Abstract
The green phototrophic bacteria contain a unique complement of chlorophyll pigments, which self-assemble efficiently into antenna structures known as chlorosomes with little involvement of protein. The few proteins found in chlorosomes have previously been thought to have a primarily structural function. The biosynthetic pathway of the chlorosome pigments, bacteriochlorophylls c, d, and e, is not well understood. In this report, we used spectroscopic, proteomic, and gene expression approaches to investigate the chlorosome proteins of the green filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. Surprisingly, Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase, AcsF, was identified under anaerobic growth conditions. The AcsF protein was found in the isolated chlorosome fractions, and the proteomics analysis suggested that significant portions of the AcsF proteins are not accessible to protease digestion. Additionally, quantitative real-time PCR studies showed that the transcript level of the acsF gene is not lower in anaerobic growth than in semiaerobic growth. Since the proposed enzymatic activity of AcsF requires molecular oxygen, our studies suggest that the roles of AcsF in C. aurantiacus need to be investigated further.
The unique chlorosome antenna complexes found in green phototrophic bacteria are the most densely packed pigmented light-harvesting complexes known and contain self-assembled bacteriochlorophyll (BChl) c, d, or e aggregates (1). Chlorosomes are central to the ability of green bacteria to carry out photosynthesis under very low light conditions (2). While most of the enzymes that contribute to the biosynthesis of BChl in protein-pigment light-harvesting antenna complexes have been investigated in detail, there have been few studies of the enzymes involved in synthesis of chlorosome pigments.
Chlorosomes contain relatively few proteins compared to other photosynthetic antenna systems. The functions of most of the chlorosome proteins remain to be understood. For example, while Chloroflexus aurantiacus is one of the most investigated green filamentous anoxygenic phototrophic (FAP) bacteria, the functions of the chlorosome proteins are completely unknown, except for CsmA, which is known to function as the baseplate pigment-binding protein and to mediate energy transfer from BChl c to BChl a in the integral light-harvesting complexes (13, 25).
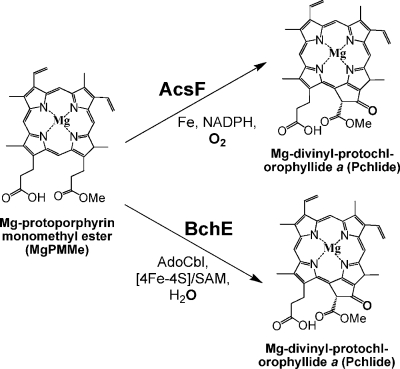
The genome of C. aurantiacus has been completely sequenced (http://genome.jgi-psf.org/finished_microbes/chlau/chlau.home.html), so combinations of biochemical and genetic studies have recently become possible. In this work, we used spectroscopic, proteomic, and gene expression approaches to investigate the chlorosome proteins and unexpectedly identified Mg-protoporphyrin IX monomethyl ester aerobic cyclase (AcsF) in chlorosomes under anaerobic growth conditions. Two cyclase enzymes are capable of forming the isocyclic ring (“E ring”) that is found in all (bacterio)chlorophylls: AcsF and Mg-protoporphyrin IX monomethyl ester anaerobic cyclase (BchE). The roles of the AcsF and BchE proteins have been suggested to be conversion of Mg-protoporphyrin IX monomethyl ester into Mg-divinyl-protochlorophyllide (PChlide) by catalyzing the isocyclic ring formation under aerobic and anaerobic conditions, respectively (Fig. 1) (21, 22). The acsF-like gene can be detected in diverse organisms, from bacteria to algae and higher plants, whereas the gene encoding BchE is found strictly in anaerobic bacteria. Interestingly, both the acsF and bchE genes exist in C. aurantiacus. As a result, alternative roles for AcsF in the chlorosomes and in anaerobic growth need to be considered, and some hypotheses are reported in this work.
FIG. 1.
Proposed isocyclic ring formation catalyzed by AcsF and BchE under aerobic and anaerobic conditions. The oxygen sources for AcsF and BchE are molecular oxygen and water molecule, respectively.
MATERIALS AND METHODS
Materials.
DNA oligomers used in this work were purchased from Integrated DNA Technologies and used without further purification. Enzymes and kits for the reported molecular biology studies are described below.
Cell cultures.
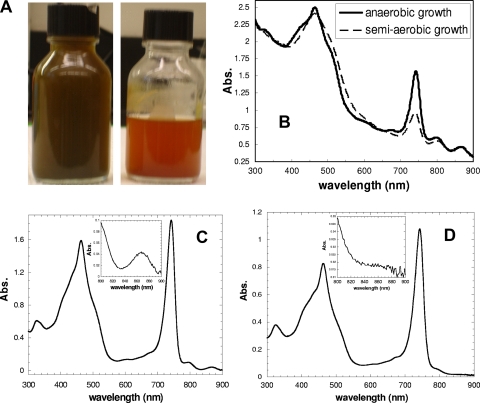
Chloroflexus aurantiacus J-10-fl cells were cultured in “D” medium as reported previously (7) under anaerobic and semiaerobic growth conditions at 48°C in low-intensity light (6 W/m2). Only one small air bubble in the incubation bottle was allowed for anaerobic growth, whereas approximately half of the volume in the 200-ml bottle was filled with medium for semiaerobic growth (see Fig. 2A). The cultures were harvested after 3 days, when the A863 and A742 values were 0.46 and 1.57 for anaerobic growth and 0.48 and 0.97 for the semiaerobic growth, respectively.
FIG. 2.
The appearance and spectra of C. aurantiacus cultures. The cultures were grown under semiaerobic (right) or anaerobic (left) conditions in low-intensity light (6 W/m2) (A). The spectra of C. aurantiacus cultures grown under anaerobic (solid line) or semiaerobic (dash line) conditions are shown (B). The spectrum of unfractionated chlorosomes and membrane (C) or the isolated chlorosome (D) is shown. The insets in panels C and D show the 800- to 900-nm spectral regions of the unfractionated chlorosome and purified chlorosome fraction, respectively. Abs., absorbance.
Isolation and characterization of chlorosomes.
C. aurantiacus cells were harvested by centrifugation at 5,471 × g for 15 min. After sonication and removal of the cell debris by centrifugation at 20,000 × g for 30 min, the membrane fraction was separated from the soluble fraction by ultracentrifugation at 200,000 × g for 2 h. The chlorosomes, located in the membrane fractions, were fractionated using a 15 to 45% sucrose density gradient as reported earlier (4, 26). The purified chlorosome fraction was characterized by the UV/visible spectrum and also subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (12% Tris-glycine).
SDS-PAGE analysis.
For SDS-PAGE analysis, the chlorosome fraction was incubated in methanol at room temperature for 10 min and centrifuged, and the supernatant liquid was removed to release the pigments and soluble components from the chlorosome envelope. The pellet (the chlorosome envelope) was then subjected to SDS-PAGE analysis.
Protein identification by MALDI-TOF fingerprinting.
In-gel protein digestion by trypsin for matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) analysis was used, following the procedure reported previously (6, 23). Briefly, the Coomassie G-250-stained SDS-PAGE (12.5% Tris-glycine) gel was destained, and bands were excised and minced into ∼1.0-mm3 pieces. After the Coomassie blue dye was removed, each gel sample was digested with ∼6 μg of proteomic-grade trypsin (Sigma) in either 500 μl of 50 mM ammonium bicarbonate or 20 mM Tris-HCl buffer (pH 8.0) at 37°C overnight. The digested peptides were extracted from the gel using 60% CH3CN and 1% trifluoroacetic acid solution and analyzed using MALDI-TOF (Applied Biosystems 4700 proteomics analyzer) measurements. The MALDI-TOF samples were prepared by mixing a 1:1 volume of sample:matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% CH3CN, 0.1% trifluoroacetic acid) on an ABI-192-AB stainless steel plate. Reflection positive mode was used for sample measurements. Each spectrum was averaged by summing 40 measurements with 50 laser shots/measurement. The proteins were identified by searching the peptide masses against the bacterial entries in the NCBI database using the Mascot software program (Matrix Science, London, United Kingdom). The oxidation of methionine, tryptophan, and histidine and the acrylamide adduct (C3H5NO) of cysteine were included as variable modifications during the database search.
In-solution trypsin digestion.
Purified chlorosomes (80 μl) were incubated with trypsin (6 μg) in 180 μl (final volume) of 20 mM Tris-HCl buffer (pH 8.0) at room temperature for 2 days. At the end of the incubation, the digested peptides were filtered by a Microcon YM-10 concentrator (Millipore) (28). The flowthrough fraction was colorless, indicating that the chlorosome envelope remains largely undisrupted after protease digestions. The digested peptides were desalted by using ZipTip C18 (Millipore), dried, and resuspended in double-distillation water. The liquid chromatography-tandem mass spectrometry (MS) studies were conducted using an LTQ-FT ICR machine (Thermo Fisher, San Jose, CA). Peptides were identified by searching the peptide accurate mass (accuracy < 10 ppm) and the tandem mass against the NCBI database using Mascot, with methionine, tryptophan, and histidine oxidation as the variable modifications.
Atomic absorption spectrum measurements.
The iron concentrations in the isolated chlorosome fraction were measured on an AA600 atomic absorption spectrometer (Perkin-Elmer, Ueberlingen, Germany) with a wavelength of 248.3 nm and Mg(NO3)2 as a modifier (9). The calibration was performed with 50, 16.6, and 5.5 μg/liter of Fe in the standard containing 100 μg/liter various metal ions. The chlorosome samples for the atomic absorption measurements were dialyzed against the metal-free distillation water before the measurements. The amount of Fe in the metal-free water is under the instrumental detection level (1.2 pg/liter or 0.0044 absorbance/s). Each measurement was carried out in triplicate, and the mean of triplicates was used.
RNA isolation and purification.
C. aurantiacus cells were harvested after 3 days' growth under semiaerobic or anaerobic conditions. RNA was isolated from the cell pellets using TRIzol reagent (Invitrogen) according to the manufacturer's protocol, and possible DNA contamination was further removed by DNase treatment. The genomic DNA contamination in the RNA samples was examined by PCR (data not shown). The RNA samples were quantified using a NanoPhotometer UV/visible spectrophotometer (Implen, Munich, Germany) and had an A260/A280 ratio of >1.95 prior to being subjected to the subsequent experiments. Two independent preparations of RNA samples under each growth condition were used to collect gene expression data (described below).
Quantitative real-time PCR (QRT-PCR).
QRT-PCR was carried out to profile the gene expression under different growth conditions of C. aurantiacus. cDNA was synthesized from 1 μg RNA and 100 μM random 9-mer DNA using Superscript III reverse transcriptase (Invitrogen). The QRT-PCRs were performed via the ABI 7500 real-time PCR system (Applied Biosystems). The primers for QRT-PCRs (shown in Table 1) were designed using the Primer Express 2.0 software program (PE Applied Biosystems) and analyzed by using the OligoAnalyzer 3.0 software program (Integrated DNA Technologies). The Power SYBR green master mix (PE Applied Biosystems) was used to amplify DNA with the following thermal cycles: an initial denaturation step (15 min at 95°C), followed by 40 amplification cycles (15 s at 95°C, 30 s at 60°C, and 45 s at 72°C) and then 1 dissociation cycle (15 s at 95°C, 1 min at 60°C, and then 15 s at 95°C). The data were analyzed as suggested in the manufacturer's protocol (Applied Biosystems). The threshold cycle (CT) was calculated as the cycle number at which ΔRn (the magnitude of the fluorescence intensity generated by the given set of PCRs) crossed the baseline. Data were normalized by calculating ΔCT = CT of the target gene − CT of the internal control gene (16S rRNA). Normalized ΔCT data from C. aurantiacus cells growing in the anaerobic condition were compared to data from C. aurantiacus cells growing in the semiaerobic condition, in which ΔΔCT = ΔCTanaerobic − ΔCTsemiaerobic. Each experiment was repeated six times for validation, and the mean value was reported (Table 2). Further, the amplified DNA fragments were verified by 1% agarose gel electrophoresis (see Fig. S1 in the supplemental material).
TABLE 1.
Primers used for QRT-PCR studies
| Gene | Forward primer(s) (5′-3′) | Reverse primer(s) (3′-5′) |
|---|---|---|
| 16S rRNA | AGATGTTGGGTTCAGTCCCG | TTACTAGCAACTCCGCCTTCAC |
| acsF | (1) TCTATGCCGAGATGGTTAAGCA | (1) CGCGGCTCATATACTTGAAAATG |
| (2) CACGCCGGCTTTCTCAA | (2) GCTTCTGCAACACCGTCAGA | |
| bchE | (1) GCCGTCGCCATGACAAC | (1) CTGCCTGACGAGAAGCTACGT |
| (2) GGTCACCCGCGTGTTGAT | (2) CACTGCCCTCTGCGAAGAAT | |
| csmM | (1) TGATGTACGCCTTTGCTTCACT | (1) CTTCCGCCCATTTCTCGAT |
| (2) TGATGTACGCCTTTGCTTCACTA | (2) CTTCCGCCCATTTCTCGAT | |
| csmN | (1) CGGGCGATGGCTGAAGT | (1) AGAGACGTTGGCCAGCTCTTT |
| (2) GTCTCTGAACGGGTGGTTGAC | (2) GGGCTGGGCAGGTTGATC |
TABLE 2.
| Gene | ΔCT
|
ΔΔCT | |
|---|---|---|---|
| Anaerobic growth | Semiaerobic growth | ||
| 16S rRNA | 0 | 0 | 0 |
| acsF | (1) 10.3 ± 0.3 | (1) 11.1 ± 0.3 | (1) −0.8 ± 0.3 |
| (2) 10.9 ± 0.2 | (2) 11.6 ± 0.2 | (2) −0.7 ± 0.2 | |
| bchE | (1) 7.8 ± 0.4 | (1) 12.3 ± 0.3 | (1) −4.5 ± 0.3 |
| (2) 7.6 ± 0.3 | (2) 11.8 ± 0.2 | (2) −4.2 ± 0.2 | |
| csmM | (1) 6.4 ± 0.2 | (1) 11.6 ± 0.3 | (1) −5.2 ± 0.3 |
| (2) 6.0 ± 0.2 | (2) 11.6 ± 0.5 | (2) −5.6 ± 0.4 | |
| csmN | (1) 5.1 ± 0.3 | (1) 10.5 ± 0.3 | (1) −5.4 ± 0.3 |
| (2) 3.6 ± 0.5 | (2) 11.2 ± 0.6 | (2) −7.6 ± 0.6 | |
ΔCT = CT of target gene − CT of the internal control gene (16S rRNA).
ΔΔCT = ΔCTanaerobic − ΔCtsemiaerobic.
The values for ΔCT and ΔΔCT in Table 2 were obtained from the various primer sets listed in Table 1.
RESULTS
Growth of C. aurantiacus cultures under various conditions.
In contrast to the strictly anaerobic environment required for growing green sulfur bacteria (GSB), green FAP bacteria, such as C. aurantiacus, are more tolerant of oxygen and are known to grow under anaerobic, semiaerobic, and aerobic conditions (7). The FAP bacteria function as phototrophs under anaerobic and semiaerobic growth conditions in the presence of light and as chemotrophs in the dark under aerobic growth conditions. In low-intensity light (6 W/m2), C. aurantiacus cultures are dark green and are orange-red under anaerobic and semiaerobic conditions, respectively (Fig. 2A). The orange-red appearance in the semiaerobic culture can be partially explained by the presence of various isoforms of carotenoids, since C. aurantiacus is known to contain large amounts of carotenoids (8).
The formation of chlorosomes in green phototrophic bacteria (both GSB and FAP) has been routinely detected by the A740 absorption of BChl c in the cell cultures. Figure 2B indicates that less chlorosome pigment is produced in semiaerobic growth (optical density at 740 nm [OD740] of 0.97) than in anaerobic growth (OD740 of 1.57), indicating that chlorosome formation is sensitive to the oxygen level in the cultures. In contrast to the decrease of BChl c and chlorosomes, the reaction center BChl a (with absorbance at 865 nm and OD865s in the anaerobic and semiaerobic cultures of 046 and 0.47, respectively) and the carotenoid level (shown in the blue-green region [400 to 500 nm] of the spectrum) are comparable in the semiaerobic cultures to those in the anaerobic cultures (Fig. 2B).
Characterization of the chlorosome.
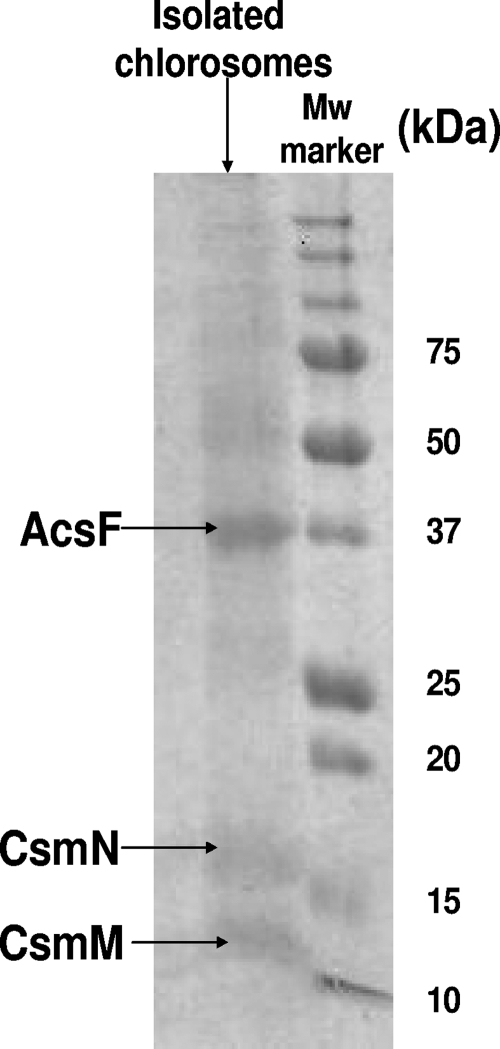
The chlorosomes described in this work were isolated from the anaerobic cultures grown in low-intensity light, while the spectral features are similar between the chlorosomes purified from anaerobic and semiaerobic cultures (data not shown). Figures 2C and D show that the reaction center BChl a, with absorbance at 865 nm, embedded in the cytoplasmic membrane (Fig. 2C), was clearly absent in the purified chlorosome fraction of C. aurantiacus (Fig. 2D). The disappearance of the 865-nm peak, along with the absorption at 740 nm, has been commonly used as a spectroscopic probe for identifying the isolated chlorosomes of C. aurantiacus (4). The purified chlorosomes were also analyzed by SDS-PAGE (Fig. 3), in which the most dominant bands on the SDS-PAGE gel were excised and subjected to in-gel digestion followed by MS analysis. In contrast to the GSB Chlorobium tepidum, in which 10 chlorosome proteins have been identified (10), only three chlorosome proteins, CsmA (∼6 kDa), CsmM (∼11 kDa), and CsmN (∼18 kDa), in C. aurantiacus were reported previously by Fuller, Feick, and coworkers (4, 16). As expected, our proteomics data confirmed that the low-molecular-mass 11- and 16-kDa bands in SDS-PAGE (Fig. 3) are the CsmM and CsmN proteins, respectively (Fig. 4; see also Tables S1 and S2 in the supplemental material). The smallest chlorosome protein reported previously, CsmA, is not shown in the 12% SDS-PAGE gel in Fig. 3 but was detected in an 18% SDS-PAGE gel instead (data not shown). Since CsmA is arguably the chlorosome protein whose structure and function have been most investigated (3, 11, 20, 29), studies described in this report focused on other relatively unknown chlorosome proteins in C. aurantiacus.
FIG. 3.
SDS-PAGE analysis of the isolated chlorosome fraction. Mw, molecular size.
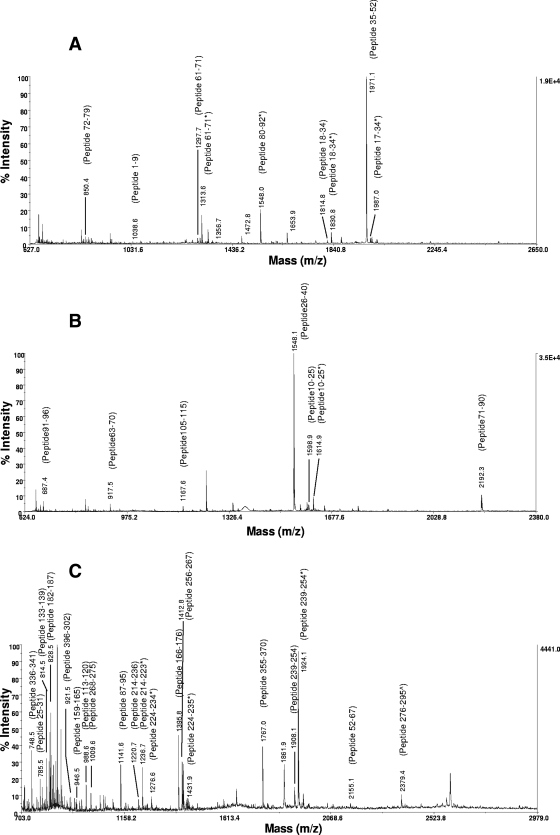
FIG. 4.
Mass spectra of CsmM (A), CsmN (B), and AcsF (C) after in-gel digestions. The data were collected by MALDI-TOF mass spectrometry. For clarity, not all of the identified peptides are labeled. The peptides including oxidation of methionine, tryptophan, or histidine (*) and with the acrylamide adduct of cysteine (∧) are shown.
Identification of the AcsF protein in the chlorosome preparation.
In contrast to the low-molecular-mass peptides CsmA, CsmM, and CsmN, no higher-molecular-mass proteins in the chlorosome were reported by Sprague and coworkers (26), so we decided to identify the dominant ∼40-kDa band on our SDS-PAGE gel by MS. Although a 0.95-kb RNA resulting from a cotranscribed csmM-csmN gene was found in Northern blot analysis by Niedermeier et al. (17), the molecular mass of the possible cotranslated product would have been around 30 kDa. However, to our surprise, the dominant 40-kDa band was identified by MS to be the magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase, AcsF (Fig. 4C; see also Table S3 in the supplemental material). Twenty-one digested fragments (sequence coverage, 49%) were found to match the predicted trypsin-digested patterns, with a score of 108 reported by Mascot (http://www.matrixscience.com/), which defines the sequence coverage as significant when the score is >71. The acsF gene in C. aurantiacus encodes a 45-kDa peptide (375 amino acid residues). Several secondary structure prediction programs were used to provide the structure information of AcsF, CsmM, and CsmN (see Fig. S4 in the supplemental material), and most portions of these proteins are predicted to have either an α-helix or random coil conformation.
To acquire the topology and more structural information for the three chlorosome proteins, we also subjected the isolated chlorosomes to in-solution trypsin digestion. The chlorosome envelope is a special lipid monolayer, in contrast to the lipid bilayer in other cellular membranes, so the transmembrane prediction programs available at the website www.expasy.org are not suitable for predicting the topology of the chlorosome proteins. All of the chlorosome proteins are predicted to be soluble proteins. The MS for the in-solution digestion can identify only a few peptide fragments from AcsF in the isolated chlorosome samples (Table 3), suggesting that a significant portion of the AcsF protein is not accessible to protease. In contrast, more peptide fragments for the chlorosome proteins CsmM and CsmN can be identified by in-solution digestion (Table 3), indicating that a large part of CsmM and CsmN is exposed to the solvent. Together, our studies suggest that the detected AcsF protein is associated with the chlorosome envelope. This observation is consistent with findings in previous studies that AcsF homologs and aerobic cyclases are found in the membranes of Chlamydomonas reinhardtii (14, 15), Arabidopsis thaliana (30), barley (24), and Synechocystis sp. strain PCC 6803 (12).
TABLE 3.
In-solution trypsin digestion of chlorosome samples
| Protein name | % Sequence coverage | Peptide fragment sequencesa |
|---|---|---|
| CsmM | 54 | MTESEGEVRV RSVPVRRNDS FVESAMEFGG GIVRLGFSIF TLPLALLPPE |
| SRQHMHNATK ELMYAFASLP RDFAEIAGKS IEKWAEEGEE PKGEAK | ||
| CsmN | 70 | MSNETTNERD GLFEMAAGFV GGATRIGLTV ASVPLVLLPR NSRRRVRRAM |
| AEVAMAVVAF PKELANVSER VVDDIFAADP PQINLPSPQR VGEQVRSFTE | ||
| RLARAAEELG TSFSRAAGRA ADAVEQGAAK VDEWVETPPK TPPAP | ||
| AcsF | 16 | MFWINPLMME MPGEFSRTTR HALKEAILSP RFYTTDFKAI DRLEIDRNGL |
| RDEFDWIRDE FAYDYNRTHF RRTDEFLQDF DDMPERDSFI DFLERSCTAE | ||
| FSGCLLYAEM VKHLHDPTLR AIFKYMSRDE GRHAGFLNRT MADLNVALDL | ||
| TVLQKRKKYT YFQPKFIFYS VYLSEKIGYS RYITIYRHLE RNPQYCIHPI | ||
| FKWFEQWCND EYRHGEFFAL LMRSQPEMLE GVNRRWIRFF LLAVYATMYL | ||
| NDARRSNFYA ALGLDWRDYD QTVIRLCNDI ATQVYPETMN LDDPRFFAYM | ||
| DKLVEYDRAL RALESRKDPI AMAHSTRLKA AIALRLLAIY RLPTRKTTEA | ||
| IRWKGLPGFP NYPGPNRPQR VVAAD |
Matched peptides are shown in bold. The peptides were identified by peptide accurate mass (accuracy, <10 ppm) and tandem mass with methionine, histidine, and tryptophan oxidations included.
Iron detected in the chlorosome fraction.
A significant amount of Fe, 1.5 μg/ml in the chlorosome fraction with an OD of ∼20 per cm at 740 nm, was detected in the isolated chlorosomes by atomic absorption spectroscopy (see Materials and Methods). Although Li and coworkers suggested that a putative iron-sulfur cluster protein, CsmY, is likely to locate in the chlorosome of C. aurantiacus (10), our SDS-PAGE data showed that even if CsmY does exist in our chlorosome fractions, the protein expression level is minor at best. Our data suggest that the detected Fe ions are most likely associated with abundant AcsF proteins in the chlorosome, since AcsF is proposed to contain the putative binuclear iron motif by sequence alignments and no Fe-binding motif can be identified in other reported chlorosome proteins from C. aurantiacus.
Gene expression of acsF, bchE, csmM, and csmN.
In addition to biochemical studies, we also applied gene expression analysis to investigate the genes encoding the proteins described above. RNA was extracted from the anaerobic and semiaerobic cultures grown in low-intensity light and was analyzed using reverse transcription followed by QRT-PCRs. The data analyses for gene expression for acsF (caur_2590), bchE (caur_3676), csmM (caur_0139) and csmN (caur_0140) were performed using the 16S rRNA gene (caur_R0008) as the internal standard. Two different primer sets were used for studying the expression of each target gene. As shown in Table 2, all of the targeted genes were upregulated under the anaerobic conditions compared to results under the semiaerobic conditions, except that the expression levels of the acsF gene are rather similar in response to different oxygen levels. The upregulation of the bchE gene in anaerobic growth is consistent with the assumption that BchE, the anaerobic cyclase, is responsible for the formation of the isocyclic ring (the “E” ring) of BChls in anaerobic bacteria. Since chlorosome formation is enhanced in anaerobic growth, upregulated gene expression is also expected for the chlorosome-associated proteins CsmM and CsmN (Table 2). It is not expected, however, that the acsF gene, encoding the “aerobic” cyclase, would be expressed under both anaerobic and semiaerobic conditions, and the gene expression level is not lower under anaerobic conditions than under semiaerobic conditions.
DISCUSSION
Significance of this work.
Although both the acsF and bchE genes can be identified in the genome of C. aurantiacus, no studies of these proteins in C. aurantiacus or any green phototrophic bacteria have been reported. Two unexpected results were discovered in our studies. First, under anaerobic growth conditions, when the bchE gene product is responsible for pigment synthesis, the AcsF proteins were detected by SDS-PAGE and the acsF gene was not downregulated. Second, the AcsF proteins were detected in our isolated chlorosome fractions.
Coexistence of acsF and bchE genes in some phototrophs.
Our studies suggested the coexistence of the acsF and bchE gene products in the FAP bacterium C. aurantiacus under anaerobic conditions. In addition to C. aurantiacus, BLAST search and previous studies suggest the coexistence of the acsF and bchE genes in several phototrophs, such as Roseiflexus sp. (FAP), Roseiflexus castenholzii (FAP), Bradyrhizobium sp. (purple bacterium), Rhodopseudomonas palustris (purple bacterium), Methylobacterium sp. (purple bacterium), Synechocystis sp. PCC 6803 (cyanobacterium) (12), Rubrivivax gelatinosus (purple bacterium) (18), Rhodobacter sphaeroides (purple bacterium), and Roseobacter denitrificans (purple bacterium). Other than Synechocystis sp. strain PCC 6803, most of the bacteria mentioned above are facultative bacteria and can be grown under both aerobic and anaerobic conditions. The AcsF and BchE proteins in those facultative bacteria have been proposed to catalyze isocyclic ring formation under aerobic or anaerobic conditions (Fig. 1), as suggested in the studies of the purple bacterium Rubrivivax gelatinosus (18).
Location of AcsF.
Previous studies of AcsF in different organisms demonstrated that AcsF is membrane associated (12, 14, 15, 24, 30). It is interesting to note that Chl27, an AcsF analogue in Arabidopsis, is found to be equally distributed in both the inner-envelope and thylakoid membranes (30). Thus, it is possible that the AcsF proteins may also be present in the cytoplasmic membrane of C. aurantiacus. Work is in progress to test this hypothesis. Nevertheless, it has been generally accepted that many plasma membrane protein complexes are absent from the chlorosome envelope, so if the AcsF protein is present in the chlorosome envelope, some special features of AcsF may lead to its location there.
Possible roles of AcsF in (the chlorosome of) C. aurantiacus.
If the roles of AcsF and BchE in C. aurantiacus are to catalyze isocyclic ring formation under different oxygen growth conditions, as proposed for some of the facultative bacteria (18), the presence of abundant AcsF in anaerobic cultures and comparable gene expression levels of acsF in anaerobic and semiaerobic growth would not be expected. The fact that the AcsF protein is abundant in anaerobic growth is puzzling, because the function of AcsF has been suggested to be catalyzing isocyclic ring formation under aerobic conditions with molecular oxygen as the oxygen source for installing the C-131 keto moiety in the isocyclic ring of (bacterio)chlorophylls (22). Further, if the AcsF proteins in the chlorosome are directly or indirectly involved in the formation of PChlide without the participation of oxygen molecules, one may expect that significant amounts of BchE would have been detected in the chlorosome. No such results were observed in our studies, even though reverse transcription-PCR data suggested that the bchE gene is upregulated under anaerobic conditions (Table 2).
Related to our work, an unexpected function was also discovered in one of the BchE homologs in Synechocystis sp. strain PCC 6803, in which three bchE-like genes were suggested from the bacterial genome (12). Minamizaki et al. demonstrated that none of the bchE genes have significant effects on PChlide formation and chlorophyll a biosynthesis under either aerobic or “micro-oxic” growth conditions and found that one of the bchE mutants, the Δsll1242 mutant, can grow only on agar plates and not in liquid medium (12). The authors suggested that that the sll1242 gene may play some important roles in Synechocystis sp. strain PCC 6803. Similarly, our presented studies may also suggest additional roles for the putative AcsF and/or BchE proteins in such facultative bacteria as C. aurantiacus. The sequence alignments indicate that in contrast to the 70% or higher sequence identity detected between the BchE protein in C. aurantiacus and those in other FAP, GSB, and purple bacteria (see Fig. S2 in the supplemental material), 50% or lower identity can be found between the AcsF protein in C. aurantiacus and those in most other organisms (except for ∼70% identity with that of Roseiflexus sp.) (see Fig. S3 in the supplemental material), implying that AcsF in C. aurantiacus may have different properties. Taken together, it is likely that AcsF in C. aurantiacus has unrevealed biological functions, irrelevant to isocyclic ring formation, in the chlorosome envelope during anaerobic growth. Here we discuss three working hypotheses based on the data presented in this report and previous studies by other groups, as follows.
(i) AcsF and BchE in C. aurantiacus may be responsible for biosynthesis of different BChls.
The absorption spectra in Fig. 2B suggest that the levels of the reaction center BChl a is almost the same in semiaerobic and anaerobic growth cultures, whereas the chlorosome level is significantly lower in the semiaerobic cultures. Table 2 indicates that the gene expression levels of acsF are comparable in anaerobic and semiaerobic growth and that gene expression of bchE is downregulated under semiaerobic conditions. Thus, it is possible that AcsF and BchE are responsible for biosynthesis of BChl a and BChl c, respectively. However, several questions cannot be resolved with this hypothesis: (i) GSB, containing only the bchE gene, can produce BChl a and BChl c, d, or e; (ii) some FAP bacteria can generate only BChl a yet contain both the acsF and bchE genes; (iii) the difference in Fig. 1B could be due to downstream gene expression being repressed for BChl c, but not BChl a, biosynthesis under semiaerobic conditions; and (iv) more importantly, if AcsF is responsible for the isocyclic ring formation of BChl a under anaerobic conditions, a different reaction mechanism needs to be used by AcsF because molecular oxygen is no longer available. Together, as with the roles suggested for other photosynthetic bacteria (5), it is likely that BchE, a putative cobalamin and oxygen-sensitive [4Fe-4S] cluster/S-adenosylmethionine-dependent enzyme, is responsible for various isoforms of BChl biosynthesis in C. aurantiacus in anaerobic growth and that AcsF and/or BchE is required for the biosynthesis of BChls under semiaerobic conditions.
(ii) The AcsF protein in C. aurantiacus may be important for electron transfer under anaerobic growth conditions.
The nonheme binuclear iron center proteins can be divided into two major types. First,the oxidoreductase-type enzymes, catalyzing the oxidation of chemical insertion hydrocarbons, include the soluble methane monooxygenase family, the stearoyl-ACP desaturase family, class I ribonucleotide reductase, etc. Electron transfer, dioxygen cleavage, and oxygen insertion are common themes in these binuclear iron proteins, and such a mechanism was proposed for the isocyclic ring formation catalyzed by AcsF in the presence of oxygen (Fig. 1) (12). None of the known binuclear iron oxidoreductases are functional without oxygen participation. Second, the ferritin-like proteins, which are responsible for iron storage, oxygen transport, phosphoryl transfer, and other unidentified biological functions, comprise hemerythrin, bacterioferritin, rubrerythrin, purple acid phosphatases, etc. In the first type, the R2 subunit of the class I ribonucleotide reductase is known to function as a proton-coupled electron transfer protein (27). It is possible that the C. aurantiacus AcsF protein may function as an electron transfer protein under anaerobic conditions and as an oxidoreductase with oxygen available. Some residues, such as three Cys residues in C. aurantiacus AcsF in the vicinity of the putative binuclear iron binding sites, may directly or indirectly be involved in electron transfer. The functions of C. aurantiacus AcsF under anaerobic and semiaerobic conditions may or may not be related.
Vassilieva and coworkers found three [2Fe-2S] cluster chlorosome proteins, CsmI, CsmJ, and CsmX, in C. tepidum and proposed that these proteins are important for the energy and electron transfer in C. tepidum chlorosomes (31, 32), because [2Fe-2S] clusters play important roles in electron transfer and cellular signaling for a variety of organisms (19). At least two of the three genes encoding [2Fe-2S] proteins have been identified for the GSB strains with sequence information available. The authors postulated that in the presence of oxygen, the excitation energy quencher (likely to be chlorobiumquinone in GSB [2]) is activated by transfer of an electron directly or indirectly to O2 and that [2Fe-2S] cluster proteins can inactivate the quencher by transferring an electron to the quencher, so the excitation energy can be transferred from BChl c to the reaction center. However, it has been demonstrated that the redox-dependent quenching effect observed in GSB is not observed in FAP bacteria (2). Thus, if AcsF functions as an electron transfer protein in the C. aurantiacus chlorosomes, the role is probably not similar to the proposed roles of [2Fe-2S] proteins in C. tepidum or other GSB.
(iii) The AcsF protein may serve as an iron transport protein.
Most of the characterized binuclear iron center proteins are involved in redox chemistry, but not the ferritin protein. A BLAST search shows that AcsF proteins belong to the ferritin-like superfamily, the class of binuclear iron center proteins responsible for iron storage and nonredox biological functions (described above). Since iron is required for the function of cytochromes in the electron transport chain, it is possible that AcsF is important for transporting the iron to the reaction center and associated proteins near the chlorosome envelope. If this hypothesis holds true, one may expect that different iron transport pathways are adopted in C. aurantiacus versus the case for the GSB and other anaerobic bacteria, in which the acsF gene is absent.
Altogether, it is clear that even for the most investigated FAP bacterium, C. aurantiacus, the role(s) of AcsF is far from being understood. Additionally, since no studies have been described previously for AcsF and BchE in any green bacteria, which have a specialized photosynthetic compartment—the chlorosome—caution is needed when comparing the properties and mechanism of action of AcsF and BchE in GSB and FAP bacteria with the studies of high plants, algae, and other bacteria. We are currently investigating these two enzymes with both in vitro and in vivo studies.
Supplementary Material
Acknowledgments
We thank the members of the Blankenship group for valuable technical advice and Himadri B. Pakrasi for assistance with atomic absorption spectroscopy. We are grateful for the assistance of the Washington University Mass Spectrometry Resource Center, supported by NIH NCRR grant no. 2P41RR000954.
This work was supported by U.S. Department of Energy Basic Energy Sciences grant DE-FG02-07ER15846 to R. E. Blankenship.
Footnotes
Published ahead of print on 3 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Blankenship, R. E. 2002. Molecular mechanisms of photosynthesis. Blackwell Science Ltd., Oxford, United Kingdom.
- 2.Blankenship, R. E., and K. Matsuura. 2003. Antenna complexes from green photosynthetic bacteria, p. 195-217. In B. R. Green and W. W. Parson (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Bryant, D. A., E. V. Vassilieva, N. U. Frigaard, and H. Li. 2002. Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 4114403-14411. [DOI] [PubMed] [Google Scholar]
- 4.Feick, R. G., M. Fitzpatrick, and R. C. Fuller. 1982. Isolation and characterization of cytoplasmic membranes and chlorosomes from the green bacterium Chloroflexus aurantiacus. J. Bacteriol. 150905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gough, S. P., B. O. Petersen, and J. O. Duus. 2000. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl. Acad. Sci. USA 976908-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granvogl, B., P. Gruber, and L. A. Eichacker. 2007. Standardisation of rapid in-gel digestion by mass spectrometry. Proteomics 7642-654. [DOI] [PubMed] [Google Scholar]
- 7.Hanada, S., and B. K. Pierson. 2006. The family chloroflexaceae. Prokaryotes 7815-842. [Google Scholar]
- 8.Imhoff, J. F., and U. Bias-Imhoff. 1995. Lipids, quinones and fatty acids of anoxygenic phototrophic bacteria, p. 179-205. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Advances in photosynthesis and respiration, vol 2. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 9.Keren, N., M. Liberton, and H. B. Pakrasi. 2005. Photochemical competence of assembled photosystem II core complex in cyanobacterial plasma membrane. J. Biol. Chem. 2806548-6553. [DOI] [PubMed] [Google Scholar]
- 10.Li, H., N. U. Frigaard, and D. A. Bryant. 2006. Molecular contacts for chlorosome envelope proteins revealed by cross-linking studies with chlorosomes from Chlorobium tepidum. Biochemistry 459095-9103. [DOI] [PubMed] [Google Scholar]
- 11.Milks, K. J., M. Danielsen, S. Persson, O. N. Jensen, R. P. Cox, and M. Miller. 2005. Chlorosome proteins studied by MALDI-TOF-MS: topology of CsmA in Chlorobium tepidum. Photosynth. Res. 86113-121. [DOI] [PubMed] [Google Scholar]
- 12.Minamizaki, K., T. Mizoguchi, T. Goto, H. Tamiaki, and Y. Fujita. 2008. Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2832684-2692. [DOI] [PubMed] [Google Scholar]
- 13.Montaño, G. A., H. M. Wu, S. Lin, D. C. Brune, and R. E. Blankenship. 2003. Isolation and characterization of the B798 light-harvesting baseplate from the chlorosomes of Chloroflexus aurantiacus. Biochemistry 4210246-10251. [DOI] [PubMed] [Google Scholar]
- 14.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 192139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn, F. Soto, S. M. Theg, M. Hippler, and S. Merchant. 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niedermeier, G., H. Scheer, and R. G. Feick. 1992. The functional role of protein in the organization of bacteriochlorophyll c in chlorosomes of Chloroflexus aurantiacus. Eur. J. Biochem. 204685-692. [DOI] [PubMed] [Google Scholar]
- 17.Niedermeier, G., J. A. Shiozawa, F. Lottspeich, and R. G. Feick. 1994. The primary structure of two chlorosome proteins from Chloroflexus aurantiacus. FEBS Lett. 34261-65. [DOI] [PubMed] [Google Scholar]
- 18.Ouchane, S., A. S. Steunou, M. Picaud, and C. Astier. 2004. Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: a strategy adopted to bypass the repressive oxygen control system. J. Biol. Chem. 2796385-6394. [DOI] [PubMed] [Google Scholar]
- 19.Page, C. C., C. C. Moser, X. Chen, and P. L. Dutton. 1999. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 40247-52. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen, M. O., L. Pham, D. B. Steensgaard, and M. Miller. 2008. A reconstituted light-harvesting complex from the green sulfur bacterium Chlorobium tepidum containing CsmA and bacteriochlorophyll a. Biochemistry 471435-1441. [DOI] [PubMed] [Google Scholar]
- 21.Pinta, V., M. Picaud, F. Reiss-Husson, and C. Astier. 2002. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 184746-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porra, R. J., W. Schafer, N. Gad'on, I. Katheder, G. Drews, and H. Scheer. 1996. Origin of the two carbonyl oxygens of bacteriochlorophyll a. Demonstration of two different pathways for the formation of ring E in Rhodobacter sphaeroides and Roseobacter denitrificans, and a common hydratase mechanism for 3-acetyl group formation. Eur. J. Biochem. 23985-92. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203173-179. [DOI] [PubMed] [Google Scholar]
- 24.Rzeznicka, K., C. J. Walker, T. Westergren, C. G. Kannangara, D. von Wettstein, S. Merchant, S. P. Gough, and M. Hansson. 2005. Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc. Natl. Acad. Sci. USA 1025886-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakuragi, Y., N. Frigaard, K. Shimada, and K. Matsuura. 1999. Association of bacteriochlorophyll a with the CsmA protein in chlorosomes of the photosynthetic green filamentous bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1413172-180. [DOI] [PubMed] [Google Scholar]
- 26.Sprague, S. G., L. A. Staehelin, M. J. DiBartolomeis, and R. C. Fuller. 1981. Isolation and development of chlorosomes in the green bacterium Chloroflexus aurantiacus. J. Bacteriol. 1471021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stubbe, J., D. G. Nocera, C. S. Yee, and M. C. Chang. 2003. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem. Rev. 1032167-2201. [DOI] [PubMed] [Google Scholar]
- 28.Tang, K. H., A. Harms, and P. A. Frey. 2002. Identification of a novel pyridoxal 5′-phosphate binding site in adenosylcobalamin-dependent lysine 5,6-aminomutase from Porphyromonas gingivalis. Biochemistry 418767-8776. [DOI] [PubMed] [Google Scholar]
- 29.Theroux, S. J., T. E. Redlinger, R. C. Fuller, and S. J. Robinson. 1990. Gene encoding the 5.7-kilodalton chlorosome protein of Chloroflexus aurantiacus: regulated message levels and a predicted carboxy-terminal protein extension. J. Bacteriol. 1724497-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tottey, S., M. A. Block, M. Allen, T. Westergren, C. Albrieux, H. V. Scheller, S. Merchant, and P. E. Jensen. 2003. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 10016119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vassilieva, E. V., M. L. Antonkine, B. L. Zybailov, F. Yang, C. U. Jakobs, J. H. Golbeck, and D. A. Bryant. 2001. Electron transfer may occur in the chlorosome envelope: the CsmI and CsmJ proteins of chlorosomes are 2Fe-2S ferredoxins. Biochemistry 40464-473. [DOI] [PubMed] [Google Scholar]
- 32.Vassilieva, E. V., V. L. Stirewalt, C. U. Jakobs, N. U. Frigaard, K. Inoue-Sakamoto, M. A. Baker, A. Sotak, and D. A. Bryant. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH, and CsmX. Biochemistry 414358-4370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.