Abstract
AIM: To investigate the role of FAT10 and mutant p53 in the pathogenesis, severity and prognosis of gastric cancer.
METHODS: FAT10, mutant p53 mRNA and protein levels were measured by reverse transcription (RT)-PCR and immunohistochemistry in gastric cancer tissue (n = 62), tumor-adjacent tissue (n = 62) and normal gastric tissue (n = 62). Relation of FAT10 and mutant p53 expression with clinicopathological features and clinical outcomes of gastric cancer patients were analyzed.
RESULTS: The FAT10, mutant p53 mRNA and protein levels were significantly higher in gastric cancer than in its adjacent and normal tissue. The FAT10 and mutant p53 levels in gastric cancer tissue were significantly correlated with lymph node metastasis and tumor, nodes, metastasis (TNM) staging. Moreover, the high FAT10 level was associated with the overall survival rate of patients. Multivariate Cox-proportional hazards model analysis showed that mRNA and protein levels of FAT10 and mutant p53, lymph node metastasis, distant metastasis and TNM stage were the independent prognostic factors for gastric cancer.
CONCLUSION: FAT10 may be involved in gastric carcinogenesis, and is a potential marker for the prognosis of gastric cancer patients. FAT10 and mutant p53 may play a common role in the carcinogenesis of gastric cancer.
Keywords: Gastric cancer, FAT10, Mutant p53, Prognosis
INTRODUCTION
FAT10, also known as diubiquitin, is a ubiquitin-like modifier (UBL) of the ubiquitin protein family, first discovered by Fan et al[1] in mapping HLA-F gene in 1996. It has been shown that FAT10 is expressed in mature B cells and dendritic cells[2]. It has been reported that FAT10 regulates cell-cycle and non-covalently binds to the human spindle assembly checkpoint protein (MAD2) that is responsible for the maintenance of spindle integrity during mitosis. Inhibition of MAD2 may lead to chromosomal instability, a common feature of tumorigenesis[3,4]. Lee et al[5] found that FAT10 is up-regulated in liver, uterine cervix, ovarian, rectal, pancreatic cancers and small intestinal adenocarcinoma, suggesting that FAT10 plays an important role in tumorigenesis.
P53 gene is located on the short arm of chromosome 17 and classified into wild p53 and mutant p53. P53 protein depletion or gene mutation has been detected in over 50% of all cancers. Under the regulation of upstream signals such as DNA damage, proto-oncogene activity, spindle damage and hypoxia, p53 is activated and functions as a modifier in the processes of cell apoptosis, cell cycle arrest, and DNA repair[6,7]. It was reported that damaged DNA enters into S stage, changes cell hereditary characteristics, and finally induces tumorigenesis when p53 is deleted or mutated[8]. There is evidence that the p53 mutation rate is higher in gastric cancer with atrophic gastritis than in that without atrophic gastritis[9]. Rugge et al[10] demonstrated that the p53 gene mutation rate is 9.7% in patients (less than 40-year old) with gastric cancer of intestinal type, and lower than in old people (40%-60%) and that the p53 mutation rate is lower in young (6.8%) than in old people (10%-25%) with gastric cancer of diffuse type. Moreover, Helicobacter pylori (H pylori) infection is an important risk factor for gastric tumorigenesis, whereas patients with gastric cancer and H pylori-related cytotoxin-associated gene (CagA) are often accompanied with p53 mutation[11], suggesting that p53 gene mutation also plays an important role in gastric tumorigenesis.
Gastric cancer, one of the most common malignant tumors, is a leading cause of cancer-related death worldwide. The mortality of male and female patients with gastric cancer is on the top of the list in China[12]. Although various genetic and molecular alterations have been found in gastric cancer that underly the malignant transformation of gastric mucosa during the multi-step process of carcinogenesis, the detailed mechanism underlying the development of gastric cancer still remains uncertain. It has recently proposed that FAT10 is a downstream target of p53, and dysregulation of FAT10 expression in p53-defective cells can contribute to carcinogenesis[13]. Therefore, it would be of importance if the function of both FAT10 and p53 and their correlation are investigated in human beings. Furthermore, although many researches are available on the structure and function of FAT10, and its inducing factors, little is known about the role of FAT10 in gastric tumorigenesis and its relation with mutant p53 and other gastric cancer biomarkers. In the present study, we analyzed the expression of FAT10 and mutant p53 in gastric cancer tissue and its adjacent tissue and normal gastric mucosa tissue, in an attempt to discover the potential role of FAT10 in the development of gastric cancer.
MATERIALS AND METHODS
Gastric cancer specimens
In this study, gastric cancer tissue and its adjacent tissue (within 2 cm next to the margin of tumor tissue) and normal gastric tissue (more than 5-10 cm next to the margin of tumor tissue) were obtained from 62 gastric cancer patients who were underwent surgical resection in the First Affiliated Hospital of Medical College, Zhejiang University, from March 2003 to May 2004. None of the patients received any preoperative therapies such as chemotherapy or radiotherapy. The patients consisted of 38 males and 24 females, their age ranged 21- 86 years (mean age: 59.62 years). The tumor, nodes, metastasis (TNM) stage of gastric cancer referring to the p-TNM stage were promulgated by the International Union against Cancer (UICC) in 1997. This study was approved by the Hospital Review Board and written consent was obtained from each involved patient.
Immunohistochemistry
FAT10 and mutant p53 protein levels were routinely measured by immuno-histochemistry. Briefly, gastric cancer tissue and its adjacent tissue and normal gastric tissue were sequentially fixed with 10% formalin, embedded in paraffin and cut into 4-μm thick sections. The sections were deparaffinaged and endogenous peroxidase was blocked with H2O2. Antigen retrieval was performed by heating the sections in a 0.01 mol/L citrate buffer in a microwave oven. Nonspecific binding was blocked by incubating the sections with normal rabbit serum for 20 min. The sections were then incubated at 37°C for 2 h with either polyclonal FAT10 antibody (Shanghai Jintai Biological Science and Technology Ltd, China) or monoclonal mutant P53 antibody (Beijing Zhongshan Biological Science and Technology Ltd, China). Controls without primary antibodies were also included. After washed three times with PBS, the sections were incubated with biotin-conjugated secondary antibody (Shanghai Jintai Biological Science and Technology Ltd, China) for 40 min at room temperature. Immunocomplexes were detected with 3, 3-diaminobenzidine (Fuzhou Maixin Biological Science and Technology Ltd, China) that acts as a chromogen and results in deposition of brown reaction products. Species with 0%, about 10%, about 50% of positively stained cells were scored as -, +, and ++, respectively.
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
RNA was extracted using Trizol reagent with one-step extraction method. In brief, 1 mL Trizol reagent was added to cultured cells or to approximately 100 mg tissue specimens, respectively, and total RNA was isolated following the manufacturer’s instructions. RNA integrity was assessed by agarose gel electrophoresis when clear 18 S and 28 S strips appeared on the gel, whereas total RNA concentration was measured with a spectrophotometer (A260/280 Ratios of 1.8-2.0) following its manufacturer’s instructions. Total RNA (5 μL) was reversed into cDNA in a 24 μL reaction system at 42°C for 60 min and at 70°C for 10 min, sequentially. The mixture contained 5 μL 5-RT-buffer, 1 μL Oligo (dT), 2 μL 10 mmol/L dNTPs mix, 1 μL ribonuclease inhibitor (20 U/μL), 1 μL M-MMLV reverse transcriptase (200 U/μL), and 9 μL DEPC water. The final RT reaction solution was used in PCR (GeneAmp 2400 PCR System, Perkin Elmer Company, USA). The primers and lengths of PCR amplifications are listed in Table 1. DNA products were run in agarose gel at 80-100 V and then analyzed by BIO-RAD image acquisition with an analysis system, followed by semi-quantitative analysis with Quantity One software. The 295 bp, 478 bp, and 492 bp long fragments were detected under UV light. Gray scale scanning was performed on electrophoresis strips using an image acquisition and analysis system.
Table 1.
Primers used in this study
| Designation | Sequences | PCR products (bp) |
| β-actin | ||
| Upstream primer | 5’-TCACCCACACCGTGCCCATCTACGA-3’ | 295 |
| Downstream primer | 5’-CAGCGGAACCGCTCATTGCCAACGG-3’ | |
| FAT10 | ||
| Upstream primer | 5’-AATGCTTCCTGCCTCTGTGT-3’ | 478 |
| Downstream primer | 5’-GCCGTAATCTGCCATCATCT-3’ | |
| Mutant p53 | ||
| Upstream primer | 5’-CCTATGGAAACTACTTCCTGAAAACAA-3’ | 492 |
| Downstream primer | 5’-ACAGCATCAAATCATCCATTGC-3’ |
Statistical analysis
Immunohistochemical expression was defined as positive when moderate to strong nuclear staining was observed in more than 10% cells. χ2 test was used to determine the difference in FAT10 and p53 protein levels between different groups and the correlation between positive FAT10 ratio and clinicopathological parameters of patients. RT-PCR semi-quantitative results were expressed as mean ± SD. Student’s t-test was employed to analyze differences in mRNA levels. Relation between FAT10 and p53 was statistically analyzed using Spearman’s rank correlation. Overall survival rate was calculated by Kaplan-Meier curves. Cox proportional hazards model was used to examine the effect of potential prognostic variables on survival. Statistical analysis was performed with SPSS software version 12.0. P < 0.05 was considered statistically significant.
RESULTS
Detection of FAT10 and mutant p53 protein in different gastric tissues by immunohistochemistry
FAT10 protein was mainly detected in nuclei of malignant and benign cells which were stained brown-yellow (Figure 1A). Positive staining of FAT10 cells was found in tumor tissue and its adjacent tissue and normal tissue samples from 32 (51.61%), 8 (12.90%) and 4 (6.45%) of the 62 cases. The FAT10 level was significantly different in gastric cancer tissue, and its adjacent tissue and normal tissue (χ2 = 40.96, P < 0.01). The rate of positive FAT10 cells in gastric cancer tissue was significantly higher than that in its adjacent tissue and normal tissue (P < 0.01) but was not significantly different between adjacent tissue and normal tissue (Table 2). Mutant P53 was expressed mainly in nuclei and cytoplasm (Figure 1B). Positive immunostaining of mutant fp53 was noted in 45.16% (28/62) of tumor tissue samples, 14.51% (9/62) of adjacent tissue samples and 9.63% (6/62) of normal tissue samples (χ2 = 25.83, P < 0.01). The positive rate of mutant p53 expression in tumor tissue was significantly higher than that in adjacent tissue and normal tissue (P < 0.01, Table 2).
Figure 1.
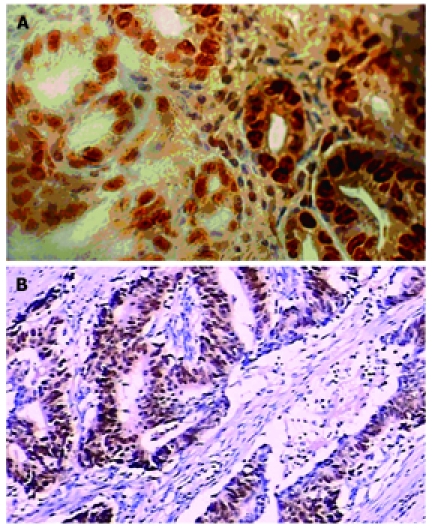
Expression of FAT10 (A) and mutant p53 (B) in gastric cancer tissue (× 400).
Table 2.
FAT10 and mutant p53 protein expression in gastric cancer tissue and its adjacent tissue and normal tissue n (%)
| Tissue |
FAT10 |
Mutant p53 |
||
| Negative | Positive | Negative | Positive | |
| Gastric tissue | 30 | 32 (51.61) | 34 | 28 (45.16) |
| Tumor-adjacent tissue | 54 | 8 (12.90) | 53 | 9 (14.51) |
| Normal gastric tissue | 58 | 4 (6.45) | 56 | 6 (9.68) |
Detection of FAT10 and mutant p53 mRNA in different gastric tissues by RT-PCR
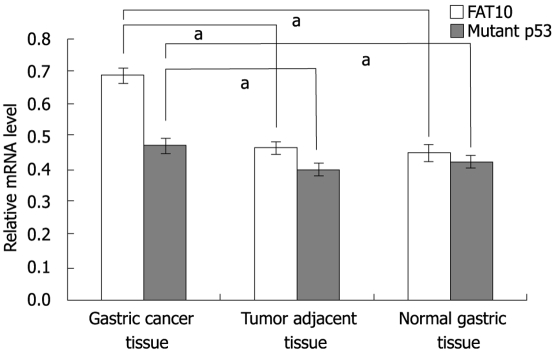
The levels of FAT10 and mutant p53-mRNA in tumor tissue and non-tumor tissue were measured. RT-PCR analysis revealed that the relative FAT10-mRNA expression in gastric cancer tissue was significantly higher than that in its adjacent tissue (t = 3.12, P < 0.01) and normal tissue (t = 4.64, P < 0.01), whereas no significant difference was detected between tumor-adjacent and normal tissues (t = 1.03, Figure 2). Mutant P53-mRNA expression was significantly higher in gastric cancer tissue than in its adjacent tissue (t = 6.79, P < 0.01) and normal tissue (t = 5.51, P < 0.01). The difference in mutant p53-mRNA expression between adjacent and normal tissue was not statistically significant (t = 1.22, Figure 2).
Figure 2.
Relative mRNA levels of FAT10 and mutant p53 in different tissues. The column represents the relative gray values of FAT10 and mutant p53 mRNA by normalizing the gray value of β-actin. aP < 0.05 (for FAT10: 0.689 ± 0.023 in gastric cancer tissue, 0.463 ± 0.019 in tumor adjacent tissue, 0.451 ± 0.028 in normal gastric tissue; for mutant p53: 0.471 ± 0.021 in gastric cancer tissue, 0.398 ± 0.017 in tumor adjacent tissue, 0.421 ± 0.019 in normal gastric tissue).
Relation between FAT10 protein and mRNA levels and clinicopathological features of gastric cancer
To test the potential value of FAT10 as a gastric cancer biomarker, we performed χ2 test to evaluate the correlation of FAT10 expression with clinicopathological features of gastric cancer (Table 3). The positive rate of FAT10 expression in gastric cancer with regional lymph node metastasis was significantly higher than that without regional lymph node metastasis (P < 0.05). Furthermore, high FAT10 expression levels were associated with advanced TNM staging (III + IV/I + II) (P < 0.05). However, there was no significant difference in FAT10 expression, age and gender of patients, tumor size, location, histological grade, and distant metastasis. The expression of FAT10-mRNA was correlated with lymph node status and TNM stage (III + IV/I + II) (P < 0.05), but not with age and sex of patients, tumor size, location, histological grade, and distant metastasis, which was similar to the immunohistochemical results (Table 4).
Table 3.
Relation between FAT10 expression and clinicopathologic factors for gastric cancer
| Clinic pathologic factors | Samples | FAT10 positive samples (%) | χ2 | P |
| Age (yr) | ||||
| < 50 | 19 | 7 (36.84) | 2.39 | > 0.05 |
| ≥ 50 | 43 | 25 (58.14) | ||
| Gender | ||||
| Male | 38 | 19 (50.00) | 0.1 | > 0.05 |
| Female | 24 | 13 (54.16) | ||
| Tumor size (cm) | ||||
| < 5 | 33 | 16 (48.48) | 0.28 | > 0.05 |
| ≥ 5 | 29 | 16 (55.17) | ||
| Location | ||||
| Antrum | 31 | 15 (48.39) | 2.81 | > 0.05 |
| Angle | 5 | 2 (40.00) | ||
| Body | 14 | 9 (64.29) | ||
| Fundus | 4 | 3 (75.00) | ||
| Cardia | 8 | 3 (37.50) | ||
| Progression degree | ||||
| Early stage | 11 | 6 (54.54) | 0.05 | > 0.05 |
| Progressive stage | 51 | 26 (50.98) | ||
| Differentiation degree | ||||
| High, middle | 15 | 7 (46.67) | 0.19 | > 0.05 |
| Low, none | 47 | 25 (53.19) | ||
| Lymph metastasis | ||||
| Positive | 39 | 25 (64.10) | 6.57 | < 0.05 |
| Negative | 23 | 7 (30.43) | ||
| Distant metastasis | ||||
| Positive | 15 | 11 (73.33) | 3.74 | > 0.05 |
| Negative | 47 | 21 (44.68) | ||
| TNM Staging | ||||
| I + II | 21 | 7 (33.33) | 4.25 | < 0.05 |
| III + IV | 41 | 25 (60.98) |
Table 4.
Relation between FAT10-mRNA level and clinicopathological factors for gastric cancer
| Clinicopathological factors | Samples (n) | Positive samples | Strap gray value (mean ± SD) | t-value | P |
| Age (yr) | |||||
| < 50 | 19 | 10 | 0.583 ± 0.036 | 1.12 | > 0.05 |
| ≥ 50 | 43 | 26 | 0.611 ± 0.026 | ||
| Sex | |||||
| Male | 38 | 21 | 0.622 ± 0.013 | 1.36 | > 0.05 |
| Female | 24 | 15 | 0.574 ± 0.024 | ||
| Tumor size (cm) | |||||
| < 5 cm | 33 | 19 | 0.594 ± 0.022 | 0.65 | > 0.05 |
| ≥ 5 cm | 29 | 17 | 0.590 ± 0.018 | ||
| Location | |||||
| Tantrum | 31 | 17 | 0.537 ± 0.019 | 1.54 | > 0.05 |
| Angle | 5 | 3 | 0.672 ± 0.037 | ||
| Body | 14 | 9 | 0.594 ± 0.021 | ||
| Fundus | 4 | 4 | 0.611 ± 0.025 | ||
| Cardia | 8 | 3 | 0.529 ± 0.032 | ||
| Progression degree | |||||
| Early | 11 | 8 | 0.623 ± 0.023 | 1.15 | > 0.05 |
| Advanced | 51 | 28 | 0.597 ± 0.020 | ||
| Differentiation degree | |||||
| Well-middle | 15 | 9 | 0.564 ± 0.031 | 1.39 | > 0.05 |
| Low-non | 47 | 27 | 0.595 ± 0.019 | ||
| Lymph node metastasis | |||||
| Positive | 39 | 27 | 0.656 ± 0.016 | 3.37 | < 0.01 |
| Negative | 23 | 9 | 0.531 ± 0.026 | ||
| Distant metastasis | |||||
| Positive | 15 | 12 | 0.623 ± 0.033 | 1.74 | > 0.05 |
| Negative | 47 | 24 | 0.598 ± 0.017 | ||
| TNM stage | |||||
| I + II | 21 | 8 | 0.667 ± 0.023 | 2.25 | < 0.05 |
| III + IV | 41 | 28 | 0.558 ± 0.015 | ||
Correlation between FAT10 and mutant p53 in gastric cancer tissue
The expression rate of FAT10 was 82.14% (23/28) and 26.47% (9/34) in positive and negative mutant p53 tumor tissues, respectively (Table 5). The correlation between FAT10 and mutant p53 was analyzed by Spearman rank correlation. The expression of FAT10 was positively correlated with mutant p53 in gastric cancer (r = 0.865, P < 0.05). Furthermore, Spearman rank correlation analysis showed that FAT10-mRNA was significantly correlated with mutant p53-mRNA (r = 0.548, P < 0.05).
Table 5.
Correlation between FAT10 and mutant p53 expressions in gastric cancer tissue
| p53 | Samples |
FAT10 expression |
|
| Positive | Negative | ||
| Positive | 28 | 23 | 5 |
| Negative | 34 | 9 | 25 |
| Total | 62 | 32 | 30 |
Predictive role of FAT10 in gastric cancer
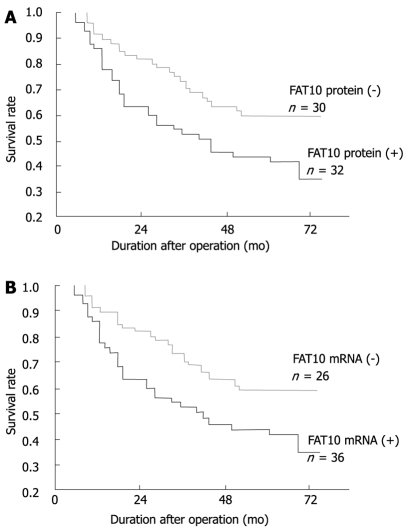
To investigate the impact of FAT10 over-expression on the clinical outcome of patients, univariate survival probability curves were plotted with respect to the immunohistochemical and RT-PCR results. Except for 4 patients (2 with positive FAT10 protein and FAT10-mRNA, and 2 with negative FAT10 protein and FAT10-mRNA), the other patients were followed up for 48-72 mo. We found that high FAT10 protein and mRNA levels in gastric cancer showed a tendency towards unfavorable prognosis regarding the overall survival rate as shown by Kaplan-Meier analysis (P < 0.05, Figure 3). In the Cox regression model, multivariate survival analyses showed that FAT10 protein and mRNA, as well as lymph node metastasis, distant metastasis, TNM stage, and mutant p53 mRNA protein, were the independent adverse prognostic factors for the overall survival rate (Table 6).
Figure 3.
Effect of FAT10 protein (A) and mRNA (B) on the survival rate of gastric cancer patients (Kaplan-Meier survival curve).
Table 6.
Cox regression model analysis results of the prognostic factors for gastric cancer
| Harzard factors | B | S χ | Wald | υ | Sig. | Exp (B) |
| Age | 0.134 | 0.144 | 0.853 | 1 | 0.533 | 1.012 |
| Tumor size | 0.243 | 0.246 | 0.804 | 1 | 0.368 | 0.763 |
| Location | 0.072 | 0.055 | 1.846 | 1 | 0.173 | 0.954 |
| Progression | 0.144 | 0.267 | 0.26 | 1 | 0.615 | 1.148 |
| Differentiation degree | 0.544 | 0.23 | 2.613 | 1 | 0.118 | 1.223 |
| Lymph node metastasis | 0.734 | 0.384 | 4.985 | 1 | 0.027 | 1.886 |
| Distant metastasis | 0.764 | 0.326 | 6.785 | 1 | 0.001 | 1.806 |
| TNM stage | 0.698 | 0.285 | 7.205 | 1 | 0.007 | 1.745 |
| FAT10 protein | 0.661 | 0.228 | 8.448 | 1 | 0.004 | 1.516 |
| FAT10 mRNA | 0.793 | 0.245 | 7.658 | 1 | 0.003 | 1.854 |
| p53 protein | 0.669 | 0.239 | 6.479 | 1 | 0.006 | 1.611 |
| p53 mRNA | 0.758 | 0.346 | 6.857 | 1 | 0.004 | 1.521 |
DISCUSSION
FAT10 is a member of the ubiquitin-like modifier family of proteins. Over-expression of the FAT10 gene has been observed in several epithelial cancers and high FAT10 expression can increase chromosome instability by reducing kinetochore localization of MAD2 during the prometaphase stage of cell-cycle[14]. In the present study, we measured the FAT10 protein and mRNA levels in gastric cancer tissue, its adjacent tissue and normal tissue from 62 patients. The immunohistochemical analysis suggested that FAT10 protein was mainly expressed in cell nuclei, indicating that FAT10 may participate in cell cycle regulation. The positive expression rate of FAT10 protein and mRNA in gastric cancer tissue was significantly higher than that in its adjacent tissue and normal tissue, suggesting that FAT10 may play an important role in the process of gastric carcinogenesis
Recent researches have shown that the expression of mutant p53 and FAT10 mRNAs is increased in cancer cell line[15,16]. However, the correlation between mutant p53 and FAT10 has not been analyzed in human gastric cancer. In this study, FAT10 and mutant p53 protein/mRNA were over-expressed in gastric cancer tissue whereas high FAT10 and mutant p53 expression levels in tumor tissue were positively correlated. It was reported that p53 negatively regulates the expression of FAT10 and p53 depletion, thus contributing to tumorigenesis by uncontrolled up-regulation of FAT10[13], suggesting that mutant p53 may also activate the FAT10 gene and promote gastric tumorigenesis due to the loss of its anti-carcinoma effect. Moreover, proinflammatory cytokines up-regulate FAT10 in liver and colon cancer, indicating that they play a potential role in activating FAT10 in gastric tumor and merit further investigation[16]. Although p53 binds to the 5’ half consensus sequence of p53-binding site at the FAT10 promoter[13], the exact p53-binding site is still unclear, thus further study is needed.
No report is available at present on the cor-relation between FAT10 protein expression and clinicopathological characteristics of gastric cancer patients. In the present study, FAT10 protein and mRNA levels were closely correlated with lymph node metastasis and TNM stage (III + IV/I + II) (P < 0.05), indicating that FAT10 can promote tumor invasion and metastasis, and may be a candidate prognostic factor for lymph node metastasis and tumor progression. Large scale studies are needed to further confirm our findings.
Tumor metastasis has become one of the most challenging problems in tumor therapy. Many efforts have been made to predict gastric cancer behaviors, but specific predictive markers for metastasis and recurrence are still lacking[17]. In our study, the patients with a high FAT10 expression level showed a tendency towards unfavorable prognosis. Since FAT10 and mutant p53 protein and mRNA, lymph node metastasis, distant metastasis, and TNM stage are the independent prognostic factors for poor patient survival, determination of FAT10 status may be an important step in formulating right therapeutic strategies. Moreover, FAT10 may be related with other predictive biomarkers of tumor metastasis, such as CD44v6, nm23, MTA1 and MMPs.
In conclusion, FAT10 is over-expressed in gastric cancer tissue, and positively correlated with mutant p53 expression, lymph node metastasis and tumor progression, and can promote tumor invasion. FAT10 is of prognostic value for human gastric cancer and is a potential target for cancer biotherapy.
COMMENTS
Background
FAT10 belongs to the ubiquitin-like modifiers of ubiquitin protein family, first discovered in mapping HLA-F gene in 1996, and is expressed in mature B cells and dendritic cells. It has been reported that FAT10 can regulate cell-cycle and may play an important role in tumorigenesis. P53 gene, located on the short arm of chromosome 17, can be divided into wild p53 and mutant p53. P53 protein depletion or gene mutation has been detected in over 50% of all cancers, suggesting that P53 may play an important role in gastric tumorigenesis. Gastric cancer, one of the most common malignant tumors, is a leading cause of cancer-related death worldwide, and is on the top of the list in China. The detailed mechanism underlying the development of gastric cancer still remains uncertain.
Research frontiers
It has been recently found that FAT10 can non-covalently bind to the human spindle assembly checkpoint protein (MAD2) that is responsible for the maintenance of spindle integrity during mitosis. Inhibition of MAD2 may lead to chromosomal instability, a common feature of tumorigenesis. It has been shown that damaged DNA enters into S stage, changes cell hereditary characteristics, and finally induces tumorigenesis when p53 is deleted or mutated. Moreover, there is evidence that the p53 mutation rate is higher in gastric cancer with atrophic gastritis than in gastric cancer without atrophic gastritis. FAT10 is a downstream target of p53, and down-regulation of FAT10 expression in p53-defective cells contributes to carcinogenesis.
Innovations and breakthroughs
In this study, the FAT10 and mutant p53 mRNA and protein levels were significantly higher in gastric cancer tissue than in its adjacent tissue and normal tissue. The FAT10 and mutant p53 levels in gastric cancer tissue were significantly correlated with lymph node metastasis and TNM staging. Moreover, mRNA and protein levels of FAT10 and mutant p53, lymph node metastasis, distant metastasis, and TNM stage were found to be independent prognostic factors for patients with gastric cancer.
Applications
FAT10 may be a potential marker of gastric cancer prognosis, which needs to be further verified. FAT10 is positively correlated with mutant p53, indicating that it may play a role in carcinogenesis and becomes a novel therapeutic target of gastric cancer.
Terminology
FAT10: a protein belonging to ubiquitin-like modifier (UBL) of ubiquitin protein family, is mainly expressed in mature B cells and dendritic cells and regulates cell-cycle and chromosomal instability, thus playing an important role in tumorigenesis.
Peer review
This study describes the increased FAT10 and mutant p53 mRNA and protein levels in gastric cancer and the correlation between FAT10 and mutant p53. Furthermore, the authors also found that mRNA and protein levels of FAT10 were independent prognostic factors for patients with gastric cancer. These results are innovative, showing that FAT10 may be involved in gastric carcinogenesis and in gastric cancer prognosis. However, further study is needed to further verify their findings.
Footnotes
Peer reviewer: Dusan M Jovanovic, Professor, Institute of Oncology, Institutski Put 4, Sremska Kamenica 21204, Serbia
S- Editor Tian L L- Editor Wang XL E- Editor Yin DH
References
- 1.Fan W, Cai W, Parimoo S, Schwarz DC, Lennon GG, Weissman SM. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics. 1996;44:97–103. doi: 10.1007/BF02660056. [DOI] [PubMed] [Google Scholar]
- 2.Bates EE, Ravel O, Dieu MC, Ho S, Guret C, Bridon JM, Ait-Yahia S, Brière F, Caux C, Banchereau J, et al. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur J Immunol. 1997;27:2471–2477. doi: 10.1002/eji.1830271002. [DOI] [PubMed] [Google Scholar]
- 3.Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci USA. 1999;96:4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- 5.Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, et al. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 7.Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Crit Rev Oncol Hematol. 2000;33:1–6. doi: 10.1016/s1040-8428(99)00051-7. [DOI] [PubMed] [Google Scholar]
- 8.Moll UM, Schramm LM. p53--an acrobat in tumorigenesis. Crit Rev Oral Biol Med. 1998;9:23–37. doi: 10.1177/10454411980090010101. [DOI] [PubMed] [Google Scholar]
- 9.Taguchi A, Ohmiya N, Itoh A, Hirooka Y, Niwa Y, Mori N, Goto H. Severity of atrophic gastritis related to antiparietal cell antibody and gastric carcinogenesis, including p53 mutations. J Gastroenterol Hepatol. 2006;21:545–551. doi: 10.1111/j.1440-1746.2005.03983.x. [DOI] [PubMed] [Google Scholar]
- 10.Rugge M, Shiao YH, Busatto G, Cassaro M, Strobbe C, Russo VM, Leo G, Parenti AR, Scapinello A, Arslan P, et al. The p53 gene in patients under the age of 40 with gastric cancer: mutation rates are low but are associated with a cardiac location. Mol Pathol. 2000;53:207–210. doi: 10.1136/mp.53.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata A, Parsonnet J, Longacre TA, Garcia MI, Puligandla B, Davis RE, Vogelman JH, Orentreich N, Habel LA. CagA status of Helicobacter pylori infection and p53 gene mutations in gastric adenocarcinoma. Carcinogenesis. 2002;23:419–424. doi: 10.1093/carcin/23.3.419. [DOI] [PubMed] [Google Scholar]
- 12.Sun XD, Mu R, Zhou YS, Dai XD, Zhang SW, Huangfu XM, Sun J, Li LD, Lu FZ, Qiao YL. [Analysis of mortality rate of stomach cancer and its trend in twenty years in China] Zhonghua Zhong Liu Za Zhi. 2004;26:4–9. [PubMed] [Google Scholar]
- 13.Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25:2318–2327. doi: 10.1038/sj.onc.1209220. [DOI] [PubMed] [Google Scholar]
- 14.Lim CB, Zhang D, Lee CG. FAT10, a gene up-regulated in various cancers, is cell-cycle regulated. Cell Div. 2006;1:20. doi: 10.1186/1747-1028-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oki E, Zhao Y, Yoshida R, Egashira A, Ohgaki K, Morita M, Kakeji Y, Maehara Y. The difference in p53 mutations between cancers of the upper and lower gastrointestinal tract. Digestion. 2009;79 Suppl 1:33–39. doi: 10.1159/000167864. [DOI] [PubMed] [Google Scholar]
- 16.Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P, Breuhahn K, Groettrup M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27:6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- 17.Ishii K, Kinami S, Funaki K, Fujita H, Ninomiya I, Fushida S, Fujimura T, Nishimura G, Kayahara M. Detection of sentinel and non-sentinel lymph node micrometastases by complete serial sectioning and immunohistochemical analysis for gastric cancer. J Exp Clin Cancer Res. 2008;27:7. doi: 10.1186/1756-9966-27-7. [DOI] [PMC free article] [PubMed] [Google Scholar]