Summary
Inflammatory activation of NF-κB involves the stimulus-induced degradation of the NF-κB-bound inhibitor, IκB, via the IκB kinase (IKK). In response to UV irradiation, however, the mechanism and function of NF-κB activation remain unclear. Using a combined biochemical, genetic, and computational modeling approach, we delineate a dual requirement for constitutive IKK-dependent and IKK-independent IκB degradation pathways in conjunction with UV-induced translational inhibition. Interestingly, we find that the high homeostatic turnover of IκB in resting cells renders the NF-κB system remarkably resistant to metabolic stresses, but the two degradation pathways critically and differentially tune NF-κB responsiveness to UV. Indeed, in the context of low chronic inflammation that accelerates NF-κB-bound IκB degradation, UV irradiation results in dramatic NF-κB activation. Our work suggests that the human health relevance of NF-κB activation by UV lies with cellular homeostatic states that are associated with pathology rather than with healthy physiology.
Introduction
Mammalian cells respond to UV irradiation by activating various transcription factors, which induce specific gene expression programs to mediate DNA damage repair, cell growth arrest, and induction of apoptosis. Nuclear factor-kappaB (NF-κB) has been described as one such transcription factor activated by UV irradiation (Devary et al., 1993; Simon et al., 1994; Stein et al., 1989). NF-κB is a critical mediator for cellular responses to inflammatory cytokines, developmental signals, pathogens, and cellular stresses (Hoffmann and Baltimore, 2006). In resting cells, NF-κB is held inactive in the cytoplasm through stoichiometric association with inhibitory proteins (IκBs). Activation of this latent NF-κB pool is known to be driven by inflammatory stimuli that lead to activation of the IκB kinase (IKK) complex, which mediates site-specific phosphorylation of the canonical IκB proteins, IκBα, -β, and -ε, that targets them for ubiquitination and proteasomal degradation. NF-κB subsequently accumulates in the nucleus where it mediates gene activation. In contrast to the strong, rapid, and transient NF-κB response to inflammatory signals, IκB proteolysis and NF-κB activation in response to UV irradiation appear to be weak, slow, and prolonged, and the underlying molecular mechanisms are not well understood.
Early studies determined that activation of NF-κB by UV irradiation involves two phases: an early phase that is independent of DNA-damage (Devary et al., 1993; Simon et al., 1994), and a second, later phase (>24 hours) that is dependent on DNA damage (Bender et al., 1998). The DNA-damage pathway of NF-κB activation has since been well-characterized to involve activation of IKK through association with the DNA double stranded break-activated kinase, ataxia telangiectasia mutated (ATM) (Wu et al., 2006).
The mechanisms underlying the early, DNA damage independent phase of UV-induced IκB degradation, however, remain unclear with reports arriving at seemingly conflicting conclusions (summarized in Figure 1A). Initially, IKK activity was proposed not to be required, as an IκBα mutant lacking the IKK phosphorylation sites transfected into HeLa cells was indeed degraded upon UV irradiation. Further, co-transfection of a dominant negative IKK kinase-defective mutant with IκBα in HeLa cells did not prevent UV-induced IκBα degradation (Bender et al., 1998; Li and Karin, 1998). Although no activation of IKK could be detected following UV irradiation (Huang et al., 2002; Li and Karin, 1998), a later study suggested that IKK activity may be required (Figure 1A, right), as expression of the IκBα mutant lacking IKK phosphorylation sites in wild-type B-cells prevented UV-induced NF-κB DNA binding activity and UV-induced NF-κB could not be detected in IKK knock-out cells (Huang et al., 2002).
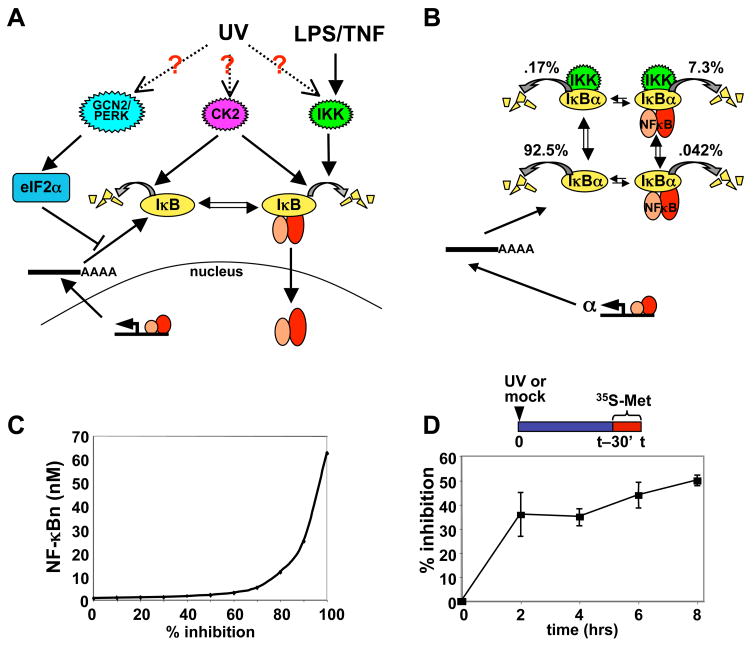
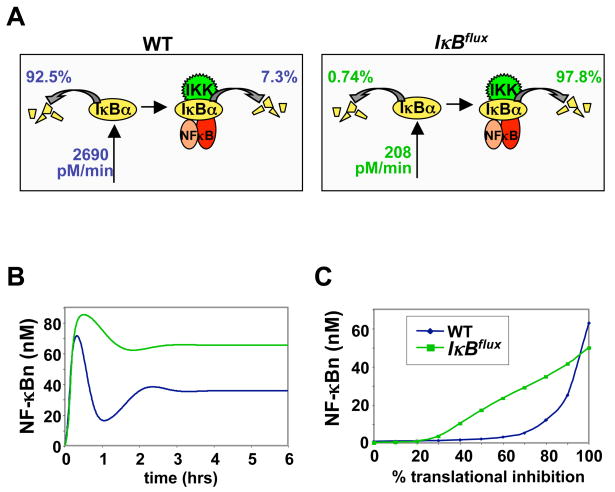
Figure 1. UV-induced translational inhibition and its effect on the IκB-NF-κB homeostasis.
(A) Proposed pathways leading to NF-κB activation by UV irradiation. Left: UV may induce GCN2 and PERK activity, which in turn phosphorylate eIF2α to inhibit translation. Center: UV may activate CK2, which may lead to degradation of free and/or bound IκB. Right: UV may also induce IKK activity, which is the primary signal transducer for inflammatory signals.
(B) Flux calculations of IκB turnover in the steady state of resting MEFs, using the computational model of the IκB-NF-κB signaling module. The schematic indicates four degradation pathways delineated in the model and described in the text. The percentages indicate the fraction of the total IκBα degradation flux that occurs within each of these pathways.
(C) Dose response curve of degrees of translational inhibition (in %) to nuclear NF-κB activity (NF-κBn in nM). A translational inhibition curve (Supplemental Figure 2) was used as an input for the computational model and the nuclear NF-κB (NF-κBn) level after 8 hours was calculated and plotted.
(D) Percentage of bulk protein synthesis inhibition in cells treated with 40 J/m2 UVC radiation. Wild-type MEFs were either mock- or UV-treated, and radiolabeled for 30 minutes prior to each respective time point with 35S-labeled methionine. Protein extracts were prepared at the indicated times and the amount of 35S incorporation was measured by scintillation count. The amount of 35S incorporation measured for each UV-treated sample was divided by the amount measured for mock-treated to calculate the percent inhibition. Error bars represent standard error of the mean.
More recently, a second mechanism was proposed wherein UV irradiation activates CK2 (casein kinase 2) via the p38/MAPK pathway (Figure 1A, center). This CK2 activity was suggested to cause degradation of IκBα through phosphorylation of a cluster of serines and threonines in the C-terminal PEST domain of IκBα (Kato et al., 2003), correlating with a prior study that indicated that the C-terminal region of IκBα is required for its degradation in response to UV (Bender et al., 1998).
In addition, a third mechanism has been proposed wherein UV-induced translational inhibition is responsible for the observed NF-κB activation. Because the half-life of IκB is shorter than that of NF-κB, treatment of cells with reagents that inhibit protein synthesis, such as cycloheximide (CHX), induce NF-κB (Sen and Baltimore, 1986). It was reported that UV irradiation causes inhibition of translation through induced phosphorylation of the eukaryotic initiation factor-2α (eIF2α) at serine 51 (Jiang and Wek, 2005; Wu et al., 2004). Stress-induced translational inhibition through eIF2α phosphorylation may also activate NF-κB (Deng et al., 2004; Jiang et al., 2003), though the degree and time of translational inhibition required has not been examined. There is strong evidence that translational inhibition by eIF2α phosphorylation, in response to UV irradiation is required for NF-κB activation (Figure 1A, left) (Jiang and Wek, 2005; Wu et al., 2004). Yet, it is unclear if translational inhibition alone is sufficient, given that UV-induced IκB degradation mechanisms have also been proposed.
Mathematical models have been constructed to investigate complex signaling pathways and are valuable in relating fundamental biochemical/biophysical properties with novel and useful predictions of cellular behavior (Aldridge et al., 2006). A mathematical model of the IKK-IκB-NF-κB signaling module was first constructed to recapitulate NF-κB activation dynamics in response to TNF treatment, and led to insights into the functions of each IκB isoform (Hoffmann et al., 2002). Subsequent refinements of the model revealed temporal control of NF-κB dynamics in responrse to TNF signaling (Kearns et al., 2006) and other inflammatory agents (Werner et al., 2005), as well as a fourth IκB protein responsible for NF-κB activation in response to developmental stimuli (Basak et al., 2007). Further, an analysis of the multiple IκB degradation mechanisms and their respective rates in resting cells was key to recapitulating homeostatic regulation of NF-κB activity (O’Dea et al., 2007).
Here, combining computational and experimental analyses, we examine the role of translational inhibition and the role of each IκB degradation pathway in NF-κB activation by UV irradiation (Figure 1A). We find that the dynamic steady-state regulation of IκB turnover critically determines the sensitivity of the NF-κB signaling pathway to UV irradiation, and suggest specific roles for the two different IκBα degradation pathways (IKK-dependent and IKK-independent) in providing powerful signaling crosstalk.
Results
Translational inhibition as an inducer of signaling
Previously, we measured the rate constants for the four possible degradation pathways that each IκB protein is subject to and found that while NF-κB-bound IκBα is extremely stable and is therefore almost exclusively degraded via the IKK-mediated degradation pathway, free, or unbound IκBα is rapidly degraded through an IKK-independent mechanism with a half-life of less than 10 minutes. These differential degradation rates were key in the construction of a computational model of the IκB-NF-κB signaling module that accurately recapitulated the steady state control of NF-κB (O’Dea et al., 2007). For example, the model recapitulated experimental results that showed that, in resting cells, approximately 85% of total cellular IκBα is present in the IκB-NF-κB complex while 15% of IκBα is not bound to NF-κB (Rice and Ernst, 1993). However, this model also predicts that in the steady state only 8% of IκBα is degraded from the NF-κB-bound form whereas almost 93% is degraded from the lower abundance free form (Figure 1B). We conclude that instability of free IκBα must be counteracted by a high synthesis rate to ensure low levels of nuclear NF-κB activity in resting cells.
Many cellular stress conditions, such as viral infection, oxidative stress, heat shock, endoplasmic reticulum (ER) stress, nutrient limitation, hypoxia, ribotoxic stress, and UV irradiation induce inhibition of protein synthesis to varying degrees (van den Beucken et al., 2006; Wek et al., 2006), which may disrupt the relationship between IκBα instability and high synthesis. To probe the sensitivity of the IκB-NF-κB homeostasis to decreased translation rates, we reduced protein synthesis parameters and calculated the resulting levels of nuclear NF-κB (Figure 1C). We found that whereas high doses of translational inhibition (<70%) induce significant levels of NF-κB after 8 hours, the system is rather resistant to lower doses, activating significantly lower levels of NF-κB.
We chose UV irradiation as a representative model system for studying cellular stress as a modulator of NF-κB activity. The level of translational inhibition induced by 40 J/m2 UVC irradiation in wild-type mouse embryonic fibroblasts (MEFs) was measured by 35S-Met/Cys incorporation. Over an 8-hour time course, UV treatment resulted in 35–50% bulk protein synthesis inhibition (Figure 1D). Our model predicted that this degree of protein synthesis inhibition would result in a modest 2.5-fold NF-κB activation (Figure 1C).
The role of IKK in UV activation of NF-κB
To determine if IKK plays a role in UV-induced NF-κB activity, we first examined IKK activity following UV irradiation. The IKK complex was immunoprecipitated from UV-treated wild-type MEFs and subjected to an in vitro kinase assay capable of detecting small changes in IKK activation following treatment with low doses of TNF (Figure 2A). In agreement with previous reports (Huang et al., 2002; Li and Karin, 1998), no activation of IKK above the basal level was seen in UV treated cells even at very high doses of UV (Figures 2B and C). Although the lack of UV-induced IKK activation suggested that IKK is dispensable for UV-induced NF-κB activity (Bender et al., 1998; Li and Karin, 1998), a more recent study proposed that IKK is required (Huang et al., 2002). We also found that in ikkβ−/− cells UV does not induce nuclear NF-κB DNA binding as measured by electrophoretic mobility shift assay (EMSA) (Figure 2D). To test whether IκB phosphorylation at the IKK-responsive serines is a requirement, iκbα−/− cells were reconstituted with an IκBα mutant in which the IKK target sites (S32/S36) were replaced with alanines (IκBαAA). We found that UV-induced NF-κB DNA binding was abolished in these cells as compared to iκbα−/− cells reconstituted with wild-type IκBα (IκBαWT) (Figure 2E), correlating with a previous report (Huang et al., 2002). These results are in contrast to transient transfection studies that reported the IκBαAA mutant to be degraded upon exposure to UV (Bender et al., 1998; Li and Karin, 1998).
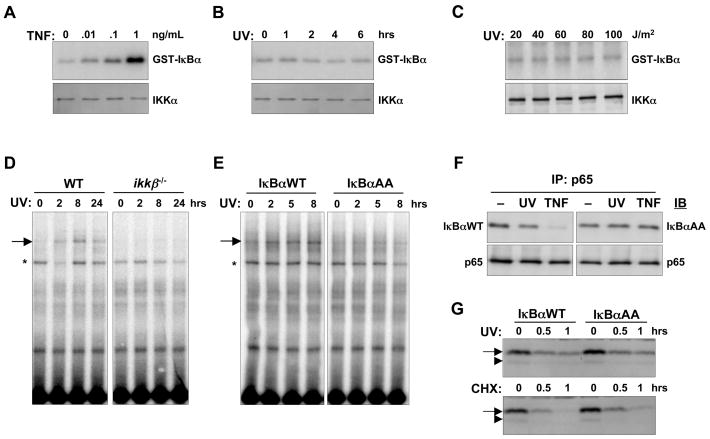
Figure 2. Different pools of IκBα are degraded with distinct pathways in response to UV irradiation.
IKK is not activated by UV:
(A) In vitro kinase assay of cytoplasmic extracts from wild-type MEFs treated with 0, 0.01, 0.1, or 1 ng/mL TNF for 10 minutes.
(B) In vitro kinase assay of cytoplasmic extracts from wild-type MEFs treated with 40 J/m2 UVC radiation for the indicated times
(C) In vitro kinase assay of cytoplasmic extracts from wild-type MEFs 8 hours after treatment with the indicated doses of UVC radiation. Immunoblots for IKKα below each kinase assay confirm equal IP-efficiency and loading of the lanes (bottom panels). UV-induced NF-κB activity requires IKK:
(D) EMSA for NF-κB activity in nuclear extracts from wild-type or ikkβ−/− cells treated with 40 J/m2 UVC for indicated times
(E) EMSA for NF-κB activity in nuclear extracts from iκbα −/− cells reconstituted with either wild-type IκBα (IκB αWT) or Ser32/36Ala mutant IκBα (IκBαAA) treated with 40 J/m2 UVC for the indicated times. An asterisk (*) indicates a non-specific band.
(F) Tracking the degradation of NF-κB-bound IκB following UV irradiation. iκbα −/− cells reconstituted with either IκBαWT or IκBαAA were mock-treated, treated with 60 J/m2 UVC, or treated with 1 ng/mL TNF. Whole cell extracts prepared 8 hours after UV treatment or after 10 minutes of TNF treatment were subject to immunoprecipitation with anti-p65 antibody. The resulting immunocomplexes were subject to immunoblot analysis for IκBα and p65, representative of multiple experiments.
(G) Tracking degradation of free IκB following UV irradiation. nfκb−/− cells (rela−/− nfkb1−/− crel−/−) stably expressing IκBαWT or IκBαAA were treated with 40 J/m2 UVC or 10 μg/mL CHX and whole cell extracts prepared at the indicated times were subject to immunoblot analysis for IκBα.
How might IKK contribute to NF-κB activation by UV if its activity is not induced? We considered that in unstimulated cells, the basal turnover of NF-κB-bound IκB involves the basal activity of IKK (Figure 1B) (O’Dea et al., 2007). To examine whether NF-κB liberation from IκB in response to UV involves the basal IKK-dependent degradation pathway, we immunoprecipitated the NF-κB subunit p65 for Western blot analysis of bound IκBαWT or IκBαAA proteins. Interestingly, UV-treatment induced a modest decrease in p65-associated IκBαWT, whereas the level of p65-associated IκBαAA remained constant (Figure 2F and Supplemental Figure 1A).
In contrast to the IKK-dependent turnover of NF-κB-bound IκBα, free IκBα turnover is regulated in a manner almost completely independent of IKK activity (Mathes et al., 2008; O’Dea et al., 2007). IκBαWT and IκBαAA were transduced into cells lacking the NF-κB subunits known to interact with IκB, RelA/p65, p50, and cRel (nfκb−/−). Treatment of these cells with UV induced rapid and sustained decreases of both IκBαWT and IκBαAA levels (Figure 2G and Supplemental Figures 1B-C), contrasting with the slow and IKK-dependent degradation of NF-κB-bound IκBα.
Together, these data demonstrate that two degradation pathways of IκBα are engaged following UV irradiation. The free IκBα degradation following UV irradiation does not require IKK activity, exposing a potentially important IκB degradation mechanism. In contrast, NF-κB-associated IκBα degradation in response to UV requires basal IKK activity.
The apparent requirement for basal IKK activity to mediate degradation of NF-κB-bound IκBα during UV-mediated induction of NF-κB suggests that the level of basal IKK activity in unstimulated cells may have a significant effect on the degree to which NF-κB can be activated. We first tested this hypothesis with the computational model. UV-induced translational inhibition was modeled by adjusting protein synthesis rates to linearly decrease to 40% inhibition by 1 hour and then linearly decrease further to 50% inhibition by 8 hours (Supplemental Figure 2B). To determine the effect of altered basal IKK activity levels on UV-induced NF-κB activation, the model was first equilibrated at different levels of basal IKK activity and was then used for simulations with the UV- induced translational inhibition curve. Decreased basal activity of IKK (0.5x) prevented NF-κB activation, while higher basal IKK (2x) activity significantly enhanced NF-κB activation (Figure 3A).
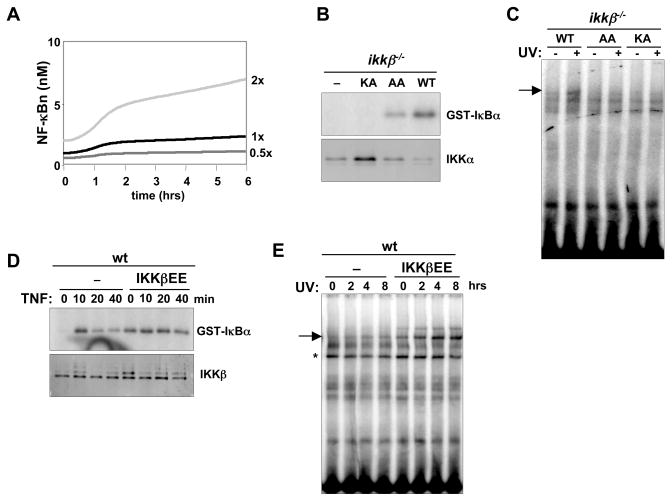
Figure 3. The level of basal IKK activity controls NF-κB responsiveness to UV irradiation.
(A) Computational simulations of NF-κB activation via 40–50% inhibition of protein synthesis over 6 hours at different equilibrium levels of basal IKK activity (0.5x, 1x, 2x). The graph shows calculated nuclear NF-κB activity plotted against time.
(B) In vitro kinase assay to measure basal IKK activity of cytoplasmic extracts from untreated ikkβ −/− cells reconstituted with IKKβWT, IKKβAA (S177A/S181A), or IKKβKA (K44A). Immunoblot of IKKα from the IP of the IKK complex (bottom panel).
(C) EMSA for NF-κB activity in nuclear extracts from ikkβ −/− cells reconstituted with IKKβWT, IKKβAA, or IKKβKA 8 hours after mock or 40 J/m2 UVC treatment.
(D) In vitro IKK kinase assay of cytoplasmic extracts from wild-type MEFs stably expressing IKKβEE (S177E/S181E) or wild-type MEFs transduced with empty vector treated with 1 ng/mL TNF for the indicated times. Immunoblot of IKKβ from the IP of the IKK complex (bottom panel).
(E) EMSA for NF-κB activity in nuclear extracts from wild-type MEFs stably expressing IKKβEE or wild-type MEFs transduced with empty vector treated with 40 J/m2 UVC for the indicated times.
To experimentally examine these computational predictions, basal activity of IKK was genetically modulated and the NF-κB activity in response to UV irradiation was measured. Reconstitution of ikkβ−/− cells with a kinase defective IKKβ mutant (IKKβKA) that cannot bind ATP or an activation defective IKKβ mutant (IKKβAA) in which the activation loop containing phospho-acceptor serines were replaced with alanines resulted in either no or decreased IKK activity in resting cells, respectively (Figure 3B). Although UV-induced NF-κB activity was recovered in ikkβ −/− cells reconstituted with wild-type IKKβ, no UV-induced NF-κB activity was detected in ikkβ−/− cells reconstituted with the mutants harboring decreased or no basal IKK activity (Figure 3C). A constitutively active IKKβ mutant (IKKβEE) expressed in wild type cells greatly enhanced IKK activity in untreated cells and could not be further activated by TNF (Figure 3D). UV irradiation of these cells resulted in significantly enhanced NF-κB activation (Figure 3E), consistent with the computational predictions.
Together, these data suggest that minimal basal IKK activity is a prerequisite for NF-κB activation in response to UV and that enhancing it sensitizes the IκB-NF-κB signaling module to UV-induced translational inhibition.
The role of IKK-independent degradation of IκB
To determine if the IKK-independent free IκB degradation pathway plays a role in UV-induced NF-κB, we simulated NF-κB activation in response to UV-induced translational inhibition with altered equilibrium rates of free IκBα degradation. The model predicted that a slower degradation rate for free IκBα (0.5x) attenuates UV-induced NF-κB activation, whereas a more unstable IκBα (2x) enhances NF-κB activation (Figure 4A).
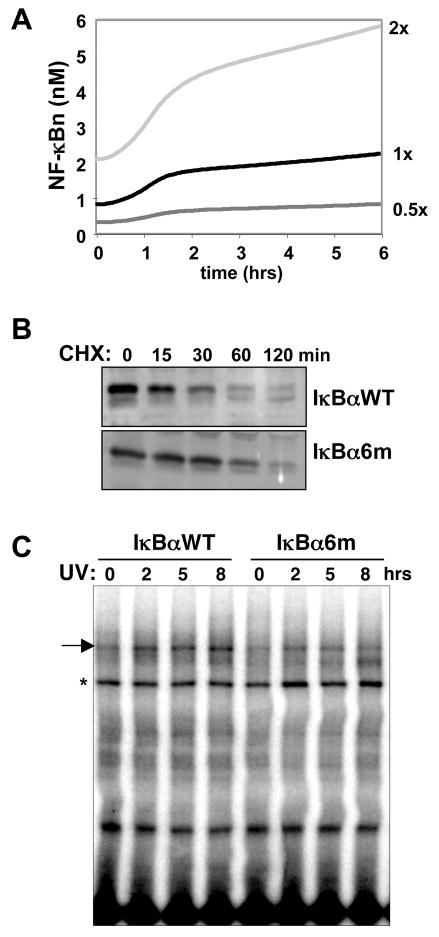
Figure 4. Free IκBα turnover is required for NF-κB activation by UV irradiation.
(A) Computational simulations of NF-κB activation by 40–50% inhibition of protein synthesis over 6 hours at different equilibrium free IκBα turnover rates (0.5x, 1x, 2x). The graph shows calculated nuclear NF-κB activity plotted against time.
(B) Immunoblots for IκBα from whole cell extracts of nfκb−/− (rela−/− nfkb1−/− crel−/−) cells stably expressing wild-type IκBα or the CK2 phospho-acceptor site mutant (IκBα6M) treated with 10 μg/mL CHX for the indicated times.
(C) EMSA for NF-κB activity in nuclear extracts from iκbα−/− cells reconstituted with either IκBαWT or IκBα6M treated with 40 J/m2 UVC.
Though the mechanism of free IκBα degradation has yet to be elucidated, some studies have implicated CK2-mediated phosphorylation of the PEST domain of free IκBα to play a role in its instability (Bren et al., 2000; Schwarz et al., 1996). An IκBα mutant with the 6 CK2 target sites in IκBα replaced with alanines (IκBα6m) was transduced into the nfκb−/− cells. CHX treatment of these cells indicated that the IκBα6M mutant has an approximately 2-fold longer half-life than the wild-type IκBα (Figure 4B and Supplemental Figure 3). Stable expression of IκBα6M in iκbα−/− cells attenuated UV-induced NF-κB activity (Figure 4C). As this mutant is still susceptible to IKK-mediated degradation of NF-κB-bound IκB (Mathes et al. 2008), our data suggests that the fast turnover rate of free IκBα is also a prerequisite for UV-induced NF-κB activity.
The observed 2-fold stabilization of the free IκBα6m mutant results in a modest increase in total cellular IκBα both experimentally (Supplemental Figure 3B) and computationally (Supplemental Figure 3C). Interestingly, simulations with a model, in which the IκBα translation rate is adjusted such that the total IκBα level is similarly increased, predict a similar attenuating effect on UV-induced NF-κB (Supplemental Figure 3D). These observations suggest that the precise steady state levels of IκBα are a determinant of NF-κB responsiveness to translational inhibition.
High IκB turnover confers resistance to ribotoxic stress
Because IκB is so rapidly degraded when unassociated from NF-κB, a high steady state synthesis rate is required to ensure low NF-κB activity in resting cells. Why might the cell have evolved such a high energy-consuming process to synthesize and degrade so much IκB protein that never associates with NF-κB?
To explore the role of IκB turnover flux on the responsiveness of the NF-κB signaling system to UV, we constructed a virtual mutant cell in which degradation and synthesis rates were both reduced. In our IκBflux mutant, IKK-independent free IκB degradation rates were decreased to match the IKK-independent degradation rates of NF-κB-bound IκB (2000-fold slower), and then the IκB synthesis rates were decreased to such a degree as to maintain the same steady state levels of free and bound IκB proteins as in the wild-type cell model (Supplemental Figure 4). Specifically, in the IκBflux mutant model, IκBα synthesis was decreased 13-fold from 2690 pM/min to 208 pM/min (Figure 5A). As a result, in the mutant the majority of degradation flux of IκBα is from the NF-κB-bound state rather than from the free state (Figure 5A). However, such a dramatic alteration in flux rates turns out to have a surprisingly small effect on NF-κB activation in response to signals functioning via IKK activation. Simulation of TNF signaling in the IκBflux mutant model results in rapid NF-κB activation that is attenuated (albeit less so) during the second phase (Figure 5B). (One may consider that in cells that do not have an excess of IκB synthesis and degradation, lower cytokine-responsive IKK activation could have co-evolved to provide identical NF-κB responsiveness to inflammatory signals.)
Figure 5. A high IκB turnover flux confers NF-κB resistance to metabolic stress.
(A) Contrasting two computational models representing virtual wild type and mutant cells. The “wild-type” model has a high IκB synthesis rate and free IκB degradation rate. In the “IκBflux” model, free IκBα degradation is decreased 2000-fold, and the synthesis rate is decreased to such a degree that free and bound IκB levels and ratios are the same as in the “wild-type” model.
(B) Computational simulation of a TNF induced NF-κB activation time course in wild-type (blue line) and IκBflux (green line) models. The graph shows calculated nuclear NF- κ B activity plotted against time.
(C) Computational simulations to determine the dose responsiveness to translational inhibition of wild-type and IκBflux mutant models. The graph shows the concentration of nuclear NF-κB activity after and 8-hour time course plotted against the degree of translational inhibition.
To determine how NF-κB is regulated in response to translational inhibition in this “low flux” model, protein synthesis was inhibited to varying degrees and the resulting NF-κB activation at 8 hours was calculated. Remarkably, these studies predicted that the IκBflux mutant would have significantly increased sensitivity to translational inhibition. While wild-type and mutant models show little difference at very low degrees of inhibition of <20%, at intermediate levels of 20–90% the mutant shows a dramatically greater response. At 50% inhibition for example, corresponding to UV irradiation conditions, the low flux mutant produces 20-fold NF-κB activation whereas the wild type model produces only 2.5-fold. Our results indicate that the high flux rates of IκB synthesis and degradation confer relative insensitivity to UV irradiation, suggesting that the energy consuming process of excess IκB synthesis and degradation may have evolved so that the NF-κB signaling module maintains resistance to ribotoxic stress.
NF-κB responsiveness to UV is differentially controlled by two homeostatic IκB degradation pathways
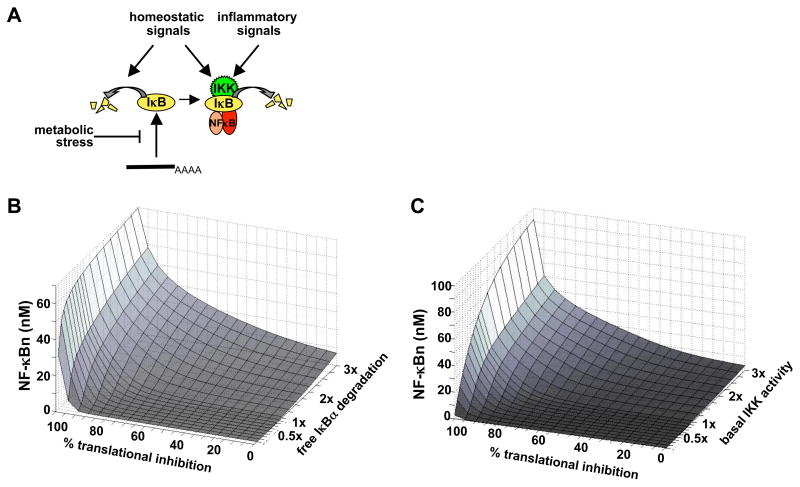
Given that our work indicated that ribotoxic stresses acting alone do not elicit strong NF-κB responses, we wondered whether altered environmental conditions may sensitize cells such that they generate higher NF-κB activation. Our experimental studies indicated that two IκB degradation pathways are important: the IKK-dependent pathway for NF-κB-bound IκB and the IKK-independent pathway for free IκB. While a host of stimuli and conditions are known to regulate IKK activity, regulation of the free IκB degradation pathway may also be possible, such that either one may impact the response to metabolic stresses that cause translational inhibition (Figure 6A).
Figure 6. Altered degradation rates of free and NF-κB-bound IκB have different effects on the NF-κB responsiveness to ribotoxic stress.
(A) Model of NF-κB activation by metabolic stress. Metabolic stress inhibits the synthesis of IκB, which is rapidly degraded in the free state (maintained and potentially altered by homeostatic signals), and bound IκB is degraded through low levels of basal IKK activity (maintained and altered by homeostatic and inflammatory signals).
(B) Computational simulations of NF-κB activation at 8 hrs in response to varying degrees of translational inhibition at different equilibrium free IκB turnover rates (in terms of a multiplier to the wild-type parameter on the z-axis).
(C) Computational simulations of NF-κB activation at 8 hrs in response to varying degrees of translational inhibition at different basal IKK activity levels (in terms of a multiplier to the wild-type parameter on the z-axis).
To examine how the apparent resistance to translational inhibition is modulated by altered turnover rates of free IκBα, simulations were run with different degrees of induced protein synthesis inhibition, each at different equilibrium rate constants of free IκB degradation (Figure 6B). As the equilibrium degradation rate constant of free IκB is increased (above 1x), NF-κB activation following 8 hours of translational inhibition moves away from a switch-like response to a more linear dose-response. In other words, decreasing the steady state level of free IκB is predicted to sensitize cells to lower doses of translational inhibition.
To examine the role of basal IKK activity (which governs the degradation rate constant of NF-κB-bound IκB), simulations were run with different degrees of induced protein synthesis inhibition, and at different equilibrium IKK activity values. The calculated NF-κB activation levels were plotted in a similar 3-D graph (Figure 6C). Similar to the effect of the free IκB degradation rate constant, elevated basal IKK activity also renders the NF-κB system more sensitive to low doses of translational inhibition. In addition, however, the maximal NF-κB activity caused by 100% translational inhibition also increased greatly with increasing basal IKK activity, although it was less sensitive to the rate of free IκBα turnover (compare Figures 6B and C at 100% translational inhibition). These predictions suggest that basal IKK regulates overall responsiveness to metabolic stress, in both low and high translational inhibition conditions. The free IκBα degradation pathway, however, provides a means for tuning the sensitivity primarily to moderate translational inhibition conditions.
UV irradiation as an amplifier of inflammation
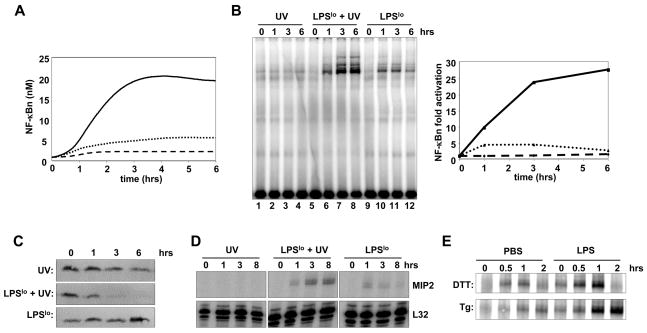
These simulation results led us to examine the potential for crosstalk between low inflammatory exposure and UV irradiation. We explored the effect of an induction of 40–50% translational inhibition in the presence of low IKK activation. A relatively low level of LPS treatment (1 ng/mL) of wild-type MEFs mildly activates IKK (Supplemental Figure 5A). When the quantitated LPS-induced IKK activity and the UV-induced translational inhibition are separately fed into the computational model, 5-fold and 2.5-fold activation of NF-κB result over a 6-hour time course, respectively (Figure 7A, dotted line and dashed line, respectively). However, when these two stimuli are fed into the model simultaneously, a surprisingly high 20-fold NF-κB activation results (Figure 7A, solid line).
Figure 7. Synergistic cross-talk between UV and inflammatory signals.
(A) Computational simulations of NF-κB activation in response to 40–50% translational inhibition (dashed line), 1 ng/mL LPS-mediated IKK activation (dotted line), or combined induction of 50% translational inhibition and 1 ng/mL LPS-mediated IKK activation (solid line).
(B) EMSA for NF-κB activity of nuclear extracts from wild-type MEFs treated with either 40 J/m2 UVC alone (lanes 1–4), 1 ng/mL LPS (“LPSlo”) alone (lanes 9–12) or co-treated with UV and LPS (lanes 5–8) for the indicated times. Signals were quantitated and graphed relative to signal levels in untreated cells (right panel).
(C) Immunoblots for IκBα from whole cell extracts of wild-type MEFs treated with either UVC, LPS, or co-treated with UV and LPS for the indicated times.
(D) RNase Protection Assay showing the mRNA levels of MIP2 in wild-type MEFs treated with either UVC alone, LPS alone, or co-treated with UV and LPS for the indicated times. mRNA levels for L32 are shown as a loading control (bottom panel).
(E) EMSA for NF-κB activity of nuclear extracts from wild-type MEFs. Cells were pretreated with PBS or 1 ng/mL LPS for 4 hours, then either 1 mM DTT (top panel) or 400 nM thapsigargin (Tg, bottom panel) was added for the indicated times.
We tested this prediction of signaling synergy between inflammation and UV exposure. Wild-type MEFs treated with 40 J/m2 UV or 1 ng/mL LPS activated nuclear NF-κB DNA binding activity about 2- and 5-fold (Figure 7B, lanes 1–4 and 9–12), consistent with the model predictions. Strikingly, co-treatment of cells with LPS and UV resulted in a greater than 25-fold induction of NF-κB (Figure 7B), even though IKK activity was unaltered (Supplemental Figure 5A). The synergy was dependent on IKK-mediated phosphorylation of IκBα (Supplemental Figure 5B) and was also observed in primary mouse keratinocytes (Supplementary Figure 5C). Whereas UV allows for the depletion of the pool of free IκBα (Figure 2G), LPS targets the IKK-mediated pathway to degrade NF-κB-bound IκBα. Consistently, co-treatment of these two stimuli resulted in a synergistic decrease in total IκBα protein levels (Figure 7C). Further, hyper-activation of NF-κB in LPS and UV co-treated cells resulted in transcriptional activation of the NF-κ B target gene MIP2 (Figure 7D).
In addition, we find that co-treatment of cells with other inflammatory signals such as IL-1 or TNF and UV also leads to synergistic activation of NF-κB (Supplemental Figure 5D), without altering the IL-1-induced IKK activity (Supplemental Figure 5E). Similarly, an intra-cellular inducer of translational inhibition is ER stress, known to activate NF-κB (Hu et al., 2006) and depletion of the free pool of IκBα (Supplementary Figure 7), also exhibited signaling synergy with inflammatory agents. We found that treating cells with the commonly used ER-stress inducing reagents DTT and thapsigargin in the presence of low LPS (1 ng/mL) resulted in drastically higher NF-κB activity in than in its absence (Figure 7E).
To determine whether the observed hyper-activation can be accounted for by UV’s translational inhibition effect, we substituted UV for a specific translational inhibitor, CHX. Titration determined that a treatment of wild-type MEFs with 0.1 μg/mL CHX blocked bulk protein synthesis by approximately 50% (data not shown). Similar to the effect of UV, when this low dose of CHX was added to cells in combination with low dose LPS, hyper-NF-κB activation resulted (Supplemental Figure 6A), as long as IκBα could be phosphorylated by IKK (Supplemental Figure 6B). Together, we surmise that by inducing translational inhibition, UV or ER stress may act as an amplifier of IKK- inducing signals that are weak and maybe constitutive. As such, the signaling effects of metabolic stress agents on NF-κB may have human health relevance in the context of inflammatory pathologies where IKK activity is elevated.
Discussion
Responsiveness to cellular stress
Intracellular signal transduction involves a stimulus-induced alteration to the steady state of a signaling network. Many intracellular signaling pathways are activated by changes to constitutive degradation rates of effector proteins. For example, in resting cells the transcription factors HIF-1α, β-catenin, and p53 are maintained at low levels through constitutive degradation, but signal-induced inhibition of the degradation pathway causes their build up to a level where they elicit gene expression responses. In these scenarios, the cellular responsiveness is determined by the high constitutive synthesis rates of the signaling effector proteins. In contrast, NF-κB protein levels are maintained through a long cellular half-life requiring only low constitutive synthesis. It is the NF-κB inhibitor, IκB, which is synthesized at a high rate and degraded very rapidly. In contrast to the cases of p53 and HIF-1α, we find in this study that high turnover of IκB confers resistance of the NF-κB signaling module to ribotoxic stress agents such as UV, which signal by inducing translational inhibition. However, the two primary degradation pathways that control IκB homeostasis critically tune the cellular responsiveness of the IκB-NF-κB signaling module, and do so with distinct effects.
Homeostatic versus signal induced regulatory mechanisms
Inflammatory signaling to NF-κB induces rapid degradation of NF-κB-bound IκB α, but does not induce the degradation of free IκBα (O’Dea et al., 2007; Mathes et al., 2008). In contrast, we find that UV irradiation causes the depletion of both free and NF-κB-bound pools of IκBα. By separately examining the NF-κB-bound and -unbound pools of IκBα we were able to clarify the previous confusion surrounding the role of IKK activity in UV-induced NF-κB, and further, determine a dual requirement of different IκB degradation pathways in UV-induced NF-κB. Previous studies that used a transiently expressed IκBαAA transgene in HeLa cells harboring NF-κB and endogenous wild-type IκBα (Bender et al., 1998; Li and Karin, 1998) were likely examining an excess of IκBα. Thus they observed UV-induced depletion of IκBα in an IKK-independent manner, just as we showed that the IκBαAA transgene in NF-κB deficient cells is degraded rapidly in response to UV. However, stably expressed IκBαAA in iκbα −/− cells showed no degradation in response to UV, indicating that the NF-κB-bound pool of IκBα absolutely requires basal IKK activity for its degradation. Indeed, this requirement is recapitulated in computational simulations, and we observe that mutations in IKK that either decrease or enhance IKK activity will either reduce or enhance UV-induced NF-κB activation (Figure 3).
Although it is evident from the present work that increasing the constitutive free IκBα degradation rate enhances NF-κB activation, it is unclear if it is increased by UV irradiation. UV-induced CK2 activity was shown to target IκBα for phosphorylation (Kato et al., 2003), but it remains unclear whether that event accelerates the degradation rate. Although CK2 phosphorylation site mutations have a modestly stabilizing effect in unstimulated cells, we were unable to detect alterations in free IκBα half-life upon UV exposure and found no effect on NF-κB activation by UV in MEFs with p38 or CK2 inhibitors (data not shown). Further, our computational simulations and experiments with low doses of CHX suggest that translational inhibition is a sufficient inducing signal to account for NF-κB activation and therefore that any contribution of UV-induced CK2 activity to IκB degradation may be less important than the constitutive degradation pathways of the NF-κB-bound and free IκB protein pools.
Physiological versus pathological relevance of NF-κB activation by UV
We find that homeostatic control of the two IκB degradation pathways regulate the sensitivity of NF-κB induction by UV irradiation. The IKK-dependent pathway regulates both the sensitivity of signaling and the maximal NF-κB activation achievable by ribotoxic stresses, whereas the IKK-independent pathway controls the former (Figure 6). As many cancerous cells have elevated IKK activity and/or elevated proteasome activities (Dahlmann, 2007), these cells have an altered homeostasis in which one or the other IκB degradation pathway is constitutively elevated. These alterations may account for the robust NF-κB activation by UV observed in previous studies in a variety of cancer cells.
In cancer cells, the functional role of metabolic stress-induced NF-κB has been thought of as being anti-apoptotic. Indeed, NF-κB inhibition potentiates anti-cancer radiation therapy. However, in primary tissue cells such as MEFs, we have not observed that NF-κB/RelA deficiency results increased apoptosis in response to UV irradiation (data not shown), consistent with our current conclusion that in such primary cells UV irradiation may not significantly activate NF-κB. Further, a recent study suggested that constitutive NF-κB may in fact have a pro-apoptotic role in UV irradiation, by controlling the expression of PKCδ which is required for UV-induced JNK activation (Liu et al., 2006). Our study resolves this apparent discrepancy by distinguishing between healthy cells and disease-associated cells with altered IκB homeostasis.
We also considered chronic inflammatory disease settings. Low levels of inflammatory agents may not activate NF-κB much, but the altered homeostasis of IκB turnover sensitizes the cells to ribotoxic stress. UV may thus be thought of primarily as an amplifier of NF-κB activation in pathology associated cells rather than an inducer of NF-κB activity in healthy cells.
This study demonstrates the value of using computational modeling as a tool for experimental analysis in signaling studies. We thereby identified rate constants that may be unaltered by an activating signal, but are critical to signal transduction. Most therapeutic strategies are aimed at inhibiting the inducing signaling pathway. However, homeostatically controlled pathways may show differential sensitivity in the signal transduction of different stimuli. Canonical signaling in response to pathogen or cytokine signals may be less critically affected by the degradation pathways identified here as being critically important for UV. Similarly, to devise therapeutic regimens that target diseased cells specifically, we suggest that the altered homeostasis of degradation pathways in diseased versus healthy cells may be considered an important specificity factor.
Experimental Procedures
Cell lines, UV radiation, plasmids, and other reagents
Primary and 3T3 immortalized MEF were generated from E12.5-14.5 embryos and maintained as previously described (Hoffmann et al., 2002; O’Dea et al., 2007). ikkβ−/− cells were a gift from Inder Verma. Primary mouse keratinocytes were cultured as described (Rheinwald and Green, 1975). Recombinant murine TNF was from Roche, LPS was from Sigma-Aldrich (L4524), and 35S-TransLabel was from MP Biomedicals. UV irradiation was performed with a UVC lamp (UVP Inc., Upland, CA). For UV irradiation as well as for mock treatment, cell media were removed and cells washed with PBS. For “mock” treatment, cells were treated just as the UV treated cells, but were not exposed to the UV light. After treatment, the original cell media were replaced. RelA/p65 (sc-372), IκBα (sc-371), IKKα (sc-7184) antibodies were from Santa Cruz Biotechnology, IKKβ antibody was from BioSource (AHO0362), and IKKγ antibody was from BD Biosciences (624084).
Retrovirus Mediated Gene Transduction
Retroviral constructs derived from pBabe-puro or pBabe-hygro were co-transfected with pCL.Eco into 293T cells, and 42 hours post-transfection filtered supernatant was used to infect MEFs. Transduced cells were selected with Puromycin Hydrochloride (Sigma) or Hygromycin B (Invitrogen). The IKKβWT-, IKKβAA-, and IKKβKA-pBabe-hygro plasmids were a kind gift from David Shultz, and the Stratagene Quickchange Site-Directed Mutagenesis Kit was used to generate the IKKβEE-pBabe-hygro plasmid.
Immunoblot and Coimmunoprecipitation
Whole cell extracts were prepared in lysis buffer and total protein concentrations were normalized to each other using Lowry assay (BioRad) before immunoblot analysis. For immunoprecipiation-western analysis, whole cell lysate was prepared in buffer containing 20% glycerol, 0.2 mM EDTA, 20 mM Tris pH 7.5, 120 mM KCl, and 0.5% NP-40, 1mM DTT, and 1 mM PMSF. For co-immunoprecipitation of p65-IκBα complexes, p65 was immunoprecipitated with 10 μg anti-p65 antibody followed by the addition of protein G sepharose beads (GE Healthcare). Beads were washed with lysis buffer and samples eluted by addition of SDS sample buffer. Proteins were resolved by SDS-PAGE and analyzed by immunoblot using ECL-plus (GE Healthcare). Band intensities were quantified using a phosphoimager (Molecular Dynamics) and ImageQuant software version 5.2 (GE Healthcare).
EMSA, IKK Kinase Assay, and RNase Protection Assay
Nuclear extracts were prepared and used for EMSA as described (Werner et al., 2005). Briefly, nuclear extract was reacted at room temperature to a 32P-labeled 38-bp double-stranded oligonucleotide containing two consensus κB-sites. Complexes were resolved on a non-denaturing acrylamide gel and visualized by autoradiography. IKK kinase assays were performed as described (Werner et al., 2005). Briefly, cytoplasmic extracts were collected and the IKK complex was immunoprecipitated with anti-IKKγ antibody, followed by addition of protein G sepharose beads. Beads were washed and subject to in vitro kinase assay containing ATPγ32, GST-IκBα(1–54). The reaction was stopped with SDS-sample buffer, proteins were resolved by SDS-PAGE and visualized by autoradiography or immunoblot. Total cellular RNA was isolated using Trizol reagent (Invitrogen), and used for RNase Protection Assay as described (Werner et al., 2005) using a RiboQuant probe set (BD Biosciences).
Metabolic labeling
Cells were either mock treated or treated with 40 J/m2 UVC. 30 minutes prior to each time point, 100μCi/mL 35S-TransLabel was added to complete cell media for a 30 minute pulse, then whole cell protein extracts were prepared. Protein concentrations for each sample were normalized to each other via Lowry assay, then spotted and dried on Whatman filter paper, and placed in scintillation fluid for counting. A sample treated with 50 μg/mL cycloheximide was treated in the same manner and included with the sample analysis to use as a background control in the scintillation counts. Experiments were performed in triplicate. To quantify the percent of translational inhibition, the scintillation count for UV-treated samples was divided by the count for mock-treated samples. This method provided for similar results as previously published (Wu et al., 2002). Measurements were confirmed by running the samples on SDS-PAGE followed by autoradiography.
Computational Modeling
A modified version (version 2.1) of a mathematical model of the IKK-IκB-NF-κB signaling module (D. Barken and A. H., unpublished results), based on the previously described version 2.0 (Werner et al., 2005), was used for all simulations. Simulations were done in Matlab version 2007a (Mathworks) using the built-in ode15s solver at default settings. To alter the free IκBα turnover rates, the rate constants governing the free IκBα degradation parameters were multiplied by the indicated variable in the equilibrium phase and the simulation phase (e.g. for 0.5x, the degradation rate was multiplied by 0.5). To alter the basal IKK activity level, the IKK activity level was multiplied by the indicated variable in the equilibrium phase and the simulation phase. To simulate the low LPS-induced IKK activity (Figure 7A), the IKK kinase assay from low LPS treated cells (Supplemental Figure 6A) was quantified and used as the IKK activity input. Simulations of translational inhibition are described in the supplement (Figure S2). Simulations involving the “slow flux” mutant are described in the supplement (Figure S4).
Supplementary Material
Acknowledgments
We thank T. Kato, M. Karin and G. Ghosh for insightful discussion, C. Lynch for excellent technical assistance, E. Mathes for IκBαAA and IκBαWT expressing iκbα −/− and nfκb−/− cells, P. Lee and C. Jamora for keratinocytes, and Santa Cruz Biotechnology for antibodies used in this study. E.L.O. is supported by the Heme Training Grant, and J.D.K by the Molecular Biophysics Training Grant (NIH GM08326-18). This study was supported by NIH grants GM72024, GM69013 and GM071573.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O’Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender K, Gottlicher M, Whiteside S, Rahmsdorf HJ, Herrlich P. Sequential DNA damage-independent and -dependent activation of NF-kappaB by UV. Embo J. 1998;17:5170–5181. doi: 10.1093/emboj/17.17.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bren GD, Pennington KN, Paya CV. PKC-zeta-associated CK2 participates in the turnover of free IkappaBalpha. J Mol Biol. 2000;297:1245–1258. doi: 10.1006/jmbi.2000.3630. [DOI] [PubMed] [Google Scholar]
- Dahlmann B. Role of proteasomes in disease. BMC Biochem . 2007;8(Suppl 1):S3. doi: 10.1186/1471-2091-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, Ron D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol. 2004;24:10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devary Y, Rosette C, DiDonato JA, Karin M. NF-kappa B activation by ultraviolet light not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Huang TT, Feinberg SL, Suryanarayanan S, Miyamoto S. The zinc finger domain of NEMO is selectively required for NF-kappa B activation by UV radiation and topoisomerase inhibitors. Mol Cell Biol. 2002;22:5813–5825. doi: 10.1128/MCB.22.16.5813-5825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, Wek RC. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Jr, Delhase M, Hoffmann A, Karin M. CK2 Is a C-Terminal IkappaB Kinase Responsible for NF-kappaB Activation during the UV Response. Mol Cell. 2003;12:829–839. doi: 10.1016/s1097-2765(03)00358-7. [DOI] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci U S A. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang D, Minemoto Y, Leitges M, Rosner MR, Lin A. NF-kappaB is required for UV-induced JNK activation via induction of PKCdelta. Mol Cell. 2006;21:467–480. doi: 10.1016/j.molcel.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Mathes E, O’Dea EL, Hoffmann A, Ghosh G. NF-kB dictates IkB degradation modes. Embo J. 2008 doi: 10.1038/emboj.2008.73. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, Hoffmann A. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. Formation of a keratinizing epithelium in culture by a cloned cell line derived from a teratoma. Cell. 1975;6:317–330. doi: 10.1016/0092-8674(75)90183-x. [DOI] [PubMed] [Google Scholar]
- Rice NR, Ernst MK. In vivo control of NF-kappa B activation by I kappa B alpha. Embo J. 1993;12:4685–4695. doi: 10.1002/j.1460-2075.1993.tb06157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EM, Van Antwerp D, Verma IM. Constitutive phosphorylation of IkappaBalpha by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IkappaBalpha. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Simon MM, Aragane Y, Schwarz A, Luger TA, Schwarz T. UVB light induces nuclear factor kappa B (NF kappa B) activity independently from chromosomal DNA damage in cell-free cytosolic extracts. J Invest Dermatol. 1994;102:422–427. doi: 10.1111/1523-1747.ep12372194. [DOI] [PubMed] [Google Scholar]
- Stein B, Rahmsdorf HJ, Steffen A, Litfin M, Herrlich P. UV-induced DNA damage is an intermediate step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol Cell Biol. 1989;9:5169–5181. doi: 10.1128/mcb.9.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Beucken T, Koritzinsky M, Wouters BG. Translational control of gene expression during hypoxia. Cancer Biol Ther. 2006;5:749–755. doi: 10.4161/cbt.5.7.2972. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- Wu S, Tan M, Hu Y, Wang JL, Scheuner D, Kaufman RJ. Ultraviolet light activates NFkappaB through translational inhibition of IkappaBalpha synthesis. J Biol Chem. 2004;279:34898–34902. doi: 10.1074/jbc.M405616200. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.