Abstract
Few molecular studies have been devoted to the finger drop process that occurs during banana fruit ripening. Recent studies revealed the involvement of changes in the properties of cell wall polysaccharides in the pedicel rupture area. In this study, the expression of cell-wall modifying genes was monitored in peel tissue during post-harvest ripening of Cavendish banana fruit, at median area (control zone) and compared with that in the pedicel rupture area (drop zone). To this end, three pectin methylesterase (PME) and seven xyloglucan endotransglycosylase/hydrolase (XTH) genes were isolated. The accumulation of their mRNAs and those of polygalaturonase, expansin, and pectate lyase genes already isolated from banana were examined. During post-harvest ripening, transcripts of all genes were detected in both zones, but accumulated differentially. MaPME1, MaPG1, and MaXTH4 mRNA levels did not change in either zone. Levels of MaPME3 and MaPG3 mRNAs increased greatly only in the control zone and at the late ripening stages. For other genes, the main molecular changes occurred 1–4 d after ripening induction. MaPME2, MaPEL1, MaPEL2, MaPG4, MaXTH6, MaXTH8, MaXTH9, MaEXP1, MaEXP4, and MaEXP5 accumulated highly in the drop zone, contrary to MaXTH3 and MaXTH5, and MaEXP2 throughout ripening. For MaPG2, MaXET1, and MaXET2 genes, high accumulation in the drop zone was transient. The transcriptional data obtained from all genes examined suggested that finger drop and peel softening involved similar mechanisms. These findings also led to the proposal of a sequence of molecular events leading to finger drop and to suggest some candidates.
Keywords: Banana, cell-wall, expansin, finger drop, Musa, pectolytic genes, quality, ripening, xyloglucan endotranglucosylase/hydrolase
Introduction
Banana is the second ranking fruit crop in the world, with an annual world production of ∼70 million metric tons (FAOstat, 2007 faostat.fao.org/site/567/DesktopDefault.aspx?). Banana fruit ripening is characterized by a burst of ethylene production and respiration concomitantly with physicochemical and biochemical changes, including chlorophyll breakdown, increased starch degradation and sugar synthesis, and fruit softening. Finger drop is one of the main features closely associated with the banana ripening process. This process, which was reported for the first time in the triploid Cavendish AAA group, involves physiological softening and weakening, thus causing individual fruit in a hand to separate from the crown (Hicks, 1934; Baldry et al., 1981; Semple and Thompson 1988). Bananas are marketed in hands of generally 4–9 fruits. Dislodgement of individual fruits from the hand considerably reduces the commercial value of the product because hands with fingers missing or fingers without pedicels cannot be sold to consumers. The finger drop process thus reduces the economic value of bananas. Despite this important economic impact, few studies have been devoted to gaining an insight into the physiological mechanisms related to banana finger drop. Susceptibility to finger drop varies widely according to cultivars and their ripening conditions, with triploid cultivars being less susceptible than tetraploids (New and Marriot, 1983). Among triploid cultivars, Valery is less susceptible than Gros Michel, and Hom Tong (Musa acuminata, AAA) shows massive drop while Namwa (Musa sp., ABB) does not (New and Marriot, 1983; Saengpook et al., 2007). In addition to cultivars, the level of fruit development ripening conditions (including ethylene, humidity, and temperature) also affect the susceptibility to banana finger drop (Semple and Thompson, 1988; Paull, 1996; Imsabai et al., 2006). Studies on the physiological mechanisms of banana finger drop have only recently been carried out and, to our knowledge, performed only at the biochemical level. In a study on two varieties that display massive (Hom Tong variety) and no finger drop (Namwa variety), respectively, Imsabai et al. (2006) showed that peel water content and thickness did not affect the finger drop process. By contrast, differential changes in the water- and CDTA-soluble fractions, and in the insoluble pectin fraction at the peel rupture area in the Hom Tong variety indicated high pectin breakdown and degradation activity in this variety. The authors also showed that this pectin breakdown was mainly the result of pectate lyase and polygalacturonase enzymes, as their corresponding activities increased concomitantly in the peel rupture area of the Hom Tong variety as compared with those of the Namwa variety. It is now assumed that cell wall modifications that occur during fruit ripening correspond to a complex process involving different mechanisms, including enzymatic and non-enzymatic degradation of cell wall components (Dumville and Fry, 2003; Bennett and Labavitch, 2008), cell wall synthesis (Mitcham et al., 1989, 1991; Greve and Labavitch, 1991; Huysamer et al., 1997; Rose and Bennett, 1999) and/or loosening of the cell wall network mediated by non-enzymatic proteins like expansin (Brummell, 2006). Enzymatic degradation of cell wall components is the result of the induction, at both molecular and biochemical levels, of pectolic and/or non-pectolitic cell wall-modifying proteins (CWMPs) according to the specific class of polysaccharides used as substrate. The pectolytic enzymes include endo- and exo-polygalacturonases (endo- EC 3.2.1.15; exo- EC 3.2.1.67), pectate lyases (PEL; EC 4.2.2.2), pectin methylesterases (PME; EC 3.1.1.11), pectin acetylesterases (PAE; EC 3.1.1.–), β-galactosidases (EC 3.2.1.23), and α-L-arabinofuranosidases (EC 3.2.1.55) (Vicente et al., 2007; Goulao and Oliveira, 2008). These enzymes are able to cleave or modify the nature of the polysaccharide backbone or to remove neutral sugars from branched side-chains. Non-pectolytic enzymes are responsible for hemicellulose modifications and include endo-1,4-β-glucanases (EGase; EC 3.2.1.4), endo-1,4-β-xylanases (EC 3.2.1.8), β-xylanases (EC 3.2.1.37), and xyloglucan endotransglycosylase/hydrolases (XTH; EC 2.4.1.207). Many of these proteins have been investigated, at both molecular and biochemical levels, in relationship with banana fruit softening. A co-ordinated increase in mRNA abundance, de novo synthesis or activity of a number of putative cell wall-modifying enzymes that may be responsible for or contribute significantly to polymer modifications has been observed (Dominguez-Puigjaner et al., 1997; Pua et al., 2001; Marin-Rodriguez et al., 2003; Trivedi and Nath, 2004; Asif and Nath, 2005; Zhuang et al., 2006, 2007; Sane et al., 2007).
As none of these genes have been examined in relation to finger drop, an attempt was made to investigate, at the molecular level, the putative relationship between the expression of some cell wall-modifying genes (CWMGs) and the finger drop process in order to identify: (i) the putative physiological mechanism of the cell wall metabolism involved in the finger drop process, and (ii) relevant genes involved for purposes of identification of functional molecular markers.
Banana is a complex species and attempts to improve the fruit quality through breeding strategies is complex and difficult to set up. Access to functional molecular markers derived from relevant genes associated with fruit quality traits should therefore provide a valuable and helpful resource for the development of new crossing strategies and the selection process. Moreover, gaining further insight into the molecular mechanisms that govern the elaboration of fruit quality traits and their regulation is a key prerequisite for the development of other strategies to improve banana quality traits like post-harvest technology and/or biotechnology approaches.
In the present study, the accumulation of mRNA genes encoding a broad array of enzymes known to hydrolyse or modify the fine structure of cell wall polysaccharides was examined, with the view of correlating particular mechanisms and genes with previously observed changes in cell wall composition during finger drop.
Materials and methods
Plant material
All tissues used in this study were harvested from three banana plants (Musa acuminata, AAA, Cavendish, cv. Grande Naine) grown at the CIRAD research station (elevation: 250 m; andosol; rainfall: 3500 mm year−1), Guadeloupe (French West Indies). During growth, bunches on banana plants were covered with blue plastic bags to hamper insect infestations, and to streamline the development of whole fruits on the bunch. Green fruits were harvested at commercial ripeness, as determined on the basis of the heat unit concept (Ganry and Meyer, 1975; Jullien et al., 2008). At each harvesting time, only internal fingers of the median hand on the bunch, considered as comparable (Liu, 1976), were taken into account for each bunch. After harvest, all fruits were kept for 24 h at 20 °C in chambers ventilated with humidified air before treatment with 10 000 ppm of azethyl (95% nitrogen/5% ethylene) for 24 h at 20 °C and ambient humidity. From 1–7 d after treatment (DAT), a sample of three fruits was taken daily for finger drop measurement. Immediately at the end of each measurement, peel tissues corresponding to the median part of the fruit (control zone) and to the rupture area of the fruit pedicel (drop zone) (Fig. 1) were sampled, separately frozen in liquid nitrogen, and stored at –80 °C until use.
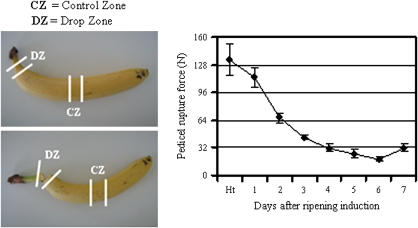
Fig. 1.
Measurement of finger drop variations during post-harvest ripening of Cavendish banana through pedicel rupture force measurement. Banana fruits harvested at the mature green stage were stored overnight at 20 °C before treatment with 10 000 ppm of azethyl (5% ethylene+95% nitrogen) for 24 h. Fruits were further kept at 20 °C in air for 7 d to ripen. Banana finger drop was evaluated on the basis of the pedicel rupture force expressed in Newtons (N). This force was measured daily from the harvesting point (Ht) to day 7, as described by Chillet et al. (2008). Each data point is the mean of values obtained from at least four fruits originating from four replicate bunches. Vertical bars indicate standard deviation (SD). When no bar is shown, the SD is smaller than the symbol.
Measurement of finger drop variations during post-harvest ripening of fruit
Finger drop was estimated by measuring the pedicel rupture force using a TA-XT2 penetrometer and an adapted probe as follows. Banana fruit were attached 6 cm above a table with a large clip. The probe was pressed down to a depth of 40 mm and at a constant speed of 5 mm s−1, at the pedicel until it separated from the fruit (Chillet et al., 2008). The required force was expressed in Newtons (N). All of these measurements were performed in triplicate on three separate fruits and the experiment was repeat three times.
Isolation of banana fruit genes coding for xyloglucan endotransglycosylase/hydrolase-like (XTH-like) and pectin methylesterase-like (PME-like)
Partial cDNAs encoding banana XTH-like genes were amplified using the 3′ RACE-PCR method (Frohman, 1990). Total RNA was extracted separately from peel tissue from control and drop zones of banana fruit taken at commercial ripeness, and at 1–4 DAT, using the modified hot-borate method (Wan and Wilkins, 1994; Mbéguié-A-Mbéguié et al., 2008a). According to the manufacturer's instructions, 2 μg of each RNA extract was DNAse-treated using RQI DNase (Promega, Charbonières, France) before being separately reverse-transcribed into cDNA using Oligo-dT17 primers (Frohman, 1990; see Supplementary Table S1 at JXB online) and AMV reverse transcriptase enzyme (Promega). These cDNA were pooled and 1 μl of this mixture was subjected to 3′ RACE-PCR with Xth-F1 and dT17-adapter primers in a 25 μl reaction containing 1× Taq buffer (Eurobio, Courtaboeuf, France), 150 nM of dNTP, 1.5 mM of MgCl2, 200 nM of each primer, and 1 U of Taq polymerase. Xth-F1 is a degenerated forward primer designed from a conserved HDE(I/M)DFEF region among plant XTH-like proteins (Table 1). The PCR program involved one cycle of 1 min at 94 °C for template denaturation and Taq activation followed by 40 cycles of 30 s at 94 °C for template denaturation, 1 min at 40 °C for primer annealing, and 2 min at 72 °C for template elongation. The isolated cDNA fragments (approximately 1100 bp) were cloned into pGEM-T vector using the pGEM-T Easy Cloning kit (Promega) and sequenced by Cogenics (Grenoble, France). GenBank accession numbers are FJ264506 (MaXTH3), FJ264507 (MaXTH4), FJ264508 (MaXTH5), FJ264509 (MaXTH6), FJ264510 (MaXTH7), FJ264511 (MaXTH8), and FJ264512 (MaXTH9).
Table 1.
Sequence analysis and comparison of banana PME genes
| (A) | ||||||||
| Sequence | Gene or cDNA length (bp) | Polypeptide length (aa) | MW (kDa) | pI | Promoter length | Exon 1 | Intron | Exon 2 |
| MaPME1 | 1923 | 565 | 61 | 7.8 | – | – | – | – |
| MaPME2 | 3365 | 559 | 61 | 6.7 | 1162 | 960 | 68 | 720 |
| MaPME3 | 3186 | 574 | 62 | 7.06 | 1000 | 1029 | 81 | 696 |
| (B) | ||||
| Polypeptides | MaPME1: 1845 bp (565 aa) |
|||
| Length | % identity (similarity) | Scov | Qcov | |
| MaPME1 | 565 | – | – | – |
| MaPME2 | 559 | 46 (60) | 81.06 | 81.93 |
| MaPME3 | 574 | 47 (59) | 97.17 | 95.64 |
| Nucleotides | CDS |
|||
| Length | % identity | Scov | Qcov | |
| MaPME1 | 1895 | – | – | – |
| MaPME2 | 2135 | 75 | 9.3 | 8.2 |
| MaPME3 | 2104 | 75 | 27.7 | 24.9 |
Analysis of nucleotide and polypeptide sequences of PME genes isolated in this study. The features of nucleotide and polypeptide sequences of MaPME1 cDNA and those of the two genomic sequences of MaPME2 and MaPME3 are given in (A). A comparison of nucleotide and polypeptide sequences of MaPME genes (B), including determination of Scov and Qcov values, was performed using BLAST 2 (Tatusova and Madden, 1999).
For pectin methylesterase genes, one partial cDNA and two genomic sequences coding for banana PME-like genes were isolated in this study. The partial cDNA sequence was isolated throughout EST sequencing. PME genomic sequences were identified throughout sequencing and annotation of BAC clones MAC077E20 (accession number AC226050) and MAC088K20 (accession number AC226051), isolated from a Cavendish BAC library (Piffanelli et al., 2003).
Bioinformatic sequence analysis
The sequence similarity of the genes isolated in this study was determined using the online BLAST (Altschul et al., 1997) and BLAST 2 sequence (Tatusova and Madden, 1999) programs on the National Center for Biotechnology Information BLAST website (National Library of Medicine, Bethesda, MD). For sequence comparison throughout the BLAST 2 program, two additional parameters were introduced, Qcov and Scov, which allows us to take into account the difference of the length between sequences. Query coverage (Qcov) is the ratio of length of the match by length of the query sequence. This value indicates the part of the query sequence covered by the alignment. Similarly, Subject coverage (Scov) was calculated as the ratio of length of the match by length of the subject or target length, and indicates the part of the subject or target sequence covered by the alignment.
All sequence alignments and phylogenetic tree construction were performed using the ClustalX algorithm with the Gonnet residue weights (Thompson et al., 1997). For XTHs genes, the pair-wise alignment parameters used included a gap opening penalty of 35 and a gap extension penalty of 0.75. For multiple alignments, the gap opening penalty was 15, with a gap extension penalty of 0.3 and the delay divergent sequences was set at 25%. The consensus tree was drawn using the TREE VIEW program (Page, 1996). Search motifs in the promoter regions of MA-PME genomic sequences was found by screening the PLACE (Prestridge, 1991; Higo et al., 1999) and PLANT CARE (Lescot et al., 2002) databases. In silico prediction of 5′- and 3′-UTR regions of MA-PME genes and prediction of signal peptides were performed using EuGène 3.2 with rice parameters (Foissac et al., 2008) and SignalP 3.0 (Bendtsen et al., 2004) programs, respectively.
Oligonucleotide primers for gene expression through real-time qPCR and end-point PCR analysis
The oligonucleotide primers used for RT-qPCR analysis were designed using the Primer 3 freeware program (Roten and Skaletsky, 2000) according to the default criteria and the corresponding sequences are described in Supplementary Table S1 at JXB online. Except for polygalacturonase genes MaPG3 and MaPG4, whose primers were designed within the divergent coding region, all other primers were designed within the 3′-UTR region. The lengths of all PCR products ranged from 125 bp to 247 bp. The gene specificity of all primer sets was tested using the following three independent procedures: (i) BLAST searches against GenBank databases were performed for each primer to check that no member of a primer set matched another sequence in the banana genome; (ii) the specificity of PCR amplification was examined by monitoring the dissociation curves during qPCR reactions using the ABI PRISM 7000 sequence detection system (Applied Biosystems, Courtaboeuf, France); (iii) individual PCR products were separated on 2% agarose gels and stained with ethidium bromide to examine their size and ensure that a single PCR product was detected for each primer pair, in accordance with the single peak observed in the dissociation curve. As for the MaPME2 and MaPME3 genes, the corresponding specific primers were designed based on the 3′-UTR predicted from the corresponding genomic sequence. The generated PCR fragment was further cloned and sequenced to confirm that it corresponded to a PME genomic sequence.
Real-time qPCR analysis
All qPCR experiments were performed on an ABI 7000 sequence detection apparatus (Applied Biosystems, Courtaboeuf, France) using the SYBR Green PCR Master Mix as described by Mbéguié-A-Mbéguié et al. (2008b). Two micrograms of total RNA extracted from different banana fruit tissues and organs treated with RQ1 DNase (Promega, Charbonnières, France) to remove possible contaminating genomic DNA were used. First-strand cDNA was synthesized from these RNA using AMV reverse transcriptase enzymes (Promega, Charbonnières, France) and random hexamers according to the manufacturer's instructions. To hamper the effect of putative PCR inhibitors that might be present in the reverse transcriptase reaction, as well as pipetting errors, the cDNA was 10-fold diluted with distilled water, and 5 μl of the diluted cDNA was used as a template for qPCR analysis. The qPCR program included one cycle of 5 min at 94 °C for template denaturation and Taq activation, followed by 40 cycles of 30 s at 94 °C for template denaturation, 15 s at 50 °C for primer annealing, and 30 s at 72 °C for primer elongation. Fluorescence was analysed using Sequence Detection Software (Applied Biosystems Courtaboeuf, France). A cycle threshold value (Ct) was calculated from the amplification curves by selecting the optimal ΔRn (emission of reporter dye over starting background fluorescence) in the exponential portion of the amplification plot.
The amplification efficiency of all primer sets was determined by qPCR analysis as described previously. To this end, we used 6 log of 2-fold serial dilutions made from a pool of cDNA reverse-transcribed from a mixture of RQI DNase-treated mRNA from different banana fruit tissues and organs. For each dilution, a qPCR was run in triplicate. The efficiency (slope) ranges were 83–100% (3.3–3.7), 87–100% (3.3–3.6), 73.3–77.3% (4.02–4.1), and 72–92% (3.12–4.2) for expansin, pectin methylesterase, polygalacturonase, pectate lyase, and xyloglucan endotransferase/hydrolase genes, respectively. They were measured using the Ct slope method (data not shown) and the formula Ex=10(–1/slope)–1.
The relative fold differences in expression of each gene between samples were determined using the 2−ΔΔCt formula (Livak and Schmittgen, 2001). For each gene and sample, a qPCR was performed in triplicate on two independent RNA extracts as described above. The MA-ACT gene was used as the internal control to standardize the difference between template amounts and fruit tissues taken at harvest before ripening induction was used as the calibrator.
Results
Finger drop evolution of banana fruit measured via the pedicel rupture force
Banana fruits were harvested at commercial harvest grade corresponding to approximately 90 days after flowering (DAF). According to the rupture force measurement (Fig. 1), Cavendish banana finger drop started 1 d after ripening induction and continued progressively throughout the post-harvest ripening stage. However, a marked decreased was observed between days 1 to 3 after ripening induction, with more than a 2-fold decrease in the pedicel rupture force. As the pedicel rupture force pattern is considered to be a way of measuring banana finger drop (Saengpook et al., 2007), our data suggest that our experimental conditions led to the development of the finger drop process of Cavendish banana.
Cloning of MaXTH genes from banana fruit and sequence analysis
Seven partial cDNA fragments encoding XTH-like polypeptides were isolated from of banana peel tissue. As two banana XTH homologues, MaXET1 and MaXET2, were previously isolated and registered in the GenBank database (Lu et al., 2004), the XTH cDNAs isolated in this study were named MaXTH3 to MaXTH9. The sequence homology between MaXTH cDNA ranges from 3.1% to 71% at the nucleotide level and from 12% to 81% at the amino acid level, with MaXTH5 and MaXTH6 being highly homologous pairs. Compared with MaXET1 and MaXET2 genes, a high homology was observed with MaXTH7 and MaXTH4, respectively. Analysis of the predicted MaXTH polypeptides revealed that they contained a DEIDFEFLG motif conserved within glycosyl hydrolase family 16 (GH16) enzymes and, adjacent to the putative active site, the potential N-linked glycosylation site N-X-S/T motif. For MaXTH5 and MaXTH6, this last motif was substituted by the N-V-R motif which is characteristic of group-3 XTHs (Campbell and Braam, 1999; Johanson et al., 2004; see Supplementary Fig. S1 at JXB online).
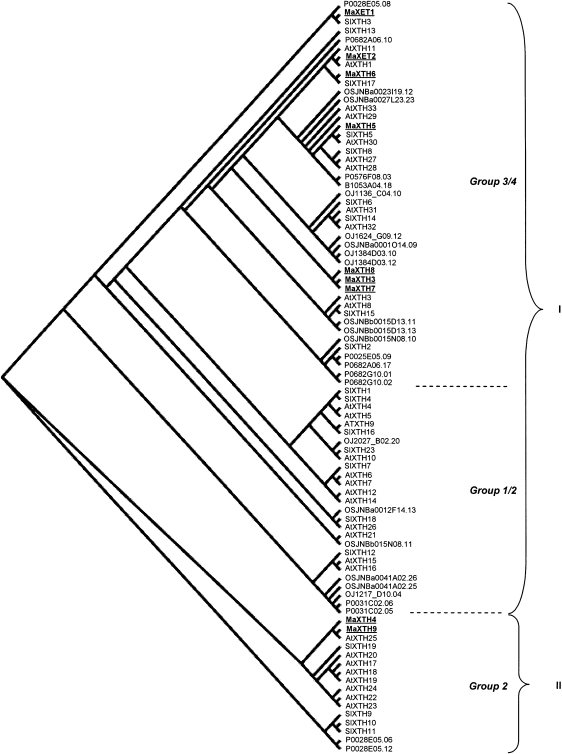
A phylogenetic tree was constructed to investigate the evolutionary relationships of XTHs polypeptides from the partial banana cDNA and those from rice, tomato, and Arabidopsis (Fig. 2). Our data showed an organization of XTH proteins into two major clusters (I and II) as observed with rice XTH proteins (Yokoyama et al., 2004), another monocotyledonous species as banana. A classification into four XTH families was observed in Arabidopsis, tomato, two dicotyledonous species, and in a broad range of plant species (Nishitani, 1997; Campbell and Braam, 1999; Rose et al., 2002; Saladié et al., 2006). Compared with this organization, our data indicated that cluster II mainly consisted of XTH proteins of family 2 of dicotyledons together with MaXTH4 and MaXTH9. Cluster I can be divided into two subclusters. The first one included MaXET1 and MaXET2, and MaXTH3, MaXTH5, MaXTH6, MaXTH7, and MaXTH8. It mainly consisted of XTH proteins of families 3 and 4 of dicotyledons. The second one mainly consisted of XTH proteins of families 1 and 2 of dicotyledons.
Fig. 2.
Phylogenetic alignment of banana, tomato, Arabidopsis, and rice xyloglucan endotransglycosylase/hydrolases (XTHs). The regions in common between the XTH polypeptide sequences of banana (MaXTHn and MaXETn), tomato (SlXTHn), Arabidopsis (AtXTHn), and rice were used to contruct a phylogenetic tree using the ClustalX program. The resulting slanted cladogram was visualized with the TREEVIEW program (Page, 1996). The sequences were grouped into two distinct groups (I–II). The numbers 1 to 4 indicate the four families groups identified in tomato and Arabidopsis and applied to a broad range of plant species (Nishitani, 1997; Campbell and Braam, 1999; Rose et al., 2002; Saladié et al., 2006). The tomato and Arabidopsis thaliana XTH sequences used in this study are available online at http://labs.plantbio.cornell.edu/XTH/genes.htm. The rice XTHs were retrieved from the Rice genome research program (http://RiceGAAS.dna.affrc.go.jp, 7 February 2009) and are indicated in the cladogram by their gene number assigned by RiceGAAS. Banana MaXET1 and MaXET2 have been registered in GenBank under accession numbers EF103137 and EF103136, respectively.
Cloning MaPME genes from banana fruit and sequences analysis
Three genes encoding a putative banana PME polypeptide have been isolated by Musa sequence data mining. Musa EST sequences produced one partial cDNA sequence (MaPME1 accession number FJ264505). A similarity search in available Musa BAC sequences produced two BAC sequences: MaC077E20 (GenBank accession number AC226050) and MaC088K20 (GenBank accession number AC226051) of 177 729 bp and 152 711 bp, respectively. Automatic and manual annotation of these BACs showed that they contained 26 and 19 predicted genes in addition to 1 and 3 repetitive elements, respectively. The two BACs sequences are schematically shown in Supplementary Fig. S2 at JXB online. They are collinear and share an overlapping region of 130 kbp. Both sequences contained one common identical putative PME gene (MaPME2). MaC077E20 contained an additional putative PME gene (MaPME3). Only the putative complete MaPME2 and MaPME3 genes from BAC MaCO77E20 were subsequently used in this study. The features of MaPME cDNAs and genomic sequences are described in Table 1A. The putative promoter regions of both MaPME2 and MaPME3 genes were analysed. They contained numerous cis-regulatory sequences that might be involved in tissue-specific or developmentally-regulated gene expression (see Supplementary Table S2 at JXB online). A sulphur-responsive element that contains the auxin-response factor (ARF) binding sequence (GAGACA) and the copper/O2-reponsive element appeared to be specific to MaPME2 and MaPME3 promoter sequences, respectively.
The predicted coding region of MaPME1 cDNA, MaPME2 and MaPME3 genomic sequences were compared at both nucleotide and polypeptide levels (Table 1B). MaPME2 and MaPME3 genes shared 7% identity at the nucleotide level, while at the amino acid level, they showed 38% and 56% identity and similarity, respectively. Compared to MaPME1 cDNA, MaPME2 and MaPME3 genomic sequences diverged at the nucleotide level. Indeed, the high percentage of identity (75%) observed between these sequences is not significant as it was obtained in a short portion of their nucleotide sequence, as shown by the low Scov and Qcov values (less than 10% with MaPME2 and 25% with MaPME3). At the polypeptide level, MaPME2 and MaPME3 shared 46% and 60% identity and similarity, respectively, with polypeptides predicted from MaPME1 cDNA. The predicted polypeptides of the three MaPME genes presented features that are conserved in all PME polypeptides. Among these, there are the pre- and pro-sequences at the N-terminal extension, and five polypeptide segments that are characteristic of the C-terminal catalytic region, which constitutes the mature PME enzyme: GxYxE, QAVAL, QDTL, DFIFG, LGRPW (Micheli, 2001; Markovic and Janecek, 2004; see Supplementary Fig. S2 at JXB online). The pre-region or signal peptide required for protein targeting to the endoplasmic reticulum is cleaved in the first step of maturation. According to the SignalP 3.0 program (Bendtsen et al., 2004); it was localized between amino acid residues 24–25 and 27–28 for MaPME2 and MaPME3, respectively. Pro-sequence cleavage of the covalently attached mature enzyme may occur in the second step of maturation close to the RR(K)LL(M) motif (Markovic and Janecek, 2004; see Supplementary Fig. S2 at JXB online), thus leading to pro-sequence lengths of 200 and 217 amino acid residues for MaPME2 and MaPME3, respectively. Based on the Arabidopsis PME sequence analysis, PME genes have been classified into two types: type I and II, having a long N-terminal pro-region and a short or absent pro-region, respectively. The role of the pro-region is currently unknown (Micheli, 2001; Markovic and Janecek, 2004; Giovane et al., 2004; Pelloux, 2007). The long N-terminal pro-sequence of MaPME2 and MaPME3 predicted polypeptides suggests that both genes belong to type I.
Expression of pectolytic genes in peel tissues from control and drop zones during banana fruit ripening
Three classes of pectolytic genes were examined in peel tissue during banana fruit ripening and in relation to finger drop. Among these, three genes encode pectin methylesterase MaPME1 to MaPME3, four encode polygalacturonase (MaPG1 to MaPG4; Asif and Nath, 2005), two encode pectate lyase (MaPEL1 and MaPEL2; Dominguez-Puigjaner et al., 1997; Pua et al., 2001), and four encode expansin protein (MaEXP1, 2, 4, and 5; Sane et al., 2007). Gene expression experiments were performed in triplicate on two independent RNA extracts and gave the same results.
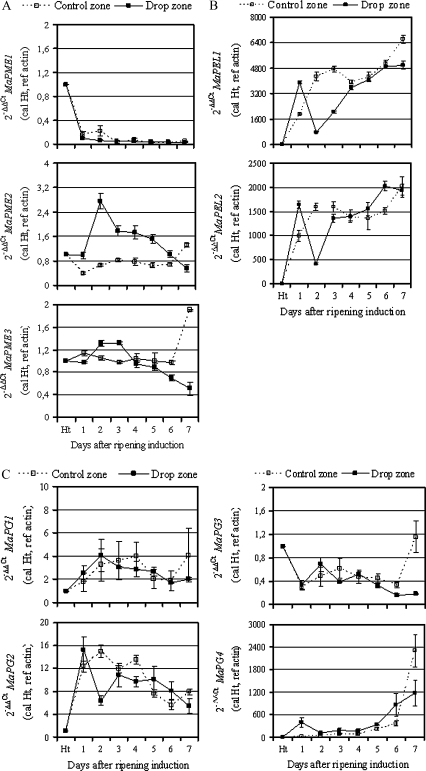
Pectolytic genes examined in his study were differentially expressed during ripening and in both the control and drop zones (Fig. 3). MaPEL1 and MaPEL2, and MaPG4 genes were the most expressed genes compared with the others. Among the three MaPME genes, MaPME1 was the least expressed in peel tissue compared with the other two. Its expression decreased during ripening in both zones (Fig. 3A). By contrast, MaPME2 was the most expressed in the drop zone. In the control zone, its mRNA level remained constant during the first 6 d after ripening induction. A slight increase was only observed in overripe fruit taken 7 d after ripening induction. In the drop zone and from day 1 after ripening induction to day 6, the MaPME2 mRNA level was approximately 2-fold increased on average, compared with the control zone, with a maximum 3-fold increase observed 2 d after ripening induction and a minimum observed in overripe fruit taken 7 d after ripening induction. During ripening, no marked changes were observed on the MaPME3 mRNA level in both control and drop zones, except at the late ripening stage when MaPME3 mRNA accumulated approximately 2-fold more in the control zone than in the drop zone.
Fig. 3.
Expression of banana pectolytic genes in banana peel tissue from the control and drop zones. Quantitative real-time PCR (RT-qPCR) was used to analyse mRNA accumulation of banana pectolytic genes, including pectin methylesterase (A), pectate lyase (B), and polygalacturonase (C) in peel tissue sampled from the middle area (control zone) and rupture area (drop zone). The y-axis represents the relative fold difference in mRNA level and was calculated using the 2–ΔΔCt formula (Livak and Schmittgen, 2001) with actin as reference. The mRNA fold difference was relative to that of fruit taken at harvest time (Ht) used as calibrator. Each data point is the mean of values obtained from qPCR reactions performed in triplicate on one cDNA sample. Each sample was prepared from four fruits from three replicate bunches. Two biological experiments were performed and they gave similar results. Vertical bars indicate standard deviation (SD). When no bar is shown, the SD is smaller than the symbol.
From day 1 to day 7 after ripening induction, both MaPEL1 and MaPEL2 mRNA levels increased drastically and continuously in the control zone (Fig. 3B). One day after ripening induction, MaPEL1 and MaPEL2 mRNA were 2-fold more expressed in the drop zone than in the control zone. The time-course of both MaPEL1 and MaPEL2 mRNA presented a transient decrease (4-fold) in the drop zone at day 2 after ripening induction. Thereafter, transcripts of both MaPEL genes increased to reach a level comparable with that of the control zone at day 3 for MaPEL2 and 4 for MaPEL1, respectively.
Concerning polygalacturonase genes, the four cDNAs previously isolated in banana present, at nucleotide level, a high percentage of identity. MaPG1 and MaPG2 were 98% identical, although MaPG1 has an additional 160 bp. MaPG3 and MaPG4 were over 90% identical while MaPG2 and MaPG3 were only 30% identical (Asif and Nath, 2005). Despite this, a set of qPCR primers was designed, as specific as possible, for each MaPG gene. For MaPG1 that spans the entire MaPG2 cDNA, primers were designed within the additional 160 bp in the 3′-UTR. For MaPG2, there was a failure to identify a divergent region with MaPG1 to design a specific MaPG2 primer. Nevertheless, MaPG1 and MaPG2 displayed quite different accumulation patterns, suggesting that putative cross amplification of the MaPG1 gene with the MaPG2 primer set would not have interfered during the gene expression analysis. No marked changes were observed on MaPG1 gene expression during fruit ripening in both the control and drop zones (Fig. 3C). In the control zone, the time-course of MaPG2 mRNA accumulation increased during ripening from day 1 to day 4 after ripening induction and decreased thereafter. A similar pattern was observed for MaPG2 in the drop zone, except for a marked but transient decrease (2-fold) observed at day 2 after ripening induction. The mRNA level of the MaPG3 gene decreased during ripening in the same manner in both the control and drop zones until day 6 after ripening induction. In overripe fruit taken at day 7, this level was approximately 10-fold higher in the control zone than in the drop zone. However, this level remained comparable to that of green fruit sampled at harvest. Finally, concerning MaPG4, the corresponding transcript was drastically induced in the control zone during the late ripening stages, namely 5, 6, and 7 d after ripening induction. In the drop zone, two peaks of MaPG4 mRNA accumulation were observed. The first one occurred 1 d after ripening induction, with approximately 400-fold more expression in the drop zone than in the control zone, and the second one, during the late ripening stage, began at day 5 after ripening induction and reached its maximum at day 7, but at a 2-fold lower level than in the control zone.
Expression of MaXTH genes in peel tissues from control and drop zones during banana fruit ripening
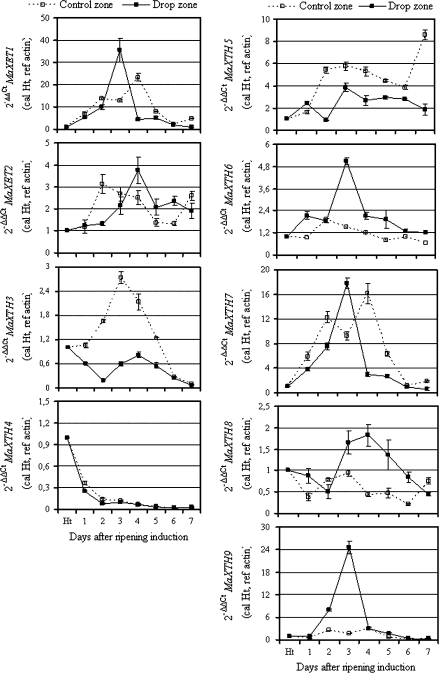
In addition to the two XET genes, namely MaXET1 and MaXET2, previously isolated from banana fruit (Lu et al., 2004), seven other cDNA sequences were isolated in this study. In accordance with the nomenclature proposed by Rose et al. (2002), these cDNAs were named MaXTH3 to MaXTH9. Specific primers designed within the 3′-UTR regions of each XET and XTH genes were used to examine the corresponding gene expression throughout qPCR analysis. Figure 4 shows the expression pattern in the drop and control zones of the nine XTH genes during banana fruit ripening. Except for MaXTH4, all other XTHs examined in this study were induced at different magnitudes in banana peel tissue, in the control and drop zones. In the control zone, MaXET1 mRNA was transiently induced during ripening with maximum expression observed 4 d after ripening induction; in line with previous Northern blot data reported in banana (Lu et al., 2004). Four XTH genes (MaXET1, MaXTH6, MaXTH8, and MaXTH9) were highly induced at the transcriptional level in the drop zone as compared to the control zone. However, this induction was transient, with a maximum of mRNA accumulation observed 3–4 d after ripening induction. MaXTH9 was the most highly and transiently expressed as compared to the four others, while MaXTH8 expression decreased slowly during the late ripening stage. By contrast, two XTH genes (MaXTH3 and MaXTH5) were more highly and differentially expressed in the control zone than in the drop zone. MaXTH3 was transiently induced in the control zone with a maximum observed 3 d after ripening induction, while in the drop zone no marked change was observed during post-harvest ripening. MaXTH5 was less expressed in the drop zone compared to the control zone. In this area, MaXTH5 presented two peaks of expression at days 3 and 7 after ripening, respectively. In the control zone, MaXET2 and MaXTH7 mRNA accumulation peaked twice at days 2 and 7, and at days 2 and 4 after ripening induction, respectively. In the drop zone, a high mRNA accumulation of MaXET2 and MaXTH7 genes was transient at days 4 and 3 after ripening induction, respectively. At this ripening stage, both genes were highly expressed in the drop zone compared to the control.
Fig. 4.
Expression of banana XTH genes in banana peel tissue from the control and drop zones. Quantitative real-time PCR (RT-qPCR) was used to analyse the mRNA accumulation of banana xyloglucan transglycosylase/hydrolase genes in peel tissue sampled from the middle area (control zone) and rupture area (drop zone). The y-axis represents the relative fold difference in mRNA level and was calculated using the 2–ΔΔCT formula (Livak and Schmittgen, 2001), with actin as reference. The mRNA fold difference was relative to that of fruit taken at harvest time (Ht) used as calibrator. Each data point is the mean of values obtained from qPCR reactions performed in triplicate on one cDNA sample. Each sample was prepared from four fruits from three replicate bunches. Two biological experiments were performed and they gave similar results. Vertical bars indicate standard deviation (SD). When no bar is shown, the SD is smaller than the symbol.
Expression of MaEXP genes in peel tissues from control- and drop zones, during banana fruit ripening
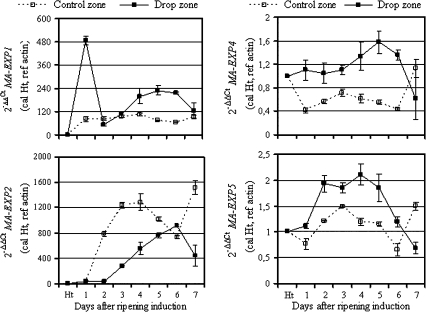
Based on five sequences of expansin genes already isolated from banana fruit (Trivedi and Nath, 2004; Sane et al., 2007), specific primers were designed from the corresponding 3′-UTR regions and used to examine expansin gene expression in both the control and drop zones (Fig. 5). Except for MaEXP3, from which there was a failure to get a single PCR product, the four other expansin genes were differentially expressed in control and drop zone tissues. By contrast with those of MaEXP2, mRNAs of MaEXP1, MaEXP4, and MaEXP5 accumulated to a greater extent in the drop zone than in the control zone. MaEXP2 mRNA presented a different accumulation pattern in both the control and drop zones as compared to the three other expansin genes analysed in this study. In the control zone, its mRNA began to accumulate 1 d after ripening induction and increased drastically (approximately 20–34-fold) until day 4 after ripening induction, when it peaked. Then this level slightly decreased at days 5 and 6, before increasing (2-fold) at day 7 after ripening induction. In the drop zone, MaEXP2 mRNA accumulated continuously from days 1 to 6 after ripening induction, where it peaked before decreasing at day 7. Compared to the pattern in the control zone, MaEXP2 was on average 1.3–20-fold less expressed in the drop zone during post-harvest ripening, except at day 6 where this expression was comparable in the two tissues. MaEXP1 mRNA sharply accumulated (480-fold) 1 d after ripening induction. This level decreased at day 2 to reach a level comparable to that of the control zone before increasing transiently, on average 2-fold more, from days 4–6. At day 7, the MaEXP1 mRNA level had reduced to a level comparable to that of the control zone. A marked increase in the MaEXP4 mRNA level was observed during the late ripening stages, namely 4, 5, and 6 d after ripening induction before decreasing approximately 3-fold at day 7 after ripening induction. Compared to the control zone and except at the last ripening stage (day 7), MaEXP4 mRNA accumulated, on average, 2-fold more in the drop zone after ripening, even during the first ripening stages (1–3 d after ripening induction), while this level was constant in the drop zone. In the drop zone, MaEXP5 gene expression was transiently induced until day 4 after ripening induction, where it peaked before decreasing during the last three ripening stages. The same pattern was observed in the control zone, except that (i) peak mRNA accumulation was observed 3 d after ripening induction, and (ii) with a 2-fold increase at the last ripening stage (day 7). Compared to the control zone, the MaEXP5 mRNA level was higher in the drop zone except at the last ripening stage.
Fig. 5.
Expression of expansin genes in banana peel tissue from the control and drop zones. Quantitative real-time PCR (RT-qPCR) was used to analyse the mRNA accumulation of banana expansin genes in peel tissue sampled from the middle area (control zone) and rupture area (drop zone). The y-axis represents the relative fold difference in mRNA level and was calculated using the 2–ΔΔCt formula (Livak and Schmittgen, 2001) with actin as reference. The mRNA fold difference was relative to that of fruit taken at harvest time (Ht) used as calibrator. Each data point is the mean of values obtained from qPCR reactions performed in triplicate on one cDNA sample. Each sample was prepared from four fruits originated from three replicate bunches. Two biological experiments were performed and they gave similar results. Vertical bars indicate standard deviation (SD). When no bar is shown, the SD is smaller than the symbol.
Discussion
In the work described here, changes in the expression of various CWMGs (cell wall modifying genes) were examined in relationship with the banana finger drop process. This study was primarily aimed at identifying genes whose expression in the drop zone could be linked to the cell wall disassembly and property changes observed during banana ripening and finger drop by Imsabai et al. (2006) and Saengpook et al. (2007).
Current evidence suggests that cellulose disassembly is not a significant contributor to cell wall disassembly during fruit ripening (Fisher and Bennett, 1991; Newman and Redgwell, 2002; Bennett and Labavitch, 2008). Therefore, our molecular studies were focused on pectolytic and hemicellulosic genes and those involved in physical cell wall properties. Taken together, our data indicated that cell wall changes that occur in the finger drop area involve major cell wall components, including xyloglucan and pectin, and also cell wall physical properties, namely cell wall loosening mediated by expansin. Overall, major changes in cell wall modifying gene expression occurred 1–4 d after ripening induction, but in a sequential manner. Firstly, there were changes in the expression of pectolytic and cell wall loosening genes, mainly during days 1–2, followed by those of xyloglucan genes, mainly during days 3–4. Moreover, within each mechanism, there was also sequential induction of the different genes involved.
The results of a comparative analysis of CWMG expression in drop and control zones allows the selection of candidate genes involved in the finger drop process. Assuming that the activity of CWMGs examined in this study such as PEL or PG are mainly regulated at the transcriptional level in banana (Pathak and Sanwal, 1998; Marin-Rodriguez et al., 2003; Lohani et al., 2004), it is considered that CWMGs expressed more in the drop zone than in the control zone were candidates.
A positive correlation was obtained between the banana cultivar prone to finger drop and PEL activity (Imsabai et al., 2006). Both MaPEL1 and MaPEL2, whose mRNA levels increased by 2-fold and 1.6-fold in the drop zone, possibly induced this increased PEL activity in the drop zone. Therefore, both MaPEL1 and MaPEL2 genes could be considered as candidates. Imsabai et al. (2006) reported that no clear change in PG activity was correlated with the finger drop process, and that PME activity does not account for the breakage of pectin associated with the finger drop process. Despite this, MaPME2 and MaPG4 could also be considered as putative candidates of finger drop based on the marked change in their mRNA accumulation in the drop zone relative to the control zone. The discrepancy between PG and PME gene expression and activities could be explained by post-transcriptional regulation of the activation of enzyme activity or subcellular localization (targeting cell walls). It is also possible that the PG and PME activities reported by Imsabai et al. (2006) might represent global activities that do not reflect a putative change in the activity of the different PG and PME isoforms. Note that the PG enzyme activity measured during banana fruit ripening suggested that more than the three identified isoforms might exist in fruit (Pathak and Sanwal, 1998; Pathak et al., 2000). For other cell wall components, MaEXP1, MaEXP4, and MaEXP5 genes appeared to be the main candidates involved in cell wall loosening related to finger drop, with MaEXP1 being the main one, while MaXET1, MaXET2, MaXTH6, MaXTH7, MaXTH8, and MaXTH9 appeared to be candidates for xyloglucan metabolism.
XTH proteins can display two distinct enzymatic activities, including transglycosylase enzymatic (XET) activity leading to xyloglycan chain synthesis or xyloglucan hydrolase activity (XEH) resulting in their degradation. These two activities are not correlated with the protein structure and subsequently with the XTH protein organization (Rose et al., 2002; Fry 2005; Saladié et al., 2006). On the other hand, the involvement of cell wall synthesis, including the xyloglucan component, throughout ripening and mainly between mature green and turning stages prior to perceptible softening, have been reported in tomato, i.e. another fleshy fruit like banana (Mitcham et al., 1989, 1991; Greve and Labavitch; 1991; Huysamer et al., 1997; Rose and Bennett, 1999). Because the XTH gene expression and phylogenetic data reported in this study do not presume the activity and the function of corresponding proteins, the involvement of both xyloglucan synthesis and degradation mediated by XTH proteins in the finger drop process cannot therefore be excluded.
Cell wall modification that occurs during the ripening process is complex. Although the data reported here highlight the involvement of the main cell wall modification enzymes, including PG, PEL, XTHs, and EXP, the possibility cannot be overlooked that, other secondary but important enzymes (β-galactosidase or α-L-arabinofuranosidase) or non-enzymatic mechanisms such as cell wall solubilization (Dumville and Fry, 2003; Zhuang et al., 2006, 2007) could be involved in the finger drop process.
Ethylene production and fruit softening are two main features that characterize the ripening process of climacteric and fleshy fruit like banana. Our data suggested that finger drop could be related to these two processes. Indeed, finger drop implies transcriptional changes in genes that are also involved in fruit softening. However, some genes seem to be specific to softening (MaXTH5, MaXTH3, and MaEXP2), finger drop (MaPME2, MaEXP1, MaEXP4, and MaEXP5, MaXTH6, MaXTH8, and MaXTH9), or both (MaPG4, MaPEL1, and MaPEL2; MaXET1, MaXET2, and MaXTH7). Moreover, the two processes seem to be quite distinct in this variety, as shown by the type of genes involved and their sequential induction, with finger drop being the main induced process followed by softening. Regarding ethylene, our data showed that Cavendish banana fruit developed finger drop mainly during the early stage of fruit ripening (days 1–4), a period corresponding to a burst of ethylene production. Concomitantly, a high level of transcription of some ripening- and ethylene-dependent genes like MaEXP1 and MaPELs (Pua et al., 2001; Trivedi and Nath, 2004) was also observed. It would be interesting to examine the relationship between ethylene production during ripening and the finger drop process. Does the specific induction of the banana finger drop process involve, at molecular level, ethylene production and/or require the ethylene transduction pathway or not?
In conclusion, the data reported here represent a substantial contribution towards understanding the post-harvest banana finger drop phenomenon at the molecular level. More specifically, the identification of several cell wall-modifying genes with expression profiles related to finger drop provided us with good candidates for functional studies in banana species. These candidate genes could be useful in molecular breeding schemes for banana improvement using cultivars differing in their proneness to develop finger drop and which are currently being assessed.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. Sequences of primers used in this study.
Table S2. Different cis-regulatory elements found within the promoter of MaPME2 and MaPME3 genomic sequences.
Fig. S1. Multiple sequence alignment of polypeptide sequences of banana XTH genes.
Fig. S2. BAC localization and structure of banana PME genes and multiple sequence alignment of polypeptide sequences.
Supplementary Material
Acknowledgments
This research was supported by the DOCUP (Document Unique de Programmation) 2000–2006 fund and The Generation Challenge Program (GCP-SP2-project no. 15).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Reseach. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M, Nath P. Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant Physiology and Biochemistry. 2005;43:177–184. doi: 10.1016/j.plaphy.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Baldry J, Coursey DG, Howard GE. The comparative consumer acceptability of triploid banana fruit. Tropical Science. 1981;23:33–66. [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bennett AB, Labavitch JM. Ethylene and ripening-regulated expression and function of fruit cell wall modifying proteins. Plant Science. 2008;175:130–136. [Google Scholar]
- Brummell DA. Cell wall disassembly in ripening fruit. Functional Plant Biology. 2006;33:103–119. doi: 10.1071/FP05234. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglucosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends in Plant Science. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Chillet M, De Lapeyre De Bellaire L, Hubert O, Mbéguié-A-Mbéguié D. Mechanical characterization of banana fruits. Fruits. 2008;63:51–52. [Google Scholar]
- Dominguez-Puigjaner E, Llop I, Vendrell M, Prat S. A cDNA clone highly expressed in ripe banana fruit shows homology to pectate lyases. Plant Physiology. 1997;114:1071–1076. doi: 10.1104/pp.114.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumville JC, Fry SC. Solubilization of tomato fruit pectins by ascorbate: a possible non-enzymatic mechanisms of fruit softening. Planta. 2003;217:951–961. doi: 10.1007/s00425-003-1061-0. [DOI] [PubMed] [Google Scholar]
- Fischer RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Annual Review of Plant Physiology and Plant Mololecular Biology. 1991;42:675–703. [Google Scholar]
- Foissac S, Gouzy J, Rombauts S, Mathé C, Amselem J, Sterck L, Van de Peer Y, Rouzé P, Schiex T. Genome annotation in plants and fungi: EuGène as a model platform. Current Bioinformatics. 2008;3:87–97. [Google Scholar]
- Frohman MA. RACE: rapid amplificatiopn of cDNA ends. In: Innis MA, Gelfrand DH, Sninsky JJ, White JJ, editors. PCR protocol: a guide to methods and applications. New York: Academic Press; 1990. pp. 28–38. [Google Scholar]
- Fry SC. Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytology. 2005;161:641–675. doi: 10.1111/j.1469-8137.2004.00980.x. [DOI] [PubMed] [Google Scholar]
- Ganry J, Meyer JP. Recherche d'une loi d'action de la température sur la croissance des fruits du bananier. Fruits. 1975;30:375–392. [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D'Avino R, Tamburrini M, Ciardiello MA, Camardella L. Pectin methylesterase inhibitor. Biochimica et Biophysica Acta Proteins Proteomics. 2004;1696:245–252. doi: 10.1016/j.bbapap.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Goulao LF, Oliveira MC. Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trend in Food Science and Technology. 2008;19:4–25. [Google Scholar]
- Greve CL, Labavitch JM. Cell wall metabolism in ripening fruit. V. Analysis of cell wall synthesis in ripening tomato pericarp tissue using a D-[U-13C]Glucose tracer and gas chromatography-mass spectrometry. Plant Physiology. 1991;97:1456–1461. doi: 10.1104/pp.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks EW. Finger dropping from bunches of Australian Cavendish bananas. Journal of Council Science of Indian Research. 1934;7:165–168. [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Place cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysamer M, Greve CL, Labavitch JM. Cell wall metabolism in ripening fruit. IX. Synthesis of pectic and hemicellulosic cell wall polymers in the outer pericarp of mature green tomato (cv. XMT-22) Plant Physiology. 1997;114:1523–1531. doi: 10.1104/pp.114.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsabai W, Ketsa S, van Doorn W. Physiological and biochemical changes during banana ripening and finger drop. Post-harvest Biology and Technology. 2006;39:211–216. [Google Scholar]
- Johansson K, El-Ahmad M, Friemann R, Jörnvall H, Markovic O, Eklund H. Crystal structure of plant pectin methylesterase. FEBS Letters. 2002;514:243–249. doi: 10.1016/s0014-5793(02)02372-4. [DOI] [PubMed] [Google Scholar]
- Jullien A, Chillet M, Malezieux E. Preharvest growth and development determine post-harvest green life of fruit in Musa (Musa sp. AAA group cv. Grande Naine (Cavendish subgroup)) Journal of Horticultural Science and Biotechnology. 2008;83:506–512. [Google Scholar]
- Lescot M, Dahais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time Quantitative PCR and the 2–ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Nakano R, Kubo Y, Inaba A, Jiang YM. Cloning and expresion analysis of an XET cDNA in the peel and pulp of banana fruit ripening and softening. Acta Botanica Sinica. 2004;46:355–362. [Google Scholar]
- Liu F. Correlation between banana storage life and minimum treatment time required for ethylene response. Journal of the American Society of Horticultural Science. 1976;101:63–65. [Google Scholar]
- Lohani S, Trivedi PK, Ntah P. Changes in activities of cell wall hydrolases during ethylene-induced ripening in banana: effect of 1-MCP, ABA and IAA. Post-harvest Biology and Technology. 2004;31:119–126. [Google Scholar]
- Marin-Rodriguez MC, Smith DL, Manning K, Orchard J, Seymour GB. Pectate lyase gene expression and enzyme activity in ripening banana fruit. Plant Molecular Biology. 2003;51:851–857. doi: 10.1023/a:1023057202847. [DOI] [PubMed] [Google Scholar]
- Markovic O, Janecek S. Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydrate Research. 2004;339:2281–2295. doi: 10.1016/j.carres.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Mbéguié-A-Mbéguié D, Fils-Lycaon B, Chillet M, Hubert O, Galas C, Gomez RM. Extraction and purification of total RNA from banana tissues (small scale) Fruit. 2008a;63:255–261. [Google Scholar]
- Mbéguié-A-Mbéguié D, Hubert O, Fils-Lycaon B, Chillet M, Baurens FC. EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande naine) Physiologia Plantarum. 2008b;133:435–448. doi: 10.1111/j.1399-3054.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- Micheli F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends in Plant Sciences. 2001;6:414–419. doi: 10.1016/s1360-1385(01)02045-3. [DOI] [PubMed] [Google Scholar]
- Mitcham EJ, Gross KC, Ng TJ. Tomato fruit cell synthasis during development and senescence. In vivo radiolabeling of wall fractions using [14C]sucrose. Plant Physiology. 1989;89:477–481. doi: 10.1104/pp.89.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitcham EJ, Gross KC, Ng TJ. Ripening and cell wall synthesis in normal and mutant tomato fruit. Phytochemistry. 1991;30:1777–1780. [Google Scholar]
- New S, Marriott J. Factors affecting the development of finger drop in bananas after ripening. Journal of Food Technology. 1983;18:241–250. [Google Scholar]
- Newman RH, Redgwell RJ. Cell wall changes in ripening kiwifruit: C-13 solid state NMR characterization of relatively rigid cell wall polymers. Carbohydrates Polymers. 2002;49:121–129. [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell wall. International Review of Cytology. 1997;173:157–206. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic tree on personal computers. Computers Applied Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pathak N, Mishra S, Sanwal GG. Purification and characterization of polygalacturonase from banana fruit. Phytochemistry. 2000;54:147–152. doi: 10.1016/s0031-9422(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Pathak N, Sanwal GG. Multiple forms of polygalacturonase from banana fruit. Phytochemistry. 1998;48:249–255. doi: 10.1016/s0031-9422(98)00005-3. [DOI] [PubMed] [Google Scholar]
- Paull RE. Ethylene, storage and ripening temperatures affect Dwarf Brazilian banana finger drop. Post-harvest Biology and Technology. 1996;8:65–74. [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin ethylesterase structure and function. Trends in Plant Sciences. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Dolezel J, Sabau X, et al. 7th international congress of plant molecular biology. Barcelona, Spain: Book of abstracts; 2003. Musagenomics: creation of a BAC-based platform for banana genomics. [Google Scholar]
- Prestridge DS. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Pua EC, Ong CK, Liu P, Liu JZ. Isolation and expression of two pectate lyase genes during fruit ripening of banana (Musa acuminata) Physiologia Plantarum. 2001;113:92–99. [Google Scholar]
- Rose JKC, Bennett AB. Cooperative disassembly of the cellulose–xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends in Plant Science. 1999;4:176–183. doi: 10.1016/s1360-1385(99)01405-3. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotranglycosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Roten S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Saengpook C, Ketsa S, van Doorn W. Effects of relative humidity on banana finger drop. Post-harvest Biology and Technology. 2007;45:151–154. [Google Scholar]
- Saladié M, Rose JCK, Cosgrove DJ, Catala C. Charaacterization of a new xyloglucan endotranglycosylases/hydrolase (XTH) from ripening tomato fruit and implications for diverse mode of enzymic action. The Plant Journal. 2006;47:282–295. doi: 10.1111/j.1365-313X.2006.02784.x. [DOI] [PubMed] [Google Scholar]
- Sane AVA, Sane AP, Nath P. Multple forms of α-expansin genes are expressed during banana fruit ripneing and development. Post-harvest Biology and Technology. 2007;45:184–192. [Google Scholar]
- Semple AJ, Thompson AK. Influence of the ripening environment on the development of finger drop in bananas. Journal of the Science of Food and Agriculture. 1988;46:139–146. [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences: a new tool for comparing protein and nucleotide sequences. FEMS Microbiology Letters. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thomson JD, Gibson TJ, Plewniak J, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: flexible strategies for multiple alignment aides by quality analysis tools. Nuclei Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi PK, Nath P. MaEXP1, an ethylene-induced expansin from ripening banana fruit. Plant Science. 2004;167:1351–1358. [Google Scholar]
- Vicente AR, Saladié M, Rose JCK, Labavith JM. The linkage between cell wall metabolism and fruit softening: looking to the future. Journal of Science of Food and Agriculture. 2007;87:1435–1448. [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton. Analytical Biochemistry. 1994;223:7–13. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JCK, Nishitani K. A suprising diversity and abundance of xyloglucan endotransglycosylase/hydrolases in rice. Classification and expression analysis. Plant Physiology. 2004;134:1025–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JP, Su J, Li XP, Chen WX. Cloning and expression analysis of beta-galactosidase gene related to softening of banana (Musa sp.) fruit. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xu Bao. 2006;32:411–419. [PubMed] [Google Scholar]
- Zhuang JP, Su J, Li XP, Chen WX. Changes in alpha-L-arabinofuranosidase activity in peel and pulp of banana (Musa sp.) fruits during ripening and softening. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xu Bao. 2007;33:131–136. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.