Abstract
Helicobacter pylori establishes a chronic infection in the human stomach, causing gastritis, peptic ulcer, or gastric cancer, and more severe diseases are associated with virulence genes such as the cag pathogenicity island (PAI). The aim of this work was to study gene content differences among H. pylori strains isolated from patients with different gastroduodenal diseases in a Mexican-Mestizo patient population. H. pylori isolates from 10 patients with nonatrophic gastritis, 10 patients with duodenal ulcer, and 9 patients with gastric cancer were studied. Multiple isolates from the same patient were analyzed by randomly amplified polymorphic DNA analysis, and strains with unique patterns were tested using whole-genome microarray-based comparative genomic hybridization (aCGH). We studied 42 isolates and found 1,319 genes present in all isolates, while 341 (20.5%) were variable genes. Among the variable genes, 127 (37%) were distributed within plasticity zones (PZs). The overall number of variable genes present in a given isolate was significantly lower for gastric cancer isolates. Thirty genes were significantly associated with nonatrophic gastritis, duodenal ulcer, or gastric cancer, 14 (46.6%) of which were within PZs and the cag PAI. Two genes (HP0674 and JHP0940) were absent in all gastric cancer isolates. Many of the disease-associated genes outside the PZs formed clusters, and some of these genes are regulated in response to acid or other environmental conditions. Validation of candidate genes identified by aCGH in a second patient cohort allowed the identification of novel H. pylori genes associated with gastric cancer or duodenal ulcer. These disease-associated genes may serve as biomarkers of the risk for severe gastroduodenal diseases.
Infection with Helicobacter pylori is one of the most common bacterial infections in humans worldwide, and like the case for many other infections, rates are higher in developing countries (80 to 90%) than in developed countries (<50%) (20, 33, 39). The infection is associated with peptic ulcers, gastric carcinoma, and mucosa-associated lymphoma (32, 36, 39). Most infected people remain asymptomatic during their lifetime, and only about 15% develop gastroduodenal illness. Environmental, host, and bacterial factors all play a role in the outcome of the infection. A number of bacterial virulence factors associated with disease have been described for H. pylori, and the most consistently reported are the cag pathogenicity island (cag PAI) (4, 40, 52) and vacA (2, 43). Outer membrane proteins such as BabA2 (18), OipA (54), and SabB (11), as well as the iceA gene, have also been reported to be associated with disease. An outstanding characteristic of H. pylori is the high level of genetic diversity among isolates from different patients, even if they belong to the same ethnic population. Several molecular typing techniques have been used to genotype H. pylori strains from different populations, demonstrating genetic differences among populations and even among isolates from the same individual, suggesting the presence of mixed infection (3, 9, 19, 26, 35, 46). The ability of this bacterium to generate such genetic diversity is due to its natural competence, high recombination and mutation rates, or the occurrence of slipped-strand synthesis and phase variation (17, 24).
H. pylori was the first bacterial species for which whole genome sequences of two independent strains were available (J99 and 26695). Their comparison showed that approximately 6 to 7% of the H. pylori genes present in one strain are absent in the other. These genes are called strain-specific genes, and almost half of them are located in hypervariable regions of the genome (1, 51). These regions contain a considerable number of restriction-modification genes and genes for transposases, topoisomerases, and outer membrane proteins. One of these regions is the cag PAI, whereas the others have been termed plasticity zones (PZs). Whole-genome DNA microarrays facilitated further analyses of the genomic contents of 15 H. pylori clinical isolates, revealing 362 genes (22% of all genes) that are not conserved among strains and represent variable or strain-specific genes (45). Similar microarray-based comparative genomic hybridization (aCGH) studies have been used to explore the genetic diversity in the H. pylori strain population colonizing the different regions of the stomach of a single host for both adults and children (46) and to correlate the genetic contents of H. pylori strains with pathogenesis in animal models (6, 25). These studies indicate that H. pylori strains gain or lose loci during chronic infection, suggesting a continuous genetic flux, mainly inside the PZs (14, 22, 26, 27, 28). The sequence of an H. pylori strain isolated from a patient with chronic atrophic gastritis was recently published, identifying additional strain-specific genes (38) and, by comparison with previous studies (22), suggesting that 121 genes are “chronic atrophic gastritis associated.”
Other studies have reported that genes located in the PZs, such as jhp0947 and jhp0949, are associated with disease (12, 37, 47). The dupA gene was associated with an increased risk for duodenal ulcer (DU) and a low risk for gastric atrophy and cancer (31). The aim of the present study was to compare the genomic contents of H. pylori strains isolated from patients with nonatrophic gastritis (NAG), DU, or gastric cancer (GC) in a Mexican-Mestizo population in order to look for genes previously associated with severe gastroduodenal diseases and to identify novel disease biomarkers.
MATERIALS AND METHODS
Patients.
Twenty-nine adult patients were selected for this study, including 10 with NAG, 10 with DU, and 9 with GC, all of whom were positive for H. pylori culture (see below). The nine GC patients presented with cancer in a noncardia location; six had diffuse and three had intestinal carcinoma types. These patients attended the Oncology Hospital of the Instituto Mexicano del Seguro Social (IMSS) and the General Hospital of the Secretaria de Salud (SS), Mexico City, Mexico; they were of medium to low socioeconomic class and a Mestizo-Mexican population. Diagnosis was based on endoscopy and histological findings. Sixty additional patients, 20 with GC, 20 with DU, and 20 with NAG, were used to validate some of the microarray results with PCR tests. These patients attended the Oncology Hospital and the Gabriel Mancera General Hospital at IMSS. Patients were informed about the nature of the study and asked to sign an informed-consent form. The study was approved by the corresponding ethical committees of the IMSS and SS.
H. pylori isolation.
All patients were subjected to endoscopy, and one biopsy specimen each from the antrum and corpus was taken for H. pylori culture. Biopsy specimens were homogenized and inoculated on horse blood agar plates under microaerobic conditions at 37°C. Colonies with characteristic morphology were confirmed by urease, catalase, and oxidase tests. From the primary growth from each biopsy site (antrum and corpus), four or five single colonies were isolated and propagated; each single isolate was suspended in saline solution for DNA isolation. DNAs were isolated using a Wizard genomic DNA purification kit (Promega Corporation) according to the manufacturer's instructions and were stored at −70°C until used. DNAs from H. pylori strains J99 and 26695 were prepared for use as competing DNAs in the microarrays.
Microarray experiments.
The microarray used represents a superset of genes present in H. pylori strains 26695 and J99, based on their whole genome sequences (45). The elements of our microarray consisted of large (mean size, 817 bp; 10th percentile, 130 bp; 90th percentile, 1,967 bp) DNA fragments corresponding to unique segments of individual open reading frames, generated by PCR using gene-specific primers. For the test, 0.5 μg each of 26695 and J99 genomic DNAs was suspended in 39 μl of H2O and denatured for 5 min at 99°C. Five microliters of 10× buffer (400 μg/ml random octamers, 0.5 M Tris-HCl, 100 mM MgSO4, and 10 mM dithiothreitol), 5 μl deoxynucleoside triphosphate-dUTP mix (0.5 mM concentration of dGTP, dATP, and dCTP, 0.2 mM aminoallyl dUTP, and 0.3 mM dTTP), and 1 μl Klenow fragment were added, and the reaction mix was incubated overnight at 37°C. Free amines were removed, and the probe (mixed J99 and 26695) was labeled with Cy3 dye (Amersham); the reaction mix was incubated for 1 h at room temperature in the dark. One microgram of genomic DNA from each test strain was labeled with Cy5 dye (Amersham Pharmacia), following the same procedures as those described above. Labeled probe and test DNAs were combined, unincorporated dye was removed, and 1 μl of 10-mg/ml yeast tRNA, 1.5 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 1.5 μl of 1% sodium dodecyl sulfate (SDS) were added. The mixture was denatured for 2 min at 99°C, applied to the microarray slide, and incubated overnight. The microarray slide was washed with 2× SSC and 0.1% SDS to remove the coverslip and then with 1× SSC for 5 min, three times. The microarray was scanned using an Axon scanner with GENEPIX 3.0 software (Axon Instruments, Redwood City, CA), and data were normalized and processed as previously described (45). The mean of the normalized red/green (R/G) ratio was calculated using data from two arrays per isolate, yielding four readings for each gene. The cutoff for absence of a gene was defined as a log2 R/G ratio of <−1.0, based on test hybridizations using the sequenced strain J99 against the 26695-J99 mixed reference. The false-positive and false-negative rates were determined to be 3.5% and 0.34%, respectively (45). Our microarray data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12932.
Validation of genes associated with disease by PCR.
The results for some variable genes found to be associated with disease by microarray analysis were confirmed using gene-specific PCR to validate the hybridization results. We selected 13 such genes (HP0025, HP0424, JHP0630, HP0713, HP0790, JHP0925, JHP0927, JHP0951, HP1426, HP1472, HP1499, HP0674, and JHP0940), and isolates from 10 patients of each disease group, i.e., the NAG, DU, and GC groups, were tested. To further validate the observation that the JHP0940 and HP0674 genes were absent in isolates from GC cases (see below), we tested these two genes by PCR for an additional group of patients; isolates from 20 patients each with NAG, DU, and distal GC were tested. Primers and conditions for each PCR were as previously described for the construction of these microarrays (45). Amplification of each gene was carried out in a total volume of 25 μl containing 1 μl of DNA template for each clinical isolate (reference strains J99 and 26695 were used as controls), 1× PCR buffer, 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim, Germany), 2 U of Taq DNA polymerase (Invitrogen, Life Technologies, Brazil), and 30 mM each primer. Each reaction mixture was amplified for 35 cycles as follows: 30 s at 92°C, 45 s of annealing (the range of annealing temperatures was 50 to 58°C, depending on the gene), and 2.5 min at 72°C. After the last cycle, extension was continued for another 5 min. Amplicons were analyzed on a 1% agarose gel stained with ethidium bromide.
Statistical analysis.
Analysis of clustering based on the variable gene content among all isolates from the different disease groups was done using Jaccard distances with the unweighted-pair group method using average linkages. For all analyses of association between the content of variable genes and disease, analysis was performed on a per-patient basis; i.e., when a gene was found in any of the isolates from that patient, the gene was considered present. To compare the frequencies of variable genes between each disease group, we used a logit model, considering disease an independent variable; P values of <0.05 were considered significant. To learn if there was any difference in the contents of genes inside the PZs among the disease groups, the Pearson correlation coefficient was used, with an r value of 1.00 meaning that the frequencies of the gene were similar between the disease groups and r values of <0.05 indicating significant differences in PZ genetic content among the disease groups. To analyze associations between the presence or absence of specific genes and disease, we used a log-linear model. Logit was used as a link function with the maximum likelihood method to estimate coefficients. The association of genes with disease was probed using the χ2 test, comparing one disease group (i.e., NAG) with the other two groups (i.e., DU and GC). Considering the exploratory nature of the study, we considered a gene significantly associated with disease when the P value was <0.1. Any possible correlation between the frequency of variable genes and age or gender was analyzed with a generalized linear model. A binomial distribution was assumed, using logit as a link function. To analyze the correlation between results of microarray hybridization and PCR tests, we used Kendall's coefficient of tau and Stuart's coefficient. We also tested if the prevalence of the JHP0940 and HP0674 genes in each disease group did not vary when tested with either microarray or PCR by using Fisher's exact test. All statistical analyses were performed using S-Plus 7.0 (Insightful Corp.).
RESULTS
Number of H. pylori isolates tested in microarrays.
Four or five single colonies of H. pylori were isolated from the antrum and corpus of each patient. Based on the finding of unique polymorphic DNA profiles obtained by random amplification of polymorphic DNA (RAPD) among isolates from the same patient, 10 of the 29 patients had mixed infections with multiple strain types, including four with NAG, five with DU, and one with GC. A representative isolate of each RAPD type found in each patient was analyzed by aCGH, giving a total of 42 different isolates (Table 1).
TABLE 1.
Content of variable genes in H. pylori isolates from patients with gastroduodenal diseases, defined using microarrays
| Group and patient | Age (yr) | Gender | No. of single isolates studieda | No. of variable genes per single isolate in patients with mixed infectiond | No. of variable genes per patiente |
|---|---|---|---|---|---|
| NAG patients | |||||
| 357b | 33 | F | 2 | A = 143; B = 112 | 152 |
| 284b | 41 | F | 2 | A = 93; B = 92 | 143 |
| 320b | 43 | F | 2 | A = 52; B = 131 | 138 |
| 103b | 37 | F | 2 | A = 81; B = 1 | 82 |
| 386c | 40 | M | 3 | A = 76; B = 72; C = 4 | 139 |
| 110 | 48 | M | 1 | 21 | |
| 370 | 44 | F | 1 | 187 | |
| 176 | 44 | M | 1 | 88 | |
| 345 | 35 | F | 1 | 85 | |
| 221 | 44 | F | 1 | 98 | |
| DU patients | |||||
| 174b | 41 | F | 2 | A = 31; B = 150 | 164 |
| 204b | 61 | M | 2 | A = 67; B = 113 | 126 |
| 372b | 46 | M | 2 | A = 43; B = 126 | 127 |
| 387b | 31 | M | 2 | A = 175; B = 127 | 181 |
| 355 | 46 | F | 1 | 30 | |
| 362 | 28 | F | 1 | 126 | |
| 263 | 55 | F | 1 | 3 | |
| 336 | 77 | F | 1 | 85 | |
| 261 | 52 | F | 1 | 100 | |
| 379 | 48 | F | 1 | 1 | |
| GC patients | |||||
| 510c | 44 | M | 4 | A = 25; B = 45; C = 13; D = 93 | 104 |
| 188 | 46 | F | 1 | 27 | |
| 206 | 61 | F | 1 | 88 | |
| 377 | 66 | M | 1 | 96 | |
| 401 | 60 | F | 1 | 30 | |
| 535 | 44 | F | 1 | 137 | |
| 456 | 57 | M | 1 | 124 | |
| 331 | 36 | F | 1 | 113 | |
| 443 | 69 | F | 1 | 77 |
Defined using RAPD fingerprinting.
Patients with two RAPD patterns.
Patients with more than two RAPD patterns.
Number of variable genes in each isolate. Each letter (A, B, C, or D) represents a single isolate with a different RAPD pattern.
Overall number of variable genes per patient, considering that in the case of mixed infection a gene was present when it was found in at least one of the isolates from this patient.
The content of variable genes varies among H. pylori isolates from each disease group.
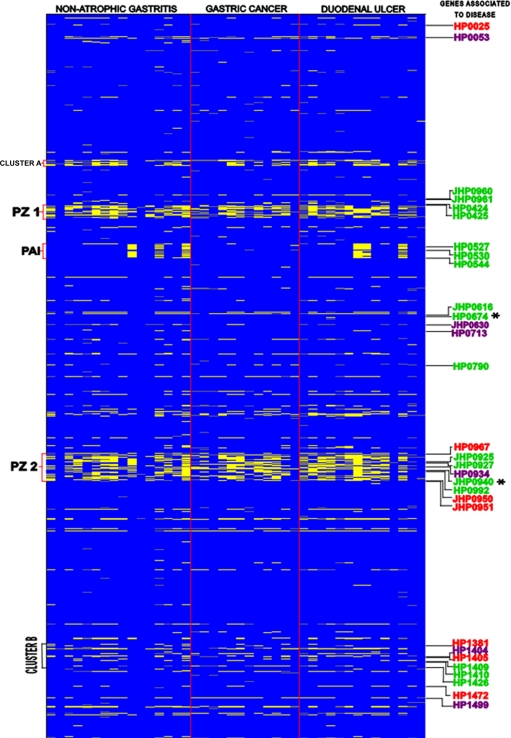
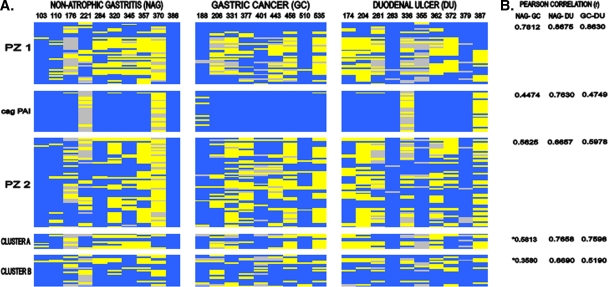
To explore the genomic content in the 42 isolates of H. pylori from patients with NAG, DU, and GC, we used aCGH. Of the 1,660 genes analyzed, 1,319 were present in all 42 isolates (genome core), whereas 341 (20.5%) were absent in one or more isolates and were considered variable genes (see Tables S1 and S2 in the supplemental material). The number of variable genes present in isolates per patient for each disease group is shown in Table 1. The range of variable genes present in isolates from patients with NAG varied from 21 to 187 genes, that for patients with DU ranged from 1 to 181, and that for patients with GC ranged from 27 to 137 genes. The overall number of variable genes present in isolates from each disease group was 291 (85.33%) for isolates from patients with NAG, 257 (75.36%) for patients with DU, and 230 (67.44%) for patients with cancer. The total number of variable genes present in the group of cancer strains was statistically lower than those for the NAG and DU strains (P < 0.01). These variable genes were classified according to their functional category, as shown in Table 2, and included genes involved in DNA metabolism, genes involved in cellular processes (including pathogenesis), transposase genes, and hypothetical genes. Of the 341 variable genes found in our studied H. pylori population, 127 (37%) were distributed within PZs, with 100 genes within the PZs and 27 genes within the cag PAI. Although most of the remaining 214 genes were scattered along the genome individually, we observed clusters of genes (clusters A and B), which are also depicted in Fig. 1. We analyzed if there were any differences in the genetic content of each PZ as well as in the clusters among H. pylori isolates from the different disease groups (Fig. 2). The cag PAI and PZ2 showed significantly different patterns of variable genes in GC isolates compared with both NAG and DU isolates, whereas PZ1 showed a different pattern for GC isolates only compared with NAG isolates (P < 0.05). The presence of a complete cag PAI was significantly more frequent for isolates from patients with GC (Fig. 2) than for isolates from NAG and ulcer patients (P < 0.05). The cag2 gene was absent in four NAG isolates, three DU isolates, and one GC isolate; this gene has been reported as nonessential for functional activity (15). Clusters A and B showed no significant differences in pattern of variable genes in a comparison of NAG and GC isolates (P > 0.05).
TABLE 2.
Classes of variable genes by functional category in isolates from Mexican patients with gastroduodenal diseases
| Functional categorya | No. of genes | % of genes |
|---|---|---|
| Hypothetical | ||
| Conserved | 24 | 59.2 |
| H. pylori specific with no known function | 178 | |
| DNA metabolism | ||
| DNA replication, recombination and repair | 9 | 13.8 |
| Restriction/modification | 38 | |
| Cellular processes | ||
| Transformation | 7 | 12.3 |
| Toxin production | 1 | |
| Pathogenesis | 32 | |
| Other | 2 | |
| Other categories | ||
| Plasmid-related functions | 3 | 5.6 |
| Transposon-related functions | 16 | |
| Cell envelope | ||
| Biosynthesis of surface polysaccharides and lipopolysaccharides | 8 | 4.7 |
| Other | 8 | |
| Unknown | ||
| General | 7 | 2.0 |
| Transport and binding proteins | ||
| Cations | 2 | 0.9 |
| Unknown substrate | 1 | |
| Energy metabolism | ||
| Fermentation | 2 | 0.6 |
| Fatty acid and phospholipid metabolism | ||
| General | 1 | 0.3 |
| Protein synthesis | ||
| tRNA aminoacylation | 1 | 0.3 |
| Protein fate | ||
| Degradation of protein, peptide and glycopeptides | 1 | 0.3 |
| Total | 341 |
Functional categories are according to http://genolist.pasteur.fr/PyloriGene/genome.cgi.
FIG. 1.
Genomotyping of H. pylori isolates from patients with gastroduodenal diseases. The figure is a representation of the entire H. pylori chromosome, which was ordered by combining the maps of strains 26695 and J99 in ascending order, where J99-specific genes are placed at the appropriate sites in the 26695 chromosomal map, starting with HP0001, in each row (top left) and the columns represent the 42 isolates analyzed from three groups of disease. The scanogram indicates genes that are present (blue) or absent (yellow), as well as missing data (gray). Genes associated with disease, i.e., NAG (purple), GC (green), and DU (red), are shown, and genes absent in GC isolates are indicated (*). The locations of PZ1, cag PAI, and PZ2, as well as the small cluster A and a disease-associated gene cluster outside the PZs (cluster B), are indicated.
FIG. 2.
Genomic comparison of H. pylori isolates in variable regions among patients with different gastroduodenal diseases. (A) The scanogram shows PZ1 (JHP0954 to HP0494), the cag PAI (HP0520 to HP0547), PZ2 (JHP0914 to HP1009), cluster A (HP0335 to HP0346), and cluster B (HP1381 to HP1426) after combination of the 26695 and J99 chromosomal maps (left). Patients with NAG, GC, and DU are indicated at the top. (B) Comparison of differences in genetic content in each region by analysis of the Pearson correlation coefficient (r). *, values are not statistically significant (P > 0.05).
Thirty variable genes are significantly associated with disease.
Hierarchical clustering based on the content of the variable genes showed no clustering of isolates by disease group (data not shown). Next, we examined the frequency of each of the 341 variable genes among patients from each disease group. We observed genes that were present in all isolates from a disease group (100% frequency) but present at lower frequencies in the other two disease groups (Table 3), including 11 in NAG, 17 in DU, and 41 in GC patient isolates. Additionally, 30 variable genes were significantly associated with one of the disease groups by being either significantly more or less frequently present in the genome. Seventeen of these genes were present at lower frequencies among isolates of one of the disease groups (Table 4). The chromosomal distribution of these genes revealed that 12 (73%) were located outside the PZs, 2 fell in PZ1, and 3 were located in PZ2. These 17 genes were distributed among the disease groups as follows: 4 were significantly less frequent in NAG isolates, 3 were significantly less frequent in DU isolates, and 10 were significantly less frequent in GC patient isolates. Two genes (HP0674 and JHP0940) were absent in all GC isolates (Table 4). Thirteen genes were significantly more frequent among isolates of one of the disease groups (Table 5), and nine of these (69%) were located inside PZs. The genes found more frequently in certain disease groups were distributed as follows: two genes in NAG isolates, four genes in DU isolates, and seven genes in GC patient isolates. Three of the genes found more frequently in GC patient isolates mapped to the cag PAI (cag7, cag10, and cag23). The distribution of the 30 genes associated with disease in the H. pylori genome is schematized in Fig. 1. Some of the genes associated with disease and located outside the PZs form a cluster (cluster B) (Fig. 1). We also searched for any correlation between age or sex and the frequency of variable genes and found that there was a significant correlation between a smaller number of variable genes present in a given strain and increasing age (P = 0.05) (data not shown).
TABLE 3.
Variable genes present in all isolates from the respective disease group (disease-specific genes)a
| Gene name and no. for indicated disease group
| ||
|---|---|---|
| Nonatrophic gastritis | Gastric cancer | Duodenal ulcer |
| N/A, HP0161, N/A | N/A, HP0025, JHP0021 | N/A, HP0059, JHP0052 |
| vapD, HP0315, N/A | N/A, HP0030, JHP0026 | N/A, HP0341, N/A |
| N/A, HP0424, N/A | N/A, HP0340, N/A | MFOKI, HP0481, JHP0433 |
| cag16, cagM, HP0537, JHP0485 | N/A, HP0446, N/A | prrB, HP0790, N/A |
| N/A, HP0987, N/A | N/A, HP0492, JHP0444 | N/A, HP0937, JHP0872 |
| hpyAIVR, HP1351, JHP1270 | N/A, HP0522, JHP0471 | N/A, HP0993, N/A |
| HINFIM, HP1352, JHP1271 | N/A, HP0523, JHP0472 | N/A, HP1144, JHP1072 |
| N/A, HP1426, N/A | virB11_1, HP0525, JHP0474 | N/A, HP1288, JHP1208 |
| N/A, HP1537, N/A | cag7, orf13 and orf14, HP0527, JHP0476 | N/A, HP1353, JHP1272 |
| N/A, N/A, JHP0933 | cag9, orf16, HP0529, JHP0478 | N/A, HP1381, N/A |
| N/A, N/A, JHP0934 | cag10, orf17, HP0530, JHP0479 | hsdS, HP1404, N/A |
| cag12, cagT, HP0532, JHP0481 | N/A, HP1405, N/A | |
| cag13, cagS, HP0534, JHP0482 | N/A, HP1518, N/A | |
| cag14, cagQ, HP0535, JHP0483 | N/A, HP1587, JHP1493 | |
| cag18, cagL, HP0539, JHP0487 | N/A, N/A, JHP0951 | |
| cag21, cagG, HP0542, JHP0490 | N/A, N/A, JHP0950 | |
| cag23, cagE, HP0544, JHP0492 | N/A, N/A, JHP0896 | |
| cag26, cagA, HP0547, JHP0495 | ||
| N/A, HP0548, N/A | ||
| N/A, HP0613, JHP0300 | ||
| N/A, HP0681, JHP0622 | ||
| N/A, HP0712, JHP0651 | ||
| N/A, HP0744, JHP0681 | ||
| N/A, HP0764, JHP0701, JHP0702 | ||
| acpP, HP0962, N/A | ||
| N/A, HP0963, JHP0897 | ||
| N/A, HP0964, JHP0898, JHP0899 | ||
| N/A, HP0965, JHP0899 | ||
| N/A, HP1396, JHP1431 | ||
| N/A, HP1397, JHP1430 | ||
| N/A, N/A, JHP0616 | ||
| N/A, N/A, JHP0961 | ||
| N/A, N/A, JHP0959 | ||
| N/A, N/A, JHP0960 | ||
| N/A, N/A, JHP0830 | ||
| N/A, N/A, JHP0540 | ||
| N/A, N/A, JHP0916 | ||
| N/A, N/A, JHP1408 | ||
| N/A, N/A, JHP0935 | ||
| N/A, N/A, JHP0814 | ||
| N/A, N/A, JHP0954 | ||
TABLE 4.
H. pylori genes significantly less frequent in isolates from one disease group than in isolates from the other two gastroduodenal disease groups
| Disease group and genesa | Description and/or function(s) (reference[s]) | Localization | P valueb |
|---|---|---|---|
| NAG | |||
| N/A, HP0713, JHP0651 | Outside PZs | 0.0291 | |
| N/A, N/A, JHP0630 | Outside PZs | 0.0570 | |
| hsdS, HP1404, N/A | Type I restriction enzyme S protein/DNA metabolism, restriction-modification/proinflammatory gene (7) | Outside PZs | 0.0920 |
| N/A, HP0053, N/A | Less frequent in cag PAI isolates (50) | Outside PZs | 0.0993 |
| GC | |||
| N/A, HP1426, N/A | Conserved hypothetical protein/hypothetical, general/less frequent in cag PAI+ isolates (45) | Outside PZs | 0.0085 |
| N/A, HP0424, N/A | PZ1 | 0.0203 | |
| prrB, HP0790, N/A | Anti-codon nuclease masking agent/DNA metabolism, DNA replication, recombination, and repair/modulation of the immune response (7) | Outside PZs | 0.0246 |
| N/A, N/A, JHP0925 | PZ2 | 0.0523 | |
| N/A, HP0992, N/A | PZ2 | 0.0605 | |
| N/A, HP1409, JHP1301, JHP1302 | Regulation of responses to environment conditions (13) | Outside PZs | 0.0812 |
| N/A, HP1410, JHP1303 | Regulation of responses to environment conditions (13) | Outside PZs | 0.0812 |
| N/A, HP0425, N/A | Regulation of response to acid (39, 40) | PZ1 | 0.0812 |
| N/A, HP0674, N/A (absent in all cancer isolates) | Outside PZs | 0.0048 | |
| N/A, N/A, JHP0940 (absent in all cancer isolates) | Proinflammatory gene, responsive to contact of H. pylori with the gastric mucosa (44) | PZ2 | 0.0636 |
| DU | |||
| omp2, HP0025, JHP0021 | Outer membrane protein, putative outer membrane protein/cell envelope, other/regulation of response to acid (8, 53) | Outside PZs | 0.0369 |
| mod, HP1472, JHP1365 | Type II S restriction enzyme M protein, putative type II DNA modification enzyme (methyltransferase)/DNA metabolism, restriction-modification | Outside PZs | 0.0437 |
| vapD, HP0967, N/A | Virulence associated protein D/other categories, adaptations and atypical conditions | Outside PZs | 0.0968 |
TABLE 5.
H. pylori genes significantly more frequent in isolates from one disease group than in isolates from the other two gastroduodenal disease groups
| Disease group and genesa | Description and/or function(s) (reference) | Localization | P valueb |
|---|---|---|---|
| NAG | |||
| N/A, HP1499, JHP1392 | Regulation of response to pH (8) | Outside PZs | 0.0332 |
| N/A, N/A, JHP0934 | PZ2 | 0.0902 | |
| GC | |||
| N/A, N/A, JHP0616 | Outside PZs | 0.0412 | |
| N/A, N/A, JHP0927 | PZ2 | 0.0593 | |
| N/A, N/A, JHP0960 | Homology with a gene in plasmids of Campylobacter sp. (5) | PZ1 | 0.0685 |
| N/A, N/A, JHP0961 | Homology with a gene in plasmids of Campylobacter sp. (5) | PZ1 | 0.0767 |
| cag7, orf13 and orf14, HP0527, JHP0476 | cag pathogenicity island protein, DNA transfer protein (Agrobacterium VirB10 homolog)/cellular processes | cag PAI | 0.0995 |
| cag10, orf17, HP0530, JHP0479 | cag pathogenicity island protein/cellular processes | PZ2 | 0.0995 |
| cag23, cagE, HP0544, JHP0492 | cag pathogenicity island protein/cellular processes | PZ2 | 0.0995 |
| DU | |||
| N/A, N/A, JHP0951 | Integrase/recombinase (xercd family)/modulation of the response to low pH and iron limitation (16) | PZ2 | 0.0374 |
| N/A, N/A, JHP0950 | PZ2 | 0.0744 | |
| N/A, HP1405, N/A | Modulation of the response to low pH (8, 30) | Outside PZs | 0.0796 |
| N/A, HP1381, N/A | Outside PZs | 0.0859 |
Variable genes in H. pylori isolates from this study which differ from variable genes in isolates from other studies.
We compared the variable genes from this study with the variable genes reported in previous works, regardless of geographic, ethnic, or disease components (Table 6). This comparison showed that the frequencies of variable genes in different populations range from 20 to 25% of the whole genome. We identified a number of genes reported as variable in previous studies which were not found as such in the isolates in our study, and conversely, there were genes identified as variable among isolates in this study which have not been reported as such in previous works. These genes were located mainly outside the PZs (see Table S2 in the supplemental material). However, it is noteworthy that among the 30 genes found to be associated with disease, 28, including the three genes of the cag PAI, have also been reported as variable in other studies. Only the HP0790 and HP1405 genes have not been reported as variable previously (Tables 4 and 5). In addition, the results of this study were analyzed together with results from a previous work that also analyzed Mexican patients (46) in comparison to reports for other populations (22, 45). We identified a number of genes found as variable exclusively in Mexican populations, with most of them (90%) located outside the PZs (Table 7). Interestingly, babB (HP0896) was one such gene.
TABLE 6.
Comparison among variable genes found in our study with variable genes reported in other studies
| No. of variable genes | Data from:
|
|||
|---|---|---|---|---|
| Gressmann et al. (22) | Salama et al. (45) | Salama et al. (46) | Our test strains | |
| Total no. of genes | 499 | 362 | 365 | 341 |
| Within the plasticity zones | 134 (27%) | 132 (36%) | 125 (34%) | 127 (37%) |
| Shared as variables with our isolates | 323 | 297 | 302 | |
| Found as core in our study and variable in other studies | 176 | 65 | 63 | |
| Found as variable in our study and as core in other studies | 18 | 44 | 39 | |
TABLE 7.
Group of genes found as variable genes in Mexican H. pylori isolates but not in isolates from other populations
| Gene name, gene no. | Functional categorya |
|---|---|
| N/A, HP0242, JHP0227 | Hypothetical |
| HPAIM, HP0263, JHP0248 | DNA metabolism (restriction-modification) |
| N/A, HP0461, N/A | Hypothetical |
| pepA, HP0570, JHP0517 | Protein fate (degradation of proteins, peptides and glycopeptides) |
| aspS, HP0617, JHP0560 | Protein synthesis (tRNA aminoacylation) |
| N/A, HP0629, JHP0572 | Hypothetical |
| N/A, HP0698, N/A | Hypothetical |
| N/A, HP0704, N/A | Hypothetical |
| fecA, fecA_2, HP0807, JHP0743 | Transport and binding proteins (cations) |
| omp19, babB, HP0896, JHP1164 | Cell envelope |
| N/A, HP1353, JHP1272 | DNA metabolism (restriction-modification) |
Functional categories are based on http://genolist.pasteur.fr/PyloriGene/genome.cgi.
The results of microarrays were validated with PCR.
To validate the microarray results, we used PCR to test 13 of the 30 disease-associated genes which had P values of <0.05. Overall, a significant correlation of 0.6278 was observed between microarray and PCR results (Kendall's tau; P < 0.0001). The relatively low value was mostly because of the number of genes present by microarray but absent by PCR, and this lack of correlation occurred mainly in four genes (HP0424, JHP0630, JHP0927, and HP1472), regardless of the disease group of the isolate. One possible explanation for this is that sequence variation in these genes occurs in the primer binding region, rendering them negative by PCR but not negative by hybridization. The absence of the HP0674 and JHP0940 genes in all GC isolates observed by aCGH was confirmed by PCR for all isolates, showing 100% concordance between the two tests. The prevalence of these genes in isolates from 60 additional patients was tested by PCR (see Table S3 in the supplemental material). Whereas HP0674 and JHP0940 were not detected in the 20 cases of GC tested, they were found in some of the 20 gastritis and 20 DU cases, and the prevalences of these two genes in each disease group were not different by either microarray or PCR test (P > 0.05; Fisher's exact test). These data support the association of the loss of these genes in isolates from GC patients.
DISCUSSION
The main goal of our study was to compare the genetic contents of H. pylori strains isolated from patients with NAG, DU, and GC, with special emphasis on genes located in PZs.
Characteristics of variable genes found in the studied isolates.
In the 42 isolates studied, we found 341 variable genes, which coded mainly for DNA metabolism components, cellular processes, transposases, and hypothetical proteins, findings similar to those of previous reports (22, 45, 46). Comparison between disease groups revealed that the overall number of variable genes present in a given strain was significantly lower in isolates from GC patients. Patient age was additionally associated with a decrease in the number of variable genes retained in the genome. As in the case of cancer patients, with age the gastric mucosa becomes increasingly atrophic, perhaps offering an environment that is less favorable for H. pylori growth and for genetic variation. Since GC patients were also the oldest group, another possibility is that with time, H. pylori strains become more adapted to the host mucosa and the presence of some genes is no longer necessary.
Characteristics of genes seen significantly less frequently in NAG, DU, and GC disease groups.
We next examined the frequency of each variable gene in H. pylori isolates from the three disease groups. Of the 341 variable genes present in our population, 30 were found to be significantly associated with a disease group. Notably, most of the genes statistically less frequently present among disease groups were located outside the PZs, whereas the genes statistically more frequently present were located inside the PZs. Among the eight genes which were less frequent in GC isolates, genes HP0425 and HP1410, both of unknown function, have homology with a RecJ exonuclease gene of Escherichia coli (38). HP0425 is transcriptionally activated in ArsS-deficient H. pylori mutants (42). The ArsRS two-component system confers urease-independent acid adaptation. The HP1409 and HP1410 genes, of unknown function, were also less frequent in cancer isolates and have been shown to be regulated by separate two-component systems (13). Thus, the above three genes may be involved in adaptive responses to acid, which during chronic atrophic gastritis may no longer be needed due to destruction of the acid-secreting compartment. Another gene that was less frequent in cancer isolates was HP0790, which encodes an anticodon nuclease. Interestingly, this gene was present in all DU isolates. HP0790 has been reported as associated with a strong inflammatory response (7). As in the case of the response to acid, after years of inducing inflammation, H. pylori leads to an atrophic mucosa and may no longer need proinflammatory genes. HP1426, of unknown function, was also found less frequently among cancer isolates, and in a previous study this gene was reported less frequently for cag PAI-positive isolates (45). In accordance with this observation, most of our cancer isolates were cag PAI positive and frequently lacked HP1426. Interestingly, there were two genes which were absent in all isolates from GC cases, namely, JHP0940 and HP0674, both of unknown function. In contrast to our results, a previous study reported JHP0940 as associated with GC (37). However, in a follow-up report, the authors did not confirm the association (47). The results of an in vivo expression study showed that the JHP0940 gene is strongly expressed in response to the interaction of H. pylori with the gerbil gastric mucosa (21), and a recent study suggested that this gene induces tumor necrosis factor alpha and interleukin-8 secretion as well as enhanced translocation of NF-κB (44). Thus, JHP0940 might be a proinflammatory gene important during the early phases of the infection, responding to contact with the gastric mucosa and eliciting the release of inflammatory mediators. During chronic infection, this gene may be downregulated or lost when the mucosa is altered by the long-lasting inflammation-induced tissue damage.
The HP0025 gene, which encodes an outer membrane protein (Omp2), was identified among the genes that were present less frequently in DU isolates. In accordance with our results, a recent report found that the protein was less frequently expressed in isolates from DU cases (10). In contrast, it is noteworthy that this gene was present in 100% of the GC isolates. HP0025 is considered part of the group of acid- responsive genes (8, 53), and it is thought to help H. pylori colonize the gastric mucosa. Among the genes less present in H. pylori isolates from gastritis patients in our study, HP0053, of unknown function, was found to be absent in cag PAI-negative strains (50). Consistent with this observation, in our study most isolates from patients with NAG had an incomplete cag PAI. Another gene that was less frequently present in gastritis isolates was HP1404, which encodes a type I restriction enzyme S protein. This gene was found in 100% of the DU isolates. It has been reported that HP1404 induces an inflammatory response in a mouse model (7). It is conceivable that after a chronic inflammatory response, this gene has to be silenced to allow the persistence of the infection.
Characteristics of genes found significantly more frequently in NAG, DU, and GC disease groups.
Seven genes were found significantly more frequently in GC isolates. Three of them are part of the cag PAI (cag7, cag10, and cag23) and have been reported as essential for translocation of CagA and induction of interleukin-8 in gastric epithelial cells (15). These results suggest that cag PAI integrity is significantly more conserved in GC isolates. Three genes of unknown function were also located in PZs, namely, JHP0927 (PZ2), JHP0960, and JHP0961 (both in PZ1). JHP0927 was also recently reported as more frequently present in isolates from GC patients in Costa Rica (37). JHP0960 and JHP0961 have shown homology with two genes from plasmids of Campylobacter species, with unknown roles (cpp24 and cpp25, respectively) (5). Thus, six of the seven genes found more frequently in isolates from patients with GC are located inside the PZs.
Among the four genes which were more frequent in DU isolates, two were inside PZ2 (JHP0950, of unknown function, and JHP0951, which encodes an integrase of the XerCD family) and two were outside the PZs (HP1381 and HP1405, both of unknown function). The JHP0951 gene is highly expressed upon mutation of the ferric uptake regulator (Fur) (16). Fur is an essential component for adaptation to low pH and iron limitation. Thus, JHP0951 might have an important role in the response to acidic environments. HP1405 shows increased expression in strains with mutations in histidine kinases HP0165 and HP1364, which function as acid sensors. These kinases are parts of two-component signal transduction systems required for acid resistance in H. pylori (30). HP1405 is also upregulated during growth of H. pylori under acidic conditions (8). Thus, two of the genes more frequently present in isolates from patients with DU are involved in the response to acid. This observation is in agreement with the fact that in DU, secretion of acid is increased and the mucosa is continuously exposed to a low pH. Interestingly, JHP0950 has been suggested as a marker for H. pylori strains from patients with gastric extranodal marginal-zone-B-cell lymphoma of the MALT type (29). Other genes in PZ2 and previously reported as associated with DU, i.e., JHP0947 and JHP0949 (12), are in the vicinity of two genes, JHP0950 and JHP0951, found by us to be associated with DU. In fact, it was recently suggested that JHP0950 may form part of an operon that includes the JHP0949 gene (37, 55).
Comparison of microarray results of this study with previous reports.
A recent study analyzed the gene contents of H. pylori isolates from patients with gastroduodenal diseases in a Chinese population (23). This study found the HP0447 gene to be absent in all DU and GC isolates, whereas in our study this gene was present in about 50% of all disease groups studied. This group also reported the JHP0918 gene as absent in GC isolates, whereas we found it in over 50% of the Mexican GC isolates. These discrepancies may be due to population differences in selective pressures of the host environment, which vary among different geographical regions, that influence whether a gene is maintained or lost. It is also possible that technical differences account for the discrepancies observed.
The number of genes found in the PZs in the Mexican isolates of this study, their locations in the genome map, and the proportion that they represent among all variable genes (around 35%) were very similar to those in previous studies (22, 45, 46). In addition, in our H. pylori isolates a number of variable genes clustered outside PZs (clusters A and B), as seen for other strains (34), and may represent novel PZs. The PZs might represent not only hot spots for variable genes but also regions where clusters of genes associated with disease are found. In our study, we found that the cag PAI and PZ2 were significantly different in isolates from patients with GC compared with those in isolates from cases of both NAG and DU. The cag2 gene was absent in four isolates from NAG patients, three from DU patients, and only one from a GC patient, which suggests that this gene is frequently absent in Mexican isolates. This gene has been reported as nonessential for function of the cag PAI (15). Of note, “cluster B” in our study also contained several of the genes which we found to be associated with GC.
Variable genes found in isolates of our study were compared with variable genes reported in previous studies (22, 45, 46), and three sets of genes were found, including genes shared between studies, genes reported as variable in previous studies but not in our isolates, and genes found as variable in our isolates but not reported as such in previous works. Most of the genes included in the last two groups were located outside the PZs. This observation suggests that whereas PZ genes are more similar among isolates from different world populations, the variable genes which differ among populations are usually genes individually scattered along the genome. The comparison among variable genes found in this study and those reported in other populations showed differences in the range of 20 to 25%. Although some of these variations may be due to genetic differences in isolates from different populations, they might also be related to differences in the numbers of isolates tested in each study as well as to the design of the microarray (i.e., the characteristics of the probes). One gene found as variable in Mexican isolates but not in other populations was babB. This finding is of interest because this gene is a paralog of babA, which codes for an adhesin reported to be a major virulence factor (18).
Concluding remarks.
In conclusion, our study allowed the identification of novel H. pylori genes associated with NAG, DU, and GC in a Mexican-Mestizo population. Many of these disease-associated genes were found within PZs or within clusters, and several coded for regulation of responses to acid or a microenvironment. These genes are candidates for future studies of the pathogenesis of GC and DU and might be useful as biomarkers of risk for GC or DU. We also identified genes which were specifically variable in our population but not in others, as well as genes reported as variable in other populations but not found as such in our isolates.
Supplementary Material
Acknowledgments
This work was supported by CONACYT, Mexico (SS; grant CO1-6957), and by grant RO1 AI0554423 from the U.S. NIH (N.R.S.). J.T. is a recipient of a Fundación IMSS exclusivity scholarship. This work was submitted as a requirement for the D.Sc. degree of Carolina Romo (Doctorado en Biomedicina Molecular, CINVESTAV, Mexico), supported by a scholarship from the CONACYT, Mexico (grant 142656), and by the Coordinación de Investigación, Instituto Mexicano del Seguro Social, Mexico.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 23 February 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. de Jonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 27017771-17777. [DOI] [PubMed] [Google Scholar]
- 3.Aviles-Jimenez, F., D. P. Letley, G. Gonzalez-Valencia, N. Salama, J. Torres, and J. C. Atherton. 2004. Evolution of the Helicobacter pylori vacuolating cytotoxin in a human stomach. J. Bacteriol. 1865182-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert, S., T. Schwarz, S. Miehlke, C. Kirsch, C. Sommer, T. Kwok, M. Gerhard, U. Goebel, N. Lehn, W. Koenig, and T. F. Meyer. 2004. Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect. Immun. 721043-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 1503507-3517. [DOI] [PubMed] [Google Scholar]
- 6.Björkholm, B., A. Lundin, A. Sillén, K. Guillemin, N. Salama, C. Rubio, J. I. Gordon, P. Falk, and L. Engstrand. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 697832-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Björkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 27734191-34197. [DOI] [PubMed] [Google Scholar]
- 8.Bury-Moné, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. de Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53623-638. [DOI] [PubMed] [Google Scholar]
- 9.Camorlinga-Ponce, M., C. Romo, G. González-Valencia, O. Muñoz, and J. Torres. 2004. Topographical localisation of cagA positive and cagA negative Helicobacter pylori strains in the gastric mucosa; an in situ hybridisation study. J. Clin. Pathol. 57822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlsohn, E., J. Nystrom, H. Karlsson, A. M. Svennerholm, and C. L. Nilsson. 2006. Characterization of the outer membrane protein profile from disease-related Helicobacter pylori isolates by subcellular fractionation and nano-LC FT-ICR MS analysis. J. Proteome Res. 53197-3204. [DOI] [PubMed] [Google Scholar]
- 11.de Jonge, R., R. G. J. Pot, R. J. L. F. Loffeld, A. H. M. van Vliet, E. J. Kuipers, and J. G. Kusters. 2004. The functional status of the Helicobacter pylori sabB adhesin gene as a putative marker for disease outcome. Helicobacter 9158-164. [DOI] [PubMed] [Google Scholar]
- 12.de Jonge, R., E. J. Kuipers, S. C. L. Langeveld, R. J. L. F. Loffeld, J. Stoof, A. H. M. van Vliet, and J. G. Kusters. 2004. The Helicobacter pylori plasticity region locus jhp0947-jhp0949 is associated with duodenal ulcer disease and interleukin-12 production in monocyte cells. FEMS Immunol. Med. Microbiol. 41161-167. [DOI] [PubMed] [Google Scholar]
- 13.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobrindt, U., and J. Hacker. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4550-557. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, W., J. Püls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 421337-1348. [DOI] [PubMed] [Google Scholar]
- 16.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Vallvé, S., P. J. Janssen, and C. A. Ouzounis. 2002. Genetic variation between Helicobacter pylori strains: gene acquisition or loss? Trends Microbiol. 10445-447. [DOI] [PubMed] [Google Scholar]
- 18.Gerhard, M., N. Lehn, N. Neumayer, T. Borén, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 9612778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghose, C., G. I. Perez-Perez, L. J. van Doorn, M. G. Domínguez-Bello, and M. J. Blaser. 2005. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J. Clin. Microbiol. 432635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go, M. F. 1996. Helicobacter pylori: its role in ulcer disease and gastric cancer and how to detect the infection. Acta Gastroenterol. Latinoam. 2645-49. [PubMed] [Google Scholar]
- 21.Graham, J. E., R. M. Peek, Jr., U. Krishna, and T. L. Cover. 2002. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology 1231637-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gressmann, H., B. Linz, R. Ghai, K. P. Pleissner, R. Schlapbach, Y. Yamaoka, C. Kraft, S. Suerbaum, T. F. Meyer, and M. Achtman. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, Y. H., W. Z. Liu, Y. Z. Shi, L. Q. Lu, S. Xiao, Q. H. Zhang, and G. P. Zhao. 2007. Comparative genomics profiling of clinical isolates of Helicobacter pylori in Chinese populations using DNA microarray. J. Microbiol. 4521-28. [PubMed] [Google Scholar]
- 24.Israel, D. A., A. S. Lou, and M. J. Blaser. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186275-280. [DOI] [PubMed] [Google Scholar]
- 25.Israel, D. A., N. Salama, C. N. Arnold, S. F. Moss, T. Ando, H. P. Wirth, K. T. Tham, M. Camorlinga, M. J. Blaser, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Investig. 107611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 9814625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen, P. J., B. Audit, and C. A. Ouzounis. 2001. Strain-specific genes of Helicobacter pylori: distribution, function and dynamics. Nucleic Acids Res. 294395-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersulyte, D., B. Velapatiño, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 1853764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehours, P., S. Dupouy, B. Bergey, A. Ruskoné-Foumestraux, J. C. Delchier, R. Rad, F. Richy, J. Tankovic, F. Zerbib, F. Mégraud, and A. Ménard. 2004. Identification of a genetic marker of Helicobacter pylori strains involved in gastric extranodal marginal zone B cell lymphoma of the MALT-type. Gut 53931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh, J. T., and T. L. Cover. 2006. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect. Immun. 743052-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, H., P. I. Hsu, D. Y. Graham, and Y. Yamaoka. 2005. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 128833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 161311-1315. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, B. J. 1994. Helicobacter pylori. Am. J. Gastroenterol. 89S116-S128. [PubMed] [Google Scholar]
- 34.Momynaliev, K. T., O. V. Smirnova, L. V. Kudryavtseva, and V. M. Govorun. 2003. Comparative genome analysis of Helicobacter pylori strains. Mol. Biol. 37529-536. [PubMed] [Google Scholar]
- 35.Morales-Espinosa, R., G. Castillo-Rojas, G. Gonzalez-Valencia, S. Ponce de León, A. Cravioto, J. C. Atherton, and Y. López-Vidal. 1999. Colonization of Mexican patients by multiple Helicobacter pylori strains with different vacA and cagA genotypes. J. Clin. Microbiol. 373001-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura, A., G. N. Stemmermann, P. H. Chyou, G. I. Perez-Perez, and M. J. Blaser. 1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med. 120977-981. [DOI] [PubMed] [Google Scholar]
- 37.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Mégraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 686240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh, J. D., H. Kling-Bäckhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 1039999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 3301310-1311. [DOI] [PubMed] [Google Scholar]
- 40.Peek, R. M., Jr., G. G. Miller, K. T. Tham, G. I. Perez-Perez, X. Zhao, J. C. Atherton, and M. J. Blaser. 1995. Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab. Investig. 73760-770. [PubMed] [Google Scholar]
- 41.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians 110531-544. [PubMed] [Google Scholar]
- 42.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 1883449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phadnis, S. H., D. Ilver, L. Janzon, S. Normark, and T. U. Westblom. 1994. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect. Immun. 621557-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizwan, M., A. Alvi, and N. Ahmed. 2007. Novel protein antigen (JHP940) from the genomic plasticity region of Helicobacter pylori induces TNF-alpha and interleukin-8 secretion by human macrophages. J. Bacteriol. 1901146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 9714668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salama, N. R., G. Gonzalez-Valencia, B. Deatherage, F. Aviles-Jimenez, J. C. Atherton, D. Y. Graham, and J. Torres. 2007. Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J. Bacteriol. 1893834-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos, A., D. M. Magalhães Queiroz, A. Ménard, A. Marais, G. Aguiar Rocha, C. Affonso Oliveira, A. M. Miguel Ferreira Nogueira, M. Uzeda, and F. Mégraud. 2003. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J. Clin. Microbiol. 411651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutera, V. A., Jr., E. S. Han, L. A. Rajman, and S. T. Lovett. 1999. Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J. Bacteriol. 1816098-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, N. S., J. G. Fox, N. S. Akopyants, D. E. Berg, N. Thompson, B. Shames, L. Yan, E. Fontham, F. Janney, and F. M. Hunter. 1995. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J. Clin. Microbiol. 33918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terry, C. E., L. M. McGinnis, K. C. Madigan, P. Cao, T. L. Cover, G. W. Liechti, R. M. Peek, Jr., and M. H. Forsyth. 2005. Genomic comparison of cag pathogenicity island (PAI)-positive and-negative Helicobacter pylori strains: identification of novel markers for cag PAI-positive strains. Infect. Immun. 733794-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 52.Weel, J. F., R. W. van der Hulst, Y. Gerrits, P. Roorda, M. Feller, J. Dankert, G. N. Tytgat, and A. van der Ende. 1996. The interrelationship between cytotoxin-associated gene A, vacuolating cytotoxin, and Helicobacter pylori-related diseases. J. Infect. Dis. 1731171-1175. [DOI] [PubMed] [Google Scholar]
- 53.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 715921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 977533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaoka, Y. 2008. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J. Med. Microbiol. 57545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.