Abstract
The host cell environment can alter bacterial pathogenicity. We employed a combination of cellular and molecular techniques to study the expression of Campylobacter jejuni polysaccharides cocultured with HCT-8 epithelial cells. After two passages, the amount of membrane-bound high-molecular-weight polysaccharide was considerably reduced. Microarray profiling confirmed significant downregulation of capsular polysaccharide (CPS) locus genes. Experiments using conditioned media showed that sugar depletion occurred only when the bacterial and epithelial cells were cocultured. CPS depletion occurred when C. jejuni organisms were exposed to conditioned media from a different C. jejuni strain but not when exposed to conditioned media from other bacterial species. Proteinase K or heat treatment of conditioned media under coculture conditions abrogated the effect on the sugars, as did formaldehyde fixation and cycloheximide treatment of host cells or chloramphenicol treatment of the bacteria. However, sugar depletion was not affected in flagellar export (fliQ) and quorum-sensing (luxS) gene mutants. Passaged C. jejuni showed reduced invasiveness and increased serum sensitivity in vitro. C. jejuni alters its surface polysaccharides when cocultured with epithelial cells, suggesting the existence of a cross talk mechanism that modulates CPS expression during infection.
The importance of the host cell environment in bacterial pathogenicity is an emerging paradigm in the study of bacterial infection. For example, human colonization creates a hyperinfectious state in Vibrio cholerae that appears to contribute to the epidemic spread of this intestinal pathogen (29). Earlier work with Campylobacter jejuni demonstrated that coculture conditions altered the protein synthesis and virulence of this organism (23). Stintzi et al. demonstrated major effects in gene expression among C. jejuni organisms exposed to pathogenic conditions (36). However, how the host cell environment alters Campylobacter pathogenesis remains largely unknown. C. jejuni is now known to produce capsular polysaccharide (CPS) (4, 21, 31). Although relatively little is known about the contribution of CPS to the pathogenicity of the organism during bacterial interaction with mucosal surfaces, existing data suggest that CPS in general plays a complex and dynamic role in the infection process (2, 33).
In a study that was carried out before CPS was identified in C. jejuni, Babakhani and Joens reported that C. jejuni cocultured with primary swine intestinal cells showed increased mucoidy when regrown on solid medium (1). In addition, passaged C. jejuni showed enhanced invasiveness in vitro. We hypothesized that the change in mucoidy observed in these experiments reflected altered CPS expression by bacteria exposed to pathogenic conditions in vitro. We further speculated that altered CPS under these conditions might have relevance to pathogenic behavior among these organisms during infection. In this study, we show that coculture of C. jejuni with human intestinal epithelial cells causes a reproducible reduction in CPS staining. The presence of viable host cells and active protein synthesis by both bacteria and host cells is necessary for the effect on CPS. The alteration in CPS appears to be dependent on a soluble factor that is both heat labile and proteinase K sensitive. Following serial passage with HCT-8 cells, C. jejuni also showed other phenotypic changes, including reduced internalization into HCT8 epithelial cells and increased serum sensitivity.
MATERIALS AND METHODS
Bacterial strains, host cell lines, and culture conditions.
The C. jejuni isolates used in these experiments were C. jejuni 81-176; a CPS-deficient 81-176 kpsM mutant (2); NCTC 11168H, which is a hypermotile derivative of strain NCTC 11168 (19); and isogenic mutants of the 11168H strain deficient in production of the flagellar fliQ protein and the quorum-sensing molecule luxS. The Campylobacter upsaliensis type strain (ATCC 43594) was obtained from the American Type Culture Collection (ATCC). All isolates were grown on Mueller-Hinton (MH) agar or in MH broth (Oxoid, United Kingdom) at 37°C for 48 h under microaerobic conditions (5% CO2, 5% O2, 90% N2). Escherichia coli DH5α and Helicobacter pylori PU4 were also used in these experiments. The E. coli strain was grown in Luria-Bertani medium at 37°C. The H. pylori PU4 strain was grown on Columbia agar supplemented with 7% (vol/vol) defibrinated horse blood. HCT-8 cells (human adenocarcinoma ileocecal cell line) were purchased from the ATCC and maintained at 37°C in 5% CO2 in RPMI 1640 medium (Bio Whittaker) containing 10% fetal bovine serum (FBS), 1% nonessential amino acids, and 2 mM l-glutamine.
Construction of fliQ and luxS mutants.
The 11168H fliQ (Cj1675) mutant was generated by insertional mutagenesis. Plasmid cam171d11 from the C. jejuni NCTC 11168 sequencing library (20, 31) was digested with the StyI restriction enzyme, blunt ended using T4 DNA polymerase, and ligated with a blunt-ended BamHI fragment containing a tagged Kanr cassette derivative (A. V. Karlyshev, unpublished). The ligation product was transformed into E. coli XL2 Blue MRF′ (Stratagene), and a recombinant plasmid with the correct orientation of the Kanr cassette was selected using restriction analysis and transformed into C. jejuni by electroporation (39) The mutants were confirmed by PCR using Kanr cassette-derived primers combined with appropriate fliQ-specific primers.
A null (deletion) luxS (Cj1198) mutant of strain C. jejuni 11168H was constructed using an inverse PCR procedure (40). luxS gene-containing plasmid cam156h10, from the C. jejuni NCTC 11168 sequencing library (31), was used as a template in inverse PCR with primers Cj1198F (TAAAAagatctCTCGAGAATGCTTAAAAAGAATCTT) and Cj1198R (TAAAagatctGCTGTCTAATAATGGCATTTTAACTC), containing a BglII restriction site (lowercase letters). The 3.5-kb PCR product was linearized with BglII, ligated with a BamHI fragment containing a Kanr cassette from plasmid pJMK30 (38), and transformed into E. coli XL2 Blue MRF′. The recombinant plasmid containing a direct orientation of the Kanr cassette was selected by restriction analysis, confirmed by sequencing, and transformed into C. jejuni 11168H. Allelic replacement leading to almost complete (94.5%) elimination of the luxS gene was confirmed by PCR using primers corresponding to the flanking genes Cj1197 and Cj1198. Both mutants were characterized using a plate motility assay. Briefly, 5 microliters of culture adjusted to an optical density at 600 nm (OD600) of 6 were inoculated into the centers of the plates, and the diameters of the resulting swarms were measured the next day. As predicted (10), both mutants showed reduced motility compared with the wild type, with the fliQ mutant substantially more attenuated (P < 0.0003 for comparison with the wild type) than the luxS mutant (P < 0.001 for comparison with the wild type).
Coculturing of C. jejuni with HCT-8 cells.
HCT-8 cells were seeded at a density of 108 cells per 25-cm2 flask and allowed to grow overnight to 60% to 70% confluence. Agar-grown bacteria were used to prepare the inoculums for the coculture experiments. Bacteria were harvested from plates and resuspended in RPMI 1640 medium. One flask containing the HCT-8 cells was inoculated with 1 ml of bacterial suspension having a bacterial density (OD600) of 0.4. Bacteria and HCT-8 cells were cocultured at 37°C under microaerobic conditions (5% O2, 5% CO2, and 90% N2) for 18 h. The HCT-8 cell culture medium containing bacteria was then removed from the flask, and bacteria (passage 1 bacteria) were pelleted by centrifugation (3,300 × g for 5 min). An aliquot of these passage 1 bacteria was resuspended in 1 ml of RPMI 1640 medium such that the OD600 was 0.4, and this was used to inoculate a fresh flask of HCT-8 cells. The remaining bacteria were used for CPS isolation (see below). The above-outlined procedure was repeated twice so that passage 2 and passage 3 bacteria were collected. Viable counts were determined for inocula at each passage (both with and without host cells) to ensure that similar numbers of live bacteria were present at each stage of coculture and for host cell-passaged organisms and controls. The above-outlined protocol was also used for coculturing of H. pylori PU4 with HCT-8 cells. For E. coli DH5α, bacteria were cocultured with the host cells for shorter periods (8 h) in order to avoid a deleterious effect on the host cells.
Inhibitor studies.
A number of different inhibitors of host cells and bacterial activity were tested for their effects on C. jejuni CPS production during coculture with HCT-8 cells. The effect of formaldehyde fixation was also tested. For formaldehyde fixation experiments, the culture medium was removed from the HCT-8 cells and the cell monolayers were washed three times with phosphate-buffered saline (PBS). Host cells were treated with 4% formaldehyde in PBS for 20 min. Monolayers were washed with PBS, and fresh tissue culture medium containing 10% FBS was added. The flasks were then inoculated with bacteria and passaged as described above. To test the effect of inhibiting host cell protein synthesis, HCT-8 cells were incubated with 10 μg ml−1 cycloheximide for 45 min prior to addition of the bacteria. Culture medium containing cycloheximide was removed, and the HCT-8 cells were washed before addition of fresh culture medium containing bacteria. In a separate experiment, the bacterial protein synthesis inhibitor chloramphenicol (100 μg ml−1) was maintained in the culture medium during coculture of bacteria and HCT-8 cells.
CPS production by C. jejuni grown in conditioned medium from HCT-8 cells.
Conditioned medium was collected from HCT-8 cells cocultured with C. jejuni 81-176 for up to two passages. The medium was centrifuged (3,951 × g for 15 min) to remove the bacteria, and the supernatant was filtered using a 0.2-μm filter (Sartorius) and stored at −20°C until use. In parallel experiments, conditioned medium from HCT-8 cells cultured without bacteria or with C. jejuni 81-176 grown in tissue culture medium (containing serum) alone was also collected. Bacteria were passaged in the differently conditioned media for up to two passages, and CPS production by the bacteria was assessed as described above. The effect of coculture of bacteria with proteinase K-treated (200 μg ml−1 for 1 h at 50°C) or heat-treated (100°C for 2 h) conditioned medium on CPS production was also tested.
CPS preparation.
CPS was prepared from bacteria by using a method described by Hitchcock (17). Bacteria were harvested by centrifugation and resuspended in 100 μl of lysis buffer containing 31.25 mM Tris-HCl (pH 6.8), 4% sodium dodecyl sulfate, 0.025% bromophenol blue, and 20% glycerol. After heating to 100°C for 5 min, 20 μl of the sample was transferred into a fresh tube, 1 μl of 20 mg ml−1 proteinase K was added to the solution, and the tubes were incubated for 1 h at 50°C. The samples were fractionated on NuPage Novex 10% bis-Tris gels (Invitrogen, United Kingdom). Following electrophoresis, gels were stained with Alcian blue (0.1% Alcian blue in 40% ethanol, 5% acetic acid) (21).
Following coculture, membrane-bound CPS was quantified using the Stains-All assay (Sigma). For this assay, 1 ml of passaged bacteria with an OD600 of approximately 0.4 was pelleted by centrifugation. Bacteria were washed twice with PBS and resuspended in 0.5 ml of water. The amount of polysaccharide in 0.5 ml of supernatant was determined by measuring the absorbance at 640 nm after the addition of 2 ml of a solution containing 20 mg of Stains-All and 60 μl of acetic acid in 100 ml of formamide. The final values were obtained by subtraction of values measured for culture medium or water alone. The experiments were done in triplicate. Additionally, the presence of detached CPS present in the culture supernatant was sought using a method described by Fournier et al. (13). Culture supernatants were autoclaved at 120°C for 1 h and examined visually for the presence of a white layer typical of polysaccharides.
Examination of coculture supernatants for released CPS and proteolytic activity.
Protease activity in the coculture supernatants was tested using a protease fluorescence detection kit (Sigma). In brief, the assay was performed with microcentrifuge tubes, and fluorescence in multiwell plates was read. Twenty microliters of incubation buffer, 20 μl of fluorescein isothiocyanate-casein substrate, and 10 μl of a test sample were added to each microcentrifuge tube. For controls, the test sample was replaced with trypsin and the blank sample with ultrapure water. Each tube was incubated at 37°C overnight in the dark. For fluorescence measurements, black 96-well plates were used. After incubation, 10 μl of a test sample was mixed with 1 ml of assay buffer. Two hundred microliters of the mixture was added to each of the 96 wells, and fluorescence was recorded with excitation at 485 nm and emission at 535 nm.
Virulence assay.
The gentamicin protection assay (9) was used to test the efficiency with which cocultured organisms entered and adhered to host epithelial cells. Briefly, HCT-8 cells were grown for 15 to 18 h in six-well tissue culture plates at a concentration of 1 × 105 cells per well. Passaged bacteria were washed and resuspended in tissue culture medium at an OD600 of 0.4. The HCT-8 cells were washed with PBS, and 2 ml of fresh culture medium was added to each well. Bacteria were added to give a multiplicity of infection of 1,000. Tissue culture plates were centrifuged at 250 × g for 5 min and incubated for 3 h at 37°C in 10% CO2. To quantify the number of cell-associated bacteria, infected monolayers were washed several times with PBS and treated with 0.1% Triton X-100 in PBS. Tenfold dilutions of each well were plated onto the appropriate agar and colonies enumerated after 3 days of incubation.
To quantify the number of bacteria that invaded HCT-8 cells, the infected monolayers were washed with PBS, tissue culture medium (2 ml) containing gentamicin (400 μg ml−1) was added to half the wells, and medium with no antibiotic was added to the remaining wells. The tissue culture plates were then incubated for a further 2 h at 37°C and washed with PBS. HCT-8 cells were lysed by the addition to each well of 100 μl of 0.1% Triton X-100 in PBS and incubated for 10 to 15 min at 37°C. Tenfold dilutions of the contents of each well were plated onto the appropriate agar and colonies enumerated after 3 days of incubation. Invasion efficiency was calculated as the average of the total number of CFU/total initial inoculum. C. jejuni 81-176 passaged in RPMI 1640 (without cells) was also tested for the ability to adhere to and invade HCT-8 cells. All assays were conducted in triplicate and repeated independently three times. The significance of differences in adhesion and invasion between samples was determined using the Student t test. A P value of <0.05 was defined as significant. Triton X-100 did not affect bacterial viability in either passaged or unpassaged cultures (data not shown).
Serum sensitivity assay.
The sensitivity of passaged bacteria to human serum was measured, as previously described (3). Cocultured and plate-grown bacteria were resuspended in fresh tissue culture medium to give an OD600 of 0.1. Five microliters of bacterial suspension was added to duplicate wells of a six-well plate containing 800 μl of Mueller-Hinton broth and 200 μl of active pooled human serum or 1 ml of Mueller-Hinton broth and heat-inactivated human serum. The plates were incubated for 1 h at 37°C under microaerobic conditions. Bacteria from each well were diluted 10-fold and plated on dry Mueller-Hinton agar plates. The plates were incubated at 37°C under microaerobic conditions. Colony counts were performed after an incubation time of 2 days. All assays were conducted in triplicate and repeated independently three times.
Microarray studies: experimental design, template labeling, and microarray hybridizations.
Gene expression profiling of C. jejuni 11168H cocultured with human intestinal epithelial cells was performed using an indirect comparison method or a type 2 experimental design (16, 41). In this experimental design, replicate test sets of Cy5-labeled C. jejuni 11168H total RNA samples were combined with a common reference sample (Cy3-labeled C. jejuni NCTC 11168 genomic DNA) as described in previous studies (11, 27). C. jejuni 11168H incubated in tissue culture medium alone was used as a comparator for analysis of coculture gene expression profiles. The microarrays used in this study were whole-genome C. jejuni NCTC 11168 arrays printed on Ultragaps glass slides (Corning), constructed by the BμG@S group (http://www.bugs.sghms.ac.uk/).
The procedures for Cy5 labeling of total RNA samples and Cy3 labeling of NCTC 11168 genomic DNA (8) were as described previously. All hybridizations were performed as described previously (8), with the following modifications. For probe hybridization, Cy5-labeled probes of C. jejuni 11168H total RNA (test) and Cy3-labeled universal reference samples of C. jejuni NCTC 11168 DNA (control) were combined and purified using a MinElute PCR purification kit (Qiagen). The final elution was then made up to a final volume of 50 μl with a final concentration of 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.3% (wt/vol) sodium dodecyl sulfate. The hybridization mixture was denatured at 98°C for 2 min and cooled slowly to room temperature. A 22- by 25-mm LifterSlip coverslip (Erie Scientific) was placed over the reporter element area on the microarray and the hybridization mixture applied underneath the coverslip. The microarray slide was placed in a humidified hybridization cassette (Telechem International) and incubated in a water bath for 18 h at 65°C without shaking.
Data acquisition and microarray data analysis.
The microarray slides were scanned with an Affymetrix 418 array scanner (MWG Biotech) according to the manufacturer's guidelines. Signal and local background intensity readings for each spot were quantified using BlueFuse software, version 3.1 (BlueGnome, Cambridge, United Kingdom). Quantified data were analyzed using GeneSpring GX software, version 7.2 (Agilent, Santa Clara, CA). Expression analysis was performed using a DNA-versus-RNA experimental setup. Statistically significantly up- and downregulated genes were selected when gene expression levels in bacteria isolated at passages 1 and 2 were compared with those in bacteria incubated in tissue culture medium alone by using analysis of variance (ANOVA). ANOVA was performed using a false discovery rate as the multiple-testing-correction method within the GeneSpring software program. Specifically, changes in the expression levels of genes involved in biosynthesis of the C. jejuni CPS were investigated (Cj1413c to Cj1448c).
Microarray data accession numbers.
The microarray design is available in BUG@Sbase (accession no. A-BUGS-9) (http://bugs.sgul.ac.uk/A-BUGS-9) and also ArrayExpress (accession no. A-BUGS-9). Fully annotated microarray data have been deposited in BUG@Sbase (accession no. E-BUGS-74) (http://bugs.sgul.ac.uk/E-BUGS-74) and also ArrayExpress (accession no. E-BUGS-74).
RESULTS
Campylobacter CPS staining is lost following serial passage with HCT-8 cells.
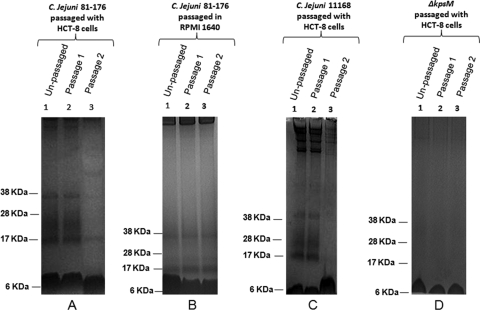
In order to explore the possibility that Campylobacter CPS changes when grown in the proximity of host cells, we isolated and stained C. jejuni 81-176 and C. jejuni 11168 CPS following coculture with HCT-8 cells. Bacteria passaged without host cells served as controls. Coculture of bacteria with HCT-8 cells resulted in dramatic loss of stainable CPS at the second passage for both strains (Fig. 1A and C). Passage of bacteria in RPMI under identical conditions without the presence of HCT-8 cells showed no decrease in CPS (Fig. 1B). In some experiments, passage 2 was associated also with a different staining pattern, with the appearance of a novel smeared band above the lipooligosaccharide region. CPS staining was also lost when other campylobacters, including C. jejuni 11168 (Fig. 1C), C. jejuni 81116, and C. upsaliensis ATCC 43954, were passaged with host cells (data not shown). These changes in polysaccharide staining were not evident when a capsule-deficient mutant (kpsM) was passaged with host cells (Fig. 1D). There was no loss of bacterial viability following coculture. When cocultured, CPS-depleted organisms were regrown on solid medium, CPS production recovered, and colonies showed enhanced mucoidy in comparison with unpassaged, plate-grown organisms.
FIG. 1.
Alcian blue-stained CPS of C. jejuni in 10% bis-Tris gels following coculture with HCT-8 cells. (A) CPS profiles from C. jejuni 81-176 following coculture with HCT-8 cells. Lane 1, unpassaged bacteria; lane 2, passage 1; lane 3, passage 2. (B) CPS isolated from C. jejuni 81-176 cocultured in RPMI medium. Lane 1, unpassaged bacteria; lane 2, passage 1; lane 3, passage 2. (C) CPS isolated from C. jejuni 11168 following coculture with HCT-8 cells. Lane 1, unpassaged bacteria, lane 2, passage 1; lane 3, passage 2. (D) Bacterial cell surface carbohydrates of capsule-deficient C. jejuni 81-176 (kpsM) following coculture with HCT-8 cells. Lane 1, unpassaged bacteria; lane 2, passage 1; lane 3, passage 2.
To ensure that the capsule structures were not being cleaved and released from organisms, we examined supernatants for the presence of CPS. Supernatants were (i) directly examined for a visible polysaccharide layer following autoclaving, (ii) run on 10% bis-Tris gels and stained using Alcian blue, and (iii) stained with Sigma Stains-All, which detects polysaccharide structures. No CPS was detected by these methods. In addition, coculture supernatants were tested for the presence of host cell-derived proteases, which could have cleaved off the CPS structures. When a protease fluorescence detection kit (Sigma) was used, no proteolytic activity was detected in coculture fluid (data not shown).
Gene expression changes in the capsule biosynthesis locus following coincubation with human intestinal epithelial cells.
Initially, microarray analysis was performed with the C. jejuni 81-176 strain. While a trend toward downregulation in expression of many of the genes in the capsule biosynthetic locus was observed during coculture, the genetic difference between the capsule biosynthetic loci in 81-176 and NCTC 11168 (represented on the microarray) made accurate interpretation of the data unfeasible. Therefore, the microarray analysis was repeated using the 11168H strain, which has been shown to be indistinguishable from NCTC 11168 by microarray analysis (19).
The expression levels of genes within the C. jejuni 11168H capsule biosynthesis locus (Cj1413c to Cj1448c) at passage 1 and passage 2 were compared with those for bacteria incubated in tissue culture medium (with FBS) alone. Analysis of the microarray data showed marked downregulation of many of the genes in the capsule biosynthesis locus at passage 1 and further downregulation at passage 2. This provides additional evidence that the loss of stainable CPS observed is due to a bacterial response to the coculture with HCT-8 cells. ANOVA of gene expression changes at passage 2 revealed seven of the capsule biosynthesis locus genes to be significantly downregulated (Table 1). Interestingly, the products of four of these genes (KpsD, KpsE, KpsF, and KpsS) are involved in the translocation of the CPS from the cytoplasm to the bacterial surface.
TABLE 1.
C. jejuni 11168H genes from the capsule biosynthesis locus (Cj1413c to Cj1448c) that are downregulated after passage 2 of coculture with HCT-8 human intestinal epithelial cells, compared to the level for passage in culture medium alonea
| Gene | Expression (fold) | Pb | Common name | Confirmed or predicted function |
|---|---|---|---|---|
| Cj1413c | 0.57 | 0.023 | kpsS | Capsule polysaccharide modification protein |
| Cj1419c | 0.47 | 0.026 | Putative methyltransferase | |
| Cj1424c | 0.68 | 0.013 | gmhA2 | Phosphoheptose isomerase |
| Cj1436c | 0.26 | 0.030 | Aminotransferase | |
| Cj1443c | 0.70 | 0.017 | kpsF | d-Arabinose 5-phosphate isomerase |
| Cj1444c | 0.79 | 0.044 | kpsD | Capsule polysaccharide export system periplasmic protein |
| Cj1445c | 0.55 | 0.012 | kpsE | Capsule polysaccharide export system inner membrane protein |
The order of the genes listed is based on capsule locus position.
Statistical verification of the data was performed with a parametric statistical test to adjust the individual P values, using the Benjamini-Hochberg false-discovery-rate multiple-testing-correction method.
Effect of conditioned medium on C. jejuni 81-176 CPS.
To further explore the origin of the effect on CPS profile under coculture conditions, the effect of conditioned medium on fresh bacteria was examined (Fig. 2). The effect on unpassaged C. jejuni 81-176 of conditioned medium taken from HCT-8 cells alone or bacteria alone was compared with that of conditioned medium under coculture conditions (following both passage 1 and passage 2). Bacteria grown in conditioned medium taken from coculture conditions after passage 1 (Fig. 2, lane 4) showed no loss of CPS. However, in keeping with the coculture experiments outlined above, for bacteria grown in conditioned medium taken from coculture conditions after passage 2 (Fig. 2, lane 5), no CPS could be detected by staining with Alcian blue (the appearance of a novel smeared band above the lipooligosaccharide region, seen in some experiments, was apparent in this gel). In contrast, there was no change in CPS profile for bacteria grown in conditioned medium taken from HCT-8 cells alone or in medium conditioned by bacteria alone (Fig. 2, lanes 2 and 3). These results suggested that a soluble factor present in the conditioned medium from HCT-8 cells cocultured with bacteria was responsible for CPS alteration.
FIG. 2.
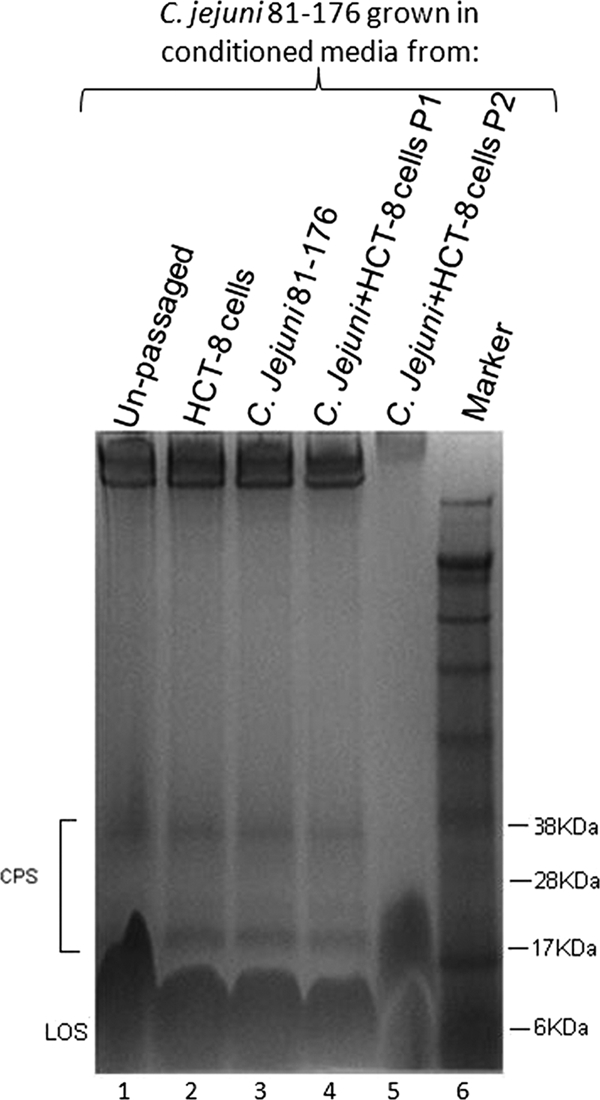
Alcian blue-stained CPS of C. jejuni 81-176 in 10% bis-Tris gels following growth in conditioned medium. CPS extracted from bacteria grown in medium taken from HCT-8 cells (lane 2), C. jejuni cultures (lane 3), or coculture conditions at passage 1 (lane 4) show unaltered profiles in comparison with CPS from organisms grown in medium from coculture conditions after two passages (lane 5). Lane 1 shows CPS taken from unpassaged, Mueller-Hinton-grown organisms, and lane 6 is a size marker. LOS, lipooligosaccharide.
To explore the nature of the signaling that might be involved in CPS alteration, the conditioned medium from passage 2 was exposed to proteinase K treatment or heat (Fig. 3). In contrast to those grown in untreated conditioned medium (Fig. 3, lane 4), the bacteria grown in conditioned medium that had been treated with proteinase K (lane 2) and in conditioned medium exposed to heat (100°C) (lane 3) showed only partial decreases in CPS staining. The conditioned media used for these experiments originated from passage 2 in an experiment where C. jejuni 81-176 was cocultured with the epithelial cell line HCT-8.
FIG. 3.
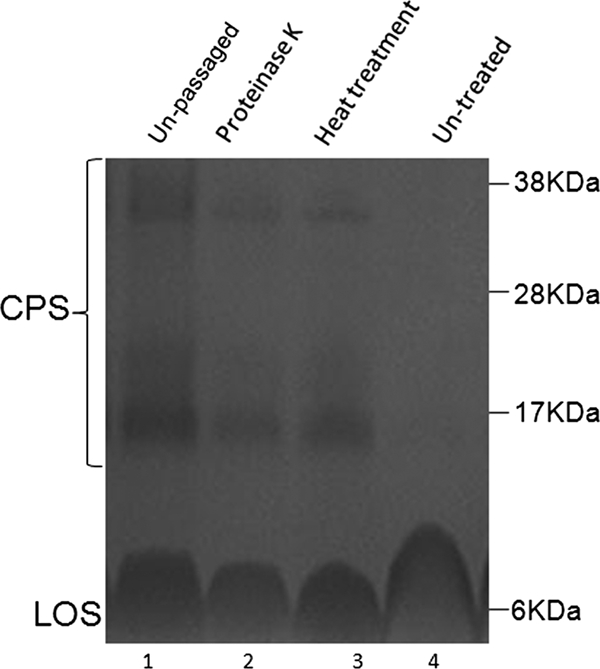
Alcian blue-stained CPS of C. jejuni 81-176 in 10% bis-Tris gels grown in conditioned medium exposed to heat or proteinase K. Shown is the CPS profile for C. jejuni 81-176 grown in proteinase K-treated (lane 2), heat-treated (lane 3), or untreated (lane 4) conditioned medium from passage 2 cocultures. Lane 1 shows the CPS profile for unpassaged bacteria. LOS, lipooligosaccharide.
To examine if the effect of conditioned medium was strain or species specific, we exposed C. jejuni 81-176 to conditioned media from C. jejuni 11168, a C. jejuni KpsM− mutant, H. pylori PU4, and E. coli DH5α. Whereas conditioned media from C. jejuni 11168 and the isogenic KpsM− strain resulted in CPS loss in exposed C. jejuni 81-176 (data not shown), no effect was observed on CPS when these organisms were exposed to conditioned medium from H. pylori or E. coli. A comparison of the effects of H. pylori PU4- and C. jejuni 81-176-conditioned media is illustrated in Fig. 4. This suggests that the effect on CPS is specific to C. jejuni and, because conditioned medium using a C. jejuni strain without capsule (KpsM−) reproduced the effect, is not dependent on signaling from capsule (i.e., to host cells).
FIG. 4.
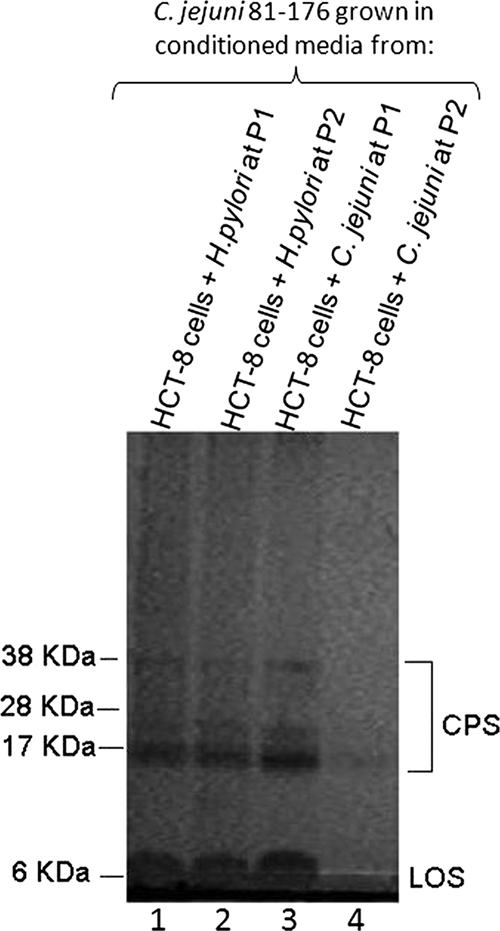
Alcian blue-stained CPS of C. jejuni 81-176 in 10% bis-Tris gels following growth in conditioned medium from HCT-8 cells cocultured with H. pylori PU4. The CPS profile for C. jejuni 81-176 following growth in conditioned medium from HCT-8 cells cocultured with H. pylori PU4 (lane 1, passage 1; lane 2, passage 2) is compared with the CPS profile for the organism following growth in conditioned medium taken from HCT-8 cells cocultured with C. jejuni 81-176 (lane 3, passage 1; lane 4, passage 2). LOS, lipooligosaccharide.
Epithelial cells actively participate in the alteration of bacterial CPS.
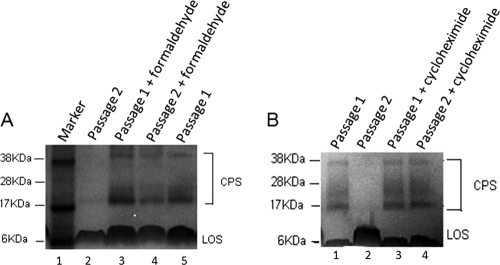
To examine the contribution of epithelial cell viability and protein synthesis to CPS alteration, we examined the effect on CPS of passaging with nonviable (fixed) host cells (Fig. 5A) and host cells treated with cycloheximide to inhibit protein synthesis (Fig. 5B). Fixation of HCT-8 cells and inhibition of protein synthesis both abrogated the effect of coculture conditions on CPS. These data suggest that the HCT-8 cells were actively participating in signaling to the organisms as part of the process of CPS alteration. An alternative explanation for these findings, following fixation, is that the observed effect is mediated through host cell surface receptors that were denatured by formaldehyde treatment.
FIG. 5.
Alcian blue-stained CPS of C. jejuni 81-176 in 10% bis-Tris gels cocultured with formaldehyde-fixed (A) and cycloheximide-treated (B) HCT-8 cells. (A) C. jejuni 81-176 CPS from conventional coculture conditions (lane 2) is compared with CPS extracted from passage 1 (lane 3) and passage 2 (lane 4) organisms cocultured with formaldehyde-fixed HCT-8 cells. Lane 5 represents CPS from passage 1 organisms under conventional coculture conditions. Lane 1 represents the standard protein marker. LOS, lipooligosaccharide. (B) CPS extracted from C. jejuni 81-176 grown with HCT-8 cells treated with cycloheximide to block host cell protein synthesis (lane 3, passage 1; lane 4, passage 2) is compared with conventional coculture conditions without cycloheximide treatment (lane 1, passage 1; lane 2, passage 2).
Bacterial protein expression, but not flagellar protein export or luxS-based quorum sensing mechanisms, is involved in CPS alteration.
To further explore the nature of the apparent communication between host cells and bacteria that underlies the alteration in CPS following coculture, we examined the roles of luxS (putative quorum-sensing process), flagellar apparatus protein secretion, and bacterial protein synthesis in the observed effects on CPS staining. These mutations were previously constructed in strain NCTC 11168H, and therefore, the parental strain, NCTC 11168H, was used as a control in these experiments. Chloramphenicol treatment either partially (as shown in Fig. 6) or completely (in other experiments) abrogated the effect of coculture on CPS depletion. Therefore, bacterial protein synthesis plays a role in the effect on CPS (suggesting that bacteria actively participate in the process). However, neither a luxS (quorum-sensing) mutant nor a fliQ (flagellar secretion) mutant was deficient in altering CPS during passage (Fig. 6). This suggests that bacteria were using mechanisms other than flagellar-apparatus-based protein secretion or luxS-based quorum sensing to mediate the effect on CPS.
FIG. 6.
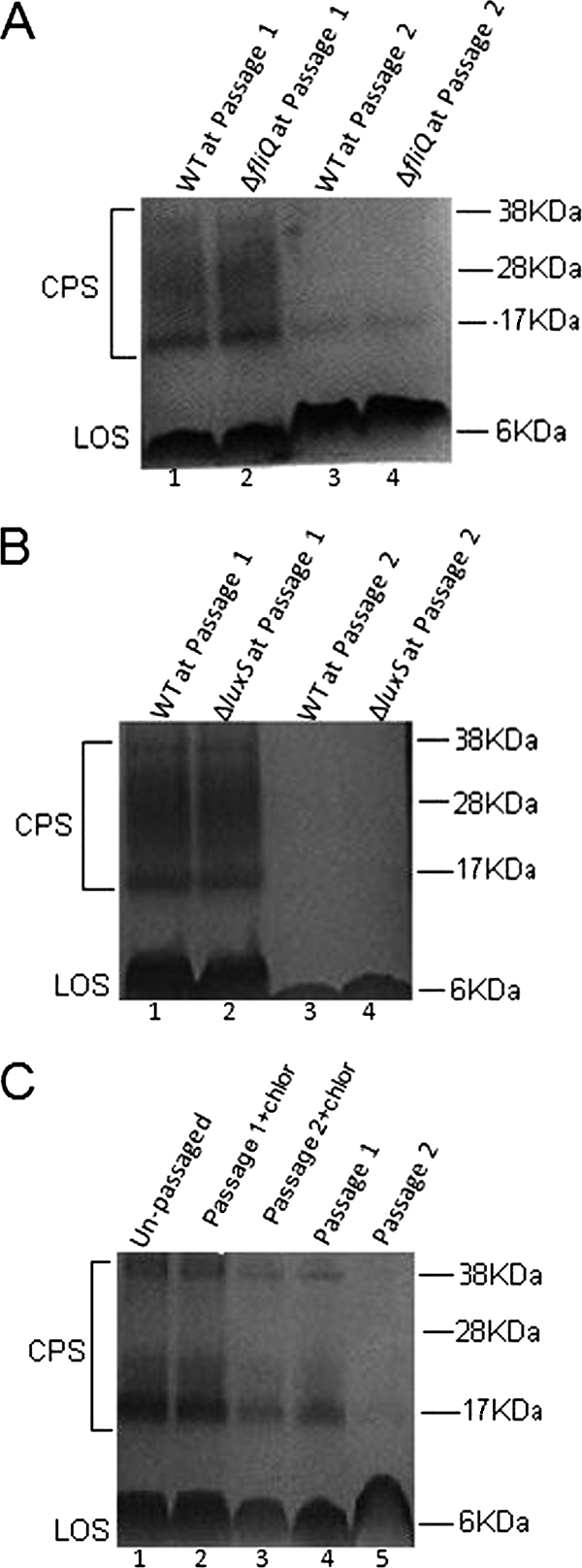
Role of flagellar apparatus protein secretion (A), luxS-based quorum sensing (B), and bacterial protein synthesis (C) on the observed effects on CPS stained with Alcian blue in 10% bis-Tris gels. (A) Unpassaged, Mueller-Hinton-grown wild-type (WT) C. jejuni 11168H (lane 1) and an isogenic fliQ mutant (lane 2) show similar CPS profiles. The parental strain (lane 3) and fliQ mutant (lane 4) both lose CPS staining following two passages with HCT-8 cells. LOS, lipooligosaccharide. (B) Unpassaged, Mueller-Hinton-grown wild-type C. jejuni 11168H (lane 1) and an isogenic luxS mutant (lane 2) show similar CPS profiles. The parental strain (lane 3) and the luxS mutant (lane 4) both lose CPS staining following two passages with HCT-8 cells. (C) Chloramphenicol-treated bacteria showed only minor reduction in CPS (lane 2, passage 1; lane 3, passage 2), compared with untreated bacteria (lane 4, passage 1; lane 5, passage 2). Lane 1 shows control CPS from unpassaged, Mueller-Hinton-grown organisms.
Coculture of C. jejuni with epithelial cells affects the pathogenicity and serum sensitivity of passaged bacteria.
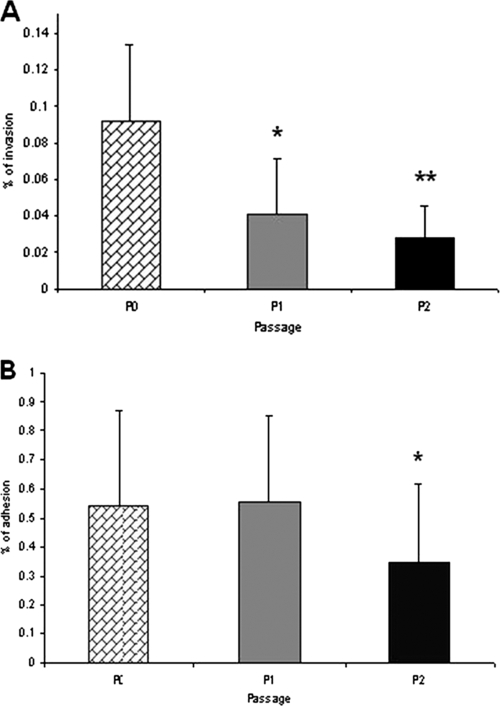
To investigate the possibility that coculture conditions resulting in altered CPS also induced other phenotypic changes in C. jejuni, we compared the adhesion and invasion (Fig. 7) and serum sensitivity (Fig. 8) of passaged bacteria with those of unpassaged bacteria. By the gentamicin protection assay, passage 1 bacteria manifested a reduction in invasion (P = 0.017), with a further reduction after passage 2 (P = 0.0016). Adhesion (Fig. 7B) was unaffected after passage 1. There was a reproducible, statistically significant reduction in numbers of bacteria attaching to host cells after passage 2. Passage in RPMI alone did not alter adhesion and invasion (data not shown).
FIG. 7.
Adhesion and invasion of C. jejuni following coculture. (A) Invasion of cocultured C. jejuni 81-176 following gentamicin protection. Statistical significance (Student's t test) relative to the level for unpassaged bacteria is indicated. *, P = 0.017; **, P = 0.0016. (B) Adhesion of cocultured C. jejuni 81-176 to HCT-8 cells. *, P = 0.17. The experiments were conducted on three separate occasions. Results for a representative experiment are presented. The error bars represent standard deviations for eight separate wells. P0, unpassaged bacteria; P1, passage 1; P2, passage 2.
FIG. 8.
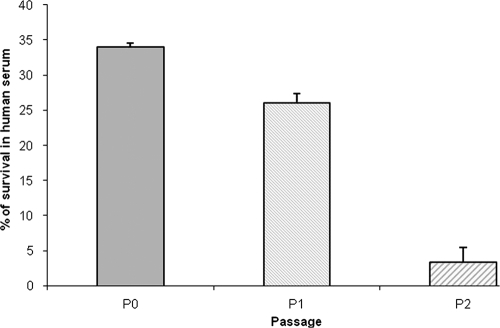
Effect of coculture on serum sensitivity of C. jejuni. The survival rate is defined as the number of C. jejuni colonies isolated following exposure to human serum divided by the number of colonies surviving in heat-inactivated serum, expressed as a percentage. The experiment was performed in triplicate. The error bars represent standard deviations. Statistical significance was assessed with Student's t test. *, P values of <0.004 for P2 versus P0. P0, agar-grown bacteria; P1, passage 1; P2, passage 2.
As encapsulation in C. jejuni is associated with resistance to complement-mediated killing and serum resistance (2, 15), we compared the sensitivities of passaged and unpassaged bacteria to human serum. Plate-grown bacteria (unpassaged) had an average rate of survival of 34%, compared to 26% survival for passage 1 bacteria. However, there was a dramatic decrease (to 3.3%) in the rate of survival of passage 2 C. jejuni in human serum (P < 0.004). These results demonstrate that following coculture with epithelial cells, C. jejuni has a decreased serum survival rate, with possible consequences for its pathogenicity.
DISCUSSION
It is known that the host environment can alter bacterial pathogenicity, and at least one other bacterial pathogen, Enterococcus faecalis, appears to actively probe the environment for target host cells (5). Although C. jejuni has been shown to alter protein synthesis under coculture conditions (23, 24), little is known of how this pathogen responds to the host environment. Herein, we report reduction in surface polysaccharides and downregulation of CPS-related genes in C. jejuni grown in the presence of epithelial cells. The alteration in surface polysaccharides requires the presence of both viable bacteria and host cells. These data suggest that some form of cross talk between C. jejuni and host epithelial cells that results in CPS depletion can occur under pathogenic conditions.
In light of earlier work suggesting increased mucoidy (possibly indicative of increased polysaccharide production) following passage with host cells (1), we were surprised to find a reduction in stainable capsule following passage in liquid culture. However, when we subsequently recultured these passaged organisms on solid medium, there was a marked increase in mucoidy, associated with strong Alcian blue staining of CPS (data not shown). These observations are in keeping with those previously reported (1), where increased mucoidy of passaged organisms was observed following growth on agar plates. The mechanisms and the potential relevance of this rebound in CPS staining remain to be explored. The other difference in our data, compared with those reported by Babakhani and Joens, relates to the change in the pathogenicity of passaged organisms. However, we suspect that the increased pathogenicity of multiply passaged organisms in this earlier study arose through repeated selection for an invasive organism phenotype that had directly infected host cells. In contrast, we did not select organisms that had invaded HCT-8 cells but examined adhesion and invasion of organisms in coculture supernatants.
C. jejuni has been shown to alter gene expression profoundly during colonization in vivo (36). Stintzi et al. have shown that C. jejuni alters stringent and heat shock responses during colonization in a rabbit ileal loop model. In addition, earlier work provided evidence that C. jejuni can respond to even subtler signals under pathogenic conditions in vitro. Konkel and Cieplak showed altered protein synthesis in C. jejuni grown under coculture conditions (23). Intriguingly, in light of our findings, these changes in protein expression occurred among bacteria in the supernatant that would not necessarily have directly interacted with the host cell model. In the present study, we show that CPS alteration and other phenotypic changes, including altered adhesion and invasion and reduced serum resistance, occur in C. jejuni following coculture with the host epithelium, even among organisms not adherent to or invading cells.
In other bacterial species, soluble factors secreted by bacteria also appear to be involved in host-pathogen communication. For example, Freitas et al. have shown that a heat-labile, low-molecular-weight compound from Bacteroides thetaiotaomicron modifies glycosylation of HT29-MTX cells (14). More recently (18), it has been shown that conditioned medium from enterohemorrhagic E. coli-infected tissue culture cells contains a soluble factor which suppresses host cell gamma interferon signaling. Indeed, it is well recognized that infection of eukaryotic cells by bacteria results in secretion of proteins of both bacterial (7) and host cell (6) origins.
The effect on CPS observed in this study was dependent on the presence of both bacteria and host cells. This observation suggests a complex interaction between C. jejuni and host cells. Recent elegant work with E. faecalis provides one model of how bacteria may actively probe the environment for target host cells (5). In the case of E. faecalis, cytolysin expression is regulated through a quorum-sensing autoinduction mechanism. Interestingly, quorum sensing appears to be important in regulation of expression of capsule in another human pathogen, Streptococcus pyogenes (28). LuxS subserves a quorum-sensing function in many prokaryotes. Its precise role in C. jejuni is uncertain. Recently, however, luxS was linked to control of biofilm formation in C. jejuni (32). A luxS C. jejuni mutant in our study did not differ from the wild type in CPS alteration following coculture with host cells. This suggests that luxS-based quorum sensing is not involved in the effect on CPS in C. jejuni that we have observed following culture with epithelial cells.
The alteration in CPS was abrogated in the setting of protein synthesis inhibition in bacteria (chloramphenicol treated) and host cells (cycloheximide treated) and following heat or proteinase K treatment of conditioned medium. These data suggest that protein production and secretion from both host cells and bacteria are required for the effect on CPS. The C. jejuni flagellar export apparatus is responsible for secretion of Campylobacter invasion antigens (Cia proteins) (24, 25). Mutation in the fliQ gene is predicted to disrupt the flagellar export apparatus. That the fliQ mutant did not differ from the wild-type strain in CPS depletion suggests that protein secretion from bacteria is occurring through a mechanism different from that reported for Cia proteins. Ongoing experiments are aimed at identifying the putative signaling molecule involved in the CPS alteration that we have observed.
The biological relevance of the alteration in CPS under coculture conditions in our experiments is not yet clear. In other bacterial species, the contribution of CPS to virulence is complex and variable, depending on the species and pathogenic model being used. For example, Klebsiella bacteria adhere better to mucus secreting host cells when the capsule is present but are impaired in adhesion to non-mucus-secreting host cells. (12). In E. coli and Klebsiella bacteria, capsule appears to act as a steric shield, which, when reduced or altered, helps expose adhesins or other virulence factors on the surfaces of bacteria (34, 35). In contrast, previous work with C. jejuni suggests that the presence of intact capsule promotes virulence. For example, in C. jejuni capsular mutants, pathogenicity is reduced in vitro and in a ferret model (2). In addition, a kpsM mutant is deficient in colonization of the avian gastrointestinal tract (19). In the present study, cocultured, capsule-deficient/altered bacteria showed significantly reduced invasion in HCT-8 cells. That the reduction in invasiveness was already seen at passage 1 suggests that coculture-induced mechanisms, in addition to the observed change in CPS, may underlie the alteration in adhesion and invasion that we observed.
That C. jejuni would alter its capsule and apparently become less invasive and more sensitive to environmental insult appears at first to be paradoxical. Despite our inability to show CPS release or proteolytic activity in medium, it is still possible that the effect on C. jejuni CPS is mediated by signaling from epithelial cells as a host-protective mechanism. However, we speculate that the change in capsule observed is a transient, niche-specific phenomenon and probably part of a complex and coordinated bacterial response to a pathogenic environment. For example, it could be in the bacterial population's interest to uncoat and sacrifice many of its number in order that a few get the opportunity to access the host cell environment (adhesion/invasion/translocation). Data from previous studies showing reduced pathogenicity for CPS mutants may be reflecting a disadvantage in permanent deletion of the capsule during the wider survival/colonization process. Similarly, the finding that invasiveness of passaged organisms is decreased using immortalized host cells that do not harbor an overlying mucous layer is likely to misrepresent conditions in vivo. A more complete understanding of this phenomenon will require insight into the mechanisms involved in the capsule alteration that we have observed.
We provide clear evidence that during coculture with epithelial cells, C. jejuni CPS transcript abundance is reduced. Other mechanisms of alteration in surface glycan structures in C. jejuni have previously been reported. For instance, the high-molecular-weight glycans of C. jejuni 81-176 and C. jejuni NCTC 11168 are variable (2, 26, 37). Comparison of cps gene clusters among various C. jejuni strains provides evidence for multiple mechanisms of structural variation (22). These include gene duplication, deletion, and fusion; horizontal transfer of gene clusters; and exchange of capsular genes. Moreover, C. jejuni polysaccharides may alter in abundance through mechanisms other than or in addition to alterations in gene expression, including perhaps transport of CPS across bacterial membranes (30). The microarray analysis suggests that downregulation of the genes responsible for the export of CPS could be a major factor in the attenuation of surface polysaccharide expression observed in this study. Further studies will be required to elucidate the exact mechanisms that result in the altered CPS expression.
In summary, we have shown that CPS abundance is reduced in C. jejuni in an environment in vitro that stimulates at least some conditions present during infection in vivo (coculture). A better understanding of the precise nature of polysaccharide alteration following passage with host cells will require characterization of the glycan structures in C. jejuni and further exploration of the mechanisms involved in CPS depletion under coculture conditions.
Acknowledgments
This work was funded by an Investigator Programme grant from Science Foundation Ireland and also by the Children's Medical and Research Foundation.
We are grateful to P. Guerry for the gift of the C. jejuni 81-176 kpsM mutant. We acknowledge the helpful advice and support of C. Szymanski. We acknowledge BUG@S (the Bacterial Microarray Group at St. George's University of London) for supply of the microarray and advice and The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resource Initiative.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 9 March 2009.
REFERENCES
- 1.Babakhani, F. K., and J. L. Joens. 1993. Primary swine intestinal cells as a model for studying Campylobacter jejuni invasiveness. Infect. Immun. 612723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40769-777. [DOI] [PubMed] [Google Scholar]
- 3.Blaser, M. J., and P. F. Kohler. 1985. Susceptibility of Campylobacter isolates to the bactericidal activity of human serum. J. Infect. Dis. 151227-235. [DOI] [PubMed] [Google Scholar]
- 4.Chart, H., D. Conway, J. A. Frost, and B. Rowe. 1996. Outer membrane characteristics of Campylobacter jejuni grown in chickens. FEMS Microbiol. Lett. 145469-472. [DOI] [PubMed] [Google Scholar]
- 5.Coburn, P. S., C. M. Pillar, B. D. Jett, W. Haas, and M. S. Gilmore. 2004. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science 3062270-2272. [DOI] [PubMed] [Google Scholar]
- 6.Coombes, B. K., and J. B. Mahony. 1999. Chlamydia pneumoniae infection of human endothelial cells induces proliferation of smooth muscle cells via an endothelial cell-derived soluble factor(s). Infect. Immun. 672909-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G. R. 2002. Yersinia type III secretion: send in the effectors. J. Cell Biol. 158401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. J. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 111706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsinghorst, E. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236405-420. [DOI] [PubMed] [Google Scholar]
- 10.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 1481475-1481. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47103-118. [DOI] [PubMed] [Google Scholar]
- 12.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, J. M., K. Hannon, M. Moreau, W. W. Karakawa, and W. F. Vann. 1987. Isolation of type 5 capsular polysaccharide from Staphylococcus aureus. Ann. Inst. Pasteur Microbiol. 138561-567. [DOI] [PubMed] [Google Scholar]
- 14.Freitas, M., C. Cayuela, J. M. Antoine, F. Piller, C. Sapin, and G. Trugnan. 2001. A heat labile soluble factor from Bacteroides thetaiotaomicron VPI-5482 specifically increases the galactosylation pattern of HT29-MTX cells. Cell. Microbiol. 3289-300. [DOI] [PubMed] [Google Scholar]
- 15.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 686656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gundogdu, O., S. D. Bentley, M. T. Holden, J. Parkhill, N. Dorrell, and B. W. Wren. 2007. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitchcock, P. J. 1983. Aberrant migration of lipopolysaccharide in sodium dodecyl sulfate/polyacrylamide gel electrophoresis. Eur. J. Biochem. 133685-688. [DOI] [PubMed] [Google Scholar]
- 18.Jandu, N., P. J. M. Ceponis, S. Kato, J. D. Riff, D. M. McKay, and P. M. Sherman. 2006. Conditioned medium from enterohemorrhagic Escherichia coli-infected T84 cells inhibits signal transducer and activator of transcription 1 activation by gamma interferon. Infect. Immun. 741809-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, M. A., K. L. Marston, C. A. Woodall, D. J. Maskell, D. Linton, A. V. Karlyshev, N. Dorrell, B. W. Wren, and P. A. Barrow. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 723769-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlyshev, A. V., J. Henderson, J. M. Ketley, and B. W. Wren. 1999. Procedure for the investigation of bacterial genomes: random shot-gun cloning, sample sequencing and muthagenesis of Campylobacter jejuni. BioTechniques 2650-56. [DOI] [PubMed] [Google Scholar]
- 21.Karlyshev, A. V., and B. W. Wren. 2001. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using Alcian blue dye. J. Clin. Microbiol. 39279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlyshev, A. V., O. L. Champion, C. Churcher, J. R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St. Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 5590-103. [DOI] [PubMed] [Google Scholar]
- 23.Konkel, M. E., and W. Cieplak, Jr. 1992. Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect. Immun. 604945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32691-701. [DOI] [PubMed] [Google Scholar]
- 25.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 1863296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linton, D., M. Gilbert, P. G. Hitchen, A. Dell, H. R. Morris, W. W. Wakarchuk, N. A. Gregson, and B. W. Wren. 2000. Phase variation of a beta;-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37501-514. [DOI] [PubMed] [Google Scholar]
- 27.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 7388-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marouni, M. J., and S. Sela. 2003. The luxS gene of Streptococcus pyogenes regulates expression of genes that affect internalization by epithelial cells. Infect. Immun. 715633-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moen, B., A. Oust, O. Langsrud, N. Dorrell, G. L. Marsden, J. Hinds, A. Kohler, B. W. Wren, and K. Rudi. 2005. Explorative multifactor approach for investigating global survival mechanisms of Campylobacter jejuni under environmental conditions. Appl. Environ. Microbiol. 712086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 32.Reeser, R. J., R. T. Medler, S. J. Billington, B. H. Jost, and L. A. Joens. 2007. Characterization of Campylobacter jejuni biofilm under defined growth conditions. Appl. Environ. Microbiol. 731908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberson, E., and W. Firestone. 1992. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl. Environ. Microbiol. 581284-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schembri, M. A., J. Blom, K. A. Krogfelt, and P. Klemm. 2005. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 734626-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 731797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szymanski, C. M., F. S. Michael, H. C. Jarrell, J. Li, M. Gilbert, S. Larocque, E. Vinogradov, and J. R. Brisson. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 27824509-24520. [DOI] [PubMed] [Google Scholar]
- 38.van Vliet, A. H. M., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1805291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassenaar, T. M., B. N. Fry, and B. A. M. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132131-135. [DOI] [PubMed] [Google Scholar]
- 40.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16994-996. [PubMed] [Google Scholar]
- 41.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3579-588. [DOI] [PubMed] [Google Scholar]