Abstract
Arsenic resistance in Synechocystis sp. strain PCC 6803 is mediated by an operon of three genes in which arsC codes for an arsenate reductase with unique characteristics. Here we describe the identification of two additional and nearly identical genes coding for arsenate reductases in Synechocystis sp. strain PCC 6803, which we have designed arsI1 and arsI2, and the biochemical characterization of both ArsC (arsenate reductase) and ArsI. Functional analysis of single, double, and triple mutants shows that both ArsI enzymes are active arsenate reductases but that their roles in arsenate resistance are essential only in the absence of ArsC. Based on its biochemical properties, ArsC belongs to a family that, though related to thioredoxin-dependent arsenate reductases, uses the glutathione/glutaredoxin system for reduction, whereas ArsI belongs to the previously known glutaredoxin-dependent family. We have also analyzed the role in arsenate resistance of the three glutaredoxins present in Synechocystis sp. strain PCC 6803 both in vitro and in vivo. Only the dithiolic glutaredoxins, GrxA (glutaredoxin A) and GrxB (glutaredoxin B), are able to donate electrons to both types of reductases in vitro, while GrxC (glutaredoxin C), a monothiolic glutaredoxin, is unable to donate electrons to either type. Analysis of glutaredoxin mutant strains revealed that only those lacking the grxA gene have impaired arsenic resistance.
Arsenic is a ubiquitous pollutant that can produce cancer and cause serious health problems in certain parts of the world (36). Because of the wide distribution of arsenic compounds, arsenic resistance is widespread among living organisms (47). Most resistance systems reduce arsenate to arsenite and export the latter to the outside of the cell or transport it to a vacuole (31). Arsenate reduction to arsenite is catalyzed by arsenate reductase, an enzyme at least three families of which have been described (25, 30, 43). The first family is exemplified by Escherichia coli ArsC, a protein that uses the glutathione (GSH)/glutaredoxin system as a reducing system and has a single catalytic cysteine. The second family is represented by Staphylococcus aureus and Bacillus subtilis arsenate reductases (also named ArsC). These enzymes are related to low-molecular-weight protein phosphotyrosine phosphatases and use thioredoxin as a reducing system through an intramolecular redox cascade requiring three cysteines for arsenate reduction (25). The last family is present only in eukaryotic organisms and was initially described in Saccharomyces cerevisae and, more recently in Leishmania major and Arabidopsis thaliana (6, 32, 53). These arsenate reductases also use the GSH/glutaredoxin system as a reductant, but they are related not to the E. coli reductase but rather to the Cdc25 family of protein phosphatases.
Arsenic resistance in Synechocystis sp. strain PCC 6803 is mediated by an operon of three genes that is regulated by an unlinked arsR homolog. The operon includes an arsenite transporter gene, arsB; an arsH homolog without a clear function in arsenic resistance; and an arsenate reductase gene, arsC (23), whose product is referred to below as ArsCsyn. ArsCsyn belongs to a new type of hybrid arsenate reductases that, though related to thioredoxin-dependent arsenate reductases, use the GSH/glutaredoxin system for reduction (19).
Glutaredoxins are small proteins first discovered as electron donors for ribonucleotide reductase but subsequently shown to be able to reduce other proteins, such as S-phosphoadenosylsulfate reductase and arsenate reductase. They are also able to reduce mixed disulfides between proteins and GSH (10). Two families of glutaredoxins have been described: dithiolic glutaredoxins (which have two cysteines in their catalytic site) and monothiolic glutaredoxins (which only have one cysteine in their catalytic site). To date, despite extensive biochemical characterization of different glutaredoxin isoforms from plants (44), little is known about glutaredoxin function in photosynthetic organisms, although recent biochemical approaches have shed some light (2, 17, 18, 38, 52).
Here we report the identification of two additional and nearly identical genes coding for arsenate reductases. We have designated them arsI1 and arsI2, because these two genes code for identical proteins belonging to the E. coli family of arsenate reductases. They are shown to play secondary roles in arsenate resistance in Synechocystis, since deletion of either of these genes has an effect only in combination with the absence of arsC. We have also performed biochemical characterization of the ArsCsyn and ArsI proteins and have found that both proteins have glutaredoxin-dependent arsenate reductase activity. Since both proteins are capable of using the GSH/glutaredoxin system, we have studied the role of this system in arsenate reduction. Synechocystis sp. strain PCC 6803 contains three genes encoding glutaredoxins. Two of these genes code for prokaryotic dithiolic glutaredoxins, while the third codes for a monothiolic glutaredoxin. In this study we have analyzed the roles of these glutaredoxins in arsenate resistance and have found that only one of the dithiolic glutaredoxins, GrxA, is essential in vivo. Nevertheless, both of the dithiolic enzymes are active in the arsenate reductase reaction in vitro.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Synechocystis sp. strain PCC 6803 was grown photoautotrophically at 30°C in BG11 medium supplemented with 1 g per liter of HCO3Na (BG11C) (40) and bubbled with a continuous stream of 1% (vol/vol) CO2 in air under continuous fluorescent illumination (50 μmol of photons per m2 per s; white light). For plate cultures, BG11C liquid medium was supplemented with 1% (wt/vol) agar. Antibiotics were added to final concentrations of 50 to 200 μg/ml for kanamycin, 20 to 40 μg/ml for chloramphenicol, or 2 to 10 μg/ml for spectinomycin when required. BG11C medium was supplemented with different concentrations of Na2HAsO4 in the presence or the absence of phosphate at the standard concentration as indicated for BG11C.
Escherichia coli DH5α (Bethesda Research Laboratories) grown in Luria-Bertani medium as described in reference 45 was used for plasmid construction and replication. E. coli BL21(DE3) and E. coli XL1-Blue grown in Luria-Bertani medium or Luria-Bertani medium supplemented with 2% glucose, respectively, were used for protein expression. E. coli cultures were supplemented with 100 μg/ml ampicillin, 40 μg/ml chloramphenicol, 50 μg/ml kanamycin, or 100 μg/ml spectinomicyn when required.
Insertional mutagenesis of Synechocystis genes.
All the oligonucleotides used for cloning and insertional or site-directed mutagenesis are described in Table S1 in the supplemental material. For the sll5104 and slr6037 insertional mutants, a 1,063-bp PCR fragment was amplified from total genomic DNA using oligonucleotides ARSI1 and ARSI2 and was cloned into pGEMT to generate pARSI1. Then an SpΩ (39) or C.C1 (8) cassette was inserted into a PflMI site in the arsI1 and arsI2 open reading frames (ORFs), generating pARSI2 and pARSI4, respectively. For double mutant construction, a 970-bp DNA fragment lacking the slr6037 ORF was constructed by two-step PCR using oligonucleotide pairs ARSI1-ARSI8 and ARSI7-NIY3, and antibiotic resistance cassettes were inserted into an NcoI site upstream of the slr6037 ORF, generating pARSI8 (SpΩ cassette) and pARSI9 (C.C1 cassette). For inactivation of grxA (ssr2061), a 1,269-bp DNA fragment was amplified using the oligonucleotide pair GRXA1-GRXA4 and cloned into pGEMT, and the C.C1 cassette was inserted into an SspI site located in the grxA ORF, generating pGRXA2. For inactivation of grxB (slr1562), an 845-bp DNA fragment was amplified using the oligonucleotide pair GRXB1-GRXB2 and cloned into pGEMT, and the SpΩ cassette was inserted into an RsaI site located in the grxB ORF, generating pGRXB2. For inactivation of grxC (slr1846), a 1,107-bp DNA fragment was amplified using the oligonucleotide pair GRXC1-GRXC2 and cloned into pGEMT, and the C.K1 cassette (8) was inserted into a BstEII site, blunt ended with Klenow enzyme, and placed in the grxC ORF, generating pGRXC2. Wild-type (WT) Synechocystis sp. strain PCC 6803 or different mutants were transformed with the plasmids listed in Table 1 to generate all the mutants used in this study.
TABLE 1.
Strains used in this work
| Strain | Genotype | Parental strain | Mutated ORF(s) | Plasmid used for transformation | Antibiotic resistance |
|---|---|---|---|---|---|
| WT | Synechocystis sp. strain PCC 6803 | ||||
| SARS4 | arsC::C.K1 | WT | slr0946 | pARSC2 | Km |
| SARS5 | arsR::C.C1 | WT | sll1957 | pARSR2 | Cm |
| SARS6 | arsI1::SpΩ | WT | sll5104 | pARSI2 | Sp |
| SARS7 | arsI2::SpΩ | WT | slr6037 | pARSI2 | Sp |
| SARS8 | ΔarsI2::SpΩ | WT | slr6037 | pARSI8 | Sp |
| SARS9 | arsI1::SpΩ ΔarsI2::C.C1 | SARS6 | slr6037, sll5104 | pARSI9 | Sp Cm |
| SARS10 | arsC::C.K1 arsI1::C.C1 | SARS4 | slr0946, sll5104 | pARSI4 | Km Cm |
| SARS11 | arsC::C.K1 ΔarsI2::SpΩ | SARS4 | slr0946, slr6037 | pARSI8 | Km Sp |
| SARS12 | arsC::C.K1 arsI1::C.C1 ΔarsI2::SpΩ | SARS10 | slr0946, slr6037, sll5104 | pARSI8 | Km Cm Sp |
| SGRXA | grxA::C.C1 | WT | ssr2061 | pGRXA2 | Cm |
| SGRXB | grxB::SpΩ | WT | slr1562 | pGRXB2 | Sp |
| SGRXC | grxC::C.K1 | WT | slr1846 | pGRXC2 | Km |
| SGRXAB | grxA::C.C1 grxB::SpΩ | SGRXA | ssr2061, slr1562 | pGRXB2 | Cm Sp |
| SGRXAC | grxA::C.C1 grxC::C.K1 | SGRXA | ssr2061, slr1846 | pGRXC2 | Cm Km |
| SGRXBC | grxB::SpΩ grxC::C.K1 | SGRXB | slr1562, slr1846 | pGRXC2 | Sp Km |
| SGRXABC | grxA::C.C1 grxB::SpΩ grxC::C.K1 | SGRXAB | ssr2061, slr1562, slr1846 | pGRX2C | Cm Sp Km |
Correct integration and complete segregation of the mutant strain were analyzed by Southern blotting. To this end, total DNA from cyanobacteria was isolated as previously described (4). DNA was digested with appropriate enzymes, electrophoresed on 0.7% agarose gels in a Tris-borate-EDTA buffer system (45), and subsequently transferred to nylon Z-probe membranes (Bio-Rad). DNA probes were 32P labeled with a random-primer kit (Amersham Biosciences) using [α-32P]dCTP (3,000 Ci/mmol).
Cloning and expression of ArsCsyn, ArsI, GrxA, GrxB, and GrxC.
Proteins were expressed with either a carboxy-terminal (ArsCsyn and ArsI) or an amino-terminal (GrxA, GrxB, and GrxC) six-histidine tag. The arsC gene was amplified from cosmid cs0223 (provided by Kazusa DNA Research Institute) using oligonucleotides ARSC3 and ARSC5, which introduced NdeI and XhoI restriction sites, respectively. The DNA fragment obtained was digested with NdeI-XhoI and cloned into NdeI-XhoI-digested pET24. The arsI gene was amplified using oligonucleotides ArsI1 and ArsI3 (which introduces an XhoI restriction site) and was cloned into NcoI-XhoI-digested pET24. The NcoI site is already included in the genomic sequence of arsI. grxA was amplified using GRXA5 (which introduces a BamHI restriction site) and GRXA4 from total genomic DNA. The corresponding DNA fragment was digested with BamHI and cloned into BamHI-digested pQE80, generating pGRXA4. grxB was amplified from total genomic DNA using oligonucleotides GRXB3 and GRXB5, which introduced BamHI and SalI restriction sites, respectively. The resulting DNA fragment was digested with BamHI and SalI and was cloned into BamHI-SalI-digested pQE80, generating pGRXB4. grxC was amplified using GRXC3 (which introduces a BamHI restriction site) and GRXC2 from total genomic DNA. The resulting DNA fragment was digested with BamHI and HindIII and was cloned into BamHI-HindIII-digested pQE80, generating pGRXC4.
ArsCsyn and ArsI were produced in E. coli BL21, and GrxA, GrxB, and GrxC in E. coli XL1-Blue, transformed with the plasmids described above. Cells were grown at 37°C to an optical density at 600 nm of 0.5; 1 mM IPTG (isopropyl-thio-β-d-galactopyranoside) was added; and the cultures were grown for 3 h at 30°C. The cells were harvested and frozen. Pellets were thawed on ice and resuspended in 20 ml of 50 mM Tris HCl (pH 8.0)-0.5 M NaCl (buffer A) per g (fresh weight). The cells were broken by sonication, and cell debris was removed by centrifugation at 50,000 × g for 20 min. The liquid supernatant was applied to a 1-ml HiTrap chelating column (Amersham Biosciences) charged with NiSO4 and was subsequently equilibrated in buffer A. The column was first washed with 100 ml of buffer A and then with 50 ml of buffer A supplemented with 25 mM imidazole, and adherent proteins were eluted by a 40-ml linear gradient from 0.025 to 0.5 M imidazole in buffer A. Imidazole and salts were removed by gel filtration using HiTrap desalting columns (Amersham Biosciences) run in 50 mM Tris-HCl pH (8.0)-150 mM NaCl.
Arsenate reductase assay.
Arsenate reductase activity was measured as described previously in a coupled assay with GSH and glutathione reductase (19). Assays for glutaredoxin Km determination were carried out in 50 mM Tris-HCl (pH 7.5), 10 mM GSH, 50 mM Na2HAsO4, 1 U of yeast glutathione reductase (Sigma), 5 μM of ArsCsyn or 1 μM of ArsI, and different amounts of glutaredoxins. Activity was monitored spectrophotometrically at 340 nm and was calculated using a ɛ of 6.22 mM for NADPH. We calculated kinetic parameters using double-reciprocal plots.
ArsCsyn site-directed mutagenesis.
The arsC gene was mutated by two overlapping PCR fragments introducing the desired changes using oligonucleotide pairs ARSCmut1-ARSC3 and ARSCmut2-ARSC5 in the first PCR and ARSC3-ARSC5 in the second. The PCR product was digested with NcoI and XhoI and was then cloned into the NcoI-XhoI-digested vector pET24. The mutant ArsCsyn protein was expressed and purified by the same procedure described for wild-type ArsCsyn.
ArsCsyn-TrxA(C35S) interaction.
Interaction assays were carried out by incubating 10 μg of WT or mutant ArsCsyn with 10 μg of TrxA(C35S), prepared as described in reference 20, in 50 mM Tris-HCl (pH 8.0) in the presence or absence of 100 mM Na2HAsO4 at room temperature for 30 min. Reactions were stopped by the addition of 1 volume of Laemmli buffer without reductant. Samples were boiled for 5 min, and 1 μg was used for Western blotting using 12% sodium dodecyl sulfate (SDS)-nonreducing polyacrylamide gel electrophoresis (PAGE) and anti-TrxA antibodies (35) at a 1:10,000 dilution.
RNA isolation and Northern blot analysis.
Total RNA was isolated from 25-ml samples of Synechocystis cultures at the mid-exponential phase (3 to 5 μg chlorophyll/ml). Extractions were performed by vortexing cells in the presence of phenol:chloroform and acid-washed baked glass beads (diameter, 0.25 to 0.3 mm; Braun, Melsungen, Germany) as previously described (12) For Northern blotting, 15 μg of total RNA was loaded per lane, electrophoresed in 1.0% agarose denaturing formaldehyde gels, and transferred to nylon membranes (Hybond N-plus; Amersham Biosciences). Prehybridization, hybridization, and washes were performed as described in the Amersham instruction manual. Probes for Northern blot hybridization were synthesized by PCR using oligonucleotide pairs ARSC1-ARSC2, ARSI3-ARSI4, GRXA250F-GRXA250F, GrxB331F-GrxB331R, and GrxC295F-GrxC295R (see Table S1 in the supplemental material) for arsC, arsI, grxA, grxB, and grxC, respectively. As a control, in all cases the filters were stripped and reprobed with a 580-bp HindIII-BamHI probe from plasmid pAV1100 containing the constitutively expressed RNase P RNA gene (rnpB) from Synechocystis sp. strain PCC 6803 (49). DNA probes were 32P labeled with a random-primer kit (Amersham Biosciences) using [α-32P]dCTP (3,000 Ci/mmol).
RESULTS
Identification and mutagenesis of two new additional arsenate reductase genes.
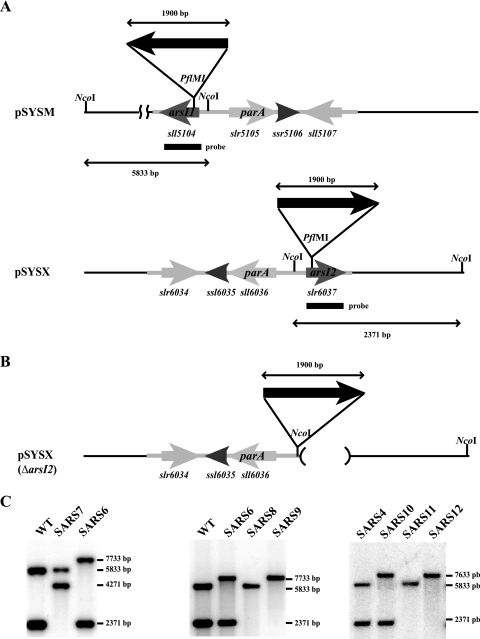
Sequence inspection of the four major Synechocystis plasmids allowed us to identify two putative genes (sll5104 and slr6037) encoding an arsenate reductase related to that of E. coli (49% identity to E. coli ArsC) (15). We designated these genes arsI1 and arsI2, respectively. The arsI genes are almost identical at the nucleotide level (99% identity) and encode identical proteins, probably reflecting a recent duplication event. They are located in a 2.5-kb region duplicated between pSYSM and pSYSX that contains three additional ORFs: one encoding a putative cytidine deaminase (sll5107 and slr6034), one small ORF encoding a protein with an unknown function (ssr5106 and ssl6035), and the third ORF encoding a ParA homolog (slr5105 and sll6036) (Fig. 1; note that although this region is duplicated, its orientation is reversed). The arsI genes are expressed at very low levels, and neither is regulated by the presence of arsenic in the medium (Fig. 2) or by arsR (data not shown).
FIG. 1.
ORF organization of arsI-containing regions in pSYSM and pSYSX. (A) Schematic representation of the pSYSM and pSYSX regions containing arsI. The repeated sequence is shaded. Arrows indicate the direction of transcription. Filled arrows represent antibiotic resistance cassettes, although for simplicity, only SpΩ is shown. (B) Schematic representation of the pSYSX ΔarsI region present in strains SARSI8, SARSI9, SARSI11, and SARS12. (C) Southern blot analysis of strains SARS4, SARS6, SARS7, SARS8, SARS9, SARS10, SARS11, and SARS12. Genomic DNA was digested with NcoI and hybridized using the same DNA fragment indicated in panel A as a probe. The sizes of the hybridizing bands are given on the right of the gels.
FIG. 2.
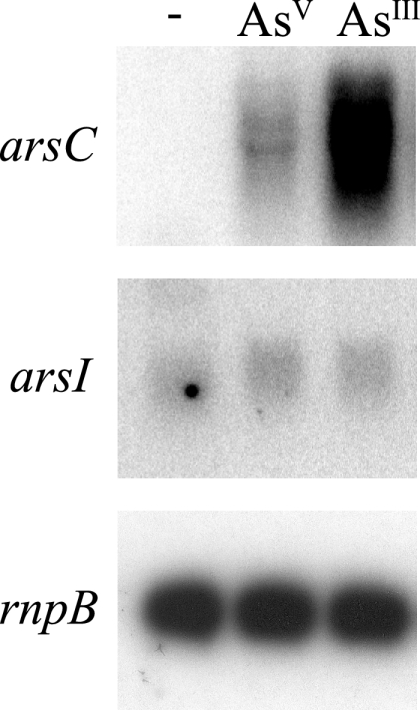
Northern blot analysis of the expression of the arsC and arsI genes. Total RNA was isolated from mid-log-phase Synechocystis cells grown in BG11C medium and exposed for 1 h either to 1 mM sodium arsenite [As(III)] or to 10 mM sodium arsenate [As(V)]. Control cells were not exposed to added compounds. Fifteen micrograms of total RNA was denatured, separated by electrophoresis on a 1% agarose gel, blotted, and hybridized with probes for arsC and arsI. The filters were stripped and rehybridized with an rnpB gene probe as a control (see Materials and Methods).
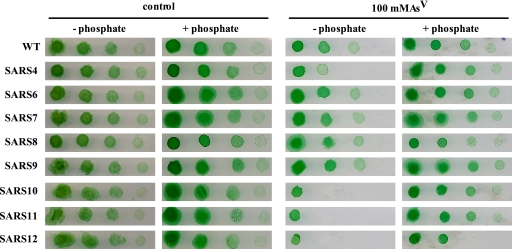
To investigate whether the presence of these additional arsenate reductases could explain the highly arsenate resistant phenotype of a Synechocystis arsC mutant, strain SARS4 (previously named arsC::CK1 [23]) (Table 1), we carried out insertional mutagenesis of the arsI1 and arsI2 genes. After transformation of the WT strain or SARS4 with plasmid pARSI2 or pARSI4, we obtained mutants with alterations in either arsI1 or arsI2 (66% arsI1 versus 33% arsI2). Subsequently, strains with arsI1 or arsI2 inactivated (SARS6, SARS7, and SARS10) were used to generate double or triple mutants using plasmids with an insertion of a different antibiotic resistance cassette in the other arsI gene. After several attempts, we still were unable to obtain mutant strains with both arsI genes inactivated, since the gene already inactivated was always partially or totally replaced by the new allele, even if both antibiotics were used in the selection step. In order to obtain double arsI mutants, we constructed plasmids in which the arsI2 downstream flanking region (not present in pSYSM [Fig. 1]) was included but the entire arsI2 ORF was deleted (from 46 bp upstream to 45 bp downstream of the arsI2 ORF) and a resistance cassette was inserted 205 bp before the starting ATG of the arsI2 ORF (pARSI8 and pARSI9). This target integration to pSYSX as an arsI2 downstream flanking region, which is included in this plasmid, is not found in pSYSM. Transformation of the WT strain, SARS4, or SARS10 with plasmid pARSI8 and of SARS6 with pARSI9 resulted in deletion of the arsI2 gene, allowing us to study the role of each arsenate reductase in arsenic resistance in Synechocystis sp. strain PCC 6803. Single mutants of any of the arsI genes or double mutants of both arsI genes, independently of the alleles (Fig. 3 and data not shown), displayed the same resistance to the presence of arsenate as the WT strain, probably due to the presence of ArsC (Fig. 3). In contrast, inactivation of either of the two arsI genes in an arsC::C.K1 background rendered the cells more sensitive to arsenate in the medium (Fig. 3, strains SARS10 and SARS11). We have previously studied arsenate toxicity in media with reduced amounts of phosphate (23) (a condition that does not affect the growth of any of the strains tested [Fig. 3]), since this compound has a protective effect against arsenate toxicity, probably affecting arsenate transport (37, 48) (Fig. 3). The strain in which all genes encoding arsenate reductases are inactivated (SARS12) was sensitive to arsenate even in the presence of the standard phosphate concentration in BG11; under the same conditions, none of the other strains were affected (Fig. 3). This indicates that the arsI genes are essential only in the absence of ArsCsyn and that both arsI genes are active in arsenate resistance.
FIG. 3.
Phenotypic characterization of arsI mutants. The tolerance of WT Synechocystis sp. strain PCC 6803 and strains SARS4, SARS6, SARS7, SARS8, SARS9, SARS10, SARS11, and SARS12 to arsenate was examined. Tenfold serial dilutions were spotted onto low-phosphate BG11C plates (− phosphate) supplemented with 100 mM sodium arsenate [As(V)], BG11C plates (+ phosphate) supplemented with 100 mM sodium arsenate [As(V)], or low-phosphate BG11C plates or BG11C plates without additions. Plates were photographed after 10 days of growth.
Biochemical characterization of ArsCsyn and ArsI.
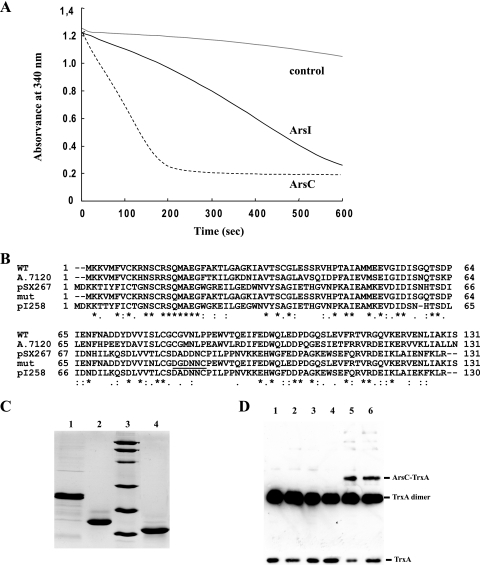
In order to study both types of arsenate reductases, we overexpressed and purified them by metal affinity chromatography. First we confirmed that ArsCsyn presents GSH-dependent arsenate reductase activity by using E. coli Grx1 as previously described (19) (Fig. 4A). ArsI displays the same activity (Fig. 4A), as predicted by its high similarity to E. coli ArsC and its conservation of residues essential for arsenate reductase activity. Although a biochemical characterization of ArsCsyn has been published previously, that study was carried out using E. coli Grx1, a heterologous electron donor (19). We aimed at characterizing the biochemical properties of ArsCsyn by using its physiological electron donor, and to that end we have used Synechocystis glutaredoxins. Synechocystis sp. strain PCC 6803 contains three ORFs coding for glutaredoxin proteins: ssr2061, slr1562, and slr1846 (16). The first two ORFs code for proteins that belong to the bacterial dithiolic family, and we have designated them GrxA and GrxB, respectively, while the third ORF encodes a protein, here designated GrxC, that belongs to the newly described monothiolic family (28, 41). We purified these glutaredoxins fused to an amino-terminal histidine tag and analyzed whether they were able to reduce ArsCsyn and ArsI. We found that only the dithiolic glutaredoxins (GrxA and GrxB) are able to reduce both arsenate reductases, while GrxC is totally inactive with both reductases. In this regard, ArsI showed a clear preference for GrxA, as reflected by a much lower Km (60 nM for GrxA versus 1.3 μM for GrxB), while ArsCsyn seemed to be able to use both GrxA and GrxB with almost the same efficiency, since it displayed similar Kms for the two glutaredoxins (0.25 μM and 0.994 μM, respectively) (Table 2). Since GrxA was the best reductant for both arsenate reductases, we used it to determine kinetic parameters for ArsCsyn and ArsI. Michaelis-Menten constants for arsenate were 5.74 mM and 11 mM for ArsCsyn and ArsI, respectively. The Vmax values were 2.81 and 1.3 μmol/min, and the kcat values were 33.72 s−1 and 79.2 s−1, for ArsCsyn and ArsI, respectively. These kinetic data for ArsCsyn and ArsI are in agreement with those previously published for ArsCsyn using a heterologous glutaredoxin (19) and for E. coli ArsC (21, 22) (Table 2).
FIG. 4.
ArsC has a unique catalytic site. (A) Glutaredoxin-dependent arsenate reductase activities of ArsC and ArsI. Portions (25 μg) of purified ArsC and ArsI were assayed with 1 μg of E. coli Grx1 following the couple reaction described in Materials and Methods. A control assay without glutaredoxin addition was carried out. (B) Sequence alignment of ArsCsyn (WT), mutant ArsCsyn (mut), Alr1105 from Anabaena sp. strain PCC 7120 (A.7120), and ArsC from Staphylococcus xylosus plasmid pSX267 or from S. aureus plasmid pI258. Identical amino acids are asterisked; conservative changes are marked with “:” or . as defined by CLUSTAL X. Changes to ArsCsyn in the mutant are underlined. (C) SDS-PAGE of purified mutant ArsCsyn (lane 1), WT ArsCsyn (lane 2), and TrxA(C35S) (lane 4). Lane 3, molecular mass markers. (D) Western blot analysis of interaction experiment between ArsCsyn and TrxA(C35S). Proteins were incubated as described in Materials and Methods. Lanes 1 and 2, reaction mixtures containing TrxA(C35S); lanes 3 and 4, reaction mixtures containing TrxA(C35S) and WT ArsCsyn; lanes 5 and 6, reaction mixtures containing TrxA(C35S) and mutant ArsCsyn. Proteins were incubated in the presence of 100 mM Na2HAsO4 (lanes 1, 3, and 5) or in the absence of Na2HAsO4 (lanes 2, 4, and 6). The membrane was incubated with anti-TrxA antibodies as described in Materials and Methods.
TABLE 2.
Catalytic properties of ArsCsyn and ArsI
| Arsenate reductase |
Km (μM)
|
Kinetic parameter determined with GrxA
|
|||
|---|---|---|---|---|---|
| GrxA | GrxB | Km for AsO32− (mM) | Vmax (μmol/min) | kcat (s−1) | |
| ArsC | 0.25 | 0.994 | 5.74 | 2.81 | 33.72 |
| ArsI | 0.063 | 1.369 | 11 | 1.32 | 79.2 |
ArsCsyn belongs to a new family of arsenate reductases.
Amino acid sequence comparison showed that ArsCsyn is related to ArsC from S. aureus and B. subtilis (42% and 37% identity, respectively). However, although they are clearly related, their biochemical properties are markedly different: S. aureus ArsC uses thioredoxin as an electron donor, while ArsCsyn uses GSH/glutaredoxin (Fig. 4A) (19, 26). The phylogenetic analysis suggests that the ArsCsyn type of arsenate reductase is restricted to freshwater cyanobacteria and that no related sequences are found in other organisms, while the group of marine cyanobacteria contains only the glutaredoxin-dependent type of arsenate reductase represented by E. coli ArsC (see Table S2 in the supplemental material). ArsCsyn contains three catalytic cysteines: C8, C80, and C82 (19), corresponding to C10, C82, and C89 of S. aureus ArsC (Fig. 4B). These three cysteines are essential for enzyme activity (3, 13, 24, 26) and are strictly conserved in all cyanobacterial homologs. We speculated that the difference in spacing between the second and the third catalytic cysteines might explain why ArsCsyn uses GSH and glutaredoxin as electron donors instead of thioredoxin, since thioredoxin would be unable to reduce the disulfide bridge formed between these two adjacent cysteines. In order to verify this hypothesis, we constructed a modified arsC gene where the spacing between active cysteines C80 and C82 was changed from 1 to 6 residues, as is the case for S. aureus ArsC (Fig. 4B). We minimized the number of changes introduced into ArsCsyn to those needed to change the spacing between the two catalytic cysteines, which led to a model with a loop, instead of an α-helix, in its oxidized state (see Discussion). The mutated protein was expressed and purified and was found to display abnormally reduced mobility in SDS-PAGE (Fig. 4C). The mutated protein was totally inactive in the coupled arsenate reductase assay using either the thioredoxin or the GSH/glutaredoxin reducing system (data not shown).
We also analyzed the capacity of ArsCsyn to interact with the most abundant thioredoxin (TrxA) in Synechocystis sp. strain PCC 6803 by using a modified version of TrxA in which the second active-site cysteine has been replaced by Ser [TrxA(C35S)], a mutation that allows the trapping of thioredoxin targets as mixed disulfides (20). We examined the abilities of WT and mutant versions of ArsCsyn to interact with TrxA(C35S) in vitro and asked if the presence of arsenate has any effect on this interaction. To this end, we incubated ArsCsyn for 15 min with or without 50 mM arsenate; then TrxA(C35S), at an equimolar concentration, was added to the reaction mixture, which was incubated for another 30 min. The reactions were stopped, and an aliquot was subjected to nonreducing SDS-PAGE and analyzed by Western blotting using anti-TrxA antibodies. While WT ArsCsyn was unable to interact with TrxA(C35S) (Fig. 4D, lanes 3 and 4), the mutated version of ArsCsyn was clearly able to form a mixed disulfide with TrxA(C35S) (Fig. 4D, lanes 5 and 6).
Glutaredoxin specificity in vivo.
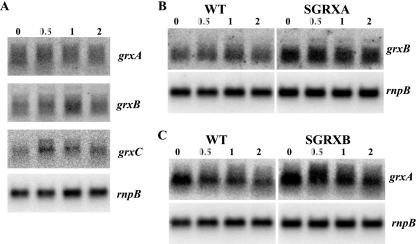
To address whether the glutaredoxins had any specificity in vivo, we studied glutaredoxin expression in response to the presence of arsenate in the medium. Total RNAs from mid-exponential-phase growing cells treated with 100 mM arsenate for 0.5, 1, and 2 h were analyzed by Northern blotting using probes against the grxA, grxB and grxC genes. While the levels of grxA and grxB transcripts did not change significantly after arsenate addition, even in the SGRXB or SGRXA mutant strain, respectively, grxC showed induction in its mRNA (Fig. 5).
FIG. 5.
Analysis of the expression of glutaredoxin genes in response to arsenate. (A) Northern blot analysis of the expression of the grxA, grxB, and grxC genes. Total RNA was isolated from mid-log-phase Synechocystis cells grown in BG11C medium and was exposed to 100 mM sodium arsenate for 0.5, 1, and 2 h. Control cells were not exposed. Fifteen micrograms of total RNA was denatured, separated by electrophoresis on a 1% agarose gel, blotted, and hybridized with probes for the grxA, grxB, and grxC genes. The filters were stripped and rehybridized with an rnpB gene probe as the control (see Materials and Methods). (B and C) Northern blot analysis of the expression of grxB in strain SGRXA (B) and of grxA in strain SGRXB (C) after arsenate exposure as described for panel A.
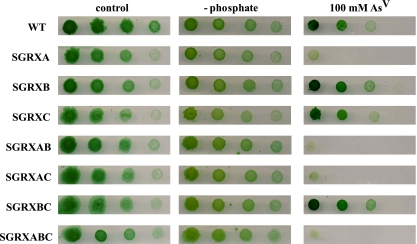
In order to further analyze the in vivo specificities of different glutaredoxins, we constructed single, double, and triple mutant strains with inactivated glutaredoxin genes. All mutants were viable, and their growth rates were comparable to those of the WT strain both in the presence and in the absence of phosphate (Fig. 6 and our unpublished results). However, analysis of their phenotypes in the presence of arsenate indicated that all strains carrying an insertion in the grxA gene were less resistant to the presence of arsenate than the WT strain (Fig. 6). Further inactivation of the grxB or grxC gene had no effect on arsenate resistance, indicating that GrxA is the main electron donor for both ArsCsyn and ArsI in vivo. These results are in agreement with our in vitro data, in which ArsI and ArsCsyn showed strong and moderate specificities for GrxA, respectively.
FIG. 6.
Phenotypic characterization of the grx mutants. The tolerance of WT Synechocystis sp. strain PCC 6803 and strains SGRXA, SGRXB, SGRXC, SGRXAB, SGRXAC, SGRXBC, and SGRXABC was tested. Tenfold serial dilutions were spotted onto low-phosphate BG11C plates supplemented with 100 mM sodium arsenate [As(V)], low-phosphate BG11C plates (− phosphate), or BG11C plates. Plates were photographed after 10 days of growth.
DISCUSSION
Here we describe the arsenate-reducing system in Synechocystis sp. strain PCC 6803, which consists of two different arsenate reductases and implies the GSH/glutaredoxin system as an electron donor. The presence of two different arsenate reductases has also been reported for other bacterial strains by genome analysis (5), but no in vivo experimental data have been reported so far. Although the main resistance determinant in Synechocystis sp. strain PCC 6803 involves the highly inducible ArsCsyn (23), ArsI is here shown to play a role in its absence. This enzyme could allow cells to tolerate small amounts of arsenate, because its gene is expressed at low and constitutive levels (Fig. 2). The fact that the differences in phenotype between SARS10 and SARS11 (double mutants with mutations of arsC and arsI), on the one hand, and SARS12 (triple mutant with all arsenate reductase genes inactivated), on the other, are observed only in media devoid of phosphate probably reflects the entry into the cells of small amounts of As(V), which are reduced by ArsI in strains SARS4, SARS10, and SARS11.
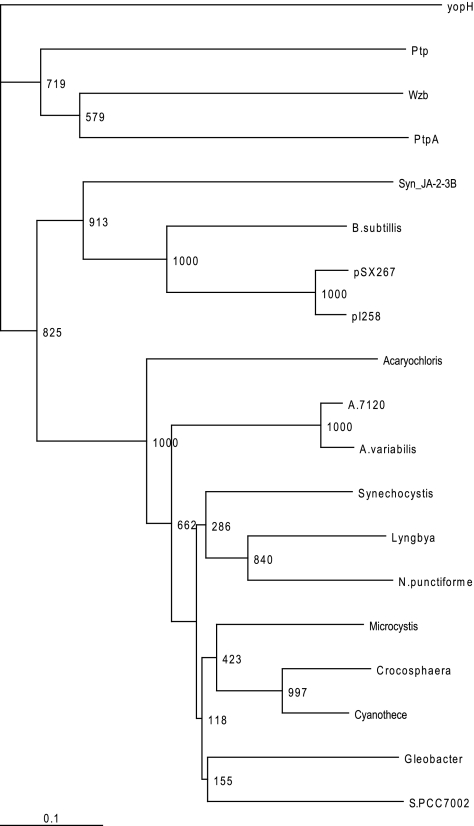
Three families of arsenate reductases have been described to date (25). Here we show that ArsCSyn, although related to thioredoxin-dependent arsenate reductases, can be considered a member of a new arsenate reductase family, since it clusters together with sequences from other cyanobacteria, forming an independent branch in phylogenetic trees (Fig. 7). The branch that includes ArsCSyn has probably shifted from a thioredoxin-dependent to a GSH-dependent reduction mechanism, and the signature for this can be found in the C80-C82 pair of cysteines (Fig. 4B). The arsenate reduction mechanism in the B. subtilis and S. aureus ArsC enzymes is well described and involves a disulfide cascade leading to conformational changes in the α-helix containing the reactive cysteines C82 and C89 (3, 24-27). These cysteines are present in an α-helix that is buried in the ArsC structure, and oxidation of these cysteines to a disulfide bridge leads to a change from an α-helix to a loop conformation. This structural reorganization exposes the disulfide bridge and allows it to be reduced by thioredoxin (3, 13, 24, 26). The change in the spacing between these two cysteines in ArsCsyn may explain why this protein is reduced by GSH but not by thioredoxin. Two reasons can be argued. The first is that a disulfide bridge between C80 and C82 probably would not cause a conformational switch from an α-helix to a loop conformation, because these cysteines are closely situated and will not disturb the α-helix. The second reason is that the spacing is not adequate for thioredoxin interaction. The site-directed mutagenesis of ArsCsyn supports the second explanation, although modeling of WT and mutant ArsCsyn enzymes using the SWISS-MODEL server with the sole constraint that the C80 and C82 cysteines form a disulfide bridge results in a model with an α-helix in the WT and a loop in the mutant (data not shown). Thus, a combination of both effects can be claimed to explain the difference in reduction mechanisms between S. aureus ArsC and ArsCsyn. Although a change in the spacing between the cysteines is essential for thioredoxin interaction (Fig. 4) and probably also for the conformational change induced upon oxidation of the cysteines, changes other than those introduced in this work may be needed to obtain thioredoxin-dependent arsenate reduction activity, since there are other residues conserved in S. aureus ArsC that are not conserved in ArsCsyn. The change from a thioredoxin- to a GSH-dependent mechanism in cyanobacterial arsenate reductase could be due to an easier availability of GSH than of reduced thioredoxin. In this regard, the more abundant thioredoxin (TrxA) is essential in Synechocystis and in other cyanobacteria (11, 33, 34), probably indicating its main role in regulating cell metabolism. Furthermore, other thioredoxins present in Synechocystis are expressed at lower levels and do not respond to the presence of arsenate in the growth medium (our unpublished observations). This is also supported by the fact that all cyanobacterial arsenate reductases use GSH as a reducing system (since they are homologs of ArsCsyn or E. coli ArsC [see Table S2 in the supplemental material), except for Synechococcus sp. strain JA-3-3Ab, which contains a gene coding for an ArsC protein of the S. aureus family (Fig. 7).
FIG. 7.
Phylogenetic analysis of ArsCsyn. Shown is a neighbor-joining tree of full-length amino acid sequences of the following arsenate reductases (sequences are from the ENTREZ protein database): ArsCSyn (Synechocystis), Alr1105 from Anabaena sp. strain PCC 7120 (A.7120; accession no. NP_485148), Ava 3712 from Anabaena variabilis (ZP_00159236), Glr0004 from Gloeobacter violaceus (NP_922950.1), and ArsC enzymes from Nostoc punctiforme (ZP_00108511), Crocosphaera watsonii WH 8501 (ZP_00349942), Lyngbya sp. strain PCC 8106 (ZP_01620002), Synechococcus sp. strain JA-2-3B′a (Syn_JA-2-3B; YP_478568), Acaryochloris marina MBIC11017 (YP_001514958), Synechococcus sp. strain PCC 7002 (S.PCC7002; YP_001733851), Microcystis aeruginosa NIES-843 (YP_001655268), Cyanothece sp. strain ATCC 51142 (YP_001804412), Bacillus subtilis (BAA12434), Staphylococcus xylosus plasmid pSX267 (Q0125), and S. aureus plasmid pI258 (P30330). Low-molecular-weight phosphatases are Wzb from E. coli (ECU38473), PtpA from Streptomyces coelicolor A3 (NP_628106), Ptp from Acintobacter johnsonii (CAA75430), and YopH from Yersinia pseudotuberculosis (P08538). YopH was used as the outgroup. The scale bar corresponds to 0.1 estimated amino acid substitution per site. Bootstrap values for 1,000 permutations are shown.
Interestingly, inactivation of all glutaredoxin genes does not seem to affect the growth of Synechocystis sp. strain PCC 6803. In most organisms, inactivation of all glutaredoxin genes is not possible (7, 9). Recently it has been reported that GrxA inactivation impairs H2O2 resistance in Synechocystis, that GrxA is reduced by a thioredoxin reductase, and that reduced GrxA is able to reduce GrxB (18). In addition, these investigators identified several putative targets that overlap with those of the thioredoxin system in Synechocystis. These data suggest that the two systems have redundant roles and that in the absence of the glutaredoxin system, thioredoxins can reduce some of its targets. Particularly striking is the lack of a clear phenotype of the SGRXC mutant, since all homologs of grxC either are essential or exhibit a drastic phenotype (2, 9, 41, 51). Synechocystis GrxC has been shown to contain an iron-sulfur group (38), like all other monothiolic glutaredoxins (9, 42), and can partially complement the grx5 mutation in Saccharomyces cerevisiae (28), suggesting that GrxC is involved in iron-sulfur assembly, as are plant and yeast monothiolic glutaredoxins (2, 42). Monothiolic glutaredoxin has been proposed to be a scaffold, storage, or regulatory protein during Fe-S cluster assembly in plant chloroplasts (2). Two Fe-S cluster assembly pathways, Suf and Isc, have been described for cyanobacteria (1, 14, 29, 50). The Suf system is involved in Fe-S cluster assembly for photosystem I (with the help of a NifU like protein), while the role of the Isc system is less well characterized. Further experiments are required to clarify if Synechocystis GrxC has roles similar to those of its plant homologs in Fe-S cluster assembly and to determine its relation to the Suf and Isc pathways.
Our in vivo experiments demonstrate that GrxA has a central role in arsenate reduction, since all strains lacking GrxA are less tolerant of the presence of arsenate in the medium than strains with GrxA. This phenotype is not due to a low level of expression of the gshB gene, encoding GSH synthetase, which is just downstream of the grxA gene and forms an operon with it (our unpublished observations). Moreover, SGRXA and WT strains contain the same levels of GSH (data not shown). Our results indicate that at least in Synechocystis sp. strain PCC 6803, and for ArsCSyn, there is a specific dependence on GrxA as an electron donor, and GrxB cannot compensate for the lack of GrxA in vivo. This is further supported by the lack of induction of the grxB or grxA gene in strain SGRXA or SGRXB, respectively, in response to the presence of arsenate (Fig. 5B and C). These data also contrast with data reported on arsenic resistance in E. coli, where resistance to arsenate was not affected in any single glutaredoxin mutant or in a triple mutant with all dithiolic glutaredoxin genes interrupted, suggesting that there is another reductant for E. coli ArsC (46). The strong specificity of ArsCsyn for GrxA in vivo contrasts with the much smaller differences observed between the specificities for GrxA and GrxB in vitro. Two reasons might explain this apparent discrepancy. First, GrxA protein may be much more abundant than GrxB. Second, the compartmentalization or spatial localization of GrxB may be different from that of ArsCsyn. Further experiments will be required to investigate these possibilities.
Supplementary Material
Acknowledgments
This research was supported by grants BMC2004-0050 and BFU2007-60300/BMC from the Ministry of Education and Science of Spain and by the Junta de Andalucia (Group BIO-0284), grant CVI-099. A.M.S.-R. is a recipient of a fellowship from the Ministerio de Educación y Ciencia (M.E.C.).
We thank the Kazusa DNA Research Institute for providing the cs02233 cosmid and Anna Lindahl for critical reading of the manuscript.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Balasubramanian, R., G. Shen, D. A. Bryant, and J. H. Golbeck. 2006. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 1883182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, S., F. Gama, M. M. Molina-Navarro, J. M. Gualberto, R. Claxton, S. G. Naik, B. H. Huynh, E. Herrero, J. P. Jacquot, M. K. Johnson, and N. Rouhier. 2008. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 271122-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, M. S., Z. Guan, M. Laurberg, and X. D. Su. 2001. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 9813577-13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1723138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cánovas, D., I. Cases, and V. de Lorenzo. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 51242-1256. [DOI] [PubMed] [Google Scholar]
- 6.Dhankher, O. P., B. P. Rosen, E. C. McKinney, and R. B. Meagher. 2006. Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2). Proc. Natl. Acad. Sci. USA 1035413-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draculic, T., I. W. Dawes, and C. M. Grant. 2000. A single glutaredoxin or thioredoxin gene is essential for viability in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 361167-1174. [DOI] [PubMed] [Google Scholar]
- 8.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68119-138. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes, A. P., M. Fladvad, C. Berndt, C. Andresen, C. H. Lillig, P. Neubauer, M. Sunnerhagen, A. Holmgren, and A. Vlamis-Gardikas. 2005. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J. Biol. Chem. 28024544-24552. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, A. P., and A. Holmgren. 2004. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 663-74. [DOI] [PubMed] [Google Scholar]
- 11.Florencio, F. J., M. E. Pérez-Pérez, L. López-Maury, A. Mata-Cabana, and M. Lindahl. 2006. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 89157-171. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Domínguez, M., and F. J. Florencio. 1997. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35723-734. [DOI] [PubMed] [Google Scholar]
- 13.Guo, X., Y. Li, K. Peng, Y. Hu, C. Li, B. Xia, and C. Jin. 2005. Solution structures and backbone dynamics of arsenate reductase from Bacillus subtilis: reversible conformational switch associated with arsenate reduction. J. Biol. Chem. 28039601-39608. [DOI] [PubMed] [Google Scholar]
- 14.Jin, Z., M. Heinnickel, C. Krebs, G. Shen, J. H. Golbeck, and D. A. Bryant. 2008. Biogenesis of iron-sulfur clusters in photosystem I: holo-NfuA from the cyanobacterium Synechococcus sp. PCC 7002 rapidly and efficiently transfers [4Fe-4S] clusters to apo-PsaC in vitro. J. Biol. Chem. 28328426-28435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, S. Sasamoto, A. Watanabe, M. Kohara, M. Matsumoto, S. Shimpo, M. Yamada, and S. Tabata. 2003. Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res. 10221-228. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3109-136. [DOI] [PubMed] [Google Scholar]
- 17.Li, M., W. Huang, Q. Yang, X. Liu, and Q. Wu. 2005. Expression and oxidative stress tolerance studies of glutaredoxin from cyanobacterium Synechocystis sp. PCC 6803 in Escherichia coli. Protein Expr. Purif. 4285-91. [DOI] [PubMed] [Google Scholar]
- 18.Li, M., Q. Yang, L. Zhang, H. Li, Y. Cui, and Q. Wu. 2007. Identification of novel targets of cyanobacterial glutaredoxin. Arch. Biochem. Biophys. 458220-228. [DOI] [PubMed] [Google Scholar]
- 19.Li, R., J. D. Haile, and P. J. Kennelly. 2003. An arsenate reductase from Synechocystis sp. strain PCC 6803 exhibits a novel combination of catalytic characteristics. J. Bacteriol. 1856780-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindahl, M., and F. J. Florencio. 2003. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 10016107-16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J., T. B. Gladysheva, L. Lee, and B. P. Rosen. 1995. Identification of an essential cysteinyl residue in the ArsC arsenate reductase of plasmid R773. Biochemistry 3413472-13476. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., and B. P. Rosen. 1997. Ligand interactions of the ArsC arsenate reductase. J. Biol. Chem. 27221084-21089. [DOI] [PubMed] [Google Scholar]
- 23.López-Maury, L., F. J. Florencio, and J. C. Reyes. 2003. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1855363-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messens, J., J. C. Martins, K. Van Belle, E. Brosens, A. Desmyter, M. De Gieter, J. M. Wieruszeski, R. Willem, L. Wyns, and I. Zegers. 2002. All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade. Proc. Natl. Acad. Sci. USA 998506-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messens, J., and S. Silver. 2006. Arsenate reduction: thiol cascade chemistry with convergent evolution. J. Mol. Biol. 3621-17. [DOI] [PubMed] [Google Scholar]
- 26.Messens, J., I. Van Molle, P. Vanhaesebrouck, M. Limbourg, K. Van Belle, K. Wahni, J. C. Martins, R. Loris, and L. Wyns. 2004. How thioredoxin can reduce a buried disulphide bond. J. Mol. Biol. 339527-537. [DOI] [PubMed] [Google Scholar]
- 27.Messens, J., I. Van Molle, P. Vanhaesebrouck, K. Van Belle, K. Wahni, J. C. Martins, L. Wyns, and R. Loris. 2004. The structure of a triple mutant of pI258 arsenate reductase from Staphylococcus aureus and its 5-thio-2-nitrobenzoic acid adduct. Acta Crystallogr. D 601180-1184. [DOI] [PubMed] [Google Scholar]
- 28.Molina-Navarro, M. M., C. Casas, L. Piedrafita, G. Belli, and E. Herrero. 2006. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 5802273-2280. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto, K., E. Yamashita, Y. Kondou, S. J. Lee, F. Arisaka, T. Tsukihara, and M. Nakai. 2006. The asymmetric IscA homodimer with an exposed [2Fe-2S] cluster suggests the structural basis of the Fe-S cluster biosynthetic scaffold. J. Mol. Biol. 360117-132. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay, R., and B. P. Rosen. 2002. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 110(Suppl. 5)745-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, R., B. P. Rosen, L. T. Phung, and S. Silver. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26311-325. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, R., J. Shi, and B. P. Rosen. 2000. Purification and characterization of ACR2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 27521149-21157. [DOI] [PubMed] [Google Scholar]
- 33.Muller, E. G., and B. B. Buchanan. 1989. Thioredoxin is essential for photosynthetic growth. The thioredoxin m gene of Anacystis nidulans. J. Biol. Chem. 2644008-4014. [PubMed] [Google Scholar]
- 34.Navarro, F., and F. J. Florencio. 1996. The cyanobacterial thioredoxin gene is required for both photoautotrophic and heterotrophic growth. Plant Physiol. 1111067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro, F., E. Martín-Figueroa, and F. J. Florencio. 2000. Electron transport controls transcription of the thioredoxin gene (trxA) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 4323-32. [DOI] [PubMed] [Google Scholar]
- 36.Nordstrom, D. K. 2002. Public health. Worldwide occurrences of arsenic in ground water. Science 2962143-2145. [DOI] [PubMed] [Google Scholar]
- 37.Oden, K. L., T. B. Gladysheva, and B. P. Rosen. 1994. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol. Microbiol. 12301-306. [DOI] [PubMed] [Google Scholar]
- 38.Picciocchi, A., C. Saguez, A. Boussac, C. Cassier-Chauvat, and F. Chauvat. 2007. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 4615018-15026. [DOI] [PubMed] [Google Scholar]
- 39.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 40.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignment, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1111-61. [Google Scholar]
- 41.Rodríguez-Manzaneque, M. T., J. Ros, E. Cabiscol, A. Sorribas, and E. Herrero. 1999. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 198180-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Manzaneque, M. T., J. Tamarit, G. Belli, J. Ros, and E. Herrero. 2002. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 131109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roos, G., L. Buts, K. Van Belle, E. Brosens, P. Geerlings, R. Loris, L. Wyns, and J. Messens. 2006. Interplay between ion binding and catalysis in the thioredoxin-coupled arsenate reductase family. J. Mol. Biol. 360826-838. [DOI] [PubMed] [Google Scholar]
- 44.Rouhier, N., E. Gelhaye, and J. P. Jacquot. 2004. Plant glutaredoxins: still mysterious reducing systems. Cell. Mol. Life Sci. 611266-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Shi, J., A. Vlamis-Gardikas, F. Aslund, A. Holmgren, and B. P. Rosen. 1999. Reactivity of glutaredoxins 1, 2, and 3 from Escherichia coli shows that glutaredoxin 2 is the primary hydrogen donor to ArsC-catalyzed arsenate reduction. J. Biol. Chem. 27436039-36042. [DOI] [PubMed] [Google Scholar]
- 47.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60107-130. [DOI] [PubMed] [Google Scholar]
- 48.Thiel, T. 1988. Phosphate transport and arsenate resistance in the cyanobacterium Anabaena variabilis. J. Bacteriol. 1701143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 206331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, T., G. Shen, R. Balasubramanian, L. McIntosh, D. A. Bryant, and J. H. Golbeck. 2004. The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J. Bacteriol. 186956-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingert, R. A., J. L. Galloway, B. Barut, H. Foott, P. Fraenkel, J. L. Axe, G. J. Weber, K. Dooley, A. J. Davidson, B. Schmid, B. H. Paw, G. C. Shaw, P. Kingsley, J. Palis, H. Schubert, O. Chen, J. Kaplan, and L. I. Zon. 2005. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 4361035-1039. [DOI] [PubMed] [Google Scholar]
- 52.Zaffagnini, M., L. Michelet, V. Massot, P. Trost, and S. D. Lemaire. 2008. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J. Biol. Chem. 2838868-8876. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, Y., N. Messier, M. Ouellette, B. P. Rosen, and R. Mukhopadhyay. 2004. Leishmania major LmACR2 is a pentavalent antimony reductase that confers sensitivity to the drug pentostam. J. Biol. Chem. 27937445-37451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.