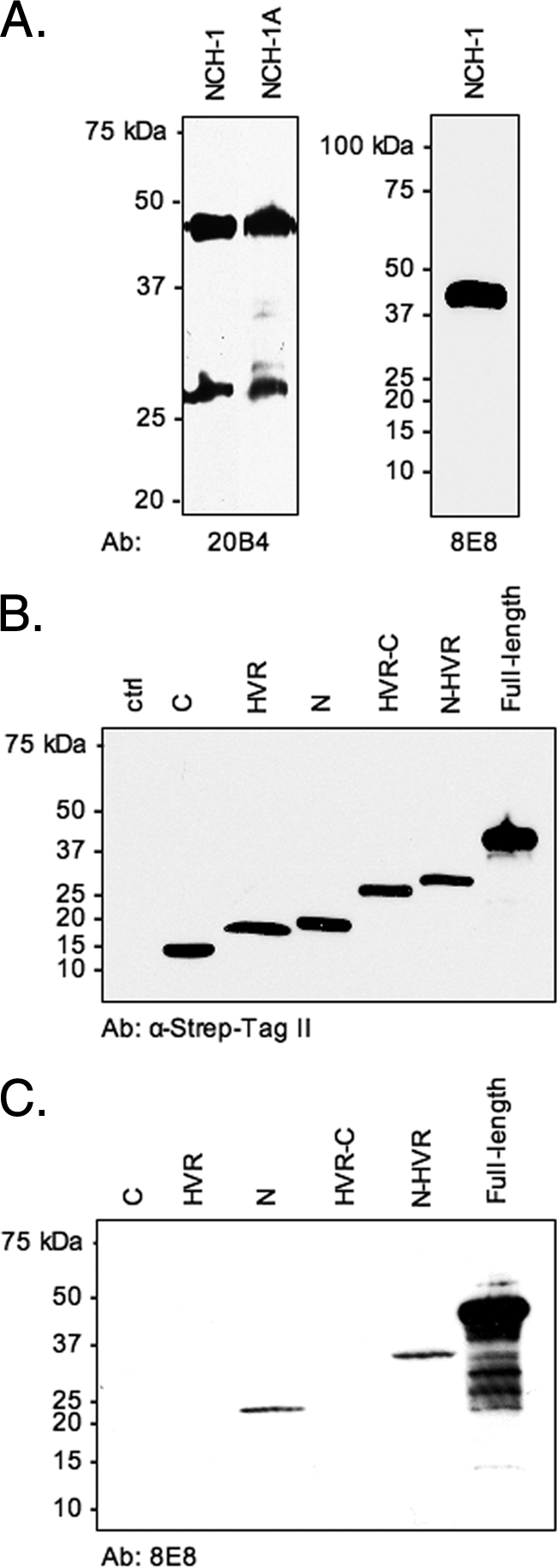
A. phagocytophilum NCH-1 and NCH-1A express both full-length and N-terminally truncated Msp2 isoforms. (A) NCH-1 and NCH-1A whole-cell lysates were screened via Western blot analysis using anti-Msp2 MAb 20B4, which recognizes full-length and truncated Msp2, or MAb 8E8, which recognizes only the full-length isoform. (B and C) Uninduced whole-cell lysates of E. coli (lane ctrl) or lysates of cells induced to express recombinant full-length Msp2 or portions of recombinant full-length Msp2, including the N-terminal conserved region (lane N), the HVR (lane HVR), the C-terminal conserved region (lane C), the N terminus through the HVR (lane N-HVR), and the HVR through the C terminus (lane HVR-C), were screened via Western blot analysis using an MAb targeting the Strep-Tag II epitope (B) or MAb 8E8 (C). Ab, antibody.
A. phagocytophilum NCH-1 expresses several Msp2(P44) and N-terminally truncated Msp2(P25) paralogs and modifies them into multiple isoforms during infection of HL-60 cells.
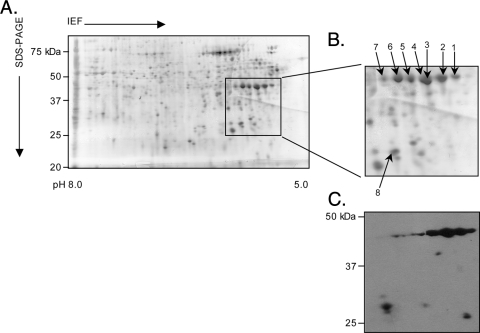
To assess whether NCH-1 Msp2(P44) is modified into multiple isoforms, we enriched for A. phagocytophilum outer membrane proteins using Triton X-114 phase partitioning (5, 36), which yields an upper aqueous phase containing hydrophilic proteins (hydrophilic liquid), a lower detergent phase (hydrophobic liquid) enriched for proteins carrying membrane anchors and one to a few transmembrane domains, and an insoluble hydrophobic pellet comprised of more complex membrane proteins. Because we previously determined that the hydrophobic pellet obtained from the HGE1 strain was the portion most enriched for Msp2(P44) proteins (38), we resolved the corresponding fraction recovered from NCH-1 by two-dimensional SDS-PAGE. High levels of several spots at apparent molecular masses of 42 to 44 kDa and at isoelectric points ranging from 5.4 to 6.0 were detected (Fig. 2A). Several MAb 20B4-reactive spots at 44 and 25 to 27 kDa were detected (Fig. 2C). Seven 42- to 44-kDa spots and one 27-kDa spot were extracted and subjected to LC-electrospray ionization ion trap MS/MS (Fig. 2B). Twenty-five different Msp2(P44)-derived peptides were recovered from the eight spots, and many of them were recovered from multiple spots (Table 1). Because the peptides recovered from the N- and C-terminal conserved domains presumably matched all Msp2(P44) paralogs and the peptides derived from the HVR were limited to a single paralog or a few paralogs, we focused on the HVR peptides to ascertain the identities of the expressed paralogs. BLAST searches of the annotated A. phagocytophilum HZ genome (www.tigrblast.tigr.org/cmr-blast) (17) and the GenBank database indicated that Msp2(P44)-23, -46, and -62 are expressed by NCH-1 and are posttranslationally modified into multiple isoforms, several of which comigrate when they are resolved by 2DE. The 27-kDa spot corresponds to an Msp2(P44)-23 paralog that has been truncated. Because the other recovered peptides each match multiple Msp2(P44) paralogs, we cannot predict which additional paralogs are expressed.
FIG. 2.
Two-dimensional SDS-PAGE and Western blot analyses of A. phagocytophilum NCH-1 Msp2(P44) proteins. Hydrophobic pellet fractions enriched for A. phagocytophilum NCH-1 outer membrane proteins were isoelectric focused (IEF) in IPG strips (pH 5.0 to 8.0), which was followed by resolution in the second dimension in SDS-polyacrylamide (4 to 20%) gels. (A) Silver-stained gel. Msp2 proteins that were excised for LC-MS/MS identification or detected by anti-Msp2(P44) antibodies are indicated by a box. (B) Enlarged view of the region of the silver-stained gel in panel A indicated by the box. Numbered spots were excised and identified by LC-MS/MS. (C) MAb 20B4 recognized spots in the predicted size range for full-length Msp2(P44) proteins, as well as multiple 25- to 27-kDa spots.
TABLE 1.
A. phagocytophilum NCH-1 strain Msp2(P44) peptide masses obtained from the hydrophobic pellet fraction
| Peptidea | Spot no.b | Msp2(P44) paralog designation(s) or no. of entriesc |
|---|---|---|
| N-terminal conserved region | ||
| IRDFSIR | 2, 4 | |
| AVYPYLKDGK | 2 | |
| AVYPYLK | 1, 2, 4 | |
| FDWNTPDPR | 2, 4 | |
| VELEIGYER | 1, 2, 3, 4, 5, 6 | |
| DSGSKEDEADTVYLLAK | 2, 4, 5, 6 | |
| -----EDEADTVYLLAK | 1, 2 | |
| ELAYDVVTGQTDNLAAAKAK | 1, 2, 4, 5 | |
| TSGKDIVQFANAVK | 1, 2, 3, 4, 5, 6, 7 | |
| ----DIVQFANAVK | 1, 2, 4 | |
| HVR | ||
| ISSPEIDGK | 1, 2, 4, 5 | 20, 23, 49, 54 |
| ISSPAIDGKd | 1, 2, 4, 5 | 46 |
| SLSGFVNTVK | 1, 2, 3, 4, 5, 6, 7, 8 | 23 |
| ASDGSSK | 1, 2, 4, 5 | 13 entries |
| ASDGSSKNIEGDPNSNAK | 5 | 23, 61 |
| -------NIEGDPNSNAK | 8 | 14 entries |
| AVAKDLVQELTPEEK | 2, 5, 6 | 4, 12, 15b, 36, 47, 48, 56 |
| ----DLVQELTPEEK | 1, 2, 3, 4, 5, 6 | 44 entries |
| AVAKDLVKELTPEEK | 2, 4, 5, 6 | 62 |
| ----DLVKELTPEEK | 1, 2, 3, 4, 5, 6 | 20 entries |
| TIVAGLLAK | 1, 4, 6 | 324 entries |
| C-terminal conserved region | ||
| TIEGGEVVEIR | 1, 2, 3, 4, 5, 6, 7, 8 | |
| VVGDGVYDDLPAQR | 1, 2, 3, 4, 5, 6 | |
| LVDDTSPAGR | 1, 2, 3, 4, 6 | |
| TKDTAIANFSMAYVGGEFGVR | 4 |
Peptide sequences are grouped for the N- or C-terminal conserved regions or the central HVR of Msp2(P44). Individual amino acids that are different in otherwise identical peptides are underlined.
The spot numbers are the numbers in Fig. 2.
The paralog designations are consistent with nomenclature used by GenBank and The Institute for Genomic Research and correspond to only the annotated paralogs with which the peptide masses exhibit 100% identity. The number of entries indicates the number of GenBank or The Institute for Genomic Research entries.
Bold type indicates peptide sequences corresponding to single Msp2(P44) paralogs and the spots from which they were derived.
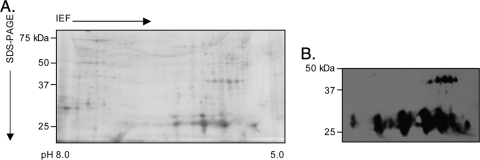
Because peptides matching Msp2(P44) paralogs were obtained from a 27-kDa spot recovered from the NCH-1 hydrophobic pellet and because a series of 25- to 27-kDa Msp2(P25) proteins were found to be enriched for in the hydrophobic liquid fraction obtained from A. phagocytophilum HGE1 (38), we resolved the NCH-1 hydrophobic liquid fraction by 2DE and assessed whether additional truncated Msp2(P25) isoforms are expressed. The full-length 44 kDa isoforms were present, but the level was lower than that in the hydrophobic pellet (Fig. 3A). Overall, this fraction contained considerably fewer proteins than the hydrophobic pellet. MAb 20B4 detected a series of 25- to 27-kDa proteins having isoelectric points of 5.3 to 6.4 (Fig. 3B), which strongly indicates that they are additional truncated Msp2(P25) isoforms.
FIG. 3.
Two-dimensional SDS-PAGE and Western blot analyses of A. phagocytophilum NCH-1 Msp2(P25) proteins. Hydrophobic liquid fractions enriched for A. phagocytophilum NCH-1 outer membrane proteins were isoelectric focused (IEF) in IPG strips (pH 5.0 to 8.0), which was followed by resolution in the second dimension in SDS-polyacrylamide (4 to 20%) gels. (A) Silver-stained gel. (B) MAb 20B4 recognized the series of full-length Msp2(P44) proteins, as well as a series of 25- to 27-kDa proteins.
A. phagocytophilum NCH-1A expresses multiple Msp2(P44) and N-terminally truncated Msp2(P25) paralogs and modifies them into different isoforms during infection of HL-60 sLex(−/low) cells.
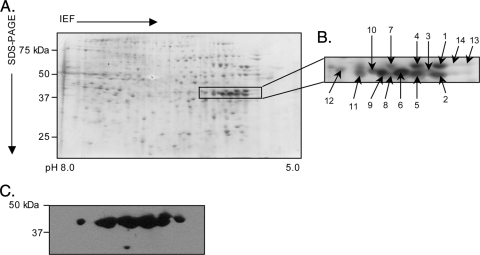
We next assessed the complement of Msp2(P44) paralogs expressed by NCH-1A during growth in sialylation-deficient HL-60 sLex(−/low) cells. A series of 14 MAb 20B4-reactive spots at 42 to 44 kDa having isoelectric points in the range expected for Msp2(P44) were subjected to LC-MS/MS analysis (Fig. 4). Twenty-nine different Msp2(P44) peptides were identified from 11 of the spots (Table 2). BLAST searches revealed that NCH-1A likely expresses Msp2(P44)-4, -8, -19, and -47 and the GenBank accession number AAO31994 sequence and modifies Msp2(P44)-4 and -8 into multiple isoforms during infection of HL-60 sLex(−/low) cells.
FIG. 4.
Two-dimensional SDS-PAGE and Western blot analyses of A. phagocytophilum NCH-1A Msp2(P44) proteins. Hydrophobic pellet fractions enriched for A. phagocytophilum NCH-1A outer membrane proteins were isoelectric focused (IEF) in IPG strips (pH 5.0 to 8.0), which was followed by resolution in the second dimension in SDS-polyacrylamide (4 to 20%) gels. (A) Silver-stained gel. Msp2(P44) proteins that were excised for LC-MS/MS identification and detected by MAb 20B4 are indicated by a box. (B) Enlarged view of the region of the silver-stained gel in panel A indicated by the box. Numbered spots were excised and identified by LC-MS/MS. (C) MAb 20B4 recognized spots in the predicted size range for full-length Msp2(P44) proteins.
TABLE 2.
A. phagocytophilum NCH-1A strain Msp2(P44) peptide masses obtained from the hydrophobic pellet fraction
| Peptidea | Spot no.b | Msp2(P44) paralog designation(s), accession no., or no. of entriesc |
|---|---|---|
| N-terminal conserved region | ||
| IRDFSIR | 4 | |
| AVYPYLKDGK | 4 | |
| AVYPYLK | 1, 4, 14 | |
| FDWNTPDPR | 1, 4 | |
| IGFKDNMLVAMEGSVGYGIGGAR | 1, 4 | |
| ----DNMLVAMEGSVGYGIGGAR | 1 | |
| VELEIGYER | 1, 4, 6, 7, 13, 14 | |
| GIRDSGSKEDEADTVYLLAK | 1, 4 | |
| ---DSGSKEDEADTVYLLAK | 1, 4, 6, 14 | |
| --------EDEADTVYLLAK | 1, 4 | |
| ELAYDVVTGQTDNLAAAKAK | 1, 2, 4, 6, 14 | |
| TSGKDIVQFAK | 1, 4, 7, 8, 13, 14 | |
| ----DIVQFAK | 1, 4 | |
| TSGKDFVQFAK | 10, 12 | |
| HVR | ||
| VALCGGAGPSDGSGSTSPQFFHDFVEKd | 1, 4 | 4 |
| TLLGNESK | 1, 4 | 4 |
| TLLENGSK | 1, 4, 7, 14 | 8 |
| NWPTSTAK | 1 | 4, 10 |
| YAVESDVK | 6 | 47 |
| DFINATMLGDGSK | 6 | 47 |
| GSNNATK | 10 | 19 |
| SGAGSTNR | 6 | AAO31994 |
| DLVQELTPEEK | 1, 4, 6, 7, 14 | 44 entries |
| DLVKELTPEEK | 1, 4, 6, 7, 14 | 20 entries |
| TIVAGLLAK | 1, 6 | 324 entries |
| C-terminal conserved region | ||
| TIEGGEVVEIR | 1, 4, 6, 7, 8, 10, 13, 14 | |
| VVGDGVYDDLPAQR | 1, 3, 4, 6, 7, 13, 14 | |
| LVDDTSPAGR | 1, 3, 6, 7, 14 | |
| TKDTAIANFSMAYVGGEFGVR | 2, 4 |
Peptide sequences are grouped for the N- or C-terminal conserved regions or the HVR of Msp2(P44). Individual amino acids that are different in otherwise identical peptides are underlined.
The spot numbers are the numbers in Fig. 4. Spots 5, 9, and 11 yielded no peptide masses.
The paralog designations are consistent with nomenclature used by GenBank and The Institute for Genomic Research and correspond to only the annotated paralogs with which the peptide masses exhibit 100% identity. GenBank accession number AAO31994 is provided because the entry is listed simply as “P44 paralog.” The number of entries indicates the number of GenBank or The Institute for Genomic Research entries.
Bold type indicates peptide sequences corresponding to single Msp2(P44) paralogs and the spots from which they were derived.
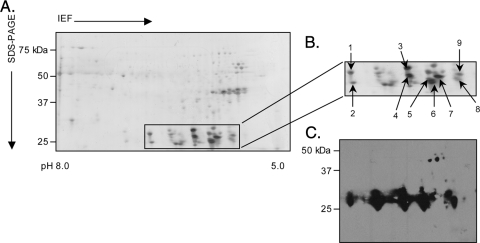
We next confirmed that NCH-1A expresses truncated Msp2(P25) isoforms during growth in HL-60 sLex(−/low) cells. Consistent with the proteins detected for NCH-1 and HGE1 (38), a series of MAb 20B4-immunoreactive full-length 44-kDa and truncated 25- to 27-kDa proteins were present in the NCH-1A hydrophobic liquid fraction, and the truncated 25- to 27-kDa proteins were more abundant (Fig. 5). LC-MS/MS analysis of nine of the 25- to 27-kDa proteins yielded a total of 18 different peptides (Table 3). Only two conserved N-terminal peptides that immediately precede the HVR were recovered. With the exception of GSSSGANYYGK and VGEGKNWPR, all the HVR peptide masses matched those recovered from the hydrophobic pellet fractions of NCH-1 and/or NCH-1A. As observed for NCH-1A Msp2(P44)-4 and -8 (Table 2), NCH-1A Msp2(P25)-8 and -23 are each modified into multiple isoforms (Table 3).
FIG. 5.
Two-dimensional SDS-PAGE and Western blot analyses of A. phagocytophilum NCH-1A Msp2(P25) proteins. Hydrophilic pellet fractions enriched for A. phagocytophilum NCH-1A outer membrane proteins were isoelectric focused (IEF) in IPG strips (pH 5.0 to 8.0), which was followed by resolution in the second dimension in SDS-polyacrylamide (4 to 20%) gels. (A) Silver-stained gel. Msp2(P25) proteins that were excised for LC-MS/MS identification are indicated by a box. (B) Enlarged view of the region of the silver-stained gel in panel A indicated by the box. Numbered spots were excised and identified by LC-MS/MS. (C) MAb 20B4 recognized spots in the predicted size range for full-length Msp2(P44) and truncated Msp2(P25) proteins.
TABLE 3.
A. phagocytophilum strain NCH-1A 27-kDa Msp2 peptide masses obtained from the hydrophobic liquid fraction
| Peptidea | Spot no.b | Msp2(P44) paralog designation(s), accession no., or no. of entriesc |
|---|---|---|
| N-terminal conserved region | ||
| TSGKDIVQFAK | 3 | |
| ----DIVQFAK | 3 | |
| HVR | ||
| GSSSGANYYGK | 9 | AAO32009 |
| ISSPAIDGKd | 5 | 46 |
| TLLENGSK | 3, 4, 7, 9 | 8 |
| SLSGFVNTVK | 5, 8 | 23 |
| VGEGKNWPR | 5 | 20, 23, 61 |
| ASDGSSK | 9 | 13 entries |
| NIEGDPNSNAK | 5, 8 | 14 entries |
| AVAKDLVQELTPEEK | 5 | 4, 12, 15b, 36, 47, 48, 56 |
| ----DLVQELTPEEK | 3, 4, 5, 7, 8, 9 | 44 entries |
| AVAKDLVKELTPEEK | 5 | 62 |
| ----DLVKELTPEEK | 3, 4, 5, 7, 9 | 20 entries |
| TIVAGLLAK | 4, 5, 6 | 324 entries |
| C-terminal conserved region | ||
| TIEGGEVVEIR | 2, 3, 4, 5, 7, 8, 9 | |
| VVGDGVYDDLPAQR | 2, 3, 4, 5, 7, 8, 9 | |
| LVDDTSPAGR | 3, 4, 6, 7 | |
| TKDTAIANFSMAYVGGEFGVR | 3, 4 |
Peptide sequences are grouped for the N- or C-terminal conserved region or the central HVR of Msp2(P44). Individual amino acids that are different in otherwise identical peptides are underlined.
The spot numbers are the numbers in Fig. 5. Spot 1 yielded no peptide masses.
The paralog designations are consistent with nomenclature used by GenBank and The Institute for Genomic Research and correspond to only the annotated paralogs with which the peptide masses exhibit 100% identity. GenBank accession number AAO32009 is provided because the entry is listed simply as “P44 paralog.” The number of entries indicates the number of GenBank or The Institute for Genomic Research entries.
Bold type indicates peptide sequences corresponding to single Msp2(P44) paralogs and the spots from which they are derived.
RT-PCR analyses of msp2(p44) paralogs expressed during A. phagocytophilum infection of HL-60, HL-60 sLex(−/low), and HL-60 A2 cells.
Because NCH-1 and NCH-1A each express multiple Msp2(P44) paralogs during infection of their host cell lines, we used RT-PCR to amplify the msp2(p44) HVR transcripts from these strains and cloned and sequenced the amplicons to determine whether a given paralog is dominant. This approach is suitable for identifying predominant transcripts, although it is not arithmetic and is likely to skew the amplicon population toward abundant messages. The msp2(p44)-23 HVR sequence was obtained from 89.5% (34 of 38; P < 0.001) of the clones derived from NCH-1-infected HL-60 cells, while msp2(p44)-4 was recovered from 75.0% (24 of 32; P = 0.007) of the clones generated from NCH-1A-infected HL-60 sLex(−/low) cells (Table 4). We next determined whether the NCH-1A msp2(p44) expression profile retains msp2(p44)-4 or reverts to being dominated by msp2(p44)-23 upon reintroduction into sialylation-competent host cells by inoculating NCH-1A into HL-60 cells. After 16 passages, msp2(p44)-4 accounted for the majority (42.0%; 21 of 50; P = 0.065) of the sequenced clones. msp2(p44)-23 was not detected. msp2(p44)-13, a paralog not previously detected during infection of HL-60 or HL-60 sLex(−/low) cells, was the second most common paralog (22.0%; 11 of 50 clones). Several low-abundance and mostly newly emerged paralogs were also recovered. To assess whether loss of not only sialylated but also α1,3-fucosylated glycans affects the msp2(p44) expression profile, we examined the complement of HVR transcripts expressed by NCH-1A2 during infection of HL-60 A2 cells. Notably, the NCH-1A2 expression profile consists almost entirely (96.6%; 57 of 59 clones; P < 0.001) of msp2(p44)-18. The predominant msp2(p44) paralogs expressed by NCH-1 and NCH-1A2 remained constant after 100 additional in vitro passages, as 91.5% (54 of 59; P < 0.001) and 85% (51 of 60; P < 0.001) of the clones obtained from NCH-1 and NCH-1A2 were msp2(p44)-23 and msp2(p44)-18, respectively (Table 5). Upon examination of the amino acid sequences predicted for the HVRs of all recovered paralogs, it became obvious that, while the Msp2(P44) expression profiles of A. phagocytophilum populations cultivated in different host cell environments are quite varied, certain HVR amino acids remain either absolutely or highly conserved (Fig. 6). These amino acids correspond to the “signature” C, C, WP, and A residues that divide the HVR into three distinct segments, as well as several individual amino acids or stretches of amino acids that have been noted by other workers (2, 27).
TABLE 4.
Msp2(P44) paralogs expressed by NCH-1, NCH-1A, and NCH-1A2a
| Paralogb | The Institute for Genomic Research designation | GenBank accession no.c | % Identityd | % of clones |
|---|---|---|---|---|
| NCH-1 in HL-60 cells | ||||
| (P44)-4 | APH_1154 | AAL86382 | 96.6-98 | 2.6 |
| (P44)-18 | APH_1194 | AAQ84091 | 100 | 2.6 |
| (P44)-20 | APH_1390 | AAL86394 | 99.3 | 2.6 |
| (P44)-23 | APH_1256 | YP_505777 | 94.3-99.3 | 89.5 |
| (P44)-56 | APH_1078 | AAP68670 | 98.5 | 2.6 |
| NCH-1A in HL-60 sLex(−/low) cells | ||||
| (P44)-4 | APH_1154 | AAL86382 | 90.5-100 | 75.0 |
| (P44)-6 | APH_1121 | AAL86384 | 100 | 3.1 |
| (P44)-11 | APH_1392 | YP_505888 | 99.3-100 | 6.2 |
| (P44)-12 | APH_1249 | YP_505773 | 100 | 3.1 |
| (P44)-43 | APH_1184 | AAO59315 | 100 | 3.1 |
| (P44)-62 | APH_1018 | AAQ75552 | 94.5 | 6.2 |
| (P44)-c20 | NAe | AAP20824 | 99.2 | 3.1 |
| NCH-1A in HL-60 cells | ||||
| (P44)-4 | APH_1154 | AAL86382 | 96.6 | 42.0 |
| (P44)-5 | APH_1312 | AAL86383 | 100 | 4.0 |
| (P44)-6 | APH_1121 | AAL86384 | 100 | 2.0 |
| (P44)-13 | APH_1238 | AAL86390, AAP14029 | 99.2-100 | 22.0 |
| (P44)-16 | APH_1269 | YP_505788 | 99.3 | 2.0 |
| (P44)-18 | APH_1194 | AAQ84091 | 100 | 2.0 |
| (P44)-19 | APH_1169 | YP_505708, ABF81554 | 68.0-76.7 | 4.0 |
| (P44)-24 | APH_1311 | YP_505824 | 100 | 10.0 |
| (P44)-43 | APH_1184 | AAO59315 | 100 | 4.0 |
| (P44)-50 | APH_1080 | AAP32153, AAL78190 | 67.1-100 | 6.0 |
| (P44)-53 | APH_1155 | AAP70037 | 100 | 2.0 |
| NCH-1A2 in HL-60 A2 cells | ||||
| (P44)-18 | APH_1194 | AAQ84091 | 98.5-99.3 | 96.6 |
| (P44)-24 | APH_1311 | YP_505824 | 100 | 1.7 |
| (P44)-c20 | NA | AAP20824 | 98.5 | 1.7 |
cDNAs corresponding to the HVRs of msp2(p44) cDNAs were TA cloned and sequenced. The nucleotide sequences were translated into amino acid sequences, which were utilized in BLAST searches.
Bold type indicates paralogs corresponding to the majority of sequenced clones and data for these paralogs.
The paralog nomenclature is the nomenclature used in the original The Institute for Genomic Research or GenBank entries. The Institute for Genomic Research designations correspond to designations for Msp2(P44) paralogs listed in the annotated A. phagocytophilum HZ genome.
Levels of amino acid identity for translated msp2(p44) HVR RT-PCR clone sequences with The Institute for Genomic Research and GenBank Msp2(P44) paralog entries.
NA, not applicable.
TABLE 5.
Msp2 (P44) paralogs expressed by NCH-1 and NCH-1A2 after 100 passagesa
| Paralogb | The Institute for Genomic Research designationc,d | GenBank accession no.c | % Identitye | % of clones |
|---|---|---|---|---|
| NCH-1 in HL-60 cells | ||||
| (P44)-6 | APH_1121 | AF412821 | 99.7 | 1.7 |
| (P44)-9 | APH_1391 | AF412824 | 99.3 | 1.7 |
| (P44)-13 | APH_1238 | AY763489 | 99.8 | 1.7 |
| (P44)-23 | APH_1256 | YP_505777 | 81.7-95 | 91.5 |
| (P44)-59 | APH_1408 | AY279321 | 98.9 | 1.7 |
| (P44)-62 | APH_1018 | AY307376 | 95 | 1.7 |
| NCH-1A2 in HL-60 A2 cells | ||||
| (P44)-18 | APH_1194 | AAQ84091 | 97.7-100.0 | 85.0 |
| (P44)-24 | APH_1311 | AY763480 | 100 | 1.7 |
| (P44)-44 | APH_0981 | AY147265 | 94.5-94.7 | 1.7 |
| (P44)-47 | APH_1152 | AY147264 | 93.7 | 1.7 |
| (P44)-48 | APH_1272 | AY147267 | 73.7-75.5 | 3.3 |
| (P44)-59 | APH_1408 | AY279321 | 71.4 | 1.7 |
| (P44)-62 | APH_1018 | AAP20824 | 98.5 | 5.0 |
NCH-1 and NCH-1A2 were cultivated in HL-60 and HL-60 A2 cells, respectively, for 100 passages from the point at which their Msp2(P44) expression profiles were examined to obtain the data shown in Table 4. cDNAs corresponding to the HVRs of msp2(p44) cDNAs were TA cloned and sequenced. The nucleotide sequences were translated into amino acid sequences, which were utilized in BLAST searches.
Bold type indicates paralogs corresponding to the majority of sequenced clones and data for these paralogs.
The paralog nomenclature is the nomenclature used in the original The Institute for Genomic Research or GenBank entries.
The Institute for Genomic Research designations correspond to designations for Msp2(P44) paralogs listed for the annotated A. phagocytophilum HZ genome.
Levels of amino acid identity for translated msp2(p44) HVR RT-PCR clone sequences with The Institute for Genomic Research and GenBank MSP2(P44) paralog entries.
FIG. 6.
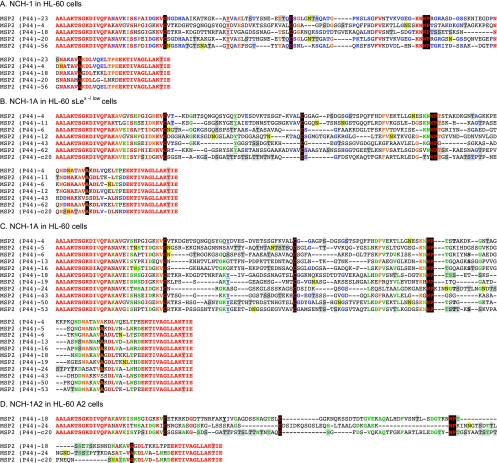
Alignment of the HVRs of Msp2(P44) paralogs expressed by NCH-1, NCH-1A, and NCH-1A2 during infection of HL-60, HL-60 sLex(−/low), and HL-60 A2 cells. The predicted amino acid sequences for the complements of msp2(p44) HVRs recovered from NCH-1 cultivated in HL-60 cells, NCH-1A cultivated in HL-60 sLex(−/low) and HL-60 cells, and NCH-1A2 cultivated in HL-60 A2 cells were aligned using Megalign. Residues that are 100% conserved in each complement of the aligned paralogs are red. Residues exhibiting 80 to 91% conservation are orange. Residues that are 64 to 73% conserved are green. Residues exhibiting 54 to 60% conservation are blue. “Signature residues” previously identified by Lin et al. (27) and Barbet et al. (2), which are very highly conserved and demarcate the most variable central portion of the HVR into three distinct segments, are highlighted with a black background. A signature tyrosine residue that may be important for intracellular replication (43) is underlined. Amino acids highlighted with a gray background are amino acids identified by NetOGlyc 3.1 (www.cbs.dtu.dk/services/NetOGlyc/) (22) and Support Vector Machines models (www.biosino.org/Oglyc) (23) as being potential O-linked glycosylation sites. Residues highlighted with a yellow background are residues predicted by NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc/) to be potential N-linked glycosylation sites.
Carbohydrate composition analyses of Msp2(P44) paralogs expressed by NCH-1, NCH-1A, and NCH-1A2.
As we have reported previously for A. phagocytophilum HGE1 Msp2(P44)-18 (38) and as has been observed for major outer membrane proteins of other Anaplasmataceae pathogens (41, 42), the multiple Msp2(P44) isoforms observed for NCH-1 and NCH-1A likely result from glycosylation or other posttranslational modifications. Because glycosylation of Msp2(P44) paralogs may contribute to their pathobiological functions, we performed in silico analyses (22, 23) to identify possible N- and O-linked glycosylation sites in the amino acid sequences predicted for the HVR of each expressed paralog. With the exception of Msp2(P44)-13, for which no glycosylation site was predicted, multiple potential glycosylation sites were identified for all paralogs (Fig. 6). The number of glycosylation sites varies widely among paralogs. For instance, although the mean number of total glycosylation sites predicted for all paralogs was 7.3 ± 4.8, Msp2(P44)-5, -53, and -c20 have 12, 13, and 22 potential glycosylation sites, respectively. Glycosylation sites were not predicted for the conserved Msp2(P44) N or C termini.
To determine if glycosyl residues decorate Msp2(P44) paralogs expressed by NCH-1, NCH-1A, and NCH-1A2 and to assess whether the Msp2(P44) carbohydrate composition varies among these isolates, the native forms from each population were analyzed en masse by gas chromatography, which has successfully determined that recombinant forms of Msp2(P44)-18 and several other Anaplasmataceae pathogen outer surface proteins are glycosylated (10, 28, 29, 38, 41, 42). As observed for the monosaccharide compositions of all recombinant forms of Anaplasmataceae outer membrane proteins analyzed to date, the Msp2(P44) proteins expressed by NCH-1 and each of its derivative populations are glycosylated and are likely glucans, as glucose is the primary sugar that decorates each of them (Table 6). The molar percentage of glucose varied among Msp2(P44) proteins recovered from NCH-1 grown in HL-60 cells, NCH-1A grown in HL-60 sLex(−/low) cells, NCH-1A organisms repassaged in HL-60 cells, and NCH-1A2 cultivated in HL-60 A2 cells. Also, several more types of sugar residues decorate Msp2(P44) proteins recovered from NCH-1A than Msp2(P44) proteins isolated from NCH-1 or NCH-1A2. Lastly, the carbohydrate composition of Msp2(P44) proteins recovered from NCH-1A2, the vast majority of which apparently is Msp2(P44)-18 (Tables 4 and 5), is 90.8 mol% glucose, and there are considerably smaller amounts of galactose, xylose, and arabinose (Table 6). This is worth noting because this glycosyl composition is nearly identical to that of recombinant Msp2(P44)-18 (38).
TABLE 6.
Carbohydrate composition analysis of Msp2(P44) proteins
| Glycosyl residue | mol%a
|
|||
|---|---|---|---|---|
| NCH-1 in HL-60 cellsb | NCH-1A in HL-60 sLex(−/low) cellsb | NCH-1A in HL-60 cellsb | NCH-1A2 in HL-60 A2 cellsb | |
| Arabinose | NDc | 9.6 | ND | ND |
| Fucose | ND | 3.8 | 11.7 | ND |
| Galactose | ND | 13.1 | 15.8 | 1.8 |
| Glucose | 93.3 | 39.0 | 28.0 | 90.8 |
| N-Acetylglucosamine | ND | 11.8 | 11.5 | ND |
| Mannose | 2.1 | 4.6 | 20.3 | 1.7 |
| Rhamnose | ND | 3.4 | ND | ND |
| Xylose | 4.7 | 14.7 | 12.6 | 5.6 |
Bold type indicates the highest molar percentages.
A. phagocytophilum strains and the host cell lines in which they were cultivated.
ND, not detected.
DISCUSSION
A. phagocytophilum NCH-1 changes not only the complement of Msp2(P44) paralogs that it expresses but also the glycosyl modifications that decorate Msp2(P44) when it is transferred from sLex-competent HL-60 cells to HL-60 cells defective for construction of sLex and other sialylated and/or α1,3-fucosylated glycans. This reinforces the ability of this bacterium to generate a large variety of surface-exposed molecules that could provide great antigenic diversity and/or provide multiple binding properties. According to our RT-PCR data, the dominant paralogs expressed by NCH-1 in HL-60 cells, by NCH-1A in HL-60 sLex(−/low) and HL-60 cells, and by NCH-1A2 in HL-60 A2 cells are Msp2(P44)-23, -4, and -18, respectively. The results observed for NCH-1 and NCH-1A are in contrast to previous observations for HGE1, HGE2, and HZ showing that Msp2(P44)-18 is the predominant, if not the only, paralog expressed during infection of HL-60 cells and thus has a fitness advantage in this cell type (3, 21, 24, 38, 46). Our data indicate that Msp2(P44)-23 and Msp2(P44)-4 confer similar fitness advantages to NCH-1 and NCH-1A in their corresponding cell lines and also indicate that there is strain-specific preferential expression of distinct paralogs in the absence of immune pressure. Because Msp2(P44)-18 confers a fitness advantage to multiple strains in HL-60 cells and to NCH-1A2 in HL-60 A2 cells, switching in the expression of specific Msp2(P44) paralogs may not be related to the host cell phenotype with respect to sLex but may instead be prompted by the introduction of the bacterium into a novel host cell milieu. In support of this hypothesis, we observed no change in the msp2(p44) transcriptional profiles of NCH-1 or its derived populations after 100 in vitro passages and observed the emergence of novel paralogs only after introduction of NCH-1 into a new host cell environment. Our data differ from those of Scorpio and colleagues, who observed the emergence of a novel msp2(p44) paralog following only 13 passages of the Webster strain in glycosylation-competent HL-60 cells (40). While this finding hints that NCH-1 is more restricted in the diversity of the msp2(p44) transcripts that it expresses when it is maintained in a stable host cell environment in the absence of immune pressure than the Webster strain, this possibility remains to be experimentally confirmed. Regardless, our data and those of Scorpio and colleagues agree that geographically diverse A. phagocytophilum strains share a propensity for generating considerable diversity in their Msp2(P44) profiles.
While the only observed differences among the host cell lines used in this study involve glycosylation, we cannot exclude the possibility that unaccounted for physiological differences among the lines may also influence A. phagocytophilum Msp2(P44) expression. Nonetheless, Msp2(P44) profile changes occurring in response to loss of one or more of the known determinants required for A. phagocytophilum to bind sLex-modified PSGL-1 hint that Msp2(P44) may facilitate adhesion to the human myeloid cell surface directly or perhaps indirectly as part of an adhesin complex. This agrees with one study which demonstrated that recombinant Msp2(P44) or MAbs targeting Msp2(P44) inhibit A. phagocytophilum propagation in HL-60 cells and partially antagonize binding to BJAB cells transfected to express sLex-modified PSGL-1 (32), as well as with a second report which showed that Msp2(P44)-targeting MAbs block A. phagocytophilum binding to or replication within HL-60 cells (43). In addition to sLex-modified PSGL-1, A. phagocytophilum is capable of utilizing an unidentified nonsialylated and nonfucosylated PSGL-1-independent receptor on human myeloid cells and a nonsialylated receptor on human endothelial cells for adhesion and entry (15, 34, 35). Therefore, if Msp2(P44) facilitates host cell adhesion and/or adaptation in vivo, then it may do so for both sLex-decorated and undecorated host cells.
If the Msp2(P44) HVR is important for adhesion and the adhesive determinants are proteinaceous, then the highly conserved signature residues would likely be critical either for providing direct points of contact for receptor recognition or for maintaining proper confirmation to facilitate binding. The adhesive epitopes would presumably be masked by flanking hypervariable amino acid segments and/or perhaps by glycosylation. Alternatively, adhesion could be glycan mediated, as has been demonstrated for Anaplasma marginale Msp1a binding to tick cells (10). In this instance, variation in the HVR would be inconsequential as long as asparagine, serine, and threonine residues were present to allow the addition of glycosyl groups needed for adhesion. Notably, Msp2(P44) glycosylation is confined to the HVR based on in silico prediction analyses. Other possible functions that have been attributed to glycosylation include maintenance of cell shape, protection from proteolysis, and protein stability (4).
Other than the signature residues, there is little homology among the HVRs of Msp2(P44)-18, -23, and -4 and the other expressed paralogs examined in this study. Interestingly, one signature residue is a tyrosine that is part of the FAKY epitope of the Msp2(P44)-18 HVR (Fig. 6) recognized by MAb 3E65, which blocks A. phagocytophilum intracellular replication (43). As this tyrosine is the only residue of the MAb 3E65 epitope that is shared by Msp2(P44)-18, -23, and -4 and many of the other expressed paralogs examined in this study, it may be a residue that is critical for intracellular development.
This study adds to the growing body of evidence demonstrating that posttranslational modification of dominant outer surface proteins into multiple isoforms is a common theme exhibited by Anaplasmataceae pathogens, and it is the first study to provide the carbohydrate composition of a native Anaplasmataceae protein. Different Msp2(P44) paralogs are glycosylated differently. For instance, P44-23 and P44-18, which are the predominant paralogs expressed by NCH-1 and NCH-1A2, respectively, are modified primarily by glucose and carry trace or undetectable amounts of other sugars. This is discernible because Msp2(P44)-23 and -18 represent 89.5% and 96.6% of the recovered RT-PCR clones, respectively, and the Msp2(P44) glucose composition is greater than 90% for both NCH-1 and NCH-1A2. The complement of NCH-1A Msp2(P44) proteins is much more diverse, and, coincidentally, the glucose composition is considerably lower, while several other sugars are detected at much higher levels in NCH-1A than in NCH-1 or NCH-1A2. To gain a better understanding of the role of differential glycosylation of Msp2(P44) paralogs, it would be of great value to determine the carbohydrate compositions of individual paralogs. Because 150 μg of glycoprotein is required for gas chromatographic analysis, determining the glycosyl composition of native paralogs is impractical. However, the recombinant (38) and native forms of Msp2(P44)-18 have highly similar glycosyl compositions, which implies that recombinant Msp2(P44) proteins undergo glycosylations in E. coli comparable to those of their native counterparts in A. phagocytophilum. This suggests that how glycosylation differs among paralogs could potentially be discerned by examining the glycosyl compositions of recombinant forms of different Msp2(P44) paralogs. The majority of Msp2(P44) isoforms can be distinguished from each other by their isoelectric points, which indicates that different paralogs may have minor side chain modifications, such as phosphorylation, sulfation, and/or acetylation.
Processing of Msp2(P44) into N-terminally truncated isoforms is not unique to NCH-1 and its derived populations as it also occurs in the HGE1 strain and is reminiscent of the processing of autotransporters (38). Members of the autotransporter superfamily are ubiquitous in the Proteobacteria and Chlamydia spp. and have roles in adhesion, invasion, cell-to-cell spread, serum resistance, and proteolysis (8, 11, 14, 18). Interestingly, their adhesive capabilities are linked to their glycosylation (11). The biological significance of Msp2(P44) N-terminal processing remains to be determined, as do the differential expression and glycosylation of Msp2(P44) paralogs. The importance of glycosylation of prokaryotic surface proteins to pathogenesis is continually becoming more apparent (4, 39), and the contribution of glycosylation to the putative pathobiological functions of Msp2(P44), particularly adhesion, warrants considerably more attention.
Acknowledgments
We thank Steve Dumler for Msp2 MAbs, Ulrike Munderloh and Curtis Nelson for HL-60 A2 cells, Leigh Ann Simmons for statistical analyses, Brian Stevenson for use of his Multiphor II electrophoresis system, Carol Beach for LC-MS/MS analyses, and Jere McBride for helpful discussions.
This work was supported by NIH grants DK065039 and AI072683 and by a grant from the National Research Fund for Tick-Borne Diseases. The University of Kentucky Center for Structural Biology Protein Core Facility is supported in part by funds from NIH National Center for Research Resources grant P20 RR020171. The University of Georgia Complex Carbohydrate Research Center is supported in part by NIH National Center for Research Resources grant P20 RR020171 and by the Department of Energy-Funded Center for Plant and Microbial Complex Carbohydrates.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 17 February 2009.
REFERENCES
- 1.Barbet, A. F., J. T. Agnes, A. L. Moreland, A. M. Lundgren, A. R. Alleman, S. M. Noh, K. A. Brayton, U. G. Munderloh, and G. H. Palmer. 2005. Identification of functional promoters in the msp2 expression loci of Anaplasma marginale and Anaplasma phagocytophilum. Gene 35389-97. [DOI] [PubMed] [Google Scholar]
- 2.Barbet, A. F., A. M. Lundgren, A. R. Alleman, S. Stuen, A. Bjoersdorff, R. N. Brown, N. L. Drazenovich, and J. E. Foley. 2006. Structure of the expression site reveals global diversity in MSP2 (P44) variants in Anaplasma phagocytophilum. Infect. Immun. 746429-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, A. F., P. F. Meeus, M. Belanger, M. V. Bowie, J. Yi, A. M. Lundgren, A. R. Alleman, S. J. Wong, F. K. Chu, U. G. Munderloh, and S. D. Jauron. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect. Immun. 711706-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45267-276. [DOI] [PubMed] [Google Scholar]
- 5.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 6.Carlyon, J. A., M. Akkoyunlu, L. Xia, T. Yago, T. Wang, R. D. Cummings, R. P. McEver, and E. Fikrig. 2003. Murine neutrophils require alpha-1,3-fucosylation but not PSGL-1 for productive infection with Anaplasma phagocytophilum. Blood 1023387-3395. [DOI] [PubMed] [Google Scholar]
- 7.Carlyon, J. A., W. T. Chan, J. Galan, D. Roos, and E. Fikrig. 2002. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J. Immunol. 1697009-7018. [DOI] [PubMed] [Google Scholar]
- 8.Dautin, N., and H. D. Bernstein. 2007. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 6189-112. [DOI] [PubMed] [Google Scholar]
- 9.Dumler, J. S., K. S. Choi, J. C. Garcia-Garcia, N. S. Barat, D. G. Scorpio, J. W. Garyu, D. J. Grab, and J. S. Bakken. 2005. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg. Infect. Dis. 111828-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Garcia, J. C., J. de la Fuente, G. Bell-Eunice, E. F. Blouin, and K. M. Kocan. 2004. Glycosylation of Anaplasma marginale major surface protein 1a and its putative role in adhesion to tick cells. Infect. Immun. 723022-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard, V., and M. Mourez. 2006. Adhesion mediated by autotransporters of Gram-negative bacteria: structural and functional features. Res. Microbiol. 157407-416. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, J. L. 2005. Human granulocytic anaplasmosis, p. 218-238. In J. L. Goodman, D. T. Dennis, and D. E. Sonenshine (ed.), Tick-borne diseases of humans. ASM Press, Washington, DC.
- 13.Goodman, J. L., C. M. Nelson, M. B. Klein, S. F. Hayes, and B. W. Weston. 1999. Leukocyte infection by the granulocytic ehrlichiosis agent is linked to expression of a selectin ligand. J. Clin. Investig. 103407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I. R., and A. C. Lam. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 9573-578. [DOI] [PubMed] [Google Scholar]
- 15.Herron, M. J., M. E. Ericson, T. J. Kurtti, and U. G. Munderloh. 2005. The interactions of Anaplasma phagocytophilum, endothelial cells, and human neutrophils. Ann. N. Y. Acad. Sci. 1063374-382. [DOI] [PubMed] [Google Scholar]
- 16.Herron, M. J., C. M. Nelson, J. Larson, K. R. Snapp, G. S. Kansas, and J. L. Goodman. 2000. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science 2881653-1656. [DOI] [PubMed] [Google Scholar]
- 17.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, H., X. Wang, T. Kikuchi, Y. Kumagai, and Y. Rikihisa. 2007. Porin activity of Anaplasma phagocytophilum outer membrane fraction and purified P44. J. Bacteriol. 1891998-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ijdo, J. W., W. Sun, Y. Zhang, L. A. Magnarelli, and E. Fikrig. 1998. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect. Immun. 663264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijdo, J. W., C. Wu, S. R. Telford III, and E. Fikrig. 2002. Differential expression of the p44 gene family in the agent of human granulocytic ehrlichiosis. Infect. Immun. 705295-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jauron, S. D., C. M. Nelson, V. Fingerle, M. D. Ravyn, J. L. Goodman, R. C. Johnson, R. Lobentanzer, B. Wilske, and U. G. Munderloh. 2001. Host cell-specific expression of a p44 epitope by the human granulocytic ehrlichiosis agent. J. Infect. Dis. 1841445-1450. [DOI] [PubMed] [Google Scholar]
- 22.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15153-164. [DOI] [PubMed] [Google Scholar]
- 23.Li, S., B. Liu, R. Zeng, Y. Cai, and Y. Li. 2006. Predicting O-glycosylation sites in mammalian proteins by using SVMs. Comput. Biol. Chem. 30203-208. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Q., and Y. Rikihisa. 2005. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect. Immun. 735106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, Q., Y. Rikihisa, N. Ohashi, and N. Zhi. 2003. Mechanisms of variable p44 expression by Anaplasma phagocytophilum. Infect. Immun. 715650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Q., C. Zhang, and Y. Rikihisa. 2006. Analysis of involvement of the RecF pathway in p44 recombination in Anaplasma phagocytophilum and in Escherichia coli by using a plasmid carrying the p44 expression and p44 donor loci. Infect. Immun. 742052-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, Q., N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Aguero-Rosenfeld, J. Raffalli, G. P. Wormser, and Y. Rikihisa. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 402981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, J. W., C. K. Doyle, X. Zhang, A. M. Cardenas, V. L. Popov, K. A. Nethery, and M. E. Woods. 2007. Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 7574-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride, J. W., X. J. Yu, and D. H. Walker. 2000. Glycosylation of homologous immunodominant proteins of Ehrlichia chaffeensis and Ehrlichia canis. Infect. Immun. 6813-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEver, R. P. 2004. Interactions of selectins with PSGL-1 and other ligands. Ernst Schering Res. Found. Workshop 44137-147. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, C. I., J. R. Storey, J. Recchia, L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, J. S. Bakken, R. T. Coughlin, and G. A. Beltz. 1998. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect. Immun. 663711-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J., K. S. Choi, and J. S. Dumler. 2003. Major surface protein 2 of Anaplasma phagocytophilum facilitates adherence to granulocytes. Infect. Immun. 714018-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, J., K. J. Kim, D. J. Grab, and J. S. Dumler. 2003. Anaplasma phagocytophilum major surface protein-2 (Msp2) forms multimeric complexes in the bacterial membrane. FEMS Microbiol. Lett. 227243-247. [DOI] [PubMed] [Google Scholar]
- 34.Reneer, D. V., S. A. Kearns, T. Yago, J. Sims, R. D. Cummings, R. P. McEver, and J. A. Carlyon. 2006. Characterization of a sialic acid- and P-selectin glycoprotein ligand-1-independent adhesin activity in the granulocytotropic bacterium Anaplasma phagocytophilum. Cell. Microbiol. 81972-1984. [DOI] [PubMed] [Google Scholar]
- 35.Reneer, D. V., M. J. Troese, B. Huang, S. A. Kearns, and J. A. Carlyon. 2008. Anaplasma phagocytophilum PSGL-1-independent infection does not require Syk and leads to less efficient AnkA delivery. Cell. Microbiol. 101827-1838. [DOI] [PubMed] [Google Scholar]
- 36.Santoni, V., S. Kieffer, D. Desclaux, F. Masson, and T. Rabilloud. 2000. Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 213329-3344. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar, M., D. V. Reneer, and J. A. Carlyon. 2007. Sialyl-Lewis x-independent infection of human myeloid cells by Anaplasma phagocytophilum strains HZ and HGE1. Infect. Immun. 755720-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar, M., M. J. Troese, S. A. Kearns, T. Yang, D. V. Reneer, and J. A. Carlyon. 2008. Anaplasma phagocytophilum MSP2(P44)-18 predominates and is modified into multiple isoforms in human myeloid cells. Infect. Immun. 762090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11554-561. [DOI] [PubMed] [Google Scholar]
- 40.Scorpio, D. G., K. Caspersen, H. Ogata, J. Park, and J. S. Dumler. 2004. Restricted changes in major surface protein-2 (msp2) transcription after prolonged in vitro passage of Anaplasma phagocytophilum. BMC Microbiol. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singu, V., H. Liu, C. Cheng, and R. R. Ganta. 2005. Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect. Immun. 7379-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singu, V., L. Peddireddi, K. R. Sirigireddy, C. Cheng, U. Munderloh, and R. R. Ganta. 2006. Unique macrophage and tick cell-specific protein expression from the p28/p30-outer membrane protein multigene locus in Ehrlichia chaffeensis and Ehrlichia canis. Cell. Microbiol. 81475-1487. [DOI] [PubMed] [Google Scholar]
- 43.Wang, X., T. Kikuchi, and Y. Rikihisa. 2006. Two monoclonal antibodies with defined epitopes of P44 major surface proteins neutralize Anaplasma phagocytophilum by distinct mechanisms. Infect. Immun. 741873-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yago, T., A. Leppanen, J. A. Carlyon, M. Akkoyunlu, S. Karmakar, E. Fikrig, R. D. Cummings, and R. P. McEver. 2003. Structurally distinct requirements for binding of P-selectin glycoprotein ligand-1 and sialyl Lewis x to Anaplasma phagocytophilum and P-selectin. J. Biol. Chem. 27837987-37997. [DOI] [PubMed] [Google Scholar]
- 45.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 27417828-17836. [DOI] [PubMed] [Google Scholar]
- 46.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 701175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhi, N., Y. Rikihisa, H. Y. Kim, G. P. Wormser, and H. W. Horowitz. 1997. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J. Clin. Microbiol. 352606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]