Abstract
Yeast ptc1 mutants are rapamycin and caffeine sensitive, suggesting a functional connection between Ptc1 and the TOR pathway that is not shared by most members of the type 2C phosphatase family. Genome-wide profiling revealed that the ptc1 mutation largely attenuates the transcriptional response to rapamycin. The lack of Ptc1 significantly prevents the nuclear translocation of Gln3 and Msn2 transcription factors to the nucleus, as well as the dephosphorylation of the Npr1 kinase, in response to rapamycin. This could explain the observed decrease in both the basal and rapamycin-induced expression of several genes subjected to nitrogen catabolite repression (GAT1, MEP1, and GLN1) and stress response element (STRE)-driven promoters. Interestingly, this decrease is abolished in the absence of the Sit4 phosphatase. Epitasis analysis indicates that the mutation of SIT4 or TIP41, encoding a Tap42-interacting protein, abolishes the sensitivity of the ptc1 strain to rapamycin and caffeine. All of these results suggest that Ptc1 is required for normal TOR signaling, possibly by regulating a step upstream of Sit4 function. According to this hypothesis, we observe that the mutation of PTC1 drastically diminishes the rapamycin-induced interaction between Tap42 and Tip41, and this can be explained by lower-than-normal levels of Tip41 in ptc1 cells. Ptc1 is not necessary for the normal expression of the TIP41 gene; instead, its absence dramatically affects the stability of Tip41. The lack of Ptc1 partially abolishes the rapamycin-induced dephosphorylation of Tip41, which may further decrease Tap42 binding. Reduced Tip41 levels contribute to the ptc1 phenotypes, although additional Ptc1 targets must exist. All of these results provide the first evidence showing that a type 2C protein phosphatase is required for the normal functioning of the TOR pathway.
Type 2C Ser/Thr protein phosphatases (PP2C) are a group of monomeric enzymes widely found in animals, plants, and fungi (35, 62, 66, 69). The function(s) and regulatory mechanisms for each specific PP2C isoform are largely unknown, and its study constitutes a major challenge, particularly because of the very important cellular roles played by this type of enzyme. PP2C activities regulate a plethora of signaling networks that control cell differentiation, proliferation, growth, survival, and metabolism. As a general rule, they act as inhibitors of cellular stress signaling (35). Because of the lack of regulatory or targeting subunits (with a few exceptions, such as the Ptc1 scaffold protein Nbp2 [40]), functional diversity most likely is achieved by the expression of multiple, probably specialized isoforms. For instance, the 16 human PP2C genes can generate at least 22 isoforms (35). In plants, PP2C genes comprise one of the largest gene families, with more than 70 members in Arabidopsis thaliana (62). In the yeast Saccharomyces cerevisiae, there are seven different PP2C isoforms (Ptc1 to Ptc7). Several of these enzymes (Ptc1 to Ptc4) have been characterized in connection with the HOG osmotic responsive pathway as Hog1 phosphatases (38, 43, 57, 71, 75), but it is now widely accepted that Ptc phosphatases play many diverse roles in yeast cells, such as tRNA splicing (52), mitochondrial and endoplasmic reticulum inheritance (20, 53), and the dephosphorylation of cyclin-dependent kinases (11). Yeast Ptc1 is the closest homolog of human Wip1, a phosphatase that may have oncogenic properties by reducing the cellular activities of p38, p53, and ATM (35). Among Ptc1 cellular tasks, a role in tolerance to lithium ions has been described. This function can be explained, at least in part, by the positive effect that Ptc1 exerts on the expression of ENA1, which encodes a major determinant for sodium/lithium efflux in yeast (56). Recent work (23) has demonstrated that, under standard growth conditions, the transcriptional profile of yeast cells lacking PTC1 is very different from those devoid of PTC2, PTC3, PTC4, or PTC5, suggesting that, from a functional point of view, Ptc1 markedly differs from other members of the family. However, in spite of the growing body of knowledge, our understanding of the functional role, isoform specificity, and regulatory properties of these enzymes still is rather limited.
The target of rapamycin (TOR) pathway plays a key role in the regulation of cell growth in eukaryotes in response to nutrient availability. In yeast cells, the TOR pathway is active when there are plenty of nutrients in the medium, whereas it is inactivated by nutrient scarcity (i.e., less favorable nitrogen sources, such as proline) or by the exposure of the cells to the antifungal macrocyclic lactone rapamycin (see reference 27 for a review). TOR controls the expression of a large number of genes that are transcribed by all three RNA polymerases. The profound effect triggered by changes in the activity of the TOR pathway was revealed by several transcriptional studies based on DNA microarray analysis using cells treated with rapamycin (8, 21, 25, 63). Rapamycin inhibits the expression of all ribosomal genes, including 35S, 5S, and tRNAs, as well as ribosomal proteins. An important set of genes whose expression is potently activated by rapamycin treatment comprises those related to nitrogen catabolite repression (NCR). These genes encode proteins that are required for the adaptation to scarce or less preferred nitrogen sources (for a review, see reference 12). The inhibition of TOR also induces genes required for the degradation of biomolecules (at the ribosome or mitochondria) in a process called autophagy (32). Similarly, treatment with rapamycin alters the expression of genes regulated by the mitochondrial signaling pathway, known as the retrograde response (RTG) (6), and affects a large number of stress-related genes by promoting entry into the nucleus of the stress-activated factors Msn2/Msn4 (3).
A well-characterized element mediating the effect of the TOR pathway on downstream NCR genes is the GATA-type transcription factor Gln3 (39). It is commonly accepted that an excess of nitrogen activates both Tor1 and Tor2 kinases, which in turn phosphorylate Tap42, thus promoting a functional association with type 2A protein phosphatases (Pph21 and Pph22) as well as with type 2A-like enzymes (Sit4, Pph3, and Ppg1) (17, 30, 70). Rapamycin treatment inactivates Tor1 and Tor2, and this triggers the disassembling of the Tap42-phosphatase complex from Tor kinases, thus allowing the dephosphorylation of Gln3. The dephosphorylated form of Gln3 dissociates from Ure2, which otherwise retains the transcription factor in the cytoplasm, and is imported into the nucleus (14, 18, 27, 74). Nuclear Gln3 mediates NCR-sensitive transcription by binding to GATA-containing promoters, such as those of GAP1 and MEP1 genes (encoding a general amino acid permease and ammonium permease, respectively), GLN1 (encoding glutamine synthetase, which incorporates ammonia into glutamate to form glutamine), or GDH1 (encoding a glutamate dehydrogenase activity that synthesizes glutamate from ammonia and α-ketoglutarate).
The ability of Tap42 to interact with Sit4 is greatly affected by the presence of the dephosphorylated form of Tip41 (29). It has been reported that in yeast cells lacking TIP41, Gln3 remains in the cytoplasm after rapamycin treatment (29). Similarly, the deletion of TIP41 prevents the nuclear migration of Msn2 in a strain expressing the temperature-sensitive tap42-106 allele (59). It has been shown that the dephosphorylation of Tip41, which would promote its binding to Tap42, also is Sit4 dependent. This would provide a feedback loop that would enhance the association of Tip41 with Tap42 (29). It has been proposed recently, on the basis of a combined experimental and mathematical approach, that the Tip41-Tap42 complex could act as a specificity factor that would trigger the rapamycin-induced and PP2A/Sit4-mediated dephosphorylation of downstream targets, such as phosphorylated Gln3 (33). Gln3 localization has been postulated to be negatively regulated by the Npr1 kinase (15), although this may be an indirect effect. It must be noted that, although useful, the models described above do not explain all experimental observations. For instance, there are discrepancies regarding the role of Sit4 in regulating the phosphorylation state of Gln3 (67) or even on the relationship between the Gln3 phosphorylation state and its nuclear localization (68).
Existing evidence indicated that the lack of Ptc1 phosphatase activity results in sensitivity to rapamycin (47, 55, 73), and we observed that ptc1 cells also were sensitive to caffeine, a compound that has been proposed to inhibit the TOR pathway (34). Interestingly, the functional mapping of the ENA1 promoter in response to salt stress in a ptc1 mutant (to be reported elsewhere) allowed linking the absence of the phosphatase with a responsive region (around nucleotide −1400 to −800) that is rich in putative GATA sequences. Since it was shown several years ago that the TOR pathway could play a role in tolerance to sodium and lithium by regulating the expression of ENA1 (13), we considered that our results may reflect a potential functional link between Ptc1 and the TOR pathway. Here, we demonstrate that Ptc1 is required for normal TOR signaling, and this could be explained, at least in part, by the requirement of functional Ptc1 to ensure normal levels of the Tap42-interacting protein Tip41. To our knowledge, this is the first report supporting a role of type 2C phosphatases in TOR-mediated signaling.
MATERIALS AND METHODS
Escherichia coli and yeast growth conditions.
Yeast cells (strains are listed in Table 1) were grown at 28°C in YPD medium (10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter dextrose) or, when carrying plasmids, in synthetic complete medium (1) containing 2% glucose and lacking the appropriate selection requirements. For the analysis of the transcriptional response to low ammonium concentrations, cells were resuspended in synthetic medium containing 2% glucose and supplemented only with 10 mM ammonium sulfate, 40 mg/liter methionine, 20 mg/liter histidine, and 100 mg/liter leucine (designated low ammonium medium). The sensitivity of yeast cells to rapamycin (Calbiochem) or caffeine (Merck) was evaluated by growth on agar plates (drop tests) or in liquid cultures as previously described in (7, 48).
TABLE 1.
Yeast strains used in this worka
| Name | Relevant genotype | Source or reference |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ met15Δ ura3Δ | 72 |
| MAR143 | BY4741 ptc1::nat1 | 56 |
| MAR146 | BY4741 sit4::kanMX4 ptc1::nat1 | This work |
| MAR163 | BY4741 tor1::kanMX4 ptc1::nat1 | This work |
| MAR164 | BY4741 tip41::kanMX4 ptc1::nat1 | This work |
| MAR165 | BY4741 ure2::kanMX4 ptc1::nat1 | This work |
| MAR166 | BY4741 gln3::kanMX4 ptc1::nat1 | This work |
| MAR167 | BY4741 gat1::kanMX4 ptc1::nat1 | This work |
| TB123 | JK9-3da GLN3-myc13-kanMX | 3 |
| AGS39 | TB123 ptc1::nat1 | This work |
| DBY746 | MATa ura3-52 leu2-3112 his3-Δ1 trp1-Δ239 | D. Bostein |
| AGS66 | DBY746 PTC1 URA3-STRE(7x)-lacZ | This work |
| AGS67 | DBY746 ptc1 URA3-STRE(7x)-lacZ | This work |
Single kanMX deletion mutants in the BY4741 background were generated in the context of the Saccharomyces Genome Deletion Project (72) and are not listed here.
E. coli DH5α cells were used as the plasmid DNA host and were grown at 37°C in Luria-Bertani broth supplemented with 50 μg/ml ampicillin, when required. Bacterial and yeast cells were transformed using standard methods (16). Standard recombinant DNA techniques were performed as described elsewhere (58).
Gene disruptions and plasmid construction.
Single kanMX deletion mutants in the BY4741 background (MATa his3Δ1 leu2Δ met15Δ ura3Δ) were generated in the context of the Saccharomyces Genome Deletion Project (72) and were kindly provided by José L. Revuelta (Universidad de Salamanca, Spain). The ptc1::nat1 disruption cassette (56) was used to transform the appropriate strains. Positive clones were selected in the presence of 100 μg/ml nourseothricin (Werner BioAgents). Strain AGS66, which contains an integrated STRE(7x)-lacZ reporter system (41) at the URA3 locus, was made as follows. Wild-type strain DBY746 was transformed with plasmid pGM18/17 (a kind gift of F. Estruch, U. Valencia), which was previously linearized at the URA3 gene marker by digestion with NotI. Positive clones were selected in synthetic medium in the absence of uracil. An identical strategy was used to generate strain AGS67, except that in this case the transformed strain was the ptc1::nat1 derivative MAR122 (56).
The construction of plasmid YEp195-PTC1 was described earlier (45). To obtain YCp33-PTC1, an EcoRI/SalI 1.56-kbp fragment that was released from YEp195-PTC1, containing the PTC1 gene, was subcloned into the same restriction sites of plasmid YCplac33 (a URA3 marker). To generate the D58N allele of PTC1, the YEp195-PTC1 construct was used as a template for PCR to change the TTG codon, encoding an Asp residue at position 58, to a TTA codon, encoding an Asn residue. The 1.56-kbp amplification fragment was digested with EcoRI/SalI and cloned into these same sites of plasmid YEplac195 to yield YEp195-PTC1[D58N]. The entire amplification fragment was sequenced to ensure the absence of unexpected mutations. For the low-level expression of the mutated version of Ptc1, the EcoRI/SalI fragment was released from YEp195-PTC1[D58N] and subcloned into the same restriction sites of plasmid YCplac33 to yield YCp33-PTC1[D58N].
To analyze the expression of GAP1, GLN1, GDH1, and MEP1 promoters, lacZ translational fusions were constructed as follows. The regions comprising nucleotides −968 to +21 (pGAP1-LacZ) and −853 to +21 (pGLN1-LacZ) were amplified by PCR with added KpnI and XbaI sites and cloned into the same sites of plasmid YEp357 (46). Regions comprising −904 to +21 (pGDH1-LacZ) and −700 to +21 (pMEP1-LacZ) were amplified by PCR with artificial EcoRI and XbaI sites and cloned into these sites of plasmid YEp357. The oligonucleotides employed are listed in Table 2. Plasmids pEJ27 (YEp195-GST-TAP42) and pEJ120 (YEp181-TIP41-HA) were a generous gift from M. Hall (University of Basel, Switzerland). Plasmid pEJ23 (YEplac181-HA-NPR1) was a generous gift from E. Jacinto (University of Medicine and Dentistry, Piscataway, NJ). All three plasmids have been described previously (29). Plasmid YCp111-TIP41-HA was generated by cloning the entire 2.1-kbp EcoRI-HindIII insert present in pEJ120 (containing the TIP41 promoter region and open reading frame, the carboxyl-terminal 3× HA tag, and the CYC1 transcription terminator) into the same sites of YCplac111 (a LEU2 marker).
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Use |
|---|---|---|
| 5′_GAP1-RT | GGTTACTAGCCTTGTCTGGG | 5′ RT-PCR for GAP1 (+1352 from ATG) |
| 3′_GAP1-RT | CGGGGAATACAGCAACGTAG | 3′ RT-PCR for GAP1 (+1561 from ATG) |
| 5′_MEP1-RT | GCAGATCGCTTACATTGCCG | 5′ RT-PCR for MEP1 (+1116 from ATG) |
| 3′_MEP1-RT | CAGAGCGTTGTGAATCTTCG | 3′ RT-PCR for MEP1 (+1317 from ATG) |
| 5′_GLN1-RT | CAGGTGGTACGAAATACATC | 5′ RT-PCR for GLN1 (+782 from ATG) |
| 3′_GLN1-RT | CCTGTAACCAAGTATGGGTC | 3′ RT-PCR for GLN1 (+1018 from ATG) |
| 5′_CPS1-RT | CTTCTGTGGCTGAAGTCTTTG | 5′ RT-PCR for CPS1 (+1268 from ATG) |
| 3′_CPS1-RT | CCAGTAGTCACATAGAACTC | 3′ RT-PCR for CPS1 (+1516 from ATG) |
| TIP41_RT_up | GCACAGAGAGCTCACCTGAG | 5′ RT-PCR for TIP41 (+638 from ATG) |
| TIP41_RT_down | CTCAGCAGCAGTAACCTTTC | 3′ RT-PCR for TIP41 (+830 from ATG) |
| Actin_fw | TGCTGTCTTCCCATCTATCG | 5′ RT-PCR for ACT1 (+400 from ATG) |
| Actin_rv | ATTGAGCTTCATCACCAAC | 3′ RT-PCR for ACT1 (+495 from ATG) |
| 5′-promGAP1 | GCGGGGTACCAAGTTGTACTATTACGAAGC | 5′ oligonucleotide (−968) for cloning GAP1 promoter in YEp357 (URA3-lacZ), contains artificial KpnI sequence |
| 3′-promGAP1 | GCGCTCTAGAGTACGAAGAAGTATTACTCA | 3′ oligonucleotide (+21) for cloning GAP1 in YEp357 (URA3-lacZ), contains artificial XbaI sequence |
| 5′-promGLN1 | GCGGGGTACCTACACATGCGCTGCACGTGC | 5′ oligonucleotide (−853) for cloning GLN1 promoter in YEp357 (URA3-lacZ), contains artificial KpnI sequence |
| 3′-promGLN1 | GCGCTCTAGATTCGATGCTTGCTTCAGCCA | 3′ oligonucleotide (+21) for cloning GLN1 promoter in YEp357 (URA3-lacZ), contains artificial XbaI sequence |
| 5′-promMEP1 | GCGGAATTCAGATAAGAAAGACTGAGCC | 5′ oligonucleotide (−700) for cloning MEP1 promoter in YEp357 (URA3-lacZ), contains artificial EcoRI sequence |
| 3′-promMEP1 | GCGCTCTAGACCCTGTAGTTCGACTCTCCA | 3′ oligonucleotide (+21) for cloning MEP1 promoter in YEp357 (URA3-lacZ), contains artificial XbaI sequence |
| 5′-promGDH1 | GCGGAATTCGACAGTAACAAACTAGC | 5′ oligonucleotide (−904) for cloning GDH1 promoter in YEp357 (URA3-lacZ), contains artificial EcoRI sequence |
| 3′-promGDH1 | GCGCTCTAGATTGAAATTCTGGCTCTGACA | 3′ oligonucleotide (+21) for cloning GDH1 promoter in YEp357 (URA3-lacZ), contains artificial XbaI sequence |
β-Galactosidase assays.
To evaluate the promoter activity of diverse NCR-sensitive genes in response to rapamycin, yeast cells were grown to saturation in the appropriate dropout medium and then inoculated into 5 ml of YPD and incubated for 4 h to reach an A660 of 0.8 to 1. Aliquots of 1 ml were centrifuged and resuspended in the same volume of YPD (noninduced cells) or YPD containing 200 ng/ml rapamycin. Growth was resumed for 60 (in the case of GAP1, GLN1, and GDH1 promoter activity assays) or 90 min (when MEP1 promoter activity was assessed). Cells were collected and processed for β-galactosidase assay as described previously (51). The same protocol was used to evaluate the stress response element (STRE) response in strains AGS66 and AGS67 after 60 min of incubation with rapamycin. The activity of the NCR-sensitive promoters in cells growing under ammonium limitation was assessed by inoculating cells (A660 of 0.005 to 0.01) in 5 ml of low ammonium medium. Cultures were grown overnight until an A660 of 0.8 to 1, cells were collected, and β-galactosidase activity was measured as described above.
RNA purification, cDNA synthesis, and DNA microarray experiments.
For RNA purification, 30 ml of yeast culture was grown at 28°C in YPD medium until an A660 of 0.6 to 0.8 and, when required, was treated with 200 ng/ml rapamycin or drug vehicle alone (90% ethanol and 10% Tween P20) for 1 h. Cells were harvested by centrifugation and washed with cold water. Dried pellets were kept at −80°C until RNA purification. Total RNA was extracted using the RiboPure-Yeast kit (Ambion) by following the manufacturer's instructions. RNA quality was assessed by electrophoresis in a denaturing 0.8% agarose gel and quantified by measuring the A260 in a BioPhotometer (Eppendorf).
Transcriptional analyses using DNA microarrays developed in our laboratory (2) were performed exactly as described previously (23). For each experimental condition (the presence versus the absence of rapamycin in both wild-type and ptc1 mutant cells), a dye swapping was performed. The scanner ScanArray 4000 (Packard Instrument Co.) was used to obtain the Cy3 and Cy5 images with a resolution of 10 μm. The fluorescent intensity of the spots was measured and processed using GenePix Pro 6.0 software (Molecular Devices). Spots with either a diameter smaller than 120 μm or fluorescence intensity for Cy3 and Cy5 of less than 150 U were not considered for further analysis. A given gene was considered to be induced when the plus/minus rapamycin signal ratio was equal or higher than 2.0-fold, whereas it was considered to be repressed when this average was equal or less than 0.50-fold. The GEPAS server (version v3.1) was used to preprocess the data (26).
RT-PCR assays.
For reverse transcription-PCR (RT-PCR), yeast cells were grown in YPD to an A660 of 0.5 to 0.8. Cultures were collected at 4°C, and total RNA was prepared as described above. RT-PCRs were performed using the Ready-To-Go RT-PCR bead kit (GE Healthcare) and 100 ng of total RNA (except for TIP41 and MEP1 amplifications, for which 200 ng was used). Specific pairs of oligonucleotides were used (Table 2) to determine the levels of TIP41 (26 amplification cycles); MEP1 (30 amplification cycles); and GAP1, GLN1, CPS1, and ACT1 (25 amplification cycles).
Microscopy techniques.
The indirect immunofluorescence detection of tagged Gln3 was accomplished as follows. TB123 and AGS39 cells were grown in YPD medium until an A660 of 0.8 to 1 was reached. Cultures then were treated for different times (10, 20, 30, and 45 min) with 200 ng/ml rapamycin or drug vehicle and fixed with 3.7% formaldehyde for 60 min. Cells were prepared for immunofluorescence as described previously (49), incubated overnight at 4°C with anti-c-myc antibody (9E10 monoclonal antibody; BabCO; a generous gift of H. Martín, University Complutense of Madrid, Spain) at a final dilution of 1:500 and subsequently with 1:100 diluted Alexa Fluor 488 goat anti-mouse immunoglobulin G antibody (Invitrogen). For Msn2 subcellular localization experiments, wild-type BY4741 or MAR143 (ptc1) was transformed with plasmid pMSN2-GFP (a generous gift of F. Estruch, University of Valencia, Spain), a YCplac111-based vector that contains a C-terminal Msn2-green fluorescent protein fusion (24). Cells were grown in YPD containing 4% glucose as the carbon source until an A660 of 0.8 to 1.0 was reached. After this, 500 μl of the culture was treated with 200 ng/ml of rapamycin or drug vehicle for 15 min and fixed for 5 min by adding 50 μl of 37% formaldehyde. Cells were harvested, washed three times with phosphate-buffered saline (PBS), and concentrated 10-fold before visualization. In all cases the cells were visualized with a fluorescein filter using a Nikon Eclipse E800 fluorescence microscope (magnification, ×1,000). Digital images were captured with an ORCA-ER 4742-80 camera (Hamamatsu) using the Wasabi software.
Preparation of cell extracts and immunoblotting.
Yeast strains were grown on YPD to an A660 of 0.8 to 1 at 28°C. For Tip41 detection, whole-cell lysates (10 ml of culture) were prepared by resuspending the cells in 200 μl of extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol, 10% glycerol, 2 mM phenylmethylsulfonyl fluoride, and complete inhibitor mixture; Roche Applied Science). For Npr1 detection, lysates were made by resuspending cells in PBS buffer (pH 7.4) containing 10 mM NaF, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1% Nonidet NP-40, and the protease inhibitor mixture mentioned above (28). One volume of acid-washed glass beads (Sigma) was added, and cells were broken at 4°C by vigorous shaking (five times for 25 s each, with intervals of 1 min on ice) in a Fast Prep cell breaker (setting 5.5; Bio 101 Inc., Vista, CA). After sedimentation at 500 × g for 10 min at 4°C, the cleared lysate was recovered and the protein concentration quantified by the Bradford assay. Total proteins (30 to 90 μg) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% polyacrylamide gels and transferred to nitrocellulose membranes (Hybond C-Extra; Amersham Biosciences).
Membranes were incubated for 2 h with anti-HA antibody (Roche Applied Science) at a 1:500 dilution, anti-glutathione S-transferase (anti-GST) (Z-5; Santa Cruz Biotechnology Inc.) at a 1:2,000 dilution, or anti-actin (I-19; Santa Cruz Biotechnology Inc.) at a 1:2,000 dilution, followed by the secondary horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G antibody (Amersham Biosciences) at a 1:20,000 dilution. The immunocomplexes were visualized using an ECL Western blotting detection kit. Chemiluminescence was detected using LAS-3000 equipment (Fuji) and quantified using Multi Gauge software, version 3.0.
GST pull-down assays.
Five milligrams of the crude protein extract, prepared as described above, was mixed with 50 μl of a slurry of glutathione-Sepharose (Amersham Biosciences) affinity matrix for 2 h at 4°C with gentle shaking. Samples were centrifuged and the supernatant was removed. Beads were resuspended in 200 μl of extraction buffer, transferred to MultiScreen filter plates (Millipore), and extensively washed with the same buffer (without protease inhibitors). Proteins retained by the affinity system were fractionated by SDS-PAGE, followed by immunoblotting using anti-HA antibody. Membranes were stripped and then incubated with anti-GST antibody.
Cycloheximide chase assays.
Yeast strains were grown on synthetic medium lacking leucine until saturation and then were inoculated (A660 of 0.2 to 0.3) into YPD and further grown for 4 h. Cultures then received 200 ng/ml of rapamycin or vehicle, and incubation was resumed for 10 min. Cycloheximide or vehicle (ethanol) (25 μg/ml) was added, and aliquots were taken at the indicated times. Cells were processed for Tip41 and actin immunodetection as described above.
Two-dimensional gel electrophoresis.
The phosphorylation state of Tip41 was monitored by two-dimensional electrophoresis according to previously published methods (28, 29), with some modifications. Cell cultures (100 ml) were grown in YPD until an A660 of 0.6 to 0.8 and treated for 30 min with 100 ng/ml of rapamycin or drug vehicle. Cells were harvested by centrifugation and resuspended in 1 ml of TNE lysis buffer (0.1 M Tris-HCl [pH 8.0], 10 mM NaF, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1% Nonidet NP-40, 1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, complete protease inhibitor mixture [Roche Applied Science], and 0.1 μg/ml RNase A) and mechanically disrupted as described above. Homogenates were centrifuged for 10 min at 500 × g, and supernatants were recovered. HA-Tip41 was immunoprecipitated from 8 (wild-type cells) or 12 mg (ptc1 cells) of crude protein extracts after incubation with 0.5 μg of anti-HA antibody and 40 μl of protein G Sepharose beads (GE Healthcare) for 3 h at 4°C. Immunoprecipitates were washed three times with 800 μl TSNE (TNE plus 0.15 M NaCl) and three times with 1 ml TNE. Samples were resuspended in 150 μl of elution buffer (8 M urea, 4 M thiourea, 0.1 M Tris-HCl [pH 8.0], 1% Nonidet NP-40, 1 mM EDTA, and 4% β-mercaptoethanol) and then incubated at 37°C for 30 min with mild agitation. Samples then were centrifuged at 400 × g for 1 min, the supernatant was recovered, and 75 μl of a solution containing 6% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 3% dithiothreitol, 3% immobilized pH gradient (IPG) buffer (pH 4 to 7) (GE Healthcare), and 0.06% bromophenol blue was added. Immobiline DryStrip IPG gel strips (GE Healthcare) (24 cm; pH 4 to 7) were rehydrated in a solution containing 8 M urea, 2% CHAPS, 0.6% DeStreak rehydration solution (GE Healthcare), and 0.5% IPG buffer and placed in the manifold ceramic tray, and 150 μl of the sample was loaded in the sample cups. First-dimension isoelectric focusing was performed on an Ettan IPGphor II unit (GE Healthcare) using the following protocol: 500 V for 1 h, 1,000 V for 1 h, and 8,000 V for 48 h (50 mA per strip). Usually, a total of 225,000 to 250,000 V/h was reached. Strips were equilibrated for 15 min in 9 ml of 50 mM Tris-HCl (pH 6.8), 6 M urea, 30 glycerol, 2% SDS, 1% dithiothreitol, and 0.002% bromophenol blue) and then for an additional 15 min with the same solution lacking dithiothreitol and supplemented with 2.5% iodoacetamide. The second dimension was performed on an SDS-10% polyacrylamide resolving gel (25 mM Tris-HCl [pH 6.8], 192 mM glycine, and 0.1% SDS as a running buffer). Proteins were detected by immunoblotting using Immobilon-P membranes (Millipore) and anti-HA antibody as described above. The position and intensity of the spots was analyzed using Melanie 7.0 software (GeneBio SA). A comparison of the relative intensity of the different spots among several experiments was done using the %vol parameter, which is a normalized value that remains independent of variations due to protein loading and/or staining, particularly for gels with a similar spot pattern.
Microarray data accession number.
Microarray data described in the manuscript have been deposited in the Gene Expression Omnibus database under accession number GSE11530.
RESULTS
Evidence for involvement of Ptc1 in the TOR pathway.
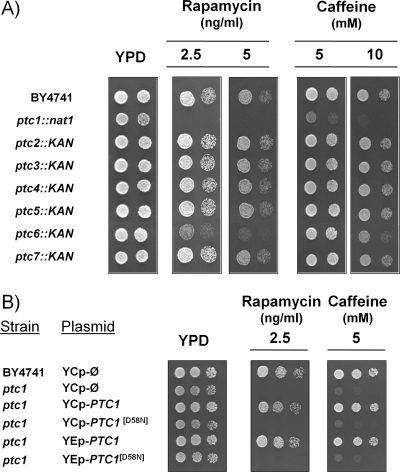
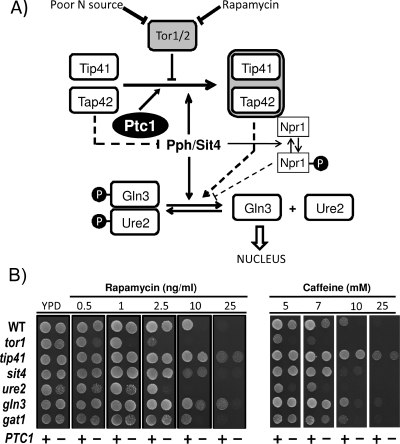
The phenotype of the sensitivity of ptc mutants to rapamycin is rather specific for the Ptc1 isoform, as deduced from growth analysis on agar plates, since it is not observed in cells lacking PTC2 to PTC5 or in the ptc7 strain (Fig. 1A). We confirm the recent observation that the lack of PTC6 confers some degree of sensitivity to rapamycin (55), although it is far less pronounced than that found in ptc1 cells. Interestingly, the same sensitivity pattern is observed (Fig. 1A) when cells are treated with different concentrations of caffeine, a likely inhibitor of the TOR pathway (34). The overexpression of PTC1, but not that of PTC2 or PTC3, conferred tolerance to toxic concentrations (20 ng/ml) of rapamycin (not shown). In addition, the growth of ptc1 mutants is somewhat impaired on media containing low concentrations of ammonium or less-preferred N sources, such as proline (data not shown). These results reinforced the notion that Ptc1 function is related to the TOR pathway. The phenotypes of ptc1 mutants in the presence of rapamycin or caffeine were not altered by the lack of the Hog1 kinase gene or the adaptor protein Nbp2 (data not shown), indicating that these effects were not mediated by the HOG pathway.
FIG. 1.
Sensitivity to rapamycin and caffeine of the diverse yeast type 2C protein phosphatase mutants. (A) Wild-type strain BY4741 and the different ptc mutants were spotted onto YPD plates containing the indicated concentrations of rapamycin or caffeine. Growth was monitored after 60 h of incubation at 28°C. (B) The BY4741 strain and its ptc1 derivative were transformed with an empty centromeric YCplac33 vector (YCp-Ø) or with the wild type or a catalytically inactive form of Ptc1 (PTC1[D58N]) cloned in either centromeric (YCp) or high-copy-number (YEp) vectors. Cells were spotted as described above and incubated for 48 h.
The regulation of the TOR pathway involves diverse protein phosphatase activities, although so far all of them belong to the type 2A family. Type 2C proteins are rather different from the PP2A or Sit4 phosphatases in primary sequence and regulatory properties. Therefore, we wanted to test whether or not the loss of Ptc1 catalytic activity could justify the phenotypes observed in the ptc1 mutant. To this end, we prepared a version of Ptc1 in which Asp58, a residue that previously has been shown to be catalytically relevant (71), was changed to Asn. As observed in Fig. 1B, the expression of the mutated Ptc1 version was unable, even at a high copy number, to rescue the rapamycin- and caffeine-sensitive phenotype of the ptc1 strain. Therefore, Ptc1 catalytic activity appears to be necessary for normal TOR function.
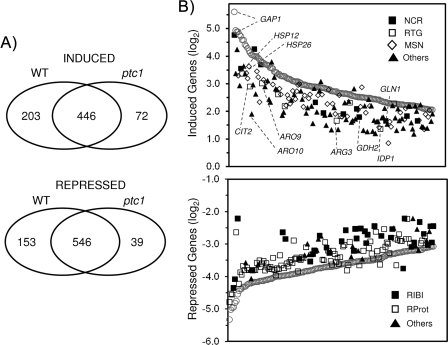
It has been shown that the inhibition of the TOR pathway (i.e., by the short-term treatment of yeast cells with rapamycin) has profound effects on the global transcriptional profile (8, 21, 25, 63). We hypothesized that if Ptc1 was required for normal TOR function, Ptc1-deficient strains may exhibit an altered transcriptional profile when challenged with rapamycin. Therefore, RNA was prepared from wild-type and ptc1 cells that had been incubated for 1 h with 200 ng/ml of rapamycin or vehicle, and DNA microarray analysis was performed. Under our working conditions, rapamycin increased by at least twofold the expression of 667 genes in wild-type cells (13.8% of genes with a measurable expression level), whereas the treatment decreased the mRNA level of 721 genes (14.9%). Gene ontology analysis shows that, as previously documented, many induced genes fall into the NCR, Msn2/Msn4-regulated stress response, or retrograde response category, whereas genes encoding cytoplasmic (but not mitochondrial) ribosomal proteins and the so-called Ribi regulon, which includes nonribosomal proteins that are involved in ribosome synthesis and maturation (31), were largely repressed. A comparison of the response to rapamycin in wild-type and ptc1 cells is shown in Fig. 2A. It can be observed that the deletion of PTC1 decreases the number of genes induced or repressed by rapamycin treatment. Remarkably, a lack of Ptc1 seems to lead to a general attenuation of changes triggered by rapamycin (see Tables S1 and S2 in the supplemental material). For instance, 91 genes were induced at least fourfold in both wild-type and ptc1 cells, but the increase in their induction averages only 6.5-fold for the ptc1 mutant and 9.2-fold for the wild-type strain. Similarly, the decrease in expression for the 251 genes repressed at least fourfold in both strains is much more pronounced in wild-type cells (9.9-fold) than in the mutant strain (7.6-fold). The attenuation of the transcriptional response to rapamycin caused by the lack of Ptc1 can clearly be observed by plotting the 150 genes showing the highest degree of induction (Fig. 2B, upper) or repression (Fig. 2B, lower) in wild-type cells and comparing their expression level to that found in ptc1 cells. It is evident that many of the highly induced genes, which include components of the NCR and the mitochondrial retrograde pathways as well as Msn2/Msn4-responsive downstream targets, decrease their expression in the absence of the phosphatase. Likewise, genes whose expression is dramatically decreased by rapamycin, including numerous ribosomal proteins as well as the Ribi regulon, clearly are less repressed in ptc1 cells. These results indicate that the lack of Ptc1 has a remarkable and general effect on the transcriptional program elicited by the inhibition of the TOR pathway and suggest that this specific type 2C phosphatase is necessary for normal signaling through the pathway.
FIG. 2.
Global transcriptional response analysis to rapamycin in wild-type (WT) and ptc1 cells. (A) Venn diagram showing the number of genes whose expression was considered to be induced (top) or repressed (bottom) by rapamycin in wild-type ptc1 cells for a set of 4,677 genes, with valid data for both strains. (B) Plots of the log2 values for the changes in the level of expression induced by rapamycin in both wild-type (open circles) and ptc1 strains for the 150 most upregulated (top) and 150 most downregulated (bottom) genes in the wild-type strain. The expression values for the ptc1 strain are shown according to the TOR-dependent regulon to which each gene belongs. For the induced genes, the following categories were used: the NCR family (closed squares), as defined previously (21); the RTG group (open squares) comprises the genes described as documented targets for the Rtg1 or Rtg3 transcription factors in YEASTRACT (44), as well as those identified elsewhere (19); and the documented targets of Msn2 or Msn4 described in YEASTRACT, plus those identified elsewhere (9), are represented as open diamonds. Genes not included in these categories are designated others (closed triangles). The localization of representative genes for each family in the plot is shown. The genes downregulated by rapamycin in the wild-type strain are classified into one of three possible families: Ribi regulon (closed squares), which include the genes described previously (31), ribosomal proteins (open squares), and others (closed triangles).
Ptc1 is required for normal Gln3- and Msn2-mediated transcriptional responses and nuclear localization.
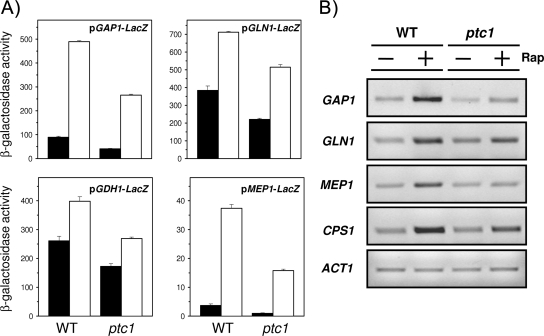
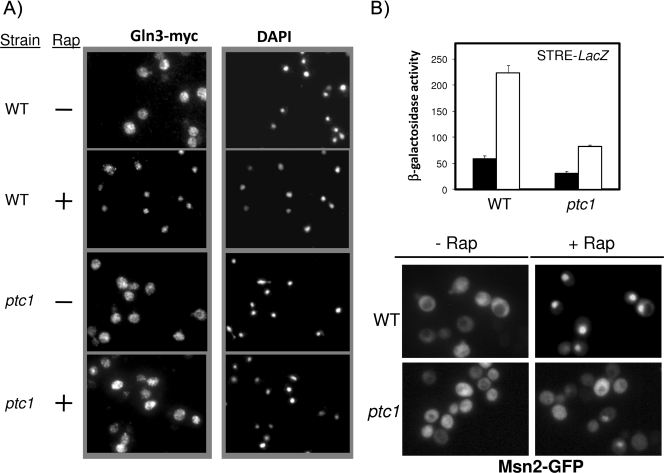
To gain further insight into the role of Ptc1 in the transcriptional response under TOR regulation, we selected for further characterization two well-known transcription factors: Gln3, which is largely responsible for the expression of NCR genes that are required for adaptation to nonpreferred nitrogen sources, and Msn2, which mediates TOR-regulated stress responses. As shown in Fig. 3A, lacZ translational fusions of the promoters of four NCR genes (GAP1, MEP1, GLN1, and GDH1) were introduced into wild-type and ptc1 strains. When cells were challenged with rapamycin, it became evident that the response to the drug was attenuated in Ptc1-deficient cells. The expression of three of these genes (plus CPS1, another NCR-regulated gene) also was investigated by RT-PCR (Fig. 3B), with similar results. Interestingly, when a Myc13-tagged version of Gln3 was expressed in wild-type and ptc1 cells and cultures were incubated with rapamycin, it was observed that the ability of Gln3 to enter into the nucleus after drug treatment was substantially impaired (Fig. 4A). Thus, after 10 min of incubation with the drug, most Gln3 was nuclear in the wild-type strain, whereas its location was largely cytosolic in the ptc1 strain. The time course monitoring of Gln3 distribution in ptc1 cells showed that Gln3 entered the nucleus only 20 to 30 min after the addition of rapamycin, although cytosolic Gln3 still was perceived all during the experiment (data not shown). This was consistent with the observed decrease in NCR-sensitive gene expression and suggests that the lack of Ptc1 impairs TOR-mediated signaling on Gln3. The effect of the ptc1 deletion on Msn2-mediated signaling was monitored by integrating an STRE-containing reporter into wild-type strain DBY746 and its isogenic ptc1 derivative. As shown in Fig. 4B, the STRE-driven response to rapamycin also is markedly decreased in ptc1 cells. As observed for Gln3, the rapamycin-triggered entry of Msn2 into the nucleus is markedly blocked in cells lacking the phosphatase. All of these observations confirm that signaling through the TOR pathway is impaired in Ptc1-deficient cells.
FIG. 3.
Decreased response of diverse NCR-sensitive genes to rapamycin treatment. (A) The indicated constructs were introduced into wild-type BY4741 (WT) and its ptc1 derivative, and cells were treated with 200 ng/ml rapamycin (Rap) (open bars) for 60 (for GAP1, GLN1, and GDH1 promoters) or 90 min (for MEP1). Control cells (closed bars) received only the solvent. β-Galactosidase activity was measured as indicated in the text. Data are means ± standard errors of the means from six independent clones. (B) RT-PCR experiments were performed using oligonucleotides specific for the indicated genes (see Materials and Methods). Amplification fragments were run on 2% agarose gels. ACT1 is included for comparison.
FIG. 4.
ptc1 mutation impairs rapamycin-induced Gln3 and Msn2 entry into the nucleus. (A) TB123 (PTC1 GLN3-myc13-kanMX) and AGS39 (ptc1 GLN3-myc13-kanMX) cells were exposed to 200 ng/ml rapamycin (Rap) or to solvent for 10 min and processed for indirect immunofluorescence using anti-Myc antibodies. Samples also were stained with DAPI to reveal the position of the nuclei (magnification, ×1,000). (B) The upper panel shows wild-type (WT) strain AGS66 and its ptc1 isogenic derivative AGS67, which contain integrated STRE-LacZ reporters. They were treated for 1 h with 200 ng/ml rapamycin (open bars) or solvent (closed bars), and β-galactosidase activity was measured. Data are means ± standard errors of the means from six independent clones. The lower panel shows strains BY4741 (WT) and MAR143 (ptc1), which were transformed with plasmid pMsn2-GFP (24) and incubated with 200 ng/ml rapamycin for 15 min. The localization of Msn2 was followed by fluorescence microscopy (magnification, ×1,000).
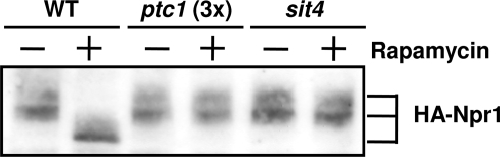
Lack of Ptc1 results in hyperphosphorylated Npr1.
The Npr1 kinase is required, under specific conditions, to maintain Gln3 in a cytosolic location, and the function of this kinase can be regulated by phosphorylation (15, 29, 61). Therefore, we considered it necessary to investigate the phosphorylation state of Npr1 in ptc1 cells. As shown in Fig. 5, in cells exposed to rapamycin for 30 min, Npr1 shows higher mobility, which corresponds to dephosphorylated forms of the protein. As reported previously (29), this shift is not observed in sit4 cells. Figure 5 also shows that the dephosphorylation of Npr1 is blocked in ptc1 cells. Remarkably, the lack of Ptc1 resulted in lower-than-normal amounts of Npr1 (note that, in ptc1 lanes, threefold more protein was loaded). In contrast, we did not observe the increase in Npr1 levels previously reported for sit4 cells (29). Therefore, our results indicate that Ptc1 is necessary for the rapamycin-induced dephosphorylation of Npr1. This observation fits with the impaired entry in the nucleus of Gln3 in Ptc1-deficient cells.
FIG. 5.
Lack of Ptc1 affects Npr1 levels and phosphorylation state. Wild-type BY4741 (WT) and its ptc1 (MAR143) or sit4 derivative were transformed with plasmid pEJ23, which carries an N-terminally HA-tagged version of Npr1. Cells were incubated for 30 min with 100 ng/ml rapamycin or vehicle, and extracts were prepared. Thirty micrograms of protein [90 μg in the case of ptc1 cells, denoted ptc1(3x)] was electrophoresed, transferred, and incubated with anti-HA monoclonal antibodies. Faster-migrating bands correspond to the dephosphorylated forms of Npr1.
Genetic interactions between the ptc1 mutation and relevant mutations in the TOR pathway.
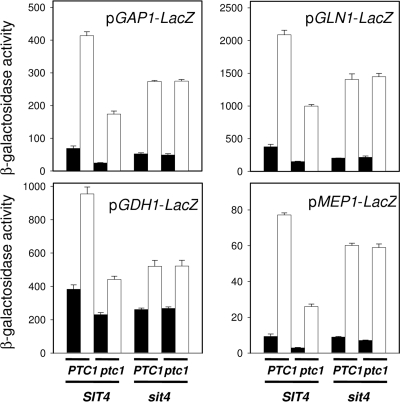
A genetic approach was taken to get insight into the role of Ptc1 in the TOR pathway. To this end, the PTC1 gene was deleted in strains carrying mutations in different components of the pathway (Fig. 6A), and the tolerance to rapamycin and caffeine was tested. As observed in Fig. 6B, the deletion of PTC1 in a tor1, ure2, gln3, gat1, or npr1 (not shown) mutant background still resulted in increased sensitivity to both drugs when tested on YPD agar plates. However, the hypertolerant phenotype of the tip41 mutant was not affected at all by a lack of Ptc1. Similarly, the sit4 ptc1 double mutant exhibited a sit4 phenotype, which was slightly more sensitive than the wild-type strain but clearly more tolerant than the single ptc1 mutant. These phenotypes were fully confirmed in liquid medium (not shown). Conversely, the transformation of the above-mentioned single mutants with plasmid YEp195-PTC1 resulted in increased rapamycin tolerance in all cases, with the only exceptions being tip41 and sit4 (data not shown). This genetic interaction also was observed by examining the response to the nitrogen starvation of the four NCR promoters described above in the ptc1, sit4, and ptc1 sit4 strains. As shown in Fig. 7, the deletion of PTC1 substantially impairs the response from GAP1, GLN1, GDH1, and MEP1 promoters (similarly to what was observed for rapamycin) (Fig. 3A). The deletion of SIT4 also decreased the promoter responses, although the effect was less prominent than in the case of the PTC1 mutation. Remarkably, the activity measured for all four promoters in the ptc1 sit4 double mutant was virtually identical to that observed in the sit4 strain. Therefore, our experiments indicate that the sit4 mutation (and also probably the tip41 mutation) is epistatic to the ptc1 mutation. This suggests that Ptc1 acts on the TOR pathway by regulating Tip41 or Sit4 function (or both).
FIG. 6.
Epistatic analysis of ptc1 and mutations affecting the TOR pathway. (A) A simplified model of signaling through the TOR pathway (focused on the regulation of NCR genes) based on previous models (15, 29, 33). (B) Rapamycin and caffeine sensitivity of diverse mutants in genes relevant in the TOR pathway in the presence (+) or the absence of PTC1 (−). Cultures were spotted on YPD plates containing the indicated concentrations of the drugs, and growth was monitored after 2 days. WT, wild type.
FIG. 7.
Effect of the ptc1 and sit4 mutations in the transcriptional response to the ammonium starvation of several NCR-sensitive genes. Strains BY4741 (PTC1 SIT4) and its ptc1, sit4, and ptc1 sit4 derivatives were transformed with the indicated NCR-sensitive reporters. Cells were grown overnight on either standard synthetic (closed bars) or low ammonium medium (open bars), and β-galactosidase activity was measured. Data are means ± standard errors of the means from three independent clones.
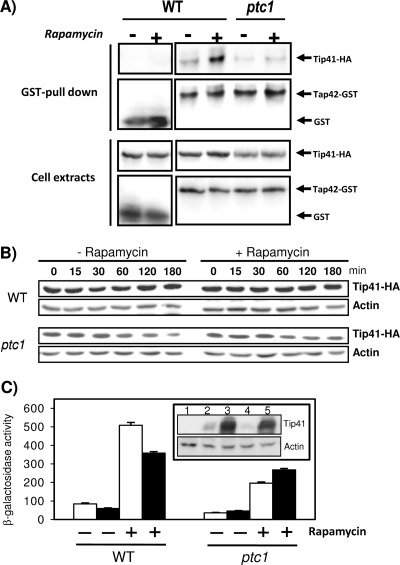
ptc1 cells display altered Tap42-Tip41 interaction, probably due to lower-than-normal Tip41 protein levels.
The interaction between Tip41 and Tap42 is an important event that regulates signaling through the TOR pathway (29). Therefore, we considered it necessary to explore whether the lack of Ptc1 affects such an interaction. To this end, wild-type and ptc1 cells were treated with rapamycin, and the drug-induced interaction between Tip41 and Tap42 was investigated by means of GST-Tap42 pull-down experiments followed by the detection of Tip41 by immunoblotting (Fig. 8A, upper). As can be observed, the ptc1 mutation greatly impaired the formation of the Tip41-Tap42 complex. This observation was in keeping with the phenotypes observed upon the deletion of the phosphatase gene. Notably, the examination of Tip41 levels in whole-cell extracts revealed that they are clearly lower in phosphatase-deficient cells than in wild-type cells, irrespective of the presence of the drug in the medium. Therefore, the failure to form a complex with Tap42, as was observed in ptc1 cells, might be caused, at least in part, by smaller-than-normal amounts of Tip41. To identify the molecular basis for this effect of the ptc1 mutation, we examined by RT-PCR the mRNA levels of TIP41 in wild-type and ptc1 cells. However, no significant differences were observed (data not shown). We considered the possibility that Ptc1 plays a role in regulating Tip41 stability. To this end, wild-type and ptc1 cells were treated with cycloheximide to arrest translation, and the amount of Tip41 was monitored at different times by immunoblotting. Figure 8B shows that in wild-type cells, Tip41 levels remained rather stable upon the blockage of translation. In sharp contrast, the amount of Tip41 decreased quite rapidly in Ptc1-deficient cells, suggesting that the lack of the phosphatase significantly affects the stability of the Tip41 protein. A similar experiment was carried out by treating the cells with 200 ng/ml of rapamycin for 10 min prior to cycloheximide addition. As shown in Fig. 8B, treatment with rapamycin does not affect Tip41 stability in wild-type cells or enhance Tip41 degradation in ptc1 cells, even after relatively long periods of incubation (180 min).
FIG. 8.
Lack of Ptc1 decreases Tip41 stability and Tip41-Tap42 rapamycin-induced interaction. (A) BY4741 and its ptc1 derivative were transformed with plasmids pEJ27 (GST-Tap42) and pEJ120 (Tip41-HA). Cultures were grown overnight on synthetic medium lacking leucine and uracil and then inoculated into YPD (A660, 0.2 to 0.3) and grown for 4 h. Cells were treated with rapamycin (100 ng/ml) or drug vehicle for 30 min, and cell extracts were prepared and GST-Tap42 affinity purified as described in Materials and Methods. Purified material (upper) or cell extracts (lower; 40 μg of proteins) were subjected to SDS-PAGE and immunoblotted with anti-HA antibodies to reveal the presence of Tip41. Membranes were stripped and incubated with anti-GST antibody to detect Tap42. (B) A BY4741 strain lacking the TIP41 gene (PTC1) was deleted for the PTC1 gene (strain MAR164; ptc1), and both strains were transformed with plasmid YCp111-TIP41-HA. Cultures received 200 ng/ml of rapamycin or vehicle, and incubation was resumed for 10 min. Cycloheximide then was added to the cultures (25 μg/ml) to halt translation, and samples were taken at the indicated times and processed for immunoblotting using anti-HA antibodies. Membranes were stripped and incubated with anti-actin antibodies as a loading and transfer control. The experiment was repeated three times, with similar results. (C) Wild-type BY4741 (WT) and MAR143 (ptc1) were transformed with plasmid pEJ120 (expressing Tip41-HA in high copy number; filled bars) or the equivalent empty plasmid (YEplac181; open bars). These strains then were transformed with plasmid pGAP1-lacZ. Cells were grown and treated with rapamycin as illustrated in the legend to Fig. 3. β-Galactosidase activity generated from the GAP1 promoter was measured as described in the text. Data are means ± standard errors of the means from nine independent clones. The inset shows the levels of Tip41-HA in tip41 cells in the presence (lanes 2 and 3) or the absence (lanes 4 and 5) of the PTC1 gene when the gene is expressed from low-copy-number (lanes 2 and 4) or high-copy-number plasmids (lanes 3 and 5). Lane 1 corresponds to tip41 cells carrying an empty plasmid. Actin levels are included as loading controls.
The results presented above clearly show that Ptc1 is required for the normal expression of Tip41. We then wondered whether the altered level of Tip41 explains some of the phenotypes caused by the ptc1 mutation. As shown in Fig. 3, a lack of Ptc1 results in the impaired expression of several NCR genes. We hypothesized that if the amount of Tip41 could be increased, the ptc1 phenotype should be alleviated. As observed in Fig. 8C, the high-copy-number expression of Tip41 in a ptc1 strain increases the rapamycin-induced expression driven from the GAP1 promoter, thus counteracting the effect of the ptc1 mutation, although the expression levels do not reach those of the wild-type strain. It is remarkable that the overexpression of Tip41 in wild-type cells does not increase but instead results in a moderate decrease of the GAP1 promoter activity. Similar results were obtained when expression from the MEP1 promoter was tested (data not shown). The expression levels of Tip41-HA in PTC1 and ptc1 cells when the protein is expressed from low-copy-number (centromeric) or high-copy-number (episomal) plasmids are shown in the inset of Fig. 8C. Interestingly, whereas the expression of TIP41 in high-copy-number plasmids is decreased by the mutation of PTC1 (Fig. 8A, lower, and C, inset, compare lanes 3 and 5), the amount of Tip41 in this case still is clearly higher than that of the protein expressed from a centromeric plasmid in PTC1 cells (which should closely mimic Tip41 wild-type levels). Since the expression of TIP41 from centromeric plasmids restores the wild-type phenotype of a tip41 mutant when Ptc1 is present (not shown), our result indicates that the restoration of Tip41 levels is not enough to fully eliminate ptc1 defects related to the signaling of the TOR pathway.
Lack of Ptc1 slightly alters Tip41 phosphorylation pattern.
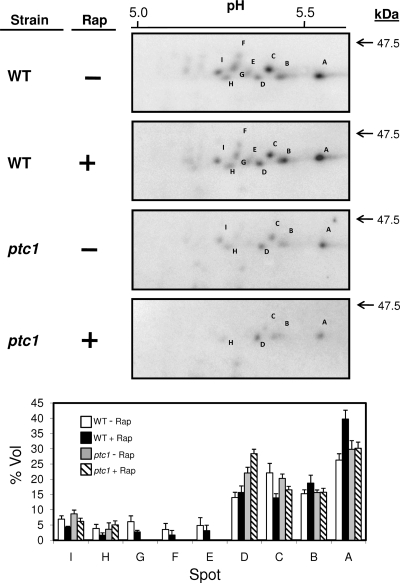
Tip41 is phosphorylated in vivo at multiple sites, and it has been reported that hyperphosphorylated Tip41 cannot properly bind Tap42. Since Ptc1 is a protein phosphatase, it was reasonable to evaluate the Tip41 phosphorylation state in ptc1 cells, both under basal conditions and in cells treated with rapamycin. HA-tagged Tip41 was immunoprecipitated, and samples were subjected to two-dimensional electrophoresis, essentially as reported previously (29). Under these conditions, Tip41 was detected in wild-type cells as nine different spots focused around pH 5.5 (Fig. 9, spots A to I). The higher number of spots observed here compared to those from a previous study (29) probably is due to the higher resolution of our methodology (i.e., larger strips and the use of a narrower pH range [4 to 7] for the first dimension). In fact, the number of species detected here approaches the number of in silico predicted phosphorylatable sites (score, >0.9) according to the NetPhos 2.0 server. As shown in Fig. 9 (lower), the major change observed after the treatment of wild-type cells with rapamycin for 30 min was an increase in spot A (focusing at higher pH and, therefore, corresponding to the most dephosphorylated form detected) and a decrease in spots G and C. These changes are compatible with the dephosphorylation of Tip41 induced by rapamycin treatment. Interestingly, when ptc1 cells are treated with the drug, the signals for the relatively highly phosphorylated spot D increased, whereas the relative intensity for spot A was unchanged. This suggests that the rapamycin-induced dephosphorylation of Tip41 is impaired in ptc1 cells.
FIG. 9.
Phosphorylation pattern of Tip41 in Ptc1-deficient cells. The upper panel shows wild-type BY4741 (WT) and ptc1 cells transformed with plasmid pEJ120, which were incubated for 30 min with 100 ng/ml rapamycin (Rap) or vehicle. Extracts were prepared for isoelectrofocusing as described in Materials and Methods. The first dimension was run using Immobiline DryStrip (pH 4 to 7) strips, and the second dimension was performed in SDS-10% polyacrylamide gels. Gels were transferred to membranes, and Tip41 was detected using anti-HA antibodies. Only the relevant region of the immunoblot is shown, and the different Tip41 forms are labeled (A to I). pHs are indicated on the top, and the molecular mass standard is on the right. The lower panel shows the vol% parameter (i.e., relative volume of a spot) for each spot, calculated using Melanie 7.0 software. The means ± standard errors from the means from three independent experiments are represented. The intensity of spots G, F, and E in the ptc1 mutant was too low to be integrated.
DISCUSSION
The results presented in this work clearly demonstrate a functional link between the type 2C phosphatase Ptc1 and the TOR pathway. We show here that cells lacking Ptc1 present a general attenuation of signaling through this pathway. This seems to be a specific feature of the Ptc1 isoform, since the mutation of other type 2C phosphatases does not cause caffeine or rapamycin sensitivity (perhaps with the exception of Ptc6), and the overexpression of other isoforms (such as PTC2 or PTC3) does not confer tolerance to rapamycin. We have observed that a ptc1 ptc6 double mutant is more sensitive to rapamycin and caffeine than the single mutants (data not shown). This suggests independent roles of both phosphatases in the TOR pathway. The role of Ptc6 in the TOR pathway currently is being investigated in our laboratory. These findings reinforce the previous notion that, from a functional point of view, Ptc1 is far different from the other members of the type 2C phosphatase family (23). In any case, we show that ptc1 defects are caused by the lack of its phosphatase activity. Therefore, it can be established that the functional alterations attributable to this mutant are caused by an imbalance in the phosphorylation state of one or more Ptc1 targets.
The requirement of Ptc1 for correct TOR signaling is evidenced by the wide effect of the ptc1 mutation on the transcriptional response to rapamycin. We show that the lack of Ptc1 causes a quite general attenuation of the transcriptional response that affects virtually all known TOR targets (8, 21, 25, 63). In some cases, such as the NCR genes, we provide a likely explanation, since the mutation of PTC1 clearly affects the nuclear entry of the Gln3 transcription factor (Fig. 4A). The inability to dephosphorylate Npr1 in response to rapamycin, as observed in ptc1 cells (Fig. 5), may contribute to this effect. In other cases, such as the effect on ribosomal proteins and the Ribi regulon, the cause might be less direct. It has been proposed that the induction by rapamycin of this set of genes involves the entry of Tor1 into the nucleus (36). However, we did not observe epistasis between the tor1 and the ptc1 mutations (Fig. 6B). It must be noted that the expression of ribosomal protein genes and the Ribi regulon also has been placed under the control of the protein kinase A (PKA) pathway (10, 42, 60, 76). Although there is no previous link between Ptc1 and PKA activity regulation, the observed effects could be explained if a lack of Ptc1 resulted in the activation of PKA. In this regard, we also observed the expression of diverse STRE-regulated genes being attenuated by the ptc1 mutation. The expression of these genes is activated upon the entry of the Msn2/Msn4 transcription factors into the nucleus. Our data indicate that a lack of Ptc1 substantially impairs the rapamycin-induced nuclear entry of Msn2 as well as STRE-driven expression (Fig. 4B). Interestingly, there is some controversy about whether or not Tap42 and Sit4 are required to direct Msn2 to the nucleus upon rapamycin stimulation (3, 21, 59, 60). Since there is long-standing evidence for regulation by PKA of stress-promoted Msn2 nuclear localization (3, 24, 64), a hypothetical regulation of the PKA pathway by Ptc1 would explain our results. This notion is reinforced by our observation that the STRE-driven response to heat shock also is decreased in ptc1 cells (data not shown).
The very wide range of effects of the ptc1 mutation on the transcriptional changes induced by rapamycin suggest that Ptc1 plays a role upstream in the TOR pathway. Indeed, the combination of the ptc1 deletion with the mutation of several genes involved in the pathway (Fig. 6B) indicated that Ptc1 could be placed at the Tip41/Sit4 level. Tip41 is known to be a regulatory component of the pathway by binding to Tap42, and it has been shown that rapamycin treatment enhances the interaction between both proteins, which appears necessary for the response to the drug (29). We show that in cells lacking Ptc1, treatment with rapamycin fails to promote the interaction between Tip41 and Tap42. Surprisingly, we also observed that whereas the level of Tap42 was normal, the amount of cellular Tip41 clearly was lower in ptc1 mutants than in wild-type cells. This effect was independent of the inhibition of the TOR pathway, as it is not observed in wild-type cells treated with rapamycin. Therefore, our data support the notion that a lack of Ptc1 affects the stability of the Tip41 protein, and this could explain the failure to observe an enhanced Tip41-Tap42 interaction in rapamycin-treated ptc1 cells. Interestingly, very recent work has shown that Wip1, the closest homolog of Ptc1 in humans (22), catalyzes the dephosphorylation of Mdm2, an E3 ubiquitin ligase involved in p53 proteosomal degradation. Dephosphorylated Mdm2 has increased stability and enhanced p53 binding, thereby facilitating p53 ubiquitination and degradation (37). The possibility that Tip41 is ubiquitinated and that Ptc1 affects this condition has been investigated in our laboratory, but no evidence supporting such a hypothesis has been found. In any case, it is apparent that the effect of Ptc1 on Tip41 stability cannot be attributed to a regulatory effect of Ptc1 on Sit4, since cells lacking Sit4 do not show significant changes in the amount of Tip41 (29 and our own data).
A key question is whether or not the low levels of Tip41 observed in Ptc1-deficient cells are sufficient to explain the interference of the ptc1 mutation with the TOR pathway. Our results indicate that, in ptc1 cells, an artificial increase of Tip41, even well above wild-type levels, can increase the expression from NCR-related gene promoters. However, the expression levels reached do not match those of wild-type cells (Fig. 8C). In addition, the sensitivity to rapamycin or caffeine of ptc1 and ptc1 sit4 mutants essentially is unaltered by the expression of high levels of Tip41 (data not shown). Therefore, the effects of a lack of Ptc1 cannot be explained exclusively by a decreased level of Tip41, indicating that the phosphatase must play additional roles in the TOR pathway. In this regard, several possibilities are worth considering. For instance, in addition to maintain proper levels of Tip41 protein, Ptc1 might be necessary to catalyze further posttranslational modifications that are required to render Tip41 a functional protein. This hypothesis is supported by the observations that Tip41 is phosphorylated in vivo at multiple sites and that it is dephosphorylated upon rapamycin treatment. It has been shown that a hyperphosphorylated form of Tip41 cannot properly bind Tap42, and Sit4 has been proposed as a likely Tip41 phosphatase acting upon the rapamycin signal. However, the lack of Sit4 does not fully abolish the dephosphorylation of Tip41 (29), suggesting that further phosphatase activities target Tip41. The evidence that a mutation of the Ptc1 catalytic site yields a functionally inactive protein (Fig. 1B), and the observation that a lack of Ptc1 partially abolishes the rapamycin-induced dephosphorylation of Tip41, support the notion that Ptc1 acts as a Tip41 phosphatase and raise the possibility that the Tip41 phosphorylation state (and, hence, its Tap42 binding ability) is controlled by a pair of phosphatases, Sit4 and Ptc1 (Fig. 6A). We wish to stress that although a lack of Ptc1 causes only a slight modification in the phosphorylation pattern of Tip41, this may still result in significant functional changes. For instance, rat liver glycogen synthase can be phosphorylated at multiple sites, and a recent report has demonstrated that the inability to phosphorylate a single Ser residue (site 2) is sufficient to constitutively activate the enzyme (54).
Our results do not rule out the possibility that Ptc1 has other targets within the TOR pathway that would be relevant for signaling, such as Tap42, which also is regulated by phosphorylation (30), or even that the phosphatase acts on other pathways functionally interacting with TOR (i.e., the PKA pathway). Interestingly, very recent work has shown that mutants in class C and D VPS genes display impaired TOR pathway signaling (50, 77), suggesting that the integrity of the Golgi body-to-endosome transport system is necessary for correct TOR function. It must be noted that ptc1 mutants display an altered vacuolar morphology that is reminiscent of class B vps mutants (4, 23, 65), and that this defect has been proposed to be responsible, at least in part, for a number of ptc1-related phenotypes (23). It is suggestive that about 50% of the class B mutants (according to the classification in reference 5) were found to be rapamycin sensitive in two recent genome-wide screens (47, 73). In addition, a recent survey for high-copy-number suppressors of diverse ptc1 phenotypes (A. González and J. Ariño, unpublished results) has shown that the overexpression of VPS70 and VPS73, encoding proteins of uncharacterized function that are required for correct vacuolar protein sorting, partially rescues the rapamycin- and caffeine-sensitive phenotype of ptc1 cells. Therefore, our findings that Ptc1 is required for correct TOR signaling may provide a new link between the TOR pathway and intracellular trafficking.
In essence, we show that Ptc1 is a specific type 2C phosphatase isoform that is necessary for normal TOR signaling. Whereas type 2A-related phosphatases have been involved in the TOR pathway for more than 10 years, as far as we know this is the first evidence for a role of type 2C enzymes in this important and conserved pathway. It is worth noting that, in this case, Ptc1 is necessary for the transmission of stress signals triggered by the inhibition of TOR, which is in contrast to the generally accepted concept that type 2C phosphatases act as inhibitors of cellular stress signaling (35).
Supplementary Material
Acknowledgments
We thank laboratory members Raquel Serrano, Carlos Casado, Anna Marco, and Maribel Marquina for support and advice. Thanks are given to Mike Hall, Estela Jacinto, Humberto Martin, and Francisco Estruch for strains and reagents. The excellent technical assistance of Anna Vilalta, María Jesús Álvarez, and Montse Robledo is acknowledged.
This work was supported by grants BFU2005-06388-C4-04-BMC and BFU2008-04188-C03-01 to J.A. and BFU2007-60342 to A.C. (Ministerio de Educación y Ciencia, Spain, and Fondo Europeo de Desarrollo Regional). J.A. is the recipient of an Ajut de Suport a les Activitats dels Grups de Recerca (2005SGR-00542; Generalitat de Catalunya).
Footnotes
Published ahead of print on 9 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Alberola, T. M., J. Garcia-Martinez, O. Antunez, L. Viladevall, A. Barcelo, J. Arino, and J. E. Perez-Ortin. 2004. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 7199-206. [PubMed] [Google Scholar]
- 3.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402689-692. [DOI] [PubMed] [Google Scholar]
- 4.Bonangelino, C. J., E. M. Chavez, and J. S. Bonifacino. 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 132486-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers, K., and T. H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744438-454. [DOI] [PubMed] [Google Scholar]
- 6.Butow, R. A., and N. G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 141-15. [DOI] [PubMed] [Google Scholar]
- 7.Calero, F., N. Gomez, J. Arino, and J. Ramos. 2000. Trk1 and Trk2 define the major K+ transport system in fission yeast. J. Bacteriol. 182394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 133271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. C., and T. Powers. 2006. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 49281-293. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, A., K. E. Ross, P. Kaldis, and M. J. Solomon. 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 132946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, T. G. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 27634441-34444. [DOI] [PubMed] [Google Scholar]
- 14.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crespo, J. L., S. B. Helliwell, C. Wiederkehr, P. Demougin, B. Fowler, M. Primig, and M. N. Hall. 2004. NPR1 kinase and RSP5-BUL1/2 ubiquitin ligase control GLN3-dependent transcription in Saccharomyces cerevisiae. J. Biol. Chem. 27937512-37517. [DOI] [PubMed] [Google Scholar]
- 16.de Nadal, E., J. Clotet, F. Posas, R. Serrano, N. Gomez, and J. Arino. 1998. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc. Natl. Acad. Sci. USA 957357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 101904-1916. [DOI] [PubMed] [Google Scholar]
- 18.Di Como, C. J., and Y. Jiang. 2006. The association of Tap42 phosphatase complexes with TORC1: another level of regulation in Tor signaling. Cell Cycle 52729-2732. [DOI] [PubMed] [Google Scholar]
- 19.Dilova, I., C. Y. Chen, and T. Powers. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 12389-395. [DOI] [PubMed] [Google Scholar]
- 20.Du, Y., L. Walker, P. Novick, and S. Ferro-Novick. 2006. Ptc1p regulates cortical ER inheritance via Slt2p. EMBO J. 254413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Düvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 111467-1478. [DOI] [PubMed] [Google Scholar]
- 22.Fiscella, M., H. Zhang, S. Fan, K. Sakaguchi, S. Shen, W. E. Mercer, G. F. Vande Woude, P. M. O'Connor, and E. Appella. 1997. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc. Natl. Acad. Sci. USA 946048-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González, A., A. Ruiz, R. Serrano, J. Arino, and A. Casamayor. 2006. Transcriptional profiling of the protein phosphatase 2C family in yeast provides insights into the unique functional roles of Ptc1. J. Biol. Chem. 28135057-35069. [DOI] [PubMed] [Google Scholar]
- 24.Görner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 9614866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, J., F. Al Shahrour, R. Diaz-Uriarte, A. Mateos, J. M. Vaquerizas, J. Santoyo, and J. Dopazo. 2003. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 313461-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoki, K., H. Ouyang, Y. Li, and K. L. Guan. 2005. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 6979-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacinto, E. 2007. Phosphatase targets in TOR signaling. Methods Mol. Biol. 365323-334. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 81017-1026. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 182782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 182491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamada, Y., T. Sekito, and Y. Ohsumi. 2004. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 27973-84. [DOI] [PubMed] [Google Scholar]
- 33.Kuepfer, L., M. Peter, U. Sauer, and J. Stelling. 2007. Ensemble modeling for analysis of cell signaling dynamics. Nat. Biotechnol. 251001-1006. [DOI] [PubMed] [Google Scholar]
- 34.Kuranda, K., V. Leberre, S. Sokol, G. Palamarczyk, and J. Francois. 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 611147-1166. [DOI] [PubMed] [Google Scholar]
- 35.Lammers, T., and S. Lavi. 2007. Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit. Rev. Biochem. Mol. Biol. 42437-461. [DOI] [PubMed] [Google Scholar]
- 36.Li, H., C. K. Tsang, M. Watkins, P. G. Bertram, and X. F. Zheng. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 4421058-1061. [DOI] [PubMed] [Google Scholar]
- 37.Lu, X., O. Ma, T. A. Nguyen, S. N. Jones, M. Oren, and L. A. Donehower. 2007. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 12342-354. [DOI] [PubMed] [Google Scholar]
- 38.Maeda, T., A. Y. Tsai, and H. Saito. 1993. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol. Cell. Biol. 135408-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2901-18. [DOI] [PubMed] [Google Scholar]
- 40.Mapes, J., and I. M. Ota. 2004. Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J. 23302-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler, G., C. Schuller, G. Adam, and H. Ruis. 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 121997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119969-979. [DOI] [PubMed] [Google Scholar]
- 43.Martín, H., M. Flandez, C. Nombela, and M. Molina. 2005. Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol. Microbiol. 586-16. [DOI] [PubMed] [Google Scholar]
- 44.Monteiro, P. T., N. D. Mendes, M. C. Teixeira, S. d'Orey, S. Tenreiro, N. P. Mira, H. Pais, A. P. Francisco, A. M. Carvalho, A. B. Lourenco, I. Sa-Correia, A. L. Oliveira, and A. T. Freitas. 2008. YEASTRACT-DISCOVERER: new tools to improve the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 36D132-D136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz, I., E. Simon, N. Casals, J. Clotet, and J. Arino. 2003. Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 20157-169. [DOI] [PubMed] [Google Scholar]
- 46.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45299-310. [DOI] [PubMed] [Google Scholar]
- 47.Parsons, A. B., R. L. Brost, H. Ding, Z. Li, C. Zhang, B. Sheikh, G. W. Brown, P. M. Kane, T. R. Hughes, and C. Boone. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2262-69. [DOI] [PubMed] [Google Scholar]
- 48.Posas, F., M. Camps, and J. Arino. 1995. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 27013036-13041. [DOI] [PubMed] [Google Scholar]
- 49.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194565-602. [DOI] [PubMed] [Google Scholar]
- 50.Puria, R., S. A. Zurita-Martinez, and M. E. Cardenas. 2008. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1057194-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds, A., V. Lundblad, D. Dorris, and M. Keaveney. 1997. Yeast vectors and assays for expression of cloned genes, p. 13.6.1-13.6.6. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, NY. [DOI] [PubMed]
- 52.Robinson, M. K., W. H. van Zyl, E. M. Phizicky, and J. R. Broach. 1994. TPD1 of Saccharomyces cerevisiae encodes a protein phosphatase 2C-like activity implicated in tRNA splicing and cell separation. Mol. Cell. Biol. 143634-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roeder, A. D., G. J. Hermann, B. R. Keegan, S. A. Thatcher, and J. M. Shaw. 1998. Mitochondrial inheritance is delayed in Saccharomyces cerevisiae cells lacking the serine/threonine phosphatase PTC1. Mol. Biol. Cell 9917-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ros, S., M. Garcia-Rocha, J. Dominguez, J. C. Ferrer, and J. J. Guinovart. 2009. Control of liver glycogen synthase activity and intracellular distribution by phosphorylation. J. Biol. Chem. 2846370-6378. [DOI] [PubMed] [Google Scholar]
- 55.Ruan, H., Z. Yan, H. Sun, and L. Jiang. 2007. The YCR079w gene confers a rapamycin-resistant function and encodes the sixth type 2C protein phosphatase in Saccharomyces cerevisiae. FEMS Yeast Res. 7209-215. [DOI] [PubMed] [Google Scholar]
- 56.Ruiz, A., A. Gonzalez, R. García-Salcedo, J. Ramos, and J. Arino. 2006. Role of protein phosphatases 2C on tolerance to lithium toxicity in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 62263-277. [DOI] [PubMed] [Google Scholar]
- 57.Saito, H., and K. Tatebayashi. 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. (Tokyo) 136267-272. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., F. E. F., and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Santhanam, A., A. Hartley, K. Duvel, J. R. Broach, and S. Garrett. 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 31261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 176924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweighofer, A., H. Hirt, and I. Meskiene. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9236-243. [DOI] [PubMed] [Google Scholar]
- 63.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 101574-1581. [DOI] [PubMed] [Google Scholar]
- 64.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 173556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahirovic, S., M. Schorr, and P. Mayinger. 2005. Regulation of intracellular phosphatidylinositol-4-phosphate by the SacI lipid phosphatase. Traffic 6116-130. [DOI] [PubMed] [Google Scholar]
- 66.Tamura, S., Li, M. G., K. Komaki, M. Sasaki, and T. Kobayashi. 2004. Roles of mammalian protein phosphatase 2C family members in the regulation of cellular functions, p. 91-105. In J. Ariño and D. Alexander (ed.), Protein phosphatases, vol. 5. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 67.Tate, J. J., A. Feller, E. Dubois, and T. G. Cooper. 2006. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J. Biol. Chem. 28137980-37992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tate, J. J., R. Rai, and T. G. Cooper. 2005. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J. Biol. Chem. 28027195-27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatebe, H., and K. Shiozaki. 2003. Protein serine/threonine-phosphatase 2C (PP2C), p. 637-640. In R. Bradshaw and E. Dennis (ed.), Handbook of cell signaling, vol. 1. Elsevier Science, New York, NY. [Google Scholar]
- 70.Wang, H., X. Wang, and Y. Jiang. 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 144342-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warmka, J., J. Hanneman, J. Lee, D. Amin, and I. Ota. 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 2151-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 73.Xie, M. W., F. Jin, H. Hwang, S. Hwang, V. Anand, M. C. Duncan, and J. Huang. 2005. Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc. Natl. Acad. Sci. USA 1027215-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan, G., X. Shen, and Y. Jiang. 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 253546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young, C., J. Mapes, J. Hanneman, S. Al Zarban, and I. Ota. 2002. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot. Cell 11032-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zurita-Martinez, S. A., and M. E. Cardenas. 2005. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 463-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zurita-Martinez, S. A., R. Puria, X. Pan, J. D. Boeke, and M. E. Cardenas. 2007. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 1762139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.