Abstract
The structure of aquaporin-0 (AQP0) has recently been determined by electron crystallography of two-dimensional (2D) crystals and by x-ray crystallography of three-dimensional (3D) crystals. The electron crystallographic structure revealed nine lipids per AQP0 monomer, which form an almost complete bilayer. The lipids adopt a wide variety of conformations and tightly fill the space between adjacent AQP0 tetramers. The conformations of the lipid acyl chains appear to be determined not only by the protein surface but also by the acyl chains of adjacent lipid molecules. In the x-ray structure, the hydrophobic region of the protein is surrounded by a detergent micelle, with two ordered detergent molecules per AQP0 monomer. Despite the different environments, the electron crystallographic and x-ray structures of AQP0 are virtually identical, but they differ in the temperature factors of the atoms that either contact the lipids in the 2D crystals or are exposed to detergents in the 3D crystals. The temperature factors are higher in the x-ray structure, suggesting that the detergent-exposed AQP0 residues are less ordered than the corresponding ones contacting lipids in the 2D crystals. An examination of ordered detergent molecules in crystal structures of other aquaporins and of lipid molecules in 2D and 3D crystals of bacteriorhodopsin suggests that the increased conformational variability of detergent-exposed residues compared to lipid-contacting residues is a general feature.
Keywords: Membrane protein, aquaporin-0, lipid-protein interactions, electron crystallography, x-ray crystallography
Introduction
Biological membranes, which separate the interior of cells from their environment, are composed of lipids and membrane proteins. The membrane proteins perform vital, more complex biological functions such as the controlled exchange of solutes, energy conversion and transmembrane signaling. The lipids form a bilayer structure, which anchors the membrane proteins in a fluid matrix, allowing both lipids and membrane proteins to move by lateral diffusion. At the same time the lipids maintain a tight seal around the membrane proteins so that proton and chemical gradients are maintained between the interior and exterior of the cell, even as membrane proteins change their shape while undergoing activity-related conformational changes. In addition, lipids also serve signaling functions [1], play a role in the correct folding of membrane proteins [2], and in some cases influence the function of membrane proteins [2].
Most structural studies of membrane processes focus on the membrane proteins, while structures of lipids are usually determined separately. Structural information on the interaction between membrane proteins and lipids is therefore difficult to obtain, and our understanding is largely based on the few crystal structures of membrane proteins with bound lipids. Most membrane protein structures have been determined by x-ray crystallography of three-dimensional (3D) crystals. These crystals are typically produced with detergent-solubilized membrane proteins, but can also be obtained in lipidic cubic phase (LCP) [3] or by inserting delipidated, detergent-solubilized proteins into perforated lipid-detergent bicelles [4]. Ordered lipids have been identified in crystal structures of several membrane proteins. These lipids have typically originated from the native membrane and have co-purified with the protein. Analysis of such tightly bound lipids has provided insight into the specific binding of lipids to membrane proteins [5].
Two-dimensional (2D) crystals, in which the membrane protein is reconstituted in an artificial lipid bilayer and which can be analyzed by electron crystallography, are an alternative way to determine membrane protein structures (reviewed in [6]). Unlike in 3D crystals of membrane proteins, which only occasionally contain lipid molecules, in 2D crystals, lipids form a continuous bilayer surrounding the membrane proteins, which could be visualized in two electron crystallographic structures determined to high resolution, namely those of bacteriorhodopsin (bR) [7] and aquaporin-0 (AQP0) [8]. The bR structure was determined using fused purple membranes, which are naturally occurring 2D arrays of bR. The lipids visualized in the bR structure are thus naturally associated with bR, and they are indeed important for its biological activity [9]. By contrast, the AQP0 structure was determined to a resolution of 1.9 Å using 2D crystals produced by reconstituting delipidated protein with the synthetically made lipid dimyrostylphosphatidylcholine (DMPC). Since the double-layered 2D crystals have the same dimensions as AQP0-mediated membrane junctions between fiber cells in the lens [10], the synthetic DMPC lipid is likely to mimic the native lipids in the in vivo junctions [8]. The lipids in the 2D crystals form an almost complete bilayer and are sandwiched in between adjacent AQP0 tetramers. Although most of the lipids are somewhat unusual in that they contact two membrane proteins rather than just one, the electron crystallographic AQP0 structure is currently the only available case that allows us to analyze non-specific lipid-protein interactions. Furthermore, the structure of AQP0 has also been determined by x-ray crystallography of 3D crystals produced with detergent-solubilized tetramers [11], enabling us to examine if detergents compensate for the lack of lipids and/or if they affect protein stability.
We provide here a detailed analysis of the non-specific lipid-protein interactions observed in the AQP0 2D crystals and compare them with specific lipid-protein interactions observed in 3D crystals of other membrane proteins. We also compare the structure of AQP0 in a lipid bilayer and a detergent micelle and extend our observations to other membrane proteins in detergent micelles or lipids.
The lipids in the double-layered AQP0 2D crystals
The structure of AQP0 determined by electron crystallography resolved in the asymmetric unit nine DMPC molecules, the lipid used for 2D crystallization [8]. These lipids form a nearly complete bilayer, one molecule wide, that separates neighboring AQP0 tetramers in the 2D array. Of the nine lipids, seven form direct contacts with the protein, thus representing a shell of annular lipids. The two remaining bulk lipids that are not in contact with the protein fill in the space at the four-fold axis between the tetramers. The modeled lipids do not completely fill the space between the tetramers, suggesting that there is probably another lipid molecule that was not detected in the density map.
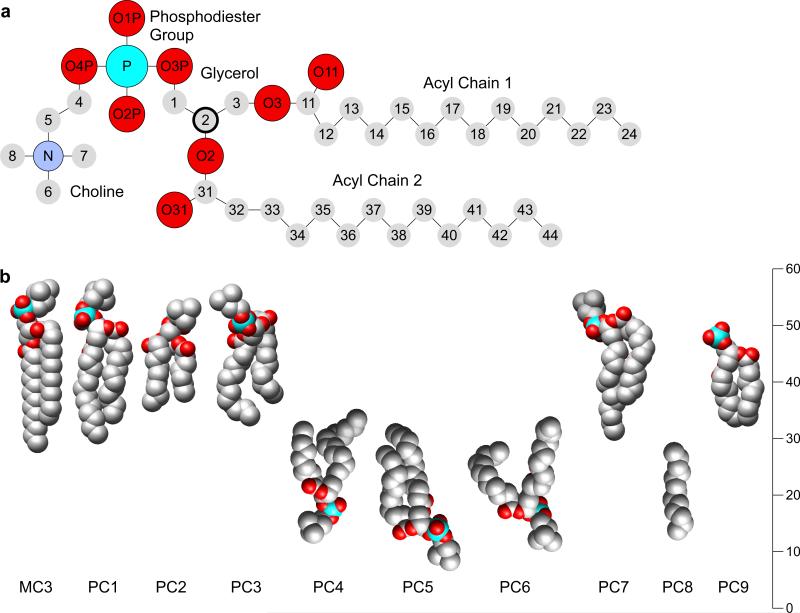
A schematic diagram of DMPC is shown in Figure 1a and annotated following the nomenclature used in the pdb file of AQP0 (PDB 2B6O) [8]. The synthetic DMPC has two saturated 14-carbon acyl chains and a phosphatidylcholine (PC) head group, linked through a glycerol backbone. It is a racemate with a single chiral center at the C2 glycerol carbon. The crystal structure of DMPC has been determined in a low hydration state (Fig. 1b, MC3) [12], in which the acyl chains adopt all-trans conformations. The tightly packed head groups in these crystals extend away from the bilayer. Figure 1b shows the models of the nine lipids found in the AQP0 2D crystals (PC1-PC9), which are placed in their correct axial positions in the membrane but are rotated to allow comparison with the DMPC crystal structure (MC3). The lipids in the AQP0 2D crystals adopt very different conformations from the extended one seen in the DMPC crystal structure. Indeed, studies measuring the positions of each of the domains in fully hydrated lipid bilayers suggest that lipids in the fluid phase are quite flexible [13]. Furthermore, the average phosphate-to-phosphate distance between DMPC molecules in the two leaflets in the bilayer in the AQP0 crystal structure is 34 Å, close to that measured for a DMPC bilayer in the gel phase (35.5 Å) [13] and substantially smaller than its 46.5 Å distance in the crystal.
Figure 1.
The structure of dimyristoylphosphatidylcholine (DMPC). (a) Schematic diagram of DMPC with atoms annotated following the nomenclature used in the PBD file of ovine junctional AQP0 (PDB 2B6O) [8]. The chiral C2 carbon of the glycerol backbone is shown bold. (b) Sphere models of the atomic structure of the B form of DMPC (MC3) [9] and the nine resolved lipids in the AQP0 structure. The lipids are positioned at their appropriate positions within the membrane but rotated to allow a comparison with the crystal structure.
Most of the lipids in the AQP0 structure are in a previously unseen environment, sandwiched between two proteins. Each DMPC molecule in the 2D crystal adopts a different conformation, presumably because of the unique local environment for each lipid. Protein surfaces contain many depressions and protrusions to which the annular lipids must adapt to ensure that the bilayer forms a tight seal. The acyl chains thus twist and bend to maximize van der Waals contacts with the protein surface, while also forming additional van der Waals contacts with adjacent lipids. PC1, PC3 and PC6 illustrate the wide variety of conformations adopted by the DMPC molecules in the AQP0 2D crystals (Fig. 1b). The acyl chains of PC1 are nearly parallel and fully extended, as in the DMPC crystal structure. Both acyl chains of PC3 are bent such that the last three methylene groups extend almost parallel to the membrane plane. The acyl chains of PC6 are also bent but splayed as well, forming a wide “V” shape. Previous structures of lipids bound to membrane proteins have shown a similar diversity of conformations to match the surface of the proteins with which they were associated [14].
Temperature factors of the lipids
While protein regions adopt rigid secondary structures, lipids are more flexible, resulting in weaker density in the experimental maps. In some cases, lipid density in the AQP0 crystal was too weak for model building, as in the case of the acyl-chain carbons beyond C17 and C40 of PC2 and beyond C40 of PC4. For the majority of the annular lipids, however, the density was sufficient to model the entire molecule, and the disorder or spread of their atoms is described by their temperature factors. The average temperature factor of AQP0 residues 5 − 224 is 40.8 Å2, whereas the average temperature factor is 102.9 Å2 for the annular lipids and 154.9 Å2 for the two bulk lipids. Higher temperature factors have also been reported before for lipids seen in x-ray structures [15]. The temperature factors of atoms in the annular lipids vary from a minimum of 45.9 Å2 for C24 of PC1 to a maximum of 145.9 Å2 for C7 of PC4, indicating that the local environment may not only influence the conformations of the acyl chains but also their local mobility or disorder.
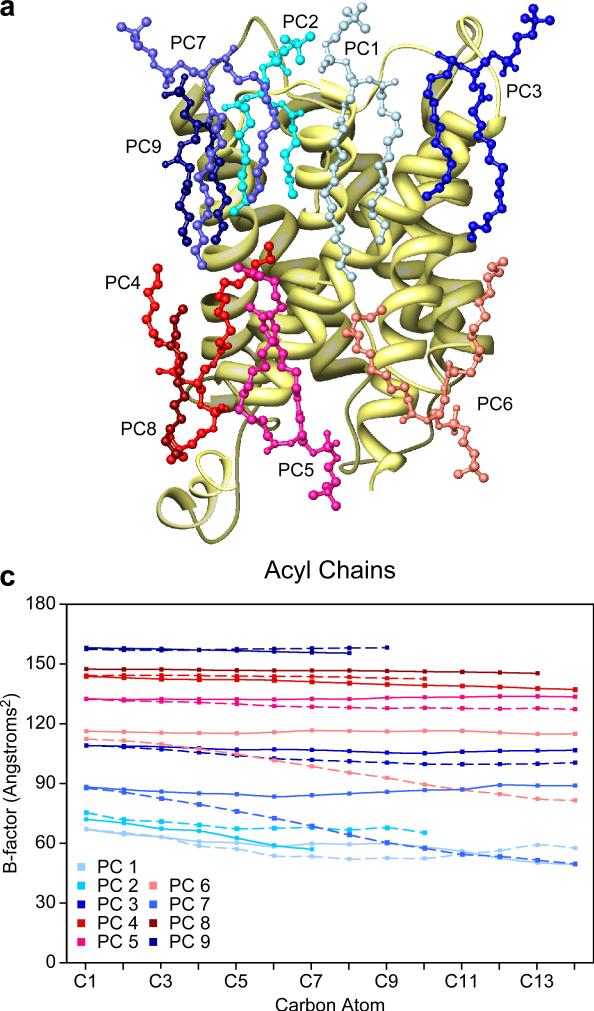
Figure 2b is a plot of the positions of the carbons in the acyl chains against their temperature factors. For some acyl chains, the temperature factors of their atoms vary considerably; in five chains the temperature factors differ by more than 15 Å2. The atoms in other chains have relatively constant temperature factors; in three chains the difference is less than 2 Å2. Overall, the temperature factors of the acyl chains tend to decrease as the distance from the head group increases, indicating that the positions of the acyl chains are more constrained in the center of the bilayer. Despite this trend, density for the ends of the acyl chains of PC2 and PC4 was missing in the map, suggesting that these particular parts of the acyl chains have few constraints on their conformations or several alternative, constrained conformations.
Figure 2.
Lipids in the AQP0 2D crystals. (a) Ribbon diagram of an AQP0 monomer with ball-and-stick models for each of the nine lipids. Lipids in the cytoplasmic leaflet are depicted in shades of red, while those in the extracellular leaflet in shades of blue. (b) The temperature factors of every atom within each of the lipid acyl chains are plotted against their position in the chain, showing that the temperature factors tend to become smaller towards the end of the chains, which are located in the center of the lipid bilayer.
The acyl chains must fill in the gaps between the membrane-embedded surfaces of the AQP0 tetramers. They interact principally with aliphatic (44.8%) and aromatic residues (37.1%) of AQP0 (Table 2). Van der Waals interactions between pairs of nonpolar atoms are non-specific, with a strength that can be estimated from the area of the interaction surface. Thus, it would be expected that variations in the temperature factors of atoms in the acyl chains would correlate with the number of contacts with the protein surface. However, when the temperature factor trends described above were compared with the number of contacts per acyl chain, the average distance to the nearest protein atom, the number of direct contacts (<4.0 Å) or the hydrophobicity of contacting residues, no correlations could be identified. In addition, no correlations could be identified between any of these metrics and the temperature factors of the individual atoms. It thus appears that the conformation and mobility of the acyl chains of annular lipids are determined not only by their interaction with the protein but also to an appreciable extent by their interactions with the acyl chains of neighboring lipid molecules.
Table 2.
Interactions of AQP0 residues with the acyl chains, glycerol backbones and phosphatidyl head groups of the lipid molecules.
| Total | Acyl Chain | Glycerol | PC Head Group | |||||
|---|---|---|---|---|---|---|---|---|
| Residue | Number | Contacts | Number | Contacts | Number | Contacts | Number | Contacts |
| ALA | 4 (6.6%) | 56 (4.5%) | 3 (5.8%) | 46 (4.7%) | 0 (0.0%) | 0 (0.0%) | 1 (4.0%) | 10 (4.5%) |
| ARG | 4 (6.6%) | 112 (9.0%) | 1 (1.9%) | 36 (3.6%) | 2 (14.3%) | 18 (27.7%) | 4 (16.0%) | 66 (29.7%) |
| ASN | 1 (1.6%) | 7 (0.6%) | 1 (1.9%) | 4 (0.4%) | 0 (0.0%) | 0 (0.0%) | 1 (4.0%) | 3 (1.4%) |
| CYS | 1 (1.6%) | 10 (0.8%) | 1 (1.9%) | 10 (1.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| GLN | 1 (1.6%) | 34 (2.7%) | 1 (1.9%) | 17 (1.7%) | 0 (0.0%) | 0 (0.0%) | 1 (4.0%) | 17 (7.7%) |
| GLY | 2 (3.3%) | 25 (2.0%) | 1 (1.9%) | 22 (2.2%) | 0 (0.0%) | 0 (0.0%) | 1 (4.0%) | 3 (1.4%) |
| HIS | 1 (1.6%) | 22 (1.8%) | 1 (1.9%) | 22 (2.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ILE | 5 (8.2%) | 106 (8.5%) | 5 (8.9%) | 106 (10.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| LEU | 11 (18.0%) | 216 (17.4%) | 10 (49.2%) | 172 (17.4%) | 4 (28.6%) | 12 (18.5%) | 5 (20.0%) | 36 (16.2%) |
| PHE | 9 (14.8%) | 185 (14.9%) | 9 (17.3%) | 155 (15.7%) | 3 (21.4%) | 16 (24.6%) | 2 (8.0%) | 23 (10.4%) |
| PRO | 2 (3.3%) | 9 (0.7%) | 1 (1.9%) | 1 (0.1%) | 0 (0.0%) | 0 (0.0%) | 2 (8.0%) | 8 (3.6%) |
| SER | 3 (4.9%) | 90 (7.3%) | 2 (3.6%) | 60 (6.1%) | 1 (7.1%) | 6 (9.2%) | 3 (12.0%) | 28 (12.6%) |
| THR | 4 (6.6%) | 24 (1.9%) | 3 (5.8%) | 12 (1.2%) | 0 (0.0%) | 0 (0.0%) | 3 (12.0%) | 12 (5.4%) |
| TRP | 3 (4.9%) | 176 (14.2%) | 3 (5.8%) | 160 (16.2%) | 2 (14.3%) | 6 (9.2%) | 0 (0.0%) | 0 (0.0%) |
| TYR | 1 (1.6%) | 59 (4.8%) | 1 (1.9%) | 57 (5.8%) | 1 (7.1%) | 5 (7.7%) | 1 (4.0%) | 15 (6.8%) |
| VAL | 9 (14.8%) | 109 (8.8%) | 9 (17.3%) | 107 (10.8%) | 1 (7.1%) | 2 (3.1%) | 1 (4.0%) | 1 (0.5%) |
| TOTAL | 61 | 1240 | 52 | 987 | 14 | 65 | 25 | 222 |
Lipid-protein interactions
In addition to the hydrophobic interactions formed by the acyl chains, lipids have polar and charged groups that can form stronger, more specific interactions with the protein. A recent study analyzed the binding specificity of the phosphodiester group using 30 unique, ordered lipids seen in x-ray structures [5]. These lipids had been co-purified with the protein, indicating that they are strongly and specifically bound. The phosphodiester group was chosen because it is frequently the best-ordered region of the lipid, indicating a tight binding interaction with the protein. A binding motif was identified, in which most phosphodiester groups were found to associate with two or more residues, of which one residue was positively charged and another one contained a polar group [5]. The positively charged residue was suggested to accept some of the electronegativity of the phosphodiester group [5]. Protein interactions with PC lipids were found to deviate from this proposed binding motif, and the authors suggested that the large, positively charged choline head group would cause steric and charge clashes [5].
The AQP0 structure contains seven DMPC molecules that contact the protein. Since DMPC is a synthetic lipid that does not occur in native membranes and was used to reconstitute fully delipidated AQP0, the lipid-protein contacts seen in the 2D crystals should represent non-specific interactions [8], which should be weaker than the specific interactions visualized previously in x-ray structures and hence less energetically favorable. The most common residue in the vicinity of the phosphodiester group is arginine, which is less than 4 Å away from four of the seven phosphodiester groups (Table 1). The remaining phosphodiester groups are located near a diverse array of residues, including several aliphatic and aromatic residues (Table 1), similar to those previously identified in interactions with PC head groups of specifically bound lipids [5].
Table 1.
Temperature factors of the phosphodiester groups of the DMPC molecules and AQP0 residues within 5 Å of the phosphodiester groups of the respective lipids.
| Lipid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 9 |
|---|---|---|---|---|---|---|---|---|
| Temperature Factor | 78.5 | 81.6 | 110.7 | 144.8 | 132.1 | 119.0 | 94.8 | 161.6 |
| Residues | R,T,L | R | Q,S,G,P,T,P,V | L,L | F,W,S | R,A | R | N/A |
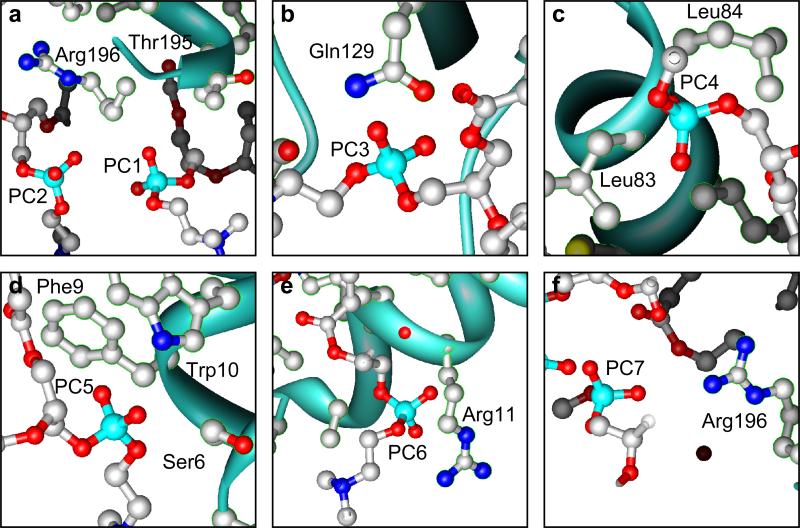
Figure 3 shows the local environments near the phosphodiester groups of the seven annular lipids. A closer examination of the phosphodiester groups suggests that some of them do not form favorable interactions with the protein. For example, the phosphate atoms of PC1 and PC2 are located only 5.1 Å from one another and the closest residue to both groups, arginine 196, is positioned such that the phosphodiester groups can only form interactions with the aliphatic carbons of its side chain rather than its positively charged amino groups. Interactions such as these are not expected to be energetically favorable, because of the strong electronegativity of the phosphodiester group. It is possible that the conformations modeled in the structure may represent the average of several different, possibly more favorable head group positions. Furthermore, because the substituted glycerol is a racemic mixture, there are two enantiomeric lipid structures, in which the head groups have different conformations and potentially different positions with respect to the acyl chains. Since the data did not suffice to refine both enantiomers for each lipid and since it was not obvious from the initial map whether a particular enantiomer was preferred for any of the lipid positions, one of the two lipid enantiomers was chosen arbitrarily during model building. The signal from the other enantiomer would thus appear as disorder during refinement, providing a possible reason for the weak densities of the phosphodiester groups in the map and their high temperature factors (Table 1). Density at C2 is also particularly weak.
Figure 3.
Interactions between the lipid phosphodiester groups of six annular lipids and AQP0. The phosphodiester groups are shown with the closest side chains of the AQP0 molecules in the 2D crystal.
To analyze further the lipid-protein interactions, we generated a list containing all pairs of lipid and protein atoms that are located less than 5 Å from one another (Table 2). The list contained 1,240 interactions between 308 protein atoms from 61 residues with 221 atoms from the 7 annular lipid molecules. The atom pairs were grouped according to whether the acyl chains, glycerol backbones or head groups of the lipids were involved in the interaction. The acyl chains make up 79.7% of the interactions with protein atoms and interact almost exclusively with hydrophobic residues (81.9%). These figures include those interactions made by the first three methylene groups of the acyl chains and the oxygens of the carboxyl and diester groups, which can also form polar interactions with the protein. The lipid head groups make many more interactions with atoms from polar (19.4%) and charged (29.7%) residues of the protein than the acyl chains. The glycerol has fewer interactions with the protein but appears to act as a barrier between the acyl chains that interact with hydrophobic protein residues and the head groups that interact with the more hydrophilic residues. The distribution of residues at the periphery of the protein is presumably the determining factor that causes the phosphate groups of the lipids to be positioned at very similar depths within the membrane despite their different conformations.
A list has previously been compiled of all residues within 5 Å of an ordered lipid or detergent molecule in x-ray structures of membrane proteins [14]. Despite the different methods used to generate the lists, the frequencies of protein residues involved in lipid interactions are quite similar in the two lists, indicating that most of the specific lipid-protein interactions seen in the 3D crystals are similar to the non-specific interactions seen in the AQP0 2D crystals. The list using the x-ray structures showed a correlation between the hydrophobicity of an amino acid and its frequency in the list. This correlation exists also for the AQP0 residues, because the vast majority of interactions involve the lipid acyl chains.
2D and 3D crystal structures of aquaporins
The structures of membrane proteins are most commonly determined in the presence of a detergent micelle, yet in the cell membrane proteins are embedded in a lipid bilayer. Only very rarely have structures been determined in both environments. Structures determined in both a detergent micelle and a lipid bilayer are often very similar (e.g., AQP1 [16, 17], photosynthetic reaction center [18,19]). The structures of EmrE, a bacterial multidrug transporter, determined in the two environments differed considerably, however, with the structure obtained in the lipid bilayer resembling the native state of the protein more closely (reviewed in [20]). The structure of AQP0 has been determined in both a detergent micelle (PDB 1YMG) [11] and a lipid bilayer [8], at the relatively high resolutions of 2.2 Å and 1.9 Å, respectively. Although the electron crystallographic structure was of junctional, ovine AQP0 and the x-ray structure of non-junctional, bovine AQP0, differences between the two structures are remarkably small, with a main chain root mean square deviation (rmsd) of 0.61 Å and a side chain rmsd of 1.18 Å. This high degree of similarity allows us to make direct comparisons between the two structures on an atom-by-atom basis.
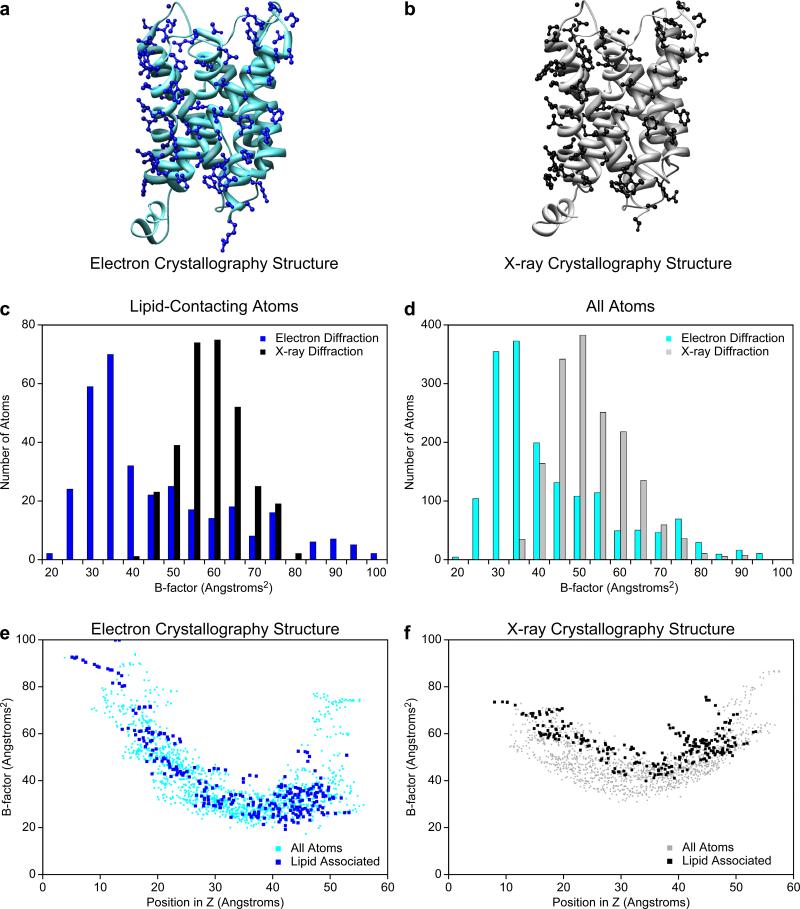
To examine the local effects of the lipids on the peripheral protein residues they contact, the atoms in the electron crystallographic structure that were within 5 Å of a lipid molecule (Fig. 4a) were compared to the corresponding atoms in the x-ray structure (Fig. 4b). Since there were no lipids present in the x-ray structure, it must be assumed that these atoms were instead exposed to the β-nonylglucoside detergent micelle. There were no gross conformational differences between the residues containing these atoms, which have a main chain rmsd of 0.51 Å and a side chain rmsd of 1.32 Å. To compare the temperature factors of the lipid-contacting or detergent-exposed atoms in the two structures (Fig. 4c), it was necessary to correct for the overall higher temperature factors in the x-ray structure compared to the electron crystallographic structure (Fig. 4d). To isolate lipid-dependent effects from the effects of different experimental parameters, we compared the lipid-contacting and detergent-exposed atoms to all the other atoms in the respective structures. A plot of the temperature factors of the lipid-contacting atoms and all the other atoms in the electron crystallographic structure is shown in Figure 4e. A corresponding plot for the x-ray structure with the detergent-exposed atoms is shown in Figure 4f. The temperature factors of the lipid-contacting atoms in the electron crystallographic structure are similar to those of the other atoms at the same z position (axial position across the bilayer) (Fig. 4e). By contrast, the temperature factors of the detergent-exposed atoms in the x-ray structure are consistently among the highest compared to the other atoms at the same z position (Fig. 4f).
Figure 4.
Effect of lipids and detergents on the AQP0 residues they contact. (a) Ribbon diagram of the AQP0 structure determined by electron crystallography. Residues that contain an atom located within 5 Å of a lipid atom are shown in ball-and-stick representations. (b) Ribbon diagram of the AQP0 structure determined by x-ray crystallography. The same residues as in (a) are shown in ball-and-stick representations. (c) Histograms of the temperature factors for the lipid-contacting atoms in the electron crystallographic structure of AQP0 (blue bars) and the x-ray structure (black). (d) Histograms of the temperature factors of all atoms in the electron crystallographic structure of AQP0 (cyan) and the x-ray structure (grey). (e) Plot of the temperature factors of all the atoms in the electron crystallographic structure of AQP0 against their position in the direction (z) normal to the membrane plane. The temperature factors of the lipid-contacting atoms are shown in blue and those of the other atoms in cyan. (f) Same plot as in (e) for the x-ray structure. The temperature factors of the detergent-exposed atoms are shown in black and those of the other atoms in grey.
The bias towards higher temperature factors of the detergent-exposed atoms in the x-ray structure indicates that the detergent molecules do not restrict their conformational variability as much as the lipid molecules do. A possible explanation for this observation is that the contacts in the 3D crystal are mainly formed by the soluble, extramembranous loops, while the residues contacting the lipids in the 2D crystals are an integral part of the crystal contacts. An alternative explanation would be lateral pressure from the lipids that could restrict the conformational flexibility of the entire transmembrane domain [2].
The conformational flexibility of the detergent-exposed atoms in the x-ray structure may also be the result of disorder in the detergent micelle that surrounds AQP0 tetramers in the 3D crystal. We therefore examined the temperature factors of the AQP0 atoms that contact the two ordered nonylglucoside molecules in the x-ray structure (Fig. 5a). More than half of the atoms that contact the two detergent molecules also contacted lipid molecules in the electron crystallographic structure. The temperature factors of the detergent-contacting atoms were again among the highest when compared to all the atoms at the same z positions in the structure (Fig. 5a). Thus, it appears that the disorder of the detergent micelle was not directly responsible for the increased temperature factors of the detergent-exposed atoms in the x-ray structure, as even the AQP0 atoms contacting ordered detergent molecules had higher temperature factors than when they were in contact with a lipid molecule.
Figure 5.
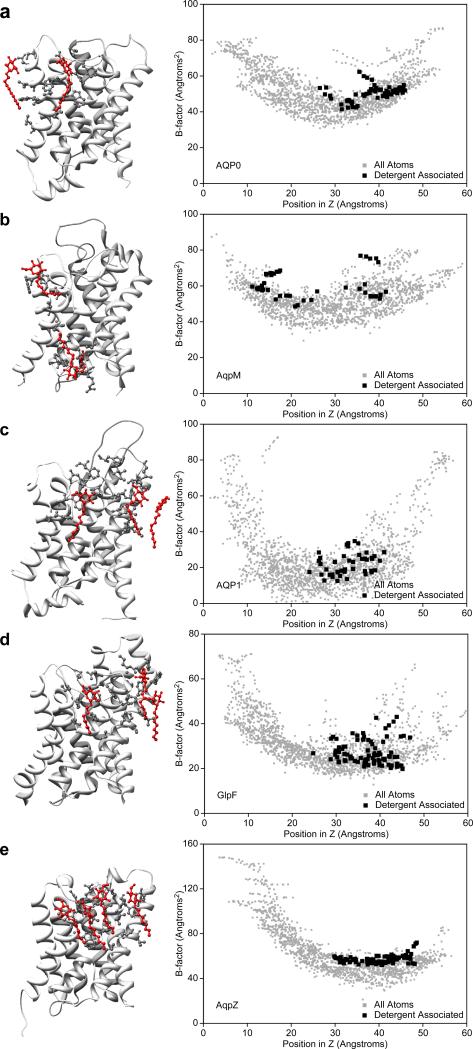
Detergent-lipid interactions in different aquaporin crystal structures. (a) Bovine AQP0, (b) AqpM from Methanobacterium thermoautotrophicum, (c) bovine AQP1, (d) GlpF from Escherichia coli, and (e) AqpZ from Escherichia coli. The left panels show the crystal structures as ribbon diagrams and the residues within 5 Å of an ordered detergent molecule (red) as ball-and-stick representations. The right panels show plots of the temperature factors of all the atoms in the structures against their position in the direction normal to the membrane plane. The temperature factors of the detergent-contacting atoms are shown in black and those of the other atoms in grey.
To corroborate the notion that detergents have little ordering effect on the protein residues they contact, we analyzed the x-ray structures of other members of the aquaporin family. None of the crystal structures revealed bound lipid molecules, indicating that none of the aquaporins of known structure have specific lipid binding sites, but six of the structures contained ordered detergent molecules. In addition to the crystal structure of AQP0 (Fig. 5a), we examined those of archaeal AqpM from Methanobacterium thermoautotrophicum (PDB 2EVU) [21] (Fig. 5b), bovine AQP1 (PDB 1J4N) [17] (Fig. 5c), and from Escherichia coli the glycerol facilitator GlpF (PDB 1FX8) [22] (Fig. 5d) and water channel AqpZ (PDB 1RC2) [23] (Fig. 5e). In all plots of the temperature factors against the z positions of the atoms, the temperature factors of the detergent-contacting atoms were among the highest (Fig. 5, left panels), corroborating the notion that ordered detergent molecules do not stabilize the protein atoms they contact. The temperature factors of the detergent-contacting atoms in the GlpF structure are least biased towards higher values (Fig. 5d). The hydrocarbon chains of the detergent molecules in the GlpF structure appear to be located in grooves between hydrophobic residues in a similar manner to the lipids in the electron crystallographic structure of AQP0, thus probably more closely mimicking the effect of lipids.
Protein-lipid interactions in bacteriorhodopsin
Bacteriorhodopsin (bR) is the only other membrane protein that has been visualized with an almost complete shell of annular lipids. Its structure was the first one determined by electron crystallography [24]. The most recent refinement yielded a 3 Å structure that contained eight modeled lipids (PDB 2AT9) [7]. 3D crystals of the purple membrane have also been obtained, the best of which diffracted to 1.55 Å using the lipidic cubic phase (LCP) technique (PDB 1C3W) [25]. Resolved in the structure were 18 phytanyl chains, but the head groups were largely missing and were thus not modeled. Another bR structure in which lipids were resolved was obtained with 3D crystals of bR in synthetic detergent-phospholipid bicelles (PDB 1XJI) [26]. These structures provide another opportunity to analyze the effects of lipids on the protein residues they contact (Fig. 6, top row).
Figure 6.
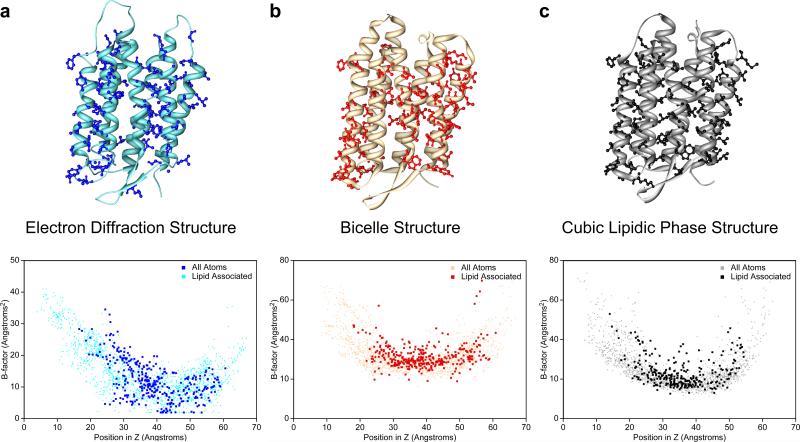
Protein-lipid interactions in bacteriorhodopsin (bR) structures. bR structures obtained with (a) 2D crystals, (b) 3D crystals produced from fused bicelles, and (c) 3D crystals obtained in lipidic cubic phase (LCP). The top panels show ribbon diagrams of bR. The residues containing an atom located within 5 Å of a lipid molecule in the LCP structure are shown in ball-and-stick representations. The lower panels show plots of the temperature factors of all the atoms in the bR structures against their position in the direction normal to the membrane plane. The temperature factors of the lipid-contacting atoms are shown in dark shades and those of the other atoms in light shades.
The LCP structure contained the most complete bilayer composed of purple membrane lipids and was used to generate a list of lipid-contacting bR atoms, which was then also used for the other two structures. Since the head groups of the lipids were not resolved, interactions of bR with the lipid head groups could not be analyzed. The list still included 380 of the 1720 resolved protein atoms, illustrating the high extent of lipid-protein interactions in purple membranes. In each of the three structures, the temperature factors of the lipid-contacting atoms have values comparable to the other atoms at the same z position (Fig. 6, bottom row).
The bR structures obtained by LCP and electron crystallography contain the native purple membrane lipids, which are intimately linked to the function of the protein [9]. By contrast, the lipids in the bR structure obtained in bicelles are entirely non-native DMPC lipids. Analysis of the temperature factors of the atoms contacting the lipid molecules actually seen in the bicelle and electron crystallographic structures showed that the values of the temperature factors of these atoms were also comparable to the other atoms at the same z position (Suppl. Fig. 1). These results from the bR structures corroborate the conclusion derived from the AQP0 structures that, unlike detergents, lipid molecules, even if they are not native, restrict the mobility of the side chains that extend into the lipid bilayer.
Outlook
X-ray structures of membrane proteins containing bound lipids have provided some insight into specific interactions of lipids with membrane proteins. Non-specific interactions between lipids and membrane proteins are more difficult to study and are thus poorly characterized. The 2D crystals of AQP0 provide a unique opportunity to examine such non-specific interactions. Analysis of the DMPC molecules in the AQP0 2D crystals shows that the acyl chains tend to be slightly better ordered towards the center of the lipid bilayer. While the forces defining the conformations of the acyl chains of the annular lipids did not become evident from our analysis, it is clear that they are not only defined by the protein surface but also by the acyl chains of the neighboring lipids. By including more diffraction patterns in the data set, it may be possible in the future to model alternative lipid conformations and thus to probe the dynamics of the acyl chains. The head groups of the synthetic DMPC molecules in the AQP0 2D crystals are less well ordered than those of specifically bound lipids seen in x-ray structures of membrane proteins. It is currently unclear whether this weak interaction is a consequence of the bulky PC head group of DMPC or whether it is a characteristic of nonspecific lipid-protein interactions more generally. Additional structures containing lipids with different head groups will be needed to resolve this question.
Detergents have long been known to be an imperfect substitution for lipids. Our comparison of detergent-exposed protein residues with lipid-contacting residues indicates that not even ordered detergents have a stabilizing effect on the protein residues they contact, while even non-native lipids do have such an effect. Aquaporins do not undergo major conformational changes as part of their biological activity and are quite stable in detergent solutions. Members of other families of membrane proteins, especially membrane transporters that have different functional states, are not as stable. For these proteins, lipids may not only order the peripheral protein side chains that they contact, as in the case of aquaporins, but may be required to stabilize the entire transmembrane domain. If this were the case, any crystallization approach in the presence of lipids would be more appropriate for structure determination of such membrane proteins than conventional crystallization in a simple detergent solution.
Supplementary Material
Supplementary Figure 1. Lipids in bacteriorhodopsin (bR) structures. bR structures obtained with (a) 2D crystals and (b) 3D crystals produced from fused bicelles. The top panels show ribbon diagrams of bR. The lipids and the bR residues of the electron crystallographic structure containing an atom located within 5 Å of a lipid molecule are shown as red and blue ball-and-stick models, respectively. The lipid and lipid contacting residues of the bicelle structure are shown as black and red ball-and-stick models, respectively. The lower panels show plots of the temperature factors of all the atoms in the bR structures against their position in the direction normal to the membrane plane. The temperature factors of the lipid-contacting atoms are shown in dark shades and those of the other atoms in light shades.
Abbreviations
- AQP0
Aquaporin-0
- 2D
two-dimensional
- 3D
three-dimensional
- LCP
lipidic cubic phase
- bR
bacteriorhodopsin
- DMPC
dimyrostylphosphatidylcholine
- PC
phosphatidylcholine
References
- 1.Eyster KM. The membrane and lipids as integral participants in signal transduction: lipid signal transduction for the non-lipid biochemist. Adv Physiol Educ. 2007;31:5–16. doi: 10.1152/advan.00088.2006. [DOI] [PubMed] [Google Scholar]
- 2.Lee AG. How lipids affect the activities of integral membrane proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Rummel G, Hardmeyer A, Widmer C, Chiu ML, Nollert P, Locher KP, Pedruzzi II, Landau EM, Rosenbusch JP. Lipidic Cubic Phases: New Matrices for the Three-Dimensional Crystallization of Membrane Proteins. J Struct Biol. 1998;121:82–91. doi: 10.1006/jsbi.1997.3952. [DOI] [PubMed] [Google Scholar]
- 4.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 5.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Hite RK, Raunser S, Walz T. Revival of electron crystallography. Curr Opin Struct Biol. 2007 doi: 10.1016/j.sbi.2007.06.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J Mol Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 8.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann R, Sickinger HD, Oesterhelt D. Quantitative aspects of energy conversion in halobacteria. FEBS Lett. 1977;82:1–6. doi: 10.1016/0014-5793(77)80873-9. [DOI] [PubMed] [Google Scholar]
- 10.Buzhynskyy N, Hite RK, Walz T, Scheuring S. The supramolecular architecture of junctional microdomains in native lens membranes. EMBO Rep. 2007;8:51–55. doi: 10.1038/sj.embor.7400858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci U S A. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson RH, Pascher I. The molecular structure of lecithin dihydrate. Nature. 1979;281:499–501. doi: 10.1038/281499a0. [DOI] [PubMed] [Google Scholar]
- 13.Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiener MC. A Census of Ordered Lipids and Detergents in X-ray Crystal Structures of Integral Membrane Proteins. In: Tamm LK, editor. Protein-Lipid Interactions: From Membrane Domains to Cellular Networks. Wiley-VCH; Weinheim: 2005. [Google Scholar]
- 15.Marsh D, Pali T. The protein-lipid interface: perspectives from magnetic resonance and crystal structures. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1666:118–141. doi: 10.1016/j.bbamem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 17.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 18.Katona G, Andreasson U, Landau EM, Andreasson LE, Neutze R. Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35Å resolution. J Mol Biol. 2003;331:681–692. doi: 10.1016/s0022-2836(03)00751-4. [DOI] [PubMed] [Google Scholar]
- 19.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. X-ray structure analysis of a membrane protein complex. Electron density map at 3 Å resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 20.Tate CG. Comparison of three structures of the multidrug transporter EmrE. Curr Opin Struct Biol. 2006;16:457–464. doi: 10.1016/j.sbi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc Natl Acad Sci U S A. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 23.Savage DF, Egea PF, Robles-Colmenares Y, O'Connell JD, 3rd, Stroud RM. Architecture and selectivity in aquaporins: 2.5 Å X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 25.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 26.Faham S, Boulting GL, Massey EA, Yohannan S, Yang D, Bowie JU. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Lipids in bacteriorhodopsin (bR) structures. bR structures obtained with (a) 2D crystals and (b) 3D crystals produced from fused bicelles. The top panels show ribbon diagrams of bR. The lipids and the bR residues of the electron crystallographic structure containing an atom located within 5 Å of a lipid molecule are shown as red and blue ball-and-stick models, respectively. The lipid and lipid contacting residues of the bicelle structure are shown as black and red ball-and-stick models, respectively. The lower panels show plots of the temperature factors of all the atoms in the bR structures against their position in the direction normal to the membrane plane. The temperature factors of the lipid-contacting atoms are shown in dark shades and those of the other atoms in light shades.