Abstract
Adenosine kinase (ADK) is a purine salvage enzyme and a typical housekeeping enzyme in eukaryotes which catalyzes the phosphorylation of adenosine to form AMP. Since prokaryotes synthesize purines de novo and no endogenous ADK activity is detectable in Escherichia coli, ADK has long been considered to be rare in bacteria. To date, only two prokaryotes, both of which are gram-positive bacteria, have been reported to contain ADK. Here we report that the gram-negative bacterium Xanthomonas campestris pathovar campestris, the causal agent of black rot of crucifers, possesses a gene (designated adkXcc) encoding an ADK (named ADKXcc), and we demonstrate genetically that the ADKXcc is involved in extracellular polysaccharide (EPS) production, cell motility, and pathogenicity of X. campestris pv. campestris. adkXcc was overexpressed as a His6-tagged protein in E. coli, and the purified His6-tagged protein exhibited ADK activity. Mutation of adkXcc did not affect bacterial growth in rich and minimal media but led to an accumulation of intracellular adenosine and diminutions of intracellular ADK activity and ATP level, as well as EPS. The adkXcc mutant displayed significant reductions in bacterial growth and virulence in the host plant.
Adenosine kinase (ADK) is a purine salvage enzyme that catalyzes the phosphorylation of adenosine to give AMP (56). ADKs belong to the PfkB family of carbohydrate and nucleoside kinases, a family of structurally related enzymes that includes ribokinase, fructokinase, and hexokinase (5, 57). ADK activity was first characterized from yeast and mammalian tissues (7, 33). Subsequently, ADKs have been investigated biochemically and/or genetically from a large number of other eukaryotes, including humans and other mammals (21, 44, 46, 53, 70), plants (20, 47), moss (66), parasitic protozoa (8, 14, 16, 23), and yeast (36, 37). The ADK enzyme has been considered to be the key metabolic regulator for maintenance of certain intra- and extracellular levels of adenosine in eukaryotes (2, 22), and there is abundant evidence to indicate that adenosine is a significant signal molecule engaged in regulation of physiology and modulation of the function of various cell types in mammals (6, 11, 35). Consequently, the identification of compounds that augment the concentrations and actions of adenosine by inhibiting the ADK has been the subject of substantial drug discovery efforts (34).
Since prokaryotes synthesize purines de novo and no detectable endogenous ADK activity was observed in Escherichia coli (44, 48), the ADK enzyme has long been considered to be rare in bacteria. To date, only two prokaryotes, both of which are gram-positive bacteria, have been reported to contain ADK. One is Mycobacterium tuberculosis, the causative agent of the human disease tuberculosis (39), and the other is Streptomyces lividans (52). M. tuberculosis possesses a unique ADK that can also catalyze the phosphorylation of the adenosine analog 2-methyladenosine, a compound that has shown selective activity against M. tuberculosis (39, 68). In S. lividans, loss of ADK activity suppresses sporulation and actinorhodin biosynthesis, while hyperproduction of undecylprodigiosin is induced (52).
The gram-negative bacterium Xanthomonas campestris pathovar campestris is the causal agent of black rot disease, one of the most destructive diseases of cruciferous crops worldwide (1). This pathogen infects almost all the members of crucifer family (Brassicaceae), including important vegetables such as broccoli, cabbage, cauliflower, mustard, and radish; the major oil crop rape; and the model plant Arabidopsis thaliana. The extracellular polysaccharide (EPS) produced by X. campestris pv. campestris is also called xanthan gum and has been widely used as a viscosifer, thickener, emulsifier, or stabilizer in both food and nonfood industries (31). Because of its agricultural and industrial importance, the molecular genetics of X. campestris pv. campestris have attracted particular attention for over two decades. The whole-genome sequences of three different X. campestris pv. campestris strains have been determined by different research groups (15, 51, 67). Although no predicted protein was annotated as ADK, a survey of the genome sequence data of the three strains revealed that the deduced proteins of the open reading frames (ORFs) XCC_3471 (GenBank accession number NP_638817.1) in strain ATCC 33913 (15), XC_0690 (GenBank accession number YP_241789.1) in strain 8004 (51), and xccb100_0723 (GenBank accession number YP_001902128.1) in strain B100 (67) display homology to the ADK of M. tuberculosis. Here we present evidence to demonstrate that the ORF XC_0690 of strain 8004 encodes an authentic ADK that is involved in cell motility, EPS production, and pathogenicity of X. campestris pv. campestris.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in LB medium (45) at 37°C. X. campestris pv. campestris strains were grown at 28°C in NYG medium (containing, per liter, 5 g peptone, 3 g yeast extract, and 20 g glycerol) (12), NY medium (NYG medium but without glycerol), or the minimal medium MMX [containing, per liter, 2. 0 g (NH4)2SO4, 4.0 g K2HPO4, 6.0 g KH2PO4, 0.2 g MgSO4·7H2O, 1.0 g citric acid, and 5.0 g glucose) (13). Antibiotics were added at the following concentrations as required: kanamycin (Kan), 25 μg ml−1; rifampin (Rif), 50 μg ml−1; ampicillin (Amp), 100 μg ml−1; spectinomycin (Spc), 50 μg ml−1; and tetracycline (Tet), 5 μg ml−1 for X. campestris pv. campestris and 15 μg ml−1 for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| JM109 | recA1 endA1 gyrA96 thi supE44 relA1 Δ(lac-proAB)/F′ [traD36 lacIqlacZ ΔM15] | 71 |
| DH5α | φ80ΔlacZM15 recA1 endA1 deoR | Gibco BRL, Life Technologies |
| M15 | lac ara gal mtl recA1 uvr1 [pREP4 lacI Kanr] | Qiagen |
| X. campestris pv. campestris strains | ||
| 8004 | Wild type, Rifr | 13 |
| 0690pk | As 8004, but XC_0690::pK18mob, Rifr Kanr | This work |
| C0690pk | 0690pk harboring pL0690; Rifr Kanr Tetr | This work |
| 010D08 | As 8004 but gumB::Tn5 gusA5, EPS deficient; Rifr Kanr Spcr Gmr | Our lab collection |
| 098A02 | As 8004 but motB::Tn5 gusA5, motility deficient; Rifr Kanr Spcr Gmr | Our lab collection |
| 8004/pL6gumGUS | 8004 harboring pL6gumGUS; Rifr Tetr | This work |
| 0690pk/pL6gumGUS | 0690pk harboring pL6gumGUS; Rifr Kanr Tetr | This work |
| 8004/pGUS0690 | 8004 harboring pGUS0690; Rifr Tetr | This work |
| Plasmids | ||
| pLAFR3 | Broad-host-range cloning vector; Tetr | 58 |
| pLAFR6 | Promoterless derivative of pLAFR3; Tetr | 27 |
| pRK2073 | Helper plasmid; Tra+ Mob+ ColE1 Spcr | 38 |
| pK18mob | pUC18 derivative, lacZα Kanr, mob site, suicide plasmid in X. campestris pv. campestris | 55 |
| pK0690 | pK18mob containing a 444-bp internal fragment of XC_0690; Kanr | This work |
| pQE-30 | Expression vector, allows production of fusion proteins containing N-terminal His6-tagged sequences; Ampr | Qiagen |
| pQE-30-0690 | pQE-30 containing a 930-bp fragment of XC_0690 coding region; Ampr | This work |
| pL0690 | pLAFR3 containing an 1,112-bp fragment including the XC_0690 gene; Tetr | This work |
| pL6gumGUS | pLAFR6 containing the promoter region of the gum operon fused to the coding region for gusA; Tetr | 65 |
| pGUS0690 | pLAFR6 containing the promoter region of the gene XC_0690 fused to the coding region for gusA; Tetr | This work |
DNA manipulations.
The methods described by Sambrook et al. (54) were used for preparation of the plasmid and chromosomal DNAs, restriction digestion, DNA ligation, agarose gel electrophoresis, and DNA transformation of E. coli. The conjugation between X. campestris pv. campestris and E. coli strains was performed as described by Turner et al. (62). The restriction endonucleases, T4 DNA ligase, and Pfu polymerase were provided by Promega (Shanghai, China) and used in accordance with the manufacturer's instructions.
Construction and complementation of an insertional mutant.
An insertional mutation of the gene XC_0690 was constructed by the homologous suicide plasmid integration method described by Windgassen et al. (69). A 444-bp internal fragment, which spans nucleotides 6 to 449 of the XC_0690 ORF sequence, was amplified by PCR using as template the total DNA of the X. campestris pv. campestris wild-type strain 8004 and as primers the oligonucleotides 5′-CGCACTGATCTGTGGTTCCCTCG-3′ and 5′-TGGATCATGCCTTCGCGCCC-3′. The amplified DNA fragment was cloned into the suicide plasmid pK18mob (55). The resulting recombinant plasmid was introduced from the E. coli strain JM109 (71) into the X. campestris pv. campestris strain 8004 by triparental conjugation using pRK2073 (38) as the helper plasmid. Transconjugants were screened on NYG medium supplemented with Rif and Kan, and the obtained transconjugants with a mutation in the XC_0690 gene were confirmed by PCR using the oligonucleotide 5′-GCCGATTCATTAATGCAGCTGGCAC-3′, which is located in pK18mob, and the oligonucleotide 5′-ACAGTTGGATCCGAGCAAGGAAAAATGCGC-3′, which is located upstream of the XC_0690 ORF, as primers. One of the confirmed mutants was named 0690pk (Table 1) and used for further study.
For complementation of the mutant 0690pk, a 1,112-bp DNA fragment containing the XC_0690 coding region and extending from 177 bp upstream of the 5′ end to 2 bp downstream of the 3′ end of the ORF was amplified using as primers oligonucleotides 5′-ACAGTTGGATCCGAGCAAGGAAAAATGCGC-3′ and 5′-ACAGTTAAGCTTGCTTACAGCGCGTAGCCG-3′. Primers were modified to give BamHI- or HindIII-compatible ends (underlined). The amplified DNA fragment was cloned into the plasmid pLAFR3 (58) to generate the recombinant plasmid pL0690 (Table 1). The recombinant plasmid was transferred into the mutant 0690pk by triparental conjugation. The transconjugants carrying the recombinant plasmid were screened on NYG medium supplemented with Rif, Kan, and Tet. The resulting complemented strain was named C0690pk (Table 1).
Determination of transcriptional start site.
Rapid amplification of cDNA 5′ ends (5′-RACE) was used to determine the transcriptional start site of the gene XC_0690. Total cellular RNA was extracted from a 10-ml culture of the X. campestris pv. campestris strain 8004 after 12 h of incubation by using the RNeasy Midi kit (Qiagen). All RNA isolation steps were performed according to the manufacturer's instructions. The isolated RNA was treated with RNase-free DNase I (Qiagen) at 25°C for 1 h, followed by a second purification using an RNeasy column. cDNA fragments were obtained using the 5′-RACE kit (Invitrogen Life Technologies). All experimental steps were performed according to the manufacturer's instructions. RNA was reverse transcribed using the XC_0690 sequence-specific primer AKP1 (5′-GTAGTGATGAACGCCTGC-3′). An anchor sequence was then added to the 3′ end of the cDNA using terminal deoxynucleotide transferase, followed by direct amplification of tailed cDNA using the nested gene-specific primers AKP2 (5′-ATCGATGATCTTCACGCGCGACAG-3′) and AKP3 (5′-GATGCCCAGCGTCTCGAAATGCTC-3′) and the anchor-specific primer provided. PCR products were then cloned into the pMD19-T vector and sequenced.
Overproduction and purification of protein.
For overproduction of XC_0690, the ORF XC_0690, which is 930 bp in length, was amplified using as primers oligonucleotides 5′-ACAGTTGGATCCATGTCCGCACTGATCTGT-3′ and 5′-ACAGTTAAGCTTCAGCGCGTAGCCGAACTG-3′. Primers were modified to give BamHI- or HindIII-compatible ends (underlined). After being confirmed by sequencing, the amplified DNA fragment was cloned into the expression vector pQE-30 (Qiagen) (Table 1) to generate the recombinant plasmid pQE-30-0690 (Table 1). In this plasmid, XC_0690 is fused N terminally in frame to the His6 tag-coding region of the plasmid pQE-30. The recombinant plasmid was transformed into the E. coli strain M15, resulting in strain M15/pQE-30-0690. For fused protein XC0690-His6 overproduction and purification, the strain M15/pQE-30-0690 was grown to an optical density at 600 nm (OD600) of 0.6 and then induced by the addition of 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The culture was grown for a further 4 h. The fused protein was purified with Ni-nitrilotriacetic acid (NTA) resin (Qiagen). The purified protein was checked by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and used for enzymatic assay.
Construction of a promoter reporter plasmid for XC_0690.
A promoter reporter plasmid for the XC_0690 gene was constructed by fusing a 471-bp DNA sequence upstream of the start codon ATG (including ATG) of the ORF XC_0690 with the promoterless β-glucuronidase (GUS)-encoding ORF (excluding the start codon ATG). The XC_0690 promoter region was amplified from the total DNA of the X. campestris pv. campestris strain 8004 by using as primers oligonucleotides 5′-ACAGTTGGATCCCATCGAGTCAGGCCTTTG-3′ and 5′-ACAGTTGAATTCCAGCATCTGCTTGGCCTC-3′. Simultaneously, the gus coding region was amplified by PCR with primers 5′-ACAGTTGGATCCTTACGTCCTGTAGAAACCCC-3′ and 5′-ACAGTTCTGCAGGGCTTTCCCCCCCCCCCCTGCAG-3′, using the transposon Tn5gusA5 DNA as template. Primers were modified to give EcoRI-, BamHI-, or PstI-compatible ends (underlined). The two fragments obtained were cloned into the promoterless EcoRI-BamHI-PstI sites of the cosmid pLAFR6 (27) to generate the reporter plasmid pGUS0690 (Table 1).
Cell extract preparation.
Bacterial cells cultured in NYG liquid medium for 18 h were harvested by centrifugation and washed in sterile water twice. The cells were then resuspended in 2.0 ml of 20-mmol/liter potassium phosphate (pH 7.5) containing 1 mmol/liter dithiothreitol and 0.1 mmol/liter EDTA and disrupted by sonication (three times for 10 s each at 350-W output). Following sonication, the cell debris and intact cells were removed by centrifugation (10 min at 18,000 × g) in a 4°C centrifuge, and the supernatant fraction was collected and used in enzymatic assay and adenosine measurement.
Enzymatic assay.
For analyzing the ability of the above-described purified protein to phosphorylate adenosine, the purified protein was incubated with 0.25 mM adenosine in 50 mM Tris-HCl (pH 7.4) containing 5 mM MgCl2, 2.5 mM ATP, and 1.0 μCi [γ-32P] ATP for 60 min at 37°C. One microliter of each reaction sample was spotted onto a polyethyleneimine-cellulose thin-layer chromatography (TLC) plate (Merck), which was developed in aqueous 1 M acetic acid and then in aqueous 1.2 M lithium chloride solution. A phosphorimager analyzer was used to monitor the radioactivity of the samples.
ADK activity from the purified protein or cell extracts was also measured using a modification of published high-pressure liquid chromatography (HPLC) assay protocols (52, 73). A standard reaction mixture (100 μl) contained 50 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol, 2.5 mM ATP, and 2.5 mM adenosine. The reaction was initiated by the addition of 0.1 or 10 μg of protein and terminated by heating in boiling water for 3 min. The reaction was conducted at 37°C. Samples were taken at 3-min intervals over the course of a 15-min assay, and the formation of AMP was monitored by HPLC on an Eclipse XDB C18 column (4.6 by 250 mm, 5 m, 80Å; Agilent Technology) at 260 nm. The mobile phase consisted of 15% methanol and 100 mM KH2PO4, and the flow rate was 1 ml/min at 30°C. According to the increase of AMP, one unit of ADK catalyzes the phosphorylation of 1 pmol adenosine per min under these conditions. The protein concentration in the samples was determined by using a bicinchoninic acid protein assay kit (Pierce Biotechnology). For characterizing the kinetic parameters of the purified protein for adenosine or ATP, a reaction mixture including a range of adenosine (1 to 100 μM) or ATP (0.1 to 20 mM) concentrations was used under standard assay conditions. The Michaelis-Menten kinetic parameter was evaluated by the double-reciprocal plot method.
For GUS activity assay, X. campestris pv. campestris strains were cultured overnight and diluted to an OD600 of 0.5, and 1.0 ml of each was inoculated into 200 ml of NY medium held in a 500-ml flask. GUS activities were determined at 8-h intervals until 48 h by measurement of the A415 using p-nitrophenyl β-d-glucuronide as the substrate, as described by Henderson et al. (26). Three independent experiments were carried out to assay the GUS activity, and each experiment was performed by determination of three samples for a given strain at every test point.
ATP determination.
ATP levels were determined by a modification of the luciferin-luciferase bioluminescence assay (32), using prepared standard reagents on a luminometer (Sirius single-tube luminometer; Berthold Detection Systems). After being cultured in NYG medium for 12, 24, or 36 h, cells were quickly harvested and immediately resuspended in Tris (100 mM)-EDTA (2 mM) buffer (pH 7.75), followed by cooling in an ice bath. Measurements were done by the addition of 80 μl of luciferase reagent (3 mg/ml) to a 20-μl sample. Each experiment was performed by determination of three samples for a given strain at every test point. The standard ATP curves were generated with standards containing known amounts of ATP in Tris-EDTA buffer.
EPS assay.
To estimate EPS production, strains were cultured in 100 ml NY liquid medium containing 2% (wt/vol) various sugars (xylose, glucose, mannose, fructose, galactose, sucrose, and maltose) at 28°C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried, and weighed as described by Tang et al. (59).
Virulence assay and determination of bacterial load in planta.
The virulence of X. campestris pv. campestris to Chinese radish (Raphanus sativus) was tested by the leaf-clipping method (18). Leaves were cut with scissors dipped in the bacterial suspensions with an OD600 of 0.1. Lesion length was measured 10 days after inoculation, and data were analyzed by the t test. The growth of bacteria in radish leaf tissue was determined as previously described (40).
Cell motility assay.
To test cell motility, 2 microliters of overnight culture (OD600 of 1.0) of each X. campestris pv. campestris strain was spotted onto NYG swarm plates containing 0.3% agarose and incubated at 28°C for 3 days. The diameter of the area occupied by strains was measured, and the values were used to indicate the motility of X. campestris pv. campestris strains. The experiment was repeated three times.
RESULTS
XC_0690 of the X. campestris pv. campestris strain 8004 encodes an ADK.
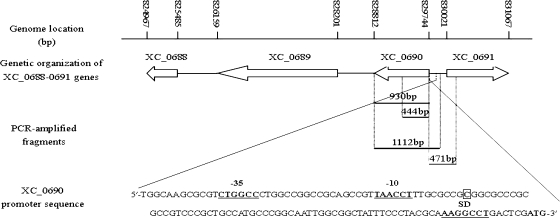
A BLAST search against the genome of the X. campestris pv. campestris strain 8004 (GenBank accession number CP000050) with the amino acid sequence of the ADK of M. tuberculosis showed that the predicted protein encoded by the ORF XC_0690 (GenBank accession number YP_241789.1), which consists of 310 amino acids and is annotated as a sugar kinase (51), has homology to the M. tuberculosis ADK. Amino acid sequence pairwise alignments using Vector NTI suite 9.0 showed that the deduced protein XC0690 shares 32.4% (46.9%) and 18.5% (29.9%) identity (similarity) to the M. tuberculosis ADK (GenBank accession number A5U4N0) and the Saccharomyces cerevisiae ADK (GenBank accession number P47143), respectively (data not shown). The ORF XC_0690 is 933 bp in length and located at positions 828812 to 829744 in the complementary strand of strain 8004's circular chromosome (Fig. 1). The nearest ORF downstream of XC_0690, XC_0689, which has the same transcriptional orientation as XC_0690 and is annotated to encode a putative transcriptional regulator, is 610 bp apart from the stop codon of XC_0690 (Fig. 1). The nearest ORF upstream of XC_0690, XC_0691, is annotated to encode a putative rod shape-determining protein and is transcribed in the opposite direction to XC_0690 (Fig. 1). Using a standard 5′-RACE method, the transcription initiation site of the gene XC_0690 was mapped at 70 nucleotides upstream of the start codon ATG (Fig. 1).
FIG. 1.
Genetic and physical map of the ADK-encoding gene XC_0690 in the genome of the X. campestris pv. campestris strain 8004. The positions and orientations of XC_0690 and other ORFs are shown. Arrows indicate lengths, locations, and orientations of the ORFs, and lines indicate the intergenic sequences. The middle elements show the PCR-amplified fragments used for expression, mutation, and complementation analysis. The transcriptional start site of XC_0690 is indicated by a square. The putative −10 and −35 sequences and the Shine-Dalgarno sequence are shown.
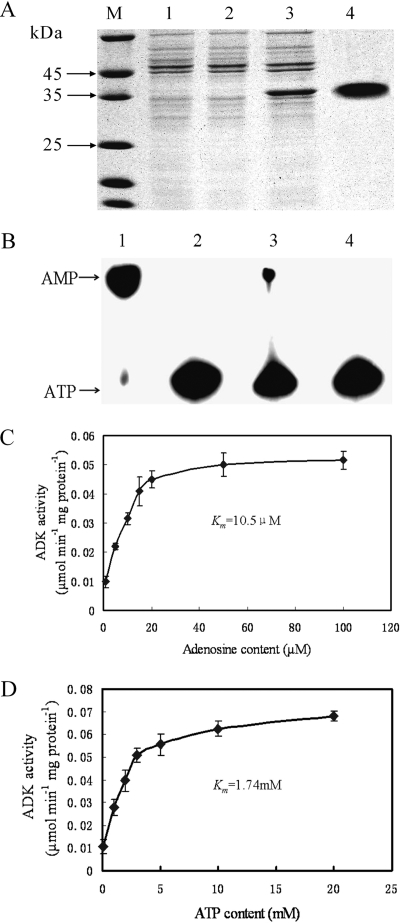
In order to investigate the biochemical characteristics of the product encoded by ORF XC_0690, XC_0690 was overexpressed with a His6 tag in the E. coli strain M15, as outlined in Materials and Methods. Separation of crude cell lysates by SDS-PAGE showed an intense protein band migrating at the expected molecular mass of 35 kDa (Fig. 2A). To obtain a preparation apparently free from contaminating proteins, the fusion protein with a His6 tag was purified using an Ni-NTA column (Fig. 2A). To verify whether the purified protein XC0690-His6 has ADK activity, its ability to phosphorylate adenosine to form AMP was examined by the radiometric TLC method. The AMP formation was detected by incubating the purified XC0690-His6 protein with 0.25 mM adenosine, 2.5 mM ATP, and 1.0 μCi [γ-32P] ATP for 1 h at 37°C before TLC separation of substrate and product. The results showed that the purified XC0690-His6 protein could direct the formation of AMP from adenosine and ATP (Fig. 2B). The ability of the protein XC0690-His6 to phosphorylate a range of sugars, including arabinose, xylose, sorbitol (glucitol), mannose, mannitol, sorbose, fructose, galactose, rhamnose, sucrose, and maltose, was also tested by the TLC method described previously (41). No detectable phosphorylation of any of these sugars by XC0690-His6 was observed (data not shown). The ADK activity of XC0690-His6 was further quantitatively measured by HPLC with 2.5 mM ATP and 1, 5, 10, 15, 20, 50, or 100 μM adenosine, and the activity values were 0.0098, 0.022, 0.0315, 0.0409, 0.045, 0.05, and 0.0516 μmol min−1 mg protein−1, respectively (Fig. 2C). The Michaelis-Menten kinetic parameters were then evaluated by the double-reciprocal plot method. The apparent Km value and the specific activity were determined to be 10.5 μM and 6.78 × 104 U (mg protein)−1, respectively. The ADK activity was also measured with a range of ATP concentrations and 2.5 mM adenosine (Fig. 2D), and the Km value was determined to be 1.74 mM. These results demonstrate that ORF XC_0690 of the X. campestris pv. campestris strain 8004 encodes an ADK. Thus, we rename the gene XC_0690 as adkXcc.
FIG. 2.
Purification and characterization of the His6-tagged recombinant protein XC0690 from E. coli. (A) SDS-PAGE analysis of XC0690-His6 expression and purification on Ni-NTA resin (Qiagen). Lanes: 1, crude extract of E. coli M15; 2, crude extract of E. coli M15/pQE-30-0690 before induction with IPTG; 3, crude extract of E. coli M15/pQE-30-0690 after induction with IPTG; 4, affinity-purified XC0690-His6 protein; M, molecular mass markers. (B) Radiometric assay result of adenosine phosphorylation by XC0690-His6. A 5-μl sample of reaction mixture was spotted onto TLC plates. Lanes: 1, AMP standard; 2, reaction without XC0690-His6; 3, reaction with 0.5 mg/ml of XC0690-His6; 4, reaction without adenosine. (C) Rate of XC0690-His6-catalyzed conversion of adenosine to AMP at different adenosine concentrations. The data shown are from a representative experiment, and values are the means ± standard deviations from triplicate measurements. (D) Rate of XC0690-His6-catalyzed conversion of adenosine to AMP at different ATP concentrations. The data shown are from a representative experiment, and values are the means ± standard deviations from triplicate measurements.
Mutation of adkXcc resulted in an accumulation of intracellular adenosine and reductions of intracellular ADK activity and ATP level.
To explore the physiological role of adkXcc, we constructed a mutant with a single-crossover insertion mutation of ORF XC_0690 by the homologous suicide plasmid integration method (69) (see Materials and Methods for details), which was designated as 0690pk (Table 1). Simultaneously, a complemented strain, named C0690pk (Table 1), was also created by introducing the recombinant plasmid pL0690 (Table 1), in which a 1,112-bp DNA fragment from 177 bp upstream to 2 bp downstream of the XC_0690 ORF sequence (Fig. 1) was cloned into the vector pLAFR3, into the mutant strain 0690pk. To clarify whether a mutation in adkXcc has any effect on bacterial growth, the growth rates of the mutant 0690pk and the wild-type 8004 in the nutrient-rich complex medium NYG and the minimal medium MMX were compared. The results showed that the mutant grew in a fashion similar to that for the wild-type strain in both media (data not shown); the doubling times of the two strains were 2.2 h in NYG and 4.4 h in MMX, respectively, indicating that a mutation in adkXcc does not affect the growth of X. campestris pv. campestris in standard media.
The intracellular ADK activities of the mutant strain 0690pk and the wild-type strain 8004 were tested and compared. Cell extracts of the strains were prepared from cultures grown in NYG liquid medium supplemented with appropriate antibiotics (i.e., Rif for the wild type and Rif and Kan for the mutant) for 18 h, and their ADK activities were measured by the HPLC assay. As summarized in Table 2, the adkXcc mutant 0690pk exhibited a substantial reduction in intracellular ADK enzymatic activity, where the level of ADK activity was approximately 10% of the wild-type level. The complemented strain C0690pk had a level of ADK activity that was considerably elevated over that of the wild type (Table 2). The residual ADK activity measured in the mutant is probably a background activity.
TABLE 2.
ADK activity and adenosine and ATP contents in X. campestris pv. campestris strains
| Straina | ADK activity (pmol AMP min−1 mg protein−1)b | Adenosine content (nmol g−1 [dry wt] cells)b | ATP content (μmol g−1 [dry wt] cells)b at:
|
||
|---|---|---|---|---|---|
| 12 h | 24 h | 36 h | |||
| 8004 (wild type) | 59.1 ± 2.2 a | 14.3 ± 2.1 a | 3.4 ± 0.18 a | 2.52 ± 0.12 a | 1.75 ± 0.14 a |
| 0690pk (adkXcc mutant) | 5.5 ± 0.8 b | 142 ± 17 b | 2.82 ± 0.13 c | 2.01 ± 0.15 c | 1.35 ± 0.11 c |
| C0690pk (complemented mutant) | 77 ± 3.8 a | 12.6 ± 3.2 a | 3.36 ± 0.15 a | 2.45 ± 0.18 a | 1.83 ± 0.15 a |
Strains were cultured in the nutrient-rich complex medium NYG.
Data presented are the means ± standard deviations of triplicate measurements from a representative experiment, and similar results were obtained in two other independent experiments. Different letters a and b indicate significant differences at a P value of 0.01 (t test); a and c indicate significant differences at a P value of 0.05.
To assess the effect of a mutation in adkXcc on the level of intracellular adenosine, adenosine was extracted from cultures grown in NYG medium for 18 h and measured by HPLC assay. The results showed that the adenosine content in the adkXcc mutant was highly enhanced. The adenosine level in the adkXcc mutant was approximately 10-fold more than that in the wild type (Table 2). In addition, the adenosine level in the complemented strain C0690pk was similar to that in the wild-type strain (Table 2). Taken together, these results reveal that a mutation in adkXcc can lead to a significant reduction of intracellular ADK activity and a substantial accumulation of the intracellular adenosine in X. campestris pv. campestris.
The effect of a mutation in adkXcc on the intracellular energy ATP level was also assessed. Strains were cultured in NYG medium, and their intracellular ATP levels were measured by the luciferin-luciferase bioluminescence assay. As shown in Table 2, the adkXcc mutant had only 82.9, 79.8, and 77.1% of the wild-type intracellular ATP level when cultured for 12, 24, and 36 h, respectively. The reduced ATP level of the mutant could be restored to the wild-type level by adkXcc in trans; i.e., the ATP levels in the complemented strain and the wild type were almost the same (Table 2). These results show that ADK deficiency can lead to a decrease of the intracellular ATP level in X. campestris pv. campestris.
Mutation in adkXcc results in attenuation of the virulence and growth of X. campestris pv. campestris in the host plant.
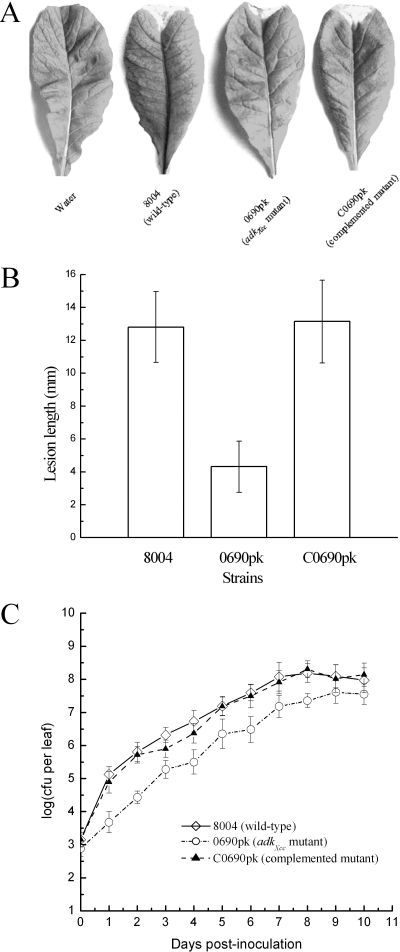
X. campestris pv. campestris is a xylem-colonizing systemic pathogen that generally invades plant leaves through hydathodes and wounds and multiplies in vascular tissues. In order to evaluate whether ADK activity in X. campestris pv. campestris plays a role in pathogenesis, we tested the virulence of the adkXcc mutant strain 0690pk on Chinese radish by the leaf-clipping method, which introduces the bacteria directly into the vascular system (18). The result showed that at 10 days after inoculation, the mutant produced disease symptoms with a mean lesion length of only 4.3 mm on the infected leaves, while the wild-type strain produced a mean lesion length of 13 mm (Fig. 3A and B). As analyzed by the t test, the mean lesion length caused by the adkXcc mutant was significantly less than that caused by the wild type (P = 0.01). The disease symptoms caused by the complemented strain were as severe as those caused by the wild type (Fig. 3A and B), indicating that the virulence of the adkXcc mutant could be restored by adkXcc in trans.
FIG. 3.
adkXcc is required for full pathogenicity of X. campestris pv. campestris for Chinese radish. (A) Symptom production on leaves 10 days after inoculation by clipping with the adkXcc mutant 0690pk, the wild-type strain 8004, and the complemented mutant C0690pk. (B) Mean lesion lengths caused by different strains. The adkXcc mutant has attenuated virulence; as analyzed by the t test, the mean lesion length caused by 0690pk was significantly less than that caused by the wild type (P = 0.01). Values given are the means ± standard deviations from 30 measurements. (C) Bacterial populations of the adkXcc mu tant, the wild type, and the complemented strain in host leaves. Inoculated leaves for each strain were taken daily and homogenized in sterile water. The homogenates were diluted and then plated on NYG plates. Bacterial CFU were counted after incubation for 3 days. Data are the means ± standard deviations from three replicates.
To verify whether a mutation in adkXcc has any effect on the growth of X. campestris pv. campestris in host tissues, the bacterial cells from the infected radish leaves were recovered and calculated as CFU. The results showed that the number of the mutant bacterial cells was more than 10-fold fewer than that of the wild type at 1 day postinoculation and afterward (Fig. 3C). The growth of the mutant in planta could be completely restored by adkXcc in trans (Fig. 3C). Taken together, these results reveal that the ADK activity is required for X. campestris pv. campestris to proliferate well and to attain full virulence in host plants.
Mutation in adkXcc negatively affects EPS production and cell motility.
X. campestris pv. campestris produces EPS and a series of extracellular enzymes, including protease, endoglucanase and amylase, which are collectively essential for pathogenesis (17). To investigate whether a mutation in adkXcc has any effect on these pathogenicity-related factors, the EPS yield and the activity of the three extracellular enzymes produced by the adkXcc mutant 0690pk were determined. The results showed that the activities of extracellular amylase, endoglucanase, and protease produced by the mutant were similar to those of the enzymes produced by the wild type (data not shown). However, the adkXcc mutant produced 30 to 60% less EPS than the wild type in NY medium supplemented with 2% of various sugars (Table 3). The EPS yields for the complemented mutant and the wild type were similar (Table 3), suggesting that the reduction in EPS production of the mutant could be restored to the wild-type level by adkXcc in trans.
TABLE 3.
EPS production in X. campestris pv. campestris strains
| Straina | EPS yield (g/liter)b with:
|
||||||
|---|---|---|---|---|---|---|---|
| Xylose | Glucose | Mannose | Fructose | Galactose | Sucrose | Maltose | |
| 8004 (wild type) | 3.32 ± 0.23 a | 9.8 ± 0.14 a | 6.56 ± 0.32 a | 6.82 ± 0.38 a | 7.17 ± 0.34 a | 9.62 ± 0.32 a | 9.59 ± 0.35 a |
| 0690pk (adkXcc mutant) | 1.77 ± 0.29 b | 4.13 ± 0.24 b | 2.85 ± 0.44 b | 3.85 ± 0.29 b | 2.76 ± 0.36 b | 4.15 ± 0.26 b | 4.16 ± 0.41 b |
| C0690pk (complemented mutant) | 3.41 ± 0.35 a | 9.85 ± 0.47 a | 6.61 ± 0.37 a | 6.96 ± 0.34 a | 7.25 ± 0.16 a | 9.71 ± 0.29 a | 9.5 ± 0.19 a |
Strains were cultured in NY medium supplemented with various sugars (2%).
Data presented are the means ± standard deviations of triplicate measurements from a representative experiment, and similar results were obtained in two other independent experiments. Different letters indicate significant differences (P = 0.01; t test).
The proteins encoded by the gum gene cluster direct assembly of the pentasaccharide repeating unit, polymerization, and the export of EPS (28, 64). To verify whether ADK deficiency in the adkXcc mutant induces changes in the expression level of the gum cluster, the reporter plasmid pL6gumGUS, which carries the promoter region of the gum operon fused to the coding region for gusA (65), was transferred to the wild type and the mutant by triparental mating, resulting in the transconjugant strains 8004/pL6gumGUS and 0690pk/pL6gumGUS (Table 1), respectively. The GUS activities produced by the transconjugant strains cultured under the conditions used for EPS production determination as described above were then assayed. The results showed that the GUS activities in the mutant background were reduced 28, 12, and 17% compared to those in the wild-type background when culture was for 24, 36, and 48 h (Table 4), respectively. As analyzed by the t test, the GUS activity in the mutant background was significantly reduced at 24 h (P = 0.01 by t test) and 48 h (P = 0.05 by t test) compared to that in the wild-type background, although the GUS activities in the mutant background at 36 h showed no significant differences (P = 0.05) by the t test compared to those in the wild-type background. This suggests that inactivation of adkXcc may negatively affect the transcription level of the gum operon.
TABLE 4.
Expression of the gum-gusA reporter in X. campestris pv. campestris strains
| Straina | GUS activity (mg para-nitrophenol produced per min per OD unit)b at:
|
||
|---|---|---|---|
| 24 h | 36 h | 48 h | |
| 8004/pL6gumGUS | 0.74 ± 0.027 a | 1.49 ± 0.15 a | 2.24 ± 0.26 a |
| 0690pk/pL6gumGUS | 0.53 ± 0.06 b | 1.31 ± 0.075 a | 1.85 ± 0.14 c |
Strains harboring the reporter plasmid pL6gumGUS were grown in NY liquid medium supplemented with 2% (wt/vol) glucose.
Values for GUS activity are the means ± standard deviations of triplicate measurements. Data presented are from a representative experiment, and similar results were obtained in two other independent experiments. Different letters a and b in the data column indicate significant differences at a P value of 0.01 (t test); a and c indicate significant differences at a P value of 0.05.
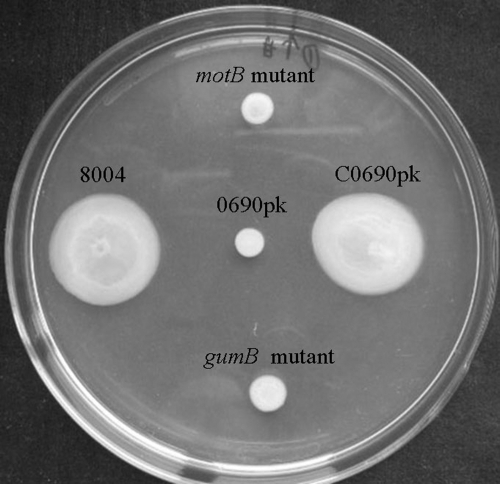
The cell motility of the mutant 0690pk was also tested. On the tested plates, the mutant displayed severely weakened motility compared to the wild type (Fig. 4). The radii of the growth zones resulting from migration away from the inoculation points on the agarose surface were about 0.5 cm for the mutant and 2.2 cm for the wild type. As analyzed by the t test, the mean radius of the mutant was significantly shorter than that of the wild type (P = 0.01). The radii of the complemented strain and the wild-type strain were not significantly different (P = 0.05), indicating that the mobility of the mutant could be restored to wild-type level by adkXcc in trans. In addition, a gumB mutant (010D08) and a motB mutant (098A02) were also included in the assay. In X. campestris pv. campestris, gumB and motB encode an EPS export protein and a putative flagellar motor component, respectively (51, 67). On the swarming test plates, the adkXcc mutant and the motB mutant showed similarly defective motility (Fig. 4). Interestingly, the gumB mutant also showed defective motility (Fig. 4). Although how EPS affects bacterial motility is unknown, these results further indicate that the ADK of X. campestris pv. campestris is implicated in bacterial motility.
FIG. 4.
Motilities of X. campestris pv. campestris strains. Cells were inoculated onto NYG plates supplemented with 0.3% agarose from overnight cultures and photographed after 3 days of incubation at 28°C. The adkXcc mutant 0690pk is deficient in cell motility, which could be restored to the wild-type level by providing a plasmid bearing the adkXcc gene (i.e., the complemented strain C0609pk).
The expression of adkXcc is enhanced by glucose and other sugars.
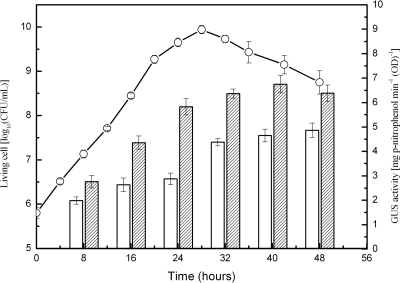
To gain understanding of the expression profile of ADK in X. campestris pv. campestris, we employed the gus reporter as an indicator to determine the expression level of adkXcc. The reporter plasmid pGUS0690, carrying the fused 471-bp adkXcc promoter region and the promoterless gus gene, was constructed and introduced into the wild-type strain 8004 to create the reporter strain 8004/pGUS0690. The GUS level in strain 8004/pGUS0690 may mirror the expression level of adkXcc. The strain 8004/pGUS0690 was cultured in NY medium and in NY containing 2% glucose, and GUS activity was measured at 8-h intervals until 48 h. As shown in Fig. 5, although the GUS activities produced by the reporter strain grown in NY medium with or without glucose rose gradually from early logarithmic phase to late decline phase, the GUS activity in the cells cultured in NY with glucose was about 30% stronger than that in cells cultured in NY alone at all of the tested points. These differences are significant (P = 0.01 by t test). The GUS activities were also tested when the reporter strain was cultured in NY medium containing 2% sucrose, fructose, mannose, or galactose. In all the cases, the values of the GUS activity were similar to those for the strain cultured in NY containing glucose (data not shown). ADK activity of the wild-type strain 8004 grown in NY supplemented with glucose was also assessed over the course of bacterial growth. The results showed that the ADK enzymatic activity profile of the wild type was similar to the GUS activity profile (data not shown). Taken together, these results imply that the expression of adkXcc may be enhanced by various sugars.
FIG. 5.
Expression of adkXcc in X. campestris pv. campestris. The wild-type strain 8004 harboring the reporter plasmid pGUS0690 was cultured in NY medium or in NY supplemented with 2% glucose. The curve represents bacterial growth in NY containing glucose, which is almost identical to that in NY only. Open and filled bars represent GUS activities of the reporter strain cultured in NY medium and NY containing glucose, respectively. Values given are the means and standard deviations of triplicate measurements. The data presented are from a representative experiment, and similar results were obtained in two other independent experiments.
DISCUSSION
ADK is a typical housekeeping enzyme in eukaryotes. Because of its essential roles in metabolism and its potential as a therapeutic target for control of parasitic protozoa and diseases such as epilepsy and stroke (4, 23, 42), the eukaryotic ADK has been intensively studied. In contrast, as described above, very little work has been done on the prokaryotic counterpart. As bacteria synthesize purines de novo and might not necessarily acquire a purine salvage enzyme, it has long been considered that ADK is not common in bacteria. To the best of our knowledge, only two bacterial ADKs have been identified from gram-positive bacteria (39, 52), and the adkXcc identified from X. campestris pv. campestris strain 8004, reported in this paper, is the first ADK to be characterized from a gram-negative bacterium. The other two X. campestris pv. campestris strains, ATCC 33913 and B100, whose genomic sequences have been determined contain a putative protein homologous to ADKXcc. Amino acid sequence pairwise alignments showed that the predicted proteins encoded by the ORF XCC_3471 of strain ATCC 33913 and the ORF xccb100_0723 of strain B100, which are annotated as a sugar kinase and a putative carbohydrate kinase, respectively (15, 67), share 100 and 99% identity to the ADKXcc of strain 8004, respectively (data not shown). In addition, our previous array-based comparative genome hybridization analyses demonstrated that the ORF encoding ADKXcc is highly conserved in all of the 18 other X. campestris pv. campestris strains tested (25). These results suggest that the ADK enzyme is widely distributed among X. campestris pv. campestris bacteria. Interestingly, a sequence comparison search showed that a number of other gram-negative bacteria, such as Xanthomonas oryzae, Xanthomonas axonopodis, Stenotrophomonas maltophilia, Nitrosococcus oceani, Methylococcus capsulatus, Burkholderia phymatum, Ralstonia solanacearum, and Bordetella petrii, contain genes encoding putative proteins that display >50% amino acid sequence homology with ADKXcc, which are annotated as sugar kinases or carbohydrate kinases (data not shown). The high sequence homology suggests that some of these proteins may have ADK activity.
The fact that ADKXcc-deficient mutant could still grow well as the wild type not only in rich medium but also in minimal medium suggests that ADKXcc is not a housekeeping enzyme in X. campestris pv. campestris and that the X. campestris pv. campestris pathogen may synthesize purines de novo. In the de novo purine synthesis pathway, there are 10 enzymatic steps that lead from 5-phosphoribosyl 1-pyrophosphate to the IMP branch point; then, IMP is converted in two steps to AMP and in two steps to GMP (19). Indeed, ORFs encoding all of the enzymes for de novo purine synthesis (i.e., purF, amidophosphoribosyltransferase [XC_3282]; purD, phosphoribosylamine-glycine ligase [XC_0509]; purT, phosphoribosylglycinamide formyltransferase [XC_1322]; purL, phosphoribosylformylglycinamidine synthase [XC_3578]; purM, phosphoribosylaminoimidazole synthetase [XC_1324]; purE, phosphoribosylaminoimidazole carboxylase catalytic subunit [XC_1613]; purK, phosphoribosylaminoimidazole carboxylase ATPase subunit [XC_1614]; purC, phosphoribosylaminoimidazole-succinocarboxamide synthase [XC_0467]; purB, adenylosuccinate lyase [XC_2744]; purH, bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase [XC_0510]; purA, adenylosuccinate synthetase [XC_3193]; and adk, adenylate kinase [XC_0873]) are present in the genome of the X. campestris pv. campestris strain 8004 (51), although the enzymatic functionality of their putative products remains to be clarified.
A mutation in adkXcc resulted in a significant attenuation of the bacterial virulence and growth in the host plant, indicating that the activity of the purine salvage enzyme ADKXcc is important for full pathogenicity of X. campestris pv. campestris. The adkXcc mutant produced significantly less EPS and displayed severely weakened motility compared to the wild type. The pathogenesis of X. campestris pv. campestris is a complex process involving several steps; these mainly include invading into host plants through hydathodes and wounds, multiplying in the intercellular spaces, spreading via the vascular system, and causing disease symptoms (9). Cell motility allows bacteria to obtain more or better nutrients, avoid toxic substances or unfavorable environments, find a host, and disperse effectively (63). Though the ability to move toward or within a potential host generally confers a significant selective advantage on bacteria that are pathogenic to animals, the role of motility in phytopathogenic bacteria is not as well understood (49, 63). In certain bacteria, e.g., Erwinia amylovora, Pseudomonas phaseolicola, Pseudomonas syringae, and R. solanacearum, motility has been suggested to make contribution to bacterial virulence in the early stages such as invasion and colonization (3, 24, 50, 60). Recently, we have demonstrated that RsmA and DsbB of X. campestris pv. campestris, which are required for full virulence, are involved in various cellular processes, including cell motility (10, 30). More recently, it has been shown that the virulence-deficient mutants derived from a mutation in the genes XC_2249 and XC_3221 of the X. campestris pv. campestris strain 8004 displayed a reduction in cell motility (43). EPS has been demonstrated to be an important virulence factor in X. campestris pv. campestris. It can enhance the susceptibility of host plants by suppressing defense responses such as callose formation (72) and can contribute to biofilm formation (18, 61) and bacterial resistance against host defenses. It may also serve to mask bacterial cells to prevent recognition by the host and to enable colonization of host tissues (1).
The biosynthesis of EPS in X. campestris pv. campestris is a complicated process, which requires a series of enzymes and a significant proportion of total cellular nicotinamide cofactors (29). The assembly and polymerization of the subunits and the export of EPS are directed by the gum cluster (28). The finding that the adkXcc mutant strain 0690pk harboring the gum promoter-gusA transcriptional fusion reporter plasmid pL6gumGUS produced lower GUS activity than the wild type carrying the same plasmid suggests that a mutation in adkXcc negatively affects the expression of the gum cluster, resulting in a reduction of EPS. EPS synthesis by X. campestris pv. campestris also requires a significant amount of ATP (29). Disruption of adkXcc led to a significant reduction of ATP in X. campestris pv. campestris. The ATP reduction may be one of the major causes that negatively affect EPS production as a result of adkXcc mutation. It is well known that ATP provides the energy that drives manifold energy-requiring activities of all living cells, including synthesis of complex biomolecules, osmosis involving in transporting substances into cells, and cell motility. In addition to reductions in EPS production and cell motility, the significant ATP diminution in the adkXcc mutant may influence other cellular processes, one (or several) of which may play roles in the pathogenicity of X. campestris pv. campestris.
Disruption of adkXcc resulted in a significant accumulation of intracellular adenosine. Whether the adenosine accumulation in X. campestris pv. campestris cells will alter the expression of certain genes or the balance of certain metabolic pathways remains to be further investigated. In eukaryotes, adenosine can act as an endogenous regulator of innate immunity and a signal regulator involved in physiology and modulation of the function of numerous cell types (6). In the gram-positive bacterium S. lividans, it has been demonstrated that loss of ADK activity results in adenosine accumulation, which in turn strongly inhibits the sporulation process, while a yet-to-be defined metabolic change(s) suppresses actinorhodin production and activates undecylprodigiosin biosynthesis (52). Further studies are required for our understanding of the full functions of ADK and adenosine in X. campestris pv. campestris and other bacteria.
Acknowledgments
This work was supported by The National Natural Science Foundation of China (30771177), the 973 Program (2006CB101902), and the 863 Program (2006AA02Z175) of the Ministry of Science and Technology of China.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Alvarez, A. M. 2000. Black rot of crucifers, p. 21-52. In A. J. Slusarenko, R. S. S. Fraser, and L. C. van Loon (ed.), Mechanisms of resistance to plant diseases. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Arch., J. R., and E. A. Newsholme. 1978. Activities and some properties of 5′-nucleotidase, adenosine kinase and adenosine deaminase in tissues from vertebrates and invertebrates in relation to the control of the concentration and physiological role of adenosine. Biochem. J. 174965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayot, R. G., and S. M. Ries. 1986. Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology 76441-445. [Google Scholar]
- 4.Boison, D. 2006. Adenosine kinase, epilepsy and stroke: mechanisms and therapies. Trends Pharmacol. Sci. 27652-658. [DOI] [PubMed] [Google Scholar]
- 5.Bork, P., C. Sander, and A. Valencia. 1993. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 231-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiec, A., K. Lechward, K. Tkacz-Stachowska, and A. C. Składanowski. 2006. Adenosine as a metabolic regulator of tissue function: production of adenosine by cytoplasmic 5′-nucleotidases. Acta Biochim. Pol. 53269-278. [PubMed] [Google Scholar]
- 7.Caputto, R. 1951. The enzymatic synthesis of adenylic acid: adenosine kinase. J. Biol. Chem. 193801-814. [PubMed] [Google Scholar]
- 8.Carret, C., S. Delbecq, G. Labesse, B. Carcy, E. Precigout, K. Moubri, T. P. Schetters, and A. Gorenflot. 1999. Characterization and molecular cloning of an adenosine kinase from Babesia canis rossi. Eur. J. Biochem. 2651015-1021. [DOI] [PubMed] [Google Scholar]
- 9.Chan, J. W., and P. H. Goodwin. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17489-508. [DOI] [PubMed] [Google Scholar]
- 10.Chao, N.-X., K. Wei, Q. Chen, Q.-L. Meng, D.-J. Tang, Y.-Q. He, G.-T. Lu, B.-L. Jiang, X.-X. Liang, J.-X. Feng, B. Chen, and J.-L. Tang. 2008. The rsmA-like gene rsmAXcc of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol. Plant-Microbe Interact. 21411-423. [DOI] [PubMed] [Google Scholar]
- 11.Cunha, R. A. 2001. Adenosine as a neuromodulator and as a homeostatic regulator in the nervous system: different roles, different sources and different receptors. Neurochem. Int. 38107-125. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, M. J., C. E. Barber, P. C. Turner, M. K. Sawczyc, R. J. W. Byrde, and A. H. Fielding. 1984. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad-host-range cosmid pLAFR1. EMBO J. 33323-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels, M. J., C. E. Barber, P. C. Turner, W. G. Cleary, and M. K. Sawczyc. 1984. Isolation of mutants of Xanthomonas campestris pathovar campestris showing altered pathogenicity. J. Gen. Microbiol. 1302447-2455. [Google Scholar]
- 14.Darling, J. A., W. J. Sullivan Jr., D. Carter, B. Ullman, and D. S. Roos. 1999. Recombinant expression, purification, and characterization of Toxoplasma gondii adenosine kinase. Mol. Biochem. Parasitol. 10315-23. [DOI] [PubMed] [Google Scholar]
- 15.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417459-463. [DOI] [PubMed] [Google Scholar]
- 16.Datta, A. K., D. Bhaumik, and R. Chatterjee. 1987. Isolation and characterization of adenosine kinase from Leishmania donovani. J. Biol. Chem. 2625515-5521. [PubMed] [Google Scholar]
- 17.Dow, J. M., and M. J. Daniels. 1994. Pathogenicity determinants and global regulation of pathogenicity in Xanthomonas campestris pv. campestris, p. 29-41. In J. L. Dangl (ed.), Molecular and cellular mechanisms in bacterial pathogenesis of plants and animals. Springer, Berlin, Germany. [DOI] [PubMed]
- 18.Dow, J. M., L. Crossman, K. Findlay, Y.-Q. He, J.-X. Feng, and J.-L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 10010995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 2628274-8287. [PubMed] [Google Scholar]
- 20.Faye, F., and F. Le Floc'h. 1997. Adenosine kinase of peach tree flower buds: purification and properties. Plant Physiol. Biochem. 3515-22. [Google Scholar]
- 21.Fisher, M. N., and E. A. Newsholme. 1984. Properties of rat heart adenosine kinase. Biochem. J. 221521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox, H. I., and N. W. Kelley. 1978. The role of adenosine and 2′-deoxyadenosine in mammalian cells. Annu. Rev. Biochem. 47655-686. [DOI] [PubMed] [Google Scholar]
- 23.Galazka, J., B. Striepen, and B. Ullman. 2006. Adenosine kinase from Crytosporidium parvum. Mol. Biochem. Parasit. 149223-230. [DOI] [PubMed] [Google Scholar]
- 24.Hatterman, D. R., and S. M. Ries. 1989. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology 79284-289. [Google Scholar]
- 25.He, Y.-Q., L. Zhang, B.-L. Jiang, Z.-C. Zhang, R.-Q. Xu, D.-J. Tang, J. Qin, W. Jiang, X. Zhang, J. Liao, J.-R. Cao, S.-S. Zhang, M.-L. Wei, X.-X. Liang, G.-T. Lu, J.-X. Feng, B. Chen, J. Cheng, and J.-L. Tang. 2007. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 8R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, R. F., J. M. Benson, F. F. Hahn, C. H. Hobbs, R. K. Jones, J. L. Mauderly, R. O. McClellan, and J. A. Pickrell. 1985. New approaches for the evaluation of pulmonary toxicity: bronchoalveolar lavage fluid analysis. Fundam. Appl. Toxicol. 5451-458. [DOI] [PubMed] [Google Scholar]
- 27.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 2451374-1377. [DOI] [PubMed] [Google Scholar]
- 28.Ielpi, L., R. O. Couso, and M. A. Dankert. 1993. Sequential assembly and polymerization of the prenol-linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris. J. Bacteriol. 1752490-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarman, T. R., and G. W. Pace. 1984. Energy requirements for microbial exopolysaccharide synthesis. Arch. Microbiol. 137231-235. [Google Scholar]
- 30.Jiang, B.-L., J. Liu, L.-F. Chen, Y.-Y. Ge, X.-H. Hang, Y.-Q. He, D.-J. Tang, G.-T. Lu, and J.-L. Tang. 2008. DsbB is required for the pathogenesis process of Xanthomonas campestris pv. campestris. Mol. Plant-Microbe Interact. 211036-1045. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy, J. F., and I. J. Bradshaw. 1984. Production, properties and applications of xanthan. Prog. Ind. Microbiol. 19319-371. [Google Scholar]
- 32.Kimmich, G. A., J. Randles, and J. S. Brand. 1975. Assay of picomole amounts of ATP, ADP, and AMP using the luciferase enzyme system. Anal. Biochem. 69187-206. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg, A., and W. E. Pricer. 1951. Enzymatic phosphorylation of adenosine and 2,6-diaminopurine riboside. J. Biol. Chem. 193481-495. [PubMed] [Google Scholar]
- 34.Kowaluk, E. A., and M. F. Jarvis. 2000. Therapeutic potential of adenosine kinase inhibitors. Expert Opin. Investig. Drugs 9551-564. [DOI] [PubMed] [Google Scholar]
- 35.Lautt, W. W., D. J. Legare, and M. S. d'Almeida. 1985. Adenosine as putative regulator of hepatic arterial flow (the buffer response). Am. J. Physiol. 248H331-H338. [DOI] [PubMed] [Google Scholar]
- 36.Lecoq, K., I. Belloc, C. Desgranges, and B. Daignan-Fornier. 2001. Role of adenosine kinase in Saccharomyces cerevisiae: identification of the ADO1 gene and study of the mutant phenotypes. Yeast 18335-342. [DOI] [PubMed] [Google Scholar]
- 37.Leibach, T. K., G. L. Spiess, T. J. Neudecker, G. J. Peschke, G. Puchwein, and G. R. Hartmann. 1971. Purification and properties of adenosine kinase from dried brewer's yeast. Hoppe-Seyler's Z. Physiol. Chem. 352328-344. [DOI] [PubMed] [Google Scholar]
- 38.Leong, S. A., G. S. Ditta, and D. R. Helinski. 1982. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 2578724-8730. [PubMed] [Google Scholar]
- 39.Long, M. C., V. Escuyer, and W. B. Parker. 2003. Identification and characterization of a unique adenosine kinase from Mycobacterium tuberculosis. J. Bacteriol. 1856548-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, G.-T., Z.-F. Ma, J.-R. Hu, D.-J. Tang, Y.-Q. He, J.-X. Feng, and J.-L. Tang. 2007. A novel locus involved in extracellular polysaccharide production and virulence of Xanthomonas campestris pathovar campestris. Microbiology 153737-746. [DOI] [PubMed] [Google Scholar]
- 41.Lu, G.-T., Z.-J. Yang, F.-Y. Peng, Y.-N. Tan, Y.-Q. Tang, J.-X. Feng, D.-J. Tang, Y.-Q. He, and J.-L. Tang. 2007. The role of glucose kinase in carbohydrate utilization and extracellular polysaccharide production in Xanthomonas campestris pathovar campestris. Microbiology 1534284-4294. [DOI] [PubMed] [Google Scholar]
- 42.Lüscher, A., P. Önal, A.-M. Schweingruber, and P. Mäser. 2007. Adenosine kinase of Trypanosoma brucei and its role in susceptibility to adenosine antimetabolites. Antimicrob. Agents Chemother. 153895-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy, Y., R. P. Ryan, K. O'Donovan, Y.-Q. He, B.-L. Jiang, J.-X. Feng, J.-L. Tang, and J. M. Dow. 2008. The role of PilZ domain proteins in the virulence of Xanthomonas campestris pathovar campestris. Mol. Plant Pathol. 9819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNally, T., R. J. Helfrich, M. Cowart, S. A. Dorwin, J. L. Meuth, K. B. Idler, K. A. Klute, R. L. Simmer, E. A. Kowaluk, and D. N. Halbert. 1997. Cloning and expression of the adenosine kinase gene from rat and human tissues. Biochem. Biophys. Res. Commun. 231645-650. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Miller, R. L., D. L. Adamczyk, and W. H. Miller. 1979. Adenosine kinase from rabbit liver. I. Purification by affinity chromatography and properties. J. Biol. Chem. 2542339-23457. [PubMed] [Google Scholar]
- 47.Moffatt, B. A., L. Wang, M. S. Allen, Y. Y. Stevens, W. Qin, J. Snider, and K. von Schwartzenberg. 2000. Adenosine kinase of Arabidopsis: kinetic properties and gene expression. Plant Physiol. 1241775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p 445-473. In F. C. Neidhardt, J. L. Ingraham, B. K. Low, B. Magasanik, M. Schaechter, H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 49.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 241109-1117. [DOI] [PubMed] [Google Scholar]
- 50.Panapoulos, N. J., and M. N. Schroth. 1974. Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 641389-1397. [Google Scholar]
- 51.Qian, W., Y. Jia, S.-X. Ren, Y.-Q. He, J.-X. Feng, L.-F. Lu, Q. Sun, G. Ying, D.-J. Tang, H. Tang, W. Wu, P. Hao, L. Wang, B.-L. Jiang, S. Zeng, W.-Y. Gu, G. Lu, L. Rong, Y. Tian, Z. Yao, G. Fu, B. Chen, R. Fang, B. Qiang, Z. Chen, G.-P. Zhao, J.-L. Tang, and C. He. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajkarnikar, A., H. J. Kwon, and J. W. Suh. 2007. Role of adenosine kinase in the control of Streptomyces differentiations: loss of adenosine kinase suppresses sporulation and actinorhodin biosynthesis while inducing hyperproduction of undecylprodigiosin in Streptomyces lividans. Biochem. Biophys. Res. Commun. 363322-328. [DOI] [PubMed] [Google Scholar]
- 53.Rotllan, P., and M. T. Miras Portugal. 1985. Adenosine kinase from bovine adrenal medulla. Eur. J. Biochem. 151365-371. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 55.Schäfer, A., A. Tauch, W. J. äger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 56.Schomberg, D., and D. Stephan. 1997. Adenosine kinase, p. 1-9. In D. Schomberg, and D. Stephan (ed.), Enzyme handbook, vol. 13. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 57.Spychala, J., N. S. Datta, K. Takabayashi, M. Datta, I. H. Fox, T. Gribbin, and B. S. Mitchell. 1996. Cloning of human adenosine kinase cDNA: sequence similarity to microbial ribokinases and fructokinases. Proc. Natl. Acad. Sci. USA 931232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 1695789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang, J.-L., Y.-N. Liu, C. E. Barber, J. M. Dow, J. C. Wootton, and M. J. Daniels. 1991. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. 226409-417. [DOI] [PubMed] [Google Scholar]
- 60.Tans-Kersten, J., H. Huang, and C. Allen. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 1833597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torres, P. S., Malamud, F., Rigano, L. A., Russo, D. M., Marano, M. R., Castagnaro, A. P., Zorreguieta, A., Bouarab, K., Dow, J. M., A. A. Vojnov. 2007. Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris. Environ. Microbiol. 92101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner, P., C. Barber, and M. J. Daniels. 1984. Behavior of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 195101-107. [Google Scholar]
- 63.VandeBroek, A., and J. Vanderleyden. 1995. The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol. Plant-Microbe Interact. 8800-810. [Google Scholar]
- 64.Vanderslice, R. W., D. H. Doherty, M. A. Capage, M. R. Betlach, R. A. Hassler, N. M. Henderson, J. Ryan-Graniero, and M. Tecklenburg. 1989. Genetic engineering of polysaccharide structure in Xanthomonas campestris, p. 145-157. In V. Crescenzi, I. C. M. Dea, S. Paoletti, S. S. Stivala, and I. W. Sutherland (ed.), Biomedical and biotechnological advances in industrial polysaccharides. Gordon and Breach Science Publishers, New York, NY.
- 65.Vojnov, A. A., H. Slater, M. J. Daniels, and J. M. Dow. 2001. Expression of the gum operon directing xanthan biosynthesis in Xanthomonas campestris and its regulation in planta. Mol. Plant-Microbe Interact. 14768-774. [DOI] [PubMed] [Google Scholar]
- 66.von Schwartzenberg, K., S. Kruse, R. Reski, B. Moffatt, and M. Laloue. 1998. Cloning and characterization of an adenosine kinase from Physcomitrella involved in cytokinin metabolism. Plant J. 13249-257. [DOI] [PubMed] [Google Scholar]
- 67.Vorhölter, F. J., S. Schneiker, A. Goesmann, L. Krause, T. Bekel, O. Kaiser, B. Linke, T. Patschkowski, C. Rückert, J. Schmid, V. K. Sidhu, V. Sieber, A. Tauch, S. A. Watt, B. Weisshaar, A. Becker, K. Niehaus, and A. Pühler. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 13433-45. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Y., M. C. Long, S. Ranganathan, V. Escuyer, W. B. Parker, and R. Li. 2005. Overexpression, purification and crystallographic analysis of a unique adenosine kinase from Mycobacterium tuberculosis. Acta Crystallogr. F 61553-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193201-205. [DOI] [PubMed] [Google Scholar]
- 70.Yamada, Y., H. Goto, and N. Ogasawara. 1982. Differences of adenosine kinases from various mammalian tissues. Comp. Biochem. Physiol. 71B367-372. [DOI] [PubMed] [Google Scholar]
- 71.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 72.Yun, M. H., P. S. Torres, M. El Oirdi, L. A. Rigano, R. Gonzalez-Lamothe, M. R. Marano, A. P. Castagnaro, M. A. Dankert, K. Bouarab, and A. A. Vojnov. 2006. Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 141178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao, J., B. Todd, and G. H. Fleet. 1994. Seperation of ribonucleotides, ribonucleosides, deoxyribonucleotides, deoxyribonucleosides and bases by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 673167-171. [Google Scholar]