Abstract
Germline mutations that inactivate BRCA1 are responsible for breast and ovarian cancer susceptibility. One possible outcome of genetic testing for BRCA1 is the finding of a genetic variant of uncertain significance for which there is no information regarding its cancer association. This outcome leads to problems in risk assessment, counseling and preventive care. The purpose of the present study was to functionally evaluate seven unclassified variants of BRCA1 including a genomic deletion that leads to the in-frame loss of exons 16/17 (Δ exons 16/17) in the mRNA, an insertion that leads to a frameshift and an extended carboxy-terminus (5673insC), and five missense variants (K1487R, S1613C, M1652I, Q1826H and V1833M). We analyzed the variants using a functional assay based on the transcription activation property of BRCA1 combined with supervised learning computational models. Functional analysis indicated that variants S1613C, Q1826H, and M1652I are likely to be neutral, whereas variants V1833M, Δ exons 16/17, and 5673insC are likely to represent deleterious variants. In agreement with the functional analysis, the results of the computational analysis also indicated that the latter three variants are likely to be deleterious. Taken together, a combined approach of functional and bioinformatics analysis, plus structural modeling, can be utilized to obtain valuable information pertaining to the effect of a rare variant on the structure and function of BRCA1. Such information can, in turn, aid in the classification of BRCA1 variants for which there is a lack of genetic information needed to provide reliable risk assessment.
Keywords: BRCA1, breast cancer, unclassified variants, functional analysis, BRCT domain
1. Introduction
BRCA1 is a tumor suppressor gene and germline mutations which disrupt its biological activity contribute to breast and ovarian cancer susceptibility [1]. Carriers of these inactivating mutations are at increased risk of developing breast and ovarian cancer with an estimated cumulative risk of breast cancer ranging from 36–71% at age 70 and up to 90% at age 80 in some populations [2–5]. The magnitude of risk remains controversial and varies according with the population studied and with study design [3, 6]. Nevertheless, it is significantly higher than the risk of breast cancer in the general population making genetic testing an important factor in the decision to undergo increased surveillance, chemoprevention, or prophylactic surgery [7]. The BRCA1 gene codes for a nuclear protein of 1863 amino acids that has been found to play a role in many cellular processes including DNA-damage repair, transcriptional activation, cell cycle regulation, apoptosis, and genomic stability [8].
Truncations and missense substitutions are two of the predominant types of BRCA1 mutations that have been identified (Breast Cancer Information Core – BIC - Database; http://research.nhgri.nih.gov/bic/). While most truncating mutations have been found to be cancer-associated, missense variants have proven more difficult to classify [9]. Over three hundred different missense variants of BRCA1 have been identified but presently their clinical significance is unknown despite intense efforts (BIC) [10, 11].
In situations in which there is a lack of clinical and genetic data to classify these variants, functional studies which assess specific biochemical properties of the protein can contribute to the classification of the variant as either deleterious or neutral [12, 13]. The BRCA1 transcription activation (TA) assay evaluates the ability of the COOH-terminus of the protein to function as a transactivation domain and has been used as a monitor of the structural integrity of the domain [12, 14–16]. Cancer-associated missense variants of BRCA1 have been found to exhibit loss of function with respect to transcriptional activity while neutral variants display activity similar to the wild type protein [12, 14]. Prediction of mutation impact on protein function by structure-based models has also been used in the classification of these rare variants [17–19].
Here, we examine functionally seven unclassified variants of BRCA1, three of which have not been reported previously. Those include a deletion of exons 16 and 17 (Δ exons 16/17), an insertion mutation (5673insC) leading to a frameshift that produces a protein which is 15 amino acids longer than the wild type, and missense variant Q1826H. Four missense variants (K1487R, S1613C, M1652I and V1833M) previously reported in the BIC database were also functionally evaluated. We provide a bioinformatics-based prediction of their impact, and for the variants that affect the BRCA1 COOH-terminal (BRCT) domains (Δ exons 16/17, 5673insC, M1652I, Q1826H and V1833M) we also provide a rationalization of their impact, based on structural modeling.
2. Materials and methods
2.1. Constructs
Constructs coding for exons 13–24 (amino acids 1396–1863) of wild-type (wt) BRCA1, positive (S1613G; neutral), and negative (M1775R and Y1853X; deleterious) controls were previously described [14–16]. Mutations were generated by splicing by overlapping extension PCR [20] using p385-BRCA1 [14] (for Δ exons 16/17, K1487R, S1613C, Q1826H, M1652I, and V1833M) or F3-BRCA1 [1] (for 5673insC) as template. PCR products were cloned into pLex9 or pGBT9 vectors. To obtain GAL4-DNA Binding Domain (DBD) fusions in a mammalian expression vector, GAL4-DBD fusion fragments were isolated from pGBT9 and subcloned into pCDNA3. Mutation nomenclature follows the nucleotide numbering found in the BIC database and uses GenBank U14680 as a reference for BRCA1 cDNA with the corresponding protein accession number AAA73985.
2.2. Yeast assays
Saccharomyces cerevisiae strain EGY48 (Matα, ura3, trp1, his3, 6 lexA operator-LEU2) has an integrated LEU2-reporter gene under the control of 6 LexA ops [21] and when the gene is activated, cells grow in the absence of leucine. EGY48 transformants expressing either wt or mutant BRCA1 constructs in pLex9 were grown overnight and saturated cultures were then used to inoculate either Synthetic drop-out (SD) medium lacking both leucine and tryptophan or only tryptophan and growth was measured by OD at 600 nm. EGY48 yeast cells were also co-transformed with the lacZ reporter plasmid pRB1840, which contains a lacZ reporter gene under the control of a single LexA operator, and a pLex9 plasmid coding for either the wt or mutant constructs [22]. β-galactosidase activity was determined using o-Nitrophenyl-β-D-galactopyranoside (ONPG) as the substrate for three independent clones each [23].
2.3. Mammalian assays
Human embryonic kidney (HEK) 293T cells were co-transfected with a reporter plasmid pG5Luc, which contains a firefly luciferase gene under the control of five GAL4 binding sites (5 × Gal4 bs), a pCDNA3 plasmid coding for either the wt or mutants fused to the GAL4 DBD, and an internal control, phRG-TK, which contains a Renilla luciferase gene under a constitutive thymidine kinase basal promoter. Transcriptional activity was determined using a dual luciferase substrate system (Promega).
2.4. Western blotting
Protein levels in yeast or in HEK-293T cells were determined by western blotting using α-LexA DBD (Upstate) or α-GAL4 DBD (Clontech) monoclonal antibodies [24]. Because of co-migration with nonspecific bands, protein levels in HEK-293T cells expressing Δ exons 16/17 or 5673insC were determined by immunoprecipitation using α-GAL4 DBD monoclonal antibody followed by blotting with the same antibody (Santa Cruz).
2.5. Structural analysis
Prediction of the impact of amino acid changes in the BRCT domains was obtained by previously described bioinformatics supervised learning computation models [18, 25–27]. To investigate the putative structural changes generated by the 5673c insertion and deletion of exons 16 and 17 (Δ exons 16/17), we built structural models with SAM-T06 and UNDERTAKER, an all-heavy-atom, fragment packing program for protein structure prediction [28].
2.6. Patients
BRCA1 mutations and sequence variants were either obtained from published data or public databases, or discovered in individuals attending oncogenetic counseling. All patients gave informed consent for the study. Besides the variants reported, no other mutation in BRCA1 (or BRCA2) was detected unless otherwise noted.
2.7. Conceptual basis of the assay
The classification used in this manuscript is based on the ability of BRCA1 constructs fused to a heterologous DNA binding domain to activate transcription of a reporter gene [12]. Previous studies have shown that constructs containing variants that disrupt transcriptional activation are likely to be disease-associated, while variants that do not affect activity and behave similarly to the wt BRCA1 construct are not likely to be disease associated [14 and references therein]. In this heterologous context, the assay is not interpreted as a measure of a bona fide biochemical function of BRCA1 but rather as a monitor of protein integrity. Thus, reduction or abrogation of activity is not only considered to indicate whether certain variants affect BRCA1 tumor suppressive activities mediated by its role in transcription [29] but also to indicate whether a variant affects other functions required for tumor suppression mediated by the COOH-terminus of BRCA1, for example phosphopeptide binding [30]. At least three lines of evidence support this conclusion. First, variants that affect transcriptional activation have been shown to also affect the integrity of the BRCT domain as measured by protease sensitivity or direct crystal structure determination [19, 31, 32]. Second, a bioinformatics structure-based analysis designed to pinpoint surface residues of the BRCT domain that affect transcription activation identified a surface patch which was independently shown to correspond to the phosphopeptide binding pocket in the BRCT domain [33, 34]. Finally, a recent study [14] showed that the transcriptional assay correctly classified all BRCA1 missense variants that have been classified by co-occurrence with other deleterious variants [35] or by integrated methods based on multifactorial likelihood models [9, 11].
2.8. Term definitions for classification and limitations of the assay
Variants subjected to functional analysis are classified as “deleterious”, “neutral”, or left unclassified. “Deleterious” is a variant for which a significant negative impact on protein function has been verified when compared to the wt control. Because inactivation of BRCA1 function has been shown to underlie cancer predisposition, a compromised activity in the assay suggests that the variant is worthy of further investigation as causally-related to cancer predisposition. Conversely, “neutral” is a variant for which no negative impact on protein function has been verified. A series of internal controls is always included to assess reproducibility and to prevent misinterpretation due to unlikely but possible technical issues (e.g. plasmid mix-ups, yeast cross-contamination, degradation of luciferase reagents). These controls have been classified by genetic and/or integrative methods: a) S1613G, a neutral polymorphic allele of BRCA1 [36, 37]; b) missense and nonsense deleterious variants M1775R and Y1853X, respectively [1, 14, 15, 37, 38].
To transform a quantitative measure, i.e. protein activity, into a qualitative classification we defined thresholds of activity (<45% of wild type activity: deleterious; >50% of wild type activity: neutral) considering the known deleterious variant that displays the highest activity and the known neutral variant displaying the lowest activity [14]. Variants whose range of percent activity (taking into account the standard deviation) falls in the intermediate range (between 45 and 50%) are left unclassified. Embedded in this classification is the simplifying notion that variants are either high or low (comparable to the risk in the general population) risk with no intermediate risk variants. For a more detailed discussion on intermediate risk see [9, 14, 39]. Lastly, the resulting classes derived from the functional assays in general (deleterious or neutral) have different degrees of certainty. For example, the disruption of function is due, in most cases, to a general effect on folding and it is likely to be deleterious to any other function dependent on that region. However, a cancer-causing surface change that does not affect folding and does not participate in the interaction being assessed in the assay may be incorrectly classified as neutral [9, 13]. Although no such case has been documented it remains a possibility.
3. Results
3.1. Rationale for choice of variants
The transcriptional assay has been widely used to classify missense variants but more dramatic changes in protein structure have not been systematically analyzed. Most nonsense and frameshift mutations are expected to completely disrupt the BRCT domain or eliminate it altogether, in a way that we can infer that they constitute loss of function mutations [24]. However, this is not the case for frameshift mutations that occur relatively close to the COOH-terminus. These variant proteins may still retain function. Thus, we decided to analyze the 5673insC variant which causes a change of only 12 amino acids.
Similarly, in-frame deletions that cause loss of complete exons but retain the frame for the remaining ones may also have activity. Because in frame deletions need to be confirmed using clinical samples (to verify that the protein product is indeed what was predicted from the DNA sequence) we chose variant Δ exons 16/17 for which these data were available.
We also analyzed five additional unclassified missense variants (Fig. 1A). Two of them (K1487R, S1613C) were chosen in order to increase coverage of the unstructured region of BRCA1 COOH-terminus and test the hypothesis that variants outside conserved domains are likely to be neutral. The remaining three (M1652I, Q1826H and V1833M) are missense variants in the BRCT domain and were chosen because they have been previously analyzed for binding to phosphopeptides (M1652I and V1833M)[31, 33], or exposed at the surface of the domain (Q1826H) which represents a class of poorly studied variants [18].
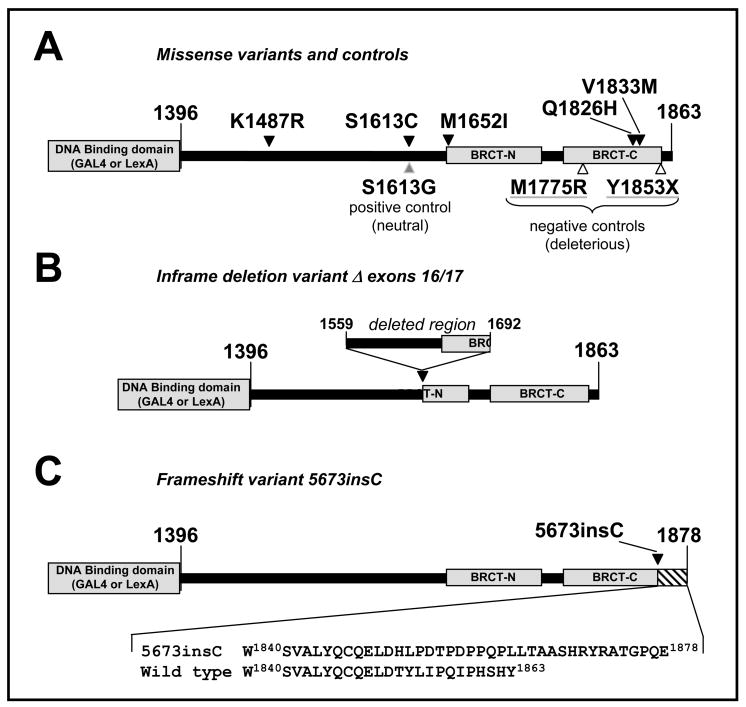
Fig. 1. Diagram of constructs containing BRCA1 variants analyzed in this study.
A. Location of missense variants (black arrowhead), positive control (grey arrowhead) and negative controls (open arrowheads). Grey boxes indicate the DNA binding domain (of GAL4 or LexA, depending on the construct) and the BRCT domains. B. Construct containing in-frame deletion Δ exons 16/17. Diagram shows the region that is left out after splicing of exon 15 to exon 18, disrupting the BRCT-N. C. Construct containing variant 5673insC with site of insertion indicated (black arrowhead). Bottom panel shows detail of wild type and mutant amino acid sequence.
3.2. Novel Variants
3.2.1.Δ exons 16/17
This variant was identified in an Australian proband in which no other mutation in BRCA1 or BRCA2 was detected (Fig. 2A). This variant is a large deletion of the genomic region including exons 16 and 17, which produces stable mRNA lacking exons 16 and 17. The net effect of the mutation is the in-frame loss of amino acids 1559–1692 (Fig. 1B). Using an RNA-based test, we confirmed the presence of the mutant mRNA transcript, lacking exons 16 and 17 (Fig. 2B & 2C), but retaining the correct frame in the splicing of exon 15 to 18 (Fig. 2D).
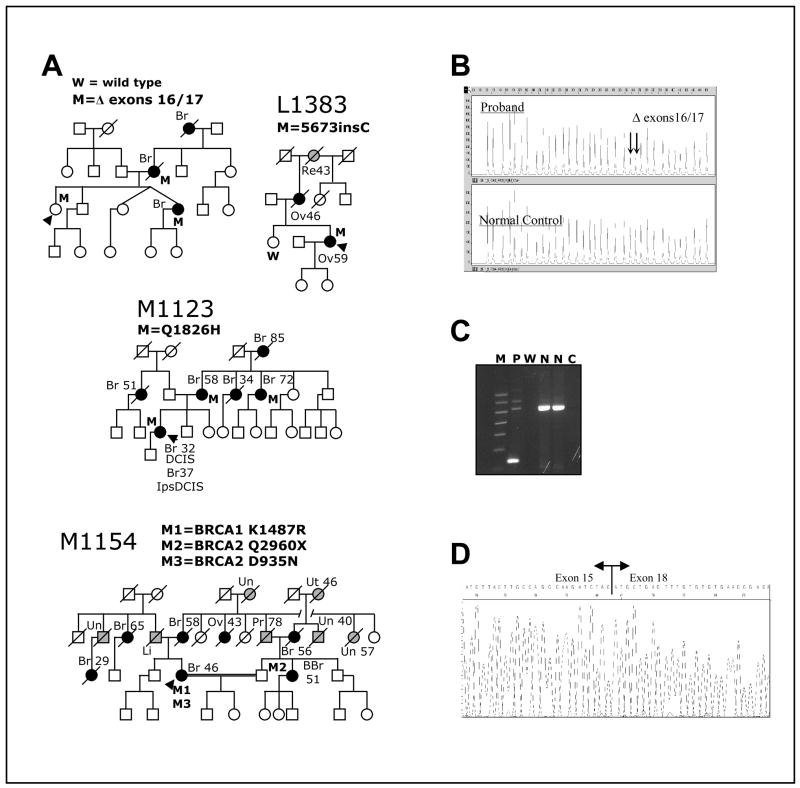
Fig. 2. Clinical data for BRCA1 variants.
A. Pedigrees of families carrying different variants. The presence (M) or absence (W) of the variants in the germ line of tested individuals is indicated. Proband is indicated by black arrowhead. Individuals affected by breast or ovarian cancer are denoted by a black circle or square and individuals affected by other cancers are denoted by a grey circle or square; site of tumor and age of diagnosis, when available is also indicated: Br, breast (individuals with two Br, reflect contralateral breast cancer); BBr, bilateral breast cancer; DCIS, ductal carcinoma in situ; Ips, ipsilateral; Li, liver; Ov, ovary; Pr, prostate; Re, rectal; Sk, skin; Un, unknown site; Ut, uterine. B. Multiplex Ligation-dependent Probe Amplification (MLPA) analysis of genomic DNA showing BRCA1 Δ exons 16/17. Note the halved intensity of the peaks indicated by arrows (corresponding to probes for exons 16 and 17, respectively) in the proband sample relative to the normal control specimen. It indicates genomic hemizygosity for BRCA1 Δ exons 16/17. C. Agarose gel of PCR products from cDNA with primers situated in exons 15 and 18 to amplify across the region containing the deletion shows BRCA1 deletion of exons 16 and 17 in proband’s RNA. M, molecular weight marker; P, Proband; W, water blank; N, normal control; C, RT blank. D. Sequencing of PCR amplicon from cDNA showing BRCA1 Δ exons 16/17 in proband’s RNA. Note that splicing of exon 15 to exon 18 preserves the reading frame.
3.2.2. 5673insC
Variant 5673insC codes for an insertion of a cytosine at nt5673 in exon 24, leading to a frameshift that changes the last 12 amino acids of the protein to a modified 27-amino acid segment (Fig. 1C). This variant was identified in a Swedish proband with ovarian cancer (at 59y) and whose mother also had ovarian cancer (at 46y) (Fig. 2A). The proband’s grandmother died of rectal cancer at age 43. LOH analysis of the proband’s tumor revealed loss of the wild-type BRCA1 allele (data not shown).
3.2.3. Q1826H
This variant was identified in family M1123 (Fig. 2A). This variant results from nucleotide change 5597G→T in BRCA1. The variant was predicted by two different computational prediction tools (ESEfinder, http://exon.cshl.edu/ESE/ and RESCUE-ESE, http://genes.mit.edu/burgelab/rescue-ese/) to destroy a potential exonic splicing enhancer (ESE) sequence in exon 24. However, an RT-PCR analysis on a lymphoblastoid cell line derived from the proband did not detect any altered cDNA in the BRCA1 C-terminal region (data not shown). Moreover, sequencing of the amplified cDNA fragment suggested that the mutant allele was expressed at a level comparable to that of the wild-type.
3.2.4. K1487R
This variant was identified in family M1154 (Fig. 2A). BRCAnalysis revealed an additional missense variant in BRCA2 (D935N), which has recently been classified in the BIC database as of no clinical importance.
3.2.5 Limitations of pedigree analysis
Because pedigree analysis was mostly uninformative and insufficient to classify the respective variants (Fig. 2A), we turned to structural-based predictions and functional analyses to characterize the BRCA1 variants (see below).
3.3. Functional and computational analysis
3.3.1 Bioinformatics analysis
We applied our previously published bioinformatics method LS-SNP [18, 25–27] to predict whether the BRCT missense variants M1652I, Q1826H, and V1833M have a deleterious or neutral effect on BRCA1. The method uses supervised learning algorithms that integrate a variety of bioinformatics-derived predictive features, including properties of amino acid sequence changes, evolutionary conservation, location in protein structure and predicted impact on protein structure. These algorithms are “black boxes” and do not explain why a variant may be deleterious. It is possible that a variant BRCA1 protein is unable to fold into a stable structure, that it partially folds and is rendered more susceptible to proteolytic degradation than the wild type, or that binding interactions with ligands or protein partners are adversely affected.
We also assess the predictions of our method, when compared to available genetic or integrative classifications, and compare them to 5 other bioinformatics missense variant prediction methods: SIFT [40], PolyPhen [41], nsSNPanalyzer [42], PMUT [43] and Panther [44] (Table 2, Table 3). We used the web servers for these five methods and submitted 20 variants to each during the first week in August, 2008. Taking into account the genetic/integrative classifications, it appears that SIFT overpredicts Tolerated class, and that PolyPhen, PMUT and Panther overpredict the Damaging, Pathological and Deleterious classes.
Table 2.
Cross validation of the transcription assay with genetic data.
Variants in bold are nonsense, frameshift or deletions.
D. Goldgar, personal communication.
Table 3.
Bioinformatics predictions of the impact of BRCA1 variants in the BRCT domains.
| Variant | LS-SNP | SIFT | PolyPhen | nsSNPanalyzer | PMUT | Panther |
|---|---|---|---|---|---|---|
| M1652I | Neutral | Tolerated | Benign | Disease | Pathological | Not deleterious |
| A1669S | Neutral | Tolerated | Benign | Neutral | Neutral | Not deleterious |
| T1685I | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| M1689R | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| R1699W | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| R1699Q | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| G1706E | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| A1708E | Deleterious | Not Tolerated | Probably Damaging | Neutral | Pathological | Deleterious |
| S1715R | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| T1720A | Neutral | Not Tolerated | Possibly Damaging | Neutral | Neutral | Not deleterious |
| V1736A | Deleterious | Tolerated | Possibly Damaging | Disease | Pathological | Deleterious |
| G1738R | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| L1764P | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| I1766S | Deleterious | Not Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| M1775K | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| M1775R | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| G1788V | Deleterious | Tolerated | Probably Damaging | Disease | Pathological | Deleterious |
| V1804D | Neutral | Not Tolerated | Benign | Neutral | Pathological | Not deleterious |
| Q1826H | Neutral | Tolerated | Probably Damaging | Neutral | Pathological | Deleterious |
| V1833M | Deleterious | Tolerated | Benign | Disease | Neutral | Deleterious |
The three BRCT missense variants analyzed in this study are shaded in gray. We also show predictions of 17 additional BRCA1 variants for which there is a genetic or integrative classification available (“Validation set”, Table 2). Red in the “Variant” column indicates that the genetic or integrative classification is deleterious; Blue that it is neutral. We compare the predictions of our LS-SNP method [18,25] with SIFT [40], PolyPhen [41], nsSNPanalyzer [42], PMUT [43] and Panther [44].
3.3.2 Functional analysis and structural rationalizations
In order to assess the functional impact of the changes in BRCA1 we determined the transcriptional activity of the C-terminal region of BRCA1 in which the variants were introduced. First, we assessed the activity of constructs containing in-frame deletion and frameshift variants (Fig. 1B–C) in yeast and mammalian cells. Yeast cells were co-transformed with a LexA-responsive β-galactosidase reporter gene (see Fig. 3) and a LexA DBD fusion to residues 1396–1863 of wild type BRCA1 (wt), or the same fragment carrying the Δ exons 16/17 or the 5673insC variants. We used the wt and the S1613G neutral polymorphism as positive controls (+). Deleterious mutations M1775R and Y1853X were used as negative controls (−). Three independent yeast clones were tested in triplicates. Mammalian cells were co-transfected with a GAL4-responsive firefly luciferase reporter gene (see Fig. 3), a Renilla luciferase driven by a constitutive promoter (internal control, not shown), and a GAL4 DNA binding domain (DBD) fusion to residues 1396–1863 of wild type BRCA1 (wt), or the same fragment carrying the Δ exons 16/17 or the 5673insC variants. Controls are the same as described above but fused to GAL4 DBD. Measurements were done in triplicates and normalized against the internal transfection controls. The activity of the wild type BRCA1:DBD fusion construct was then expressed as 100%, with the other results placed on this scale. The thresholds of activity (see Materials and Methods) used for classification are indicated by the dotted lines in Figure 3. Constructs containing the Δ exons 16/17 and 5673insC variants failed to activate transcription of the lacZ or the luciferase reporter genes in yeast as well as in mammalian cells, respectively (Fig. 3A).
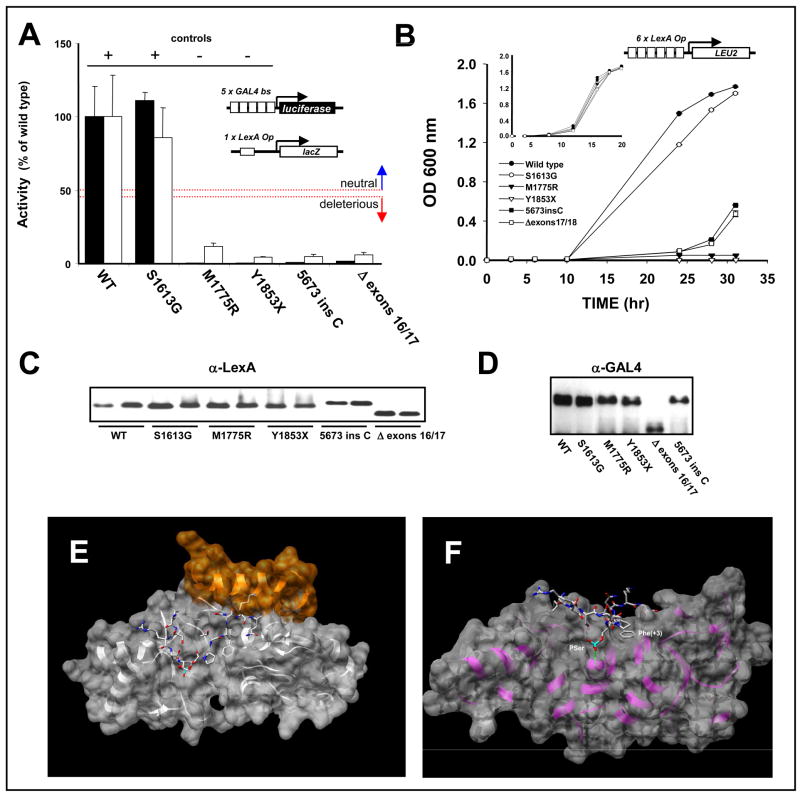
Fig. 3. Functional analysis and structural modeling of frameshift and splicing variants in BRCA1.
A. Quantitative transcriptional assay in yeast (white bars) and in mammalian cells (black bars). B. Transcriptional assay in yeast using a less stringent reporter (diagram shown above the graph) driving LEU2. EGY48 cells expressing wt or mutant BRCA1 constructs were grown in the absence of tryptophan and leucine. Inset, Control growth assay in the absence of tryptophan only. To control for possible variations in protein expression levels, samples were analyzed by western blot using α-LexA DBD polyclonal antibody in yeast extracts (C) or mouse α-GAL4 DBD monoclonal antibody in mammalian cell extracts (D). E. Model of the structural changes induced by the 5673insC variant built with SAM-T06 and UNDERTAKER [25], an all-heavy-atom fragment packing program for protein structure prediction. The insertion may form a novel 13-residue helix (shown in gold). F. Model of the structural changes induced by the Δ exons 16/17 variant built with SAM-T06 and UNDERTAKER. The image shows a transparent surface representation of the model (light gray) superimposed on a ribbon representation (purple) with BACH1 peptide (stick representation colored by element). The two key binding residues Phe(+3) and phosphoserine are labeled. The deletion of exons 16/17 does not impact the hydrophobic binding pocket of Phe(+3), but a hydrogen bond between the backbone nitrogen of GLY 1656 in BRCT-N and the phosphoserine O2P oxygen is disrupted as GLY 1656 is not present in the deletion mutant. Images generated with UCSF Chimera [25,46–47].
In order to determine whether the constructs containing the variants had any residual activity, we also assessed their activity in a less stringent growth assay. Instead of using a reporter gene driven by a single binding site, as in the previous experiment, we use a reporter with six LexA binding sites which allows growth in the absence of leucine when the reporter is activated [21]. Yeast expressing Δ exons 16/17 or 5673insC exhibited a growth pattern similar to the negative controls up to 24h, with modest growth at 28h and 32h (Fig. 3B). In non-selective medium their growth was similar to wt indicating that the expression of the variants did not result in impaired growth (Fig. 3B, inset). Both variant constructs are expressed at levels comparable to the wt in yeast cells (Fig. 3C) but display somewhat reduced levels in mammalian cells (Fig. 3D) indicating that the mutations might confer a destabilizing effect in human cells.
In order to investigate the putative structural changes generated by the 5673insC insertion we built a structural model that suggests that the insertion could generate a novel 13-residue α-helix that might modify the binding of phosphopeptide to the BRCT binding pocket (Fig. 3E). We also built a model for the putative structure of the Δ exons 16/17 variant in which the β1, β2, β3 strands, and the α1 helix are missing (Fig. 3F). Of the two key binding residues in the BACH1 peptide (Phe+3 and phosphoserine) [33], the Phe+3 binding pocket is intact in the deletion model but a hydrogen bond between BRCT-N (backbone nitrogen of GLY 1656) and the phosphoserine O2P oxygen is disrupted because GLY 1656 is missing. We predict that these changes will result in weaker phosphopeptide binding. Taken together, the functional data and the structure prediction suggest that both variants constitute deleterious mutations.
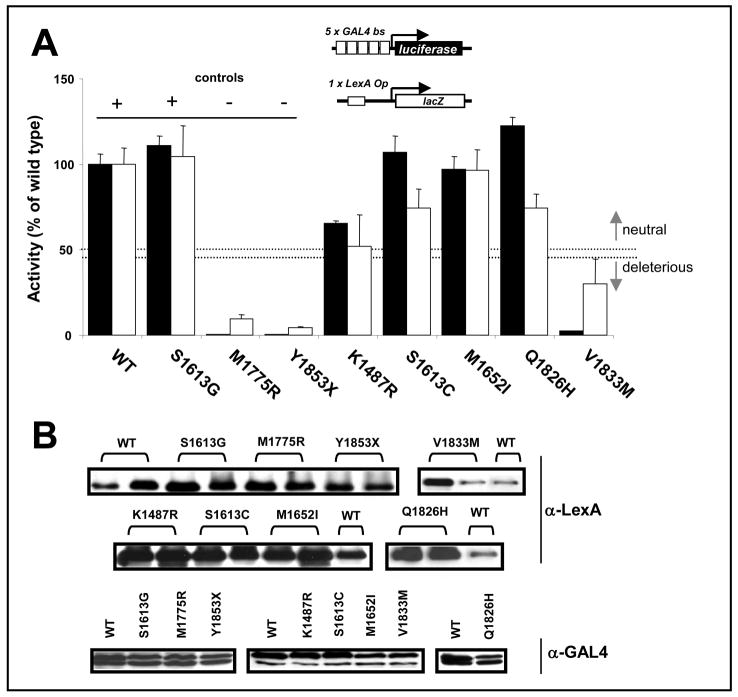
The missense variant series (K1487R, S1613C, M1652I, Q1826H, and V1833M) was evaluated for transactivation activity exclusively in the context of stringent reporters (Fig. 4A) as previously defined for missense variants [12, 14–16]. Variants S1613C, M1652I, and Q1826H exhibited activity significantly higher than 50% of the wt activity and are likely to constitute neutral variants (Fig. 4A). Variant V1833M showed reduced activity of ~30% compared to the wild type in yeast and a markedly reduced activity in mammalian cells. Thus, it is likely to be a deleterious variant (Fig. 4A). Variant K1487R displayed ~50% of the wt activity, which is considered the threshold for classification into deleterious or neutral [14] and therefore cannot be classified with the current data. Differences in activity could not be accounted for by differences in protein levels, as expression of fusion constructs did not necessarily correlate with activity (Fig. 4B).
Fig. 4. Functional analysis of missense variants in BRCA1.
A. Quantitative assay in yeast (white bars) and mammalian cells (black bars) performed as described in Fig. 3. The thresholds of activity (see Material and Methods) used for classification are indicated by dotted lines. B. Protein levels determined by western blot using α-LexA DBD polyclonal antibody for yeast extracts (top panels) or mouse α-GAL4 DBD monoclonal antibody in mammalian cell extracts (bottom panel).
Table 1 summarizes the results obtained when the missense variants located at the BRCT region (M1652I, Q1826H, and V1833M) were rationalized based on our structural models. Variant V1833M is predicted to disrupt the hydrophobic core of the BRCT domain. V1833 in BRCT-C is completely inaccessible to solvent. It lies in the β4 strand, an edge strand of a four-stranded parallel β-sheet. The side chain points inward towards the protein core. Valine is a β-branched residue that is more favored in β-strands than methionine. In addition, methionine is larger than valine, so this replacement likely disrupts the tight packing in the hydrophobic core of the BRCT-C domain. On the other hand, we found no evidence for functional impact in variants M1652I and Q1826H. Residue Q1826 in BRCT-C is on the protein surface and its side chain points outward to the solvent. There are no known or predicted binding interactions at this position. M1652 is located in BRCT-N and is completely inaccessible to solvent. It lies in the β1 strand, an interior strand of a four-stranded parallel β-sheet. Isoleucine is a hydrophobic, β-branched residue that is more favored in β-strands than methionine. We expect that this replacement is benign with respect to BRCT function and may even have a protective, stabilizing effect in the presence of other destabilizing mutations.
Table 1.
Structure-based analysis of BRCA1 variants in the BRCT domains.
| CLASS | VARIANT | NOTES |
|---|---|---|
| Hydrophobic core disruption | V1833M | V1833 in BRCT-C is completely inaccessible to solvent. It lies in the β4 strand, an edge strand of a four-stranded parallel beta sheet. The sidechain points inward towards the protein core. Valine is a β-branched residue that is more favored in β-strands than methionine and methionine is larger than valine, so this replacement likely disrupts the tight packing in the domain’s hydrophobic core. |
|
| ||
| No Evidence for Functional Impact | Q1826H | Q1826 in BRCT-C is on the protein surface and its sidechain points outward to the solvent. There are no known or predicting binding interactions at this position. |
| M1652I | M1652 in BRCT-N is completely inaccessible to solvent. It lies in the β1 strand, an interior strand of a four-stranded parallel β-sheet. Isoleucine is a hydrophobic, β-branched residue that is more favored in β-strands than methionine. We expect that this replacement is benign with respect to BRCT function and may even have a protective, stabilizing effect in the presence of other destabilizing mutations. | |
3.4. Validation
Recently we evaluated the specificity and sensitivity of the transcriptional assay using a set of variants that had been classified by genetic and integrative methods [14]. Importantly, we only considered missense variants, and in that case the assay correctly classified all 24 variants tested [14]. This resulted in 100% sensitivity (95% CI 69% - no defined upper bound) and 100% specificity (95% CI 77% - no defined upper bound)[14]. Here, we update the validation for the transcription assay taking into consideration additional variants for which genetic or integrative data has emerged and classified variants with truncations, frameshift, or in-frame deletions, totaling 31 variants (Table 2). Sensitivity for the updated set is 100% (95% CI 79.4% - no defined upper bound). Specificity was 93% (95% CI 68.1% – 99.8%) with the assay correctly classifying 14/15 neutral variants. Variant V1736A was classified as deleterious in the transcriptional assay which was supported by structure-based analysis [14]. However, integrative analysis using family data resulted in odds of 2219:1 in favor of neutrality [11]. Interestingly, this variant is also classified as deleterious by all other prediction models except SIFT (Table 2). Myriad Genetics considers this variant unclassified (Michelle Martin, Myriad Genetics, personal communication). Despite the uncertainty about V1736A classification we decided to be consistent and keep it in the validation set. Figure 5 shows a diagram of BRCA1 C-terminal region and the classification of all variants tested to date. For a complete list of variants in BRCA1 and their current classification the reader is referred to the Breast Cancer Information Core database (http://research.nhgri.nih.gov/bic/). In summary, the transcription assay is highly accurate and, despite the small number variants studied to date, can reliably classify truncations, frameshift, and in-frame deletions.
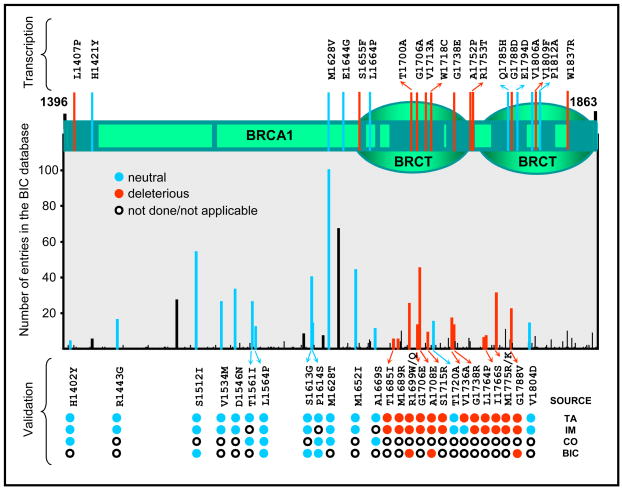
Fig. 5. Classification of missense variants in the carboxy-terminal region of BRCA1.
Top, Diagram of human BRCA1 depicting the carboxy-terminal region including the BRCT domains. Regions showing conservation above 0.75 in a multiple sequence alignment (from human to puffer fish) are shown as dark green boxes. Variants that have been tested by the transcription assay are indicated by red (deleterious) or blue (lines) but have not yet been classified by other integrative methods. Bottom, graph aligned to the top diagram shows the number of entries in the BIC database for each missense variants recorded to date. Bars record entries for all variants for a particular codon (number of entries for specific variant may be lower than what is shown because different substitutions may have been recorded). The lower part of the graph shows cross-validation of the transcription assay (TA) with integrative methods (IM), the BIC classification (BIC), or co-occurrence data (CO). Variants are shown as black (unclassified), red (deleterious), and blue (neutral) bars. All deleterious and neutral variants as classified in the BIC database are shown. Note preferential localization of deleterious variants in regions of recognized domains.
4. Discussion
Due to the low frequency of most BRCA1 unclassified variants there is a shortage of clinical data to base a decision as to whether or not they will predispose to disease. Here we have combined functional analysis and bioinformatics-based prediction models to aid in the classification of in-frame deletion, frameshift, and missense variants of BRCA1. We have recently provided validation for our functional assays for missense variants and used the validated functional data to fine tune structure-based computation models [12, 18]. Based on these methods we provide a tentative classification for the variants tested. Currently, there is no genetic or clinical data available for these variants that is conclusive and which could contribute to the classification. Although it is not possible to conclude with certainty, without additional genetic data, whether or not our classification reflects the in vivo biology, we believe it provides an important addition to our knowledge about these variants.
The data obtained in vitro indicate that frameshift (5673insC) and in-frame deletion (Δ exons 16/17) novel variants display a loss-of-function phenotype and are likely to constitute cancer-predisposing variants of BRCA1 in vivo. Deletion analysis revealed that the integrity of the most BRCA1 C-terminal hydrophobic cluster (I1855, L1854, and Y1853) is necessary for transcriptional activity [24]. The Δ exons 16/17 variant, although still retaining an intact minimal transactivation domain (aa 1796–1863) [45, 46], results in loss of function. These results are in line with the idea that the tandem BRCT repeats function as an integrated unit [17, 47, 48]. Unfortunately, there is no additional published data for these two variants.
Interestingly, modeling of the 5673insC suggested the existence of an α-helix that could disrupt binding to the BRCT phosphopeptide binding pocket (Fig. 3E). It is important to stress that structural models are inherently less reliable than experimental structures obtained through x-ray crystallography. In addition, although our functional assay has been validated [12 and the present study] we have thus far only examined missense variants, and have not yet conducted a systematic analysis of the behavior of insertion or deletion variants. Therefore, caution should be used when considering structural implications of these variants.
Recently, using a large set of variants we have determined a tentative threshold of activity below which variants are likely to be cancer-predisposing [14]. Neutral and deleterious variants display > 50% or < 45% of wild type activity, respectively. In this context, variants S1613C, M1652I and Q1826H are likely to represent neutral variants. Variant M1652I is documented as a validated cSNP (rs1799967) in NCBI’s SNP database (http://www.ncbi.nlm.nih.gov/SNP/) with 0.002 of heterozygosity. Variant S1613C failed to activate transcription in yeast using a less sensitive qualitative assay, providing a cautionary note to the interpretation of functional assays [49]. Interestingly, variant S1613C is located at the same residue as one of the most common neutral polymorphisms in BRCA1, S1613G. S1613G does not contribute to risk for breast and ovarian cancer, and in functional assays its activity is comparable to the wild type BRCA1 [14]. For this reason, it is commonly used as a neutral control (see Fig. 3 and 4).
M1652I is one of the most common missense variants with a heterozygote frequency of 4.08% in a control population [50]. Taking into account co-occurrence with other known deleterious mutations and examination of the range of amino acid variation in other BRCA1 orthologs, Tavtigian et al. arrived at an integrated likelihood ratio of <1.0 × 10−10, and was considered neutral [36]. Moreover, this variant is comparable to the wt in the yeast growth assay and in its phosphopeptide binding properties [33, 51]. Thus, taken together, the functional and computation prediction data presented here and the published data from multiple independent sources indicate that the M1652I constitutes a neutral variant. There is no additional available data for Q1826H.
The K1487R variant exhibits an amino acid change (Lys to Arg) with no alteration in charge and with large resemblance in structure in a position that allows Glycine in rodent species. In the present analysis, this variant displayed a decreased activity only slightly above 50% of the wild type, making it difficult to classify as either deleterious or neutral. To the present date, all tested variants located in this unstructured region of BRCA1, bordered by a coiled-coil region on its amino-terminus and the BRCT on its carboxy-terminus, have so far shown a pattern of activity compatible with a neutral classification [14]. Thus, although the conservative nature of the change and its location in a nonconserved and unstructured region may suggest neutrality, it is not possible to provide a classification for this variant without additional data.
On the other hand, variant V1833M presented activity comparable to that of the negative control variants (i.e., significantly below 45% of the wild type) and is therefore classified as a deleterious allele. It has also been shown to destabilize the BRCT in vitro but its thermal unfolding seems to be only moderately affected and it remains able to bind phosphopeptides when measured by isothermal titration calorimetric experiments [31, 52]. It should be noted that an early version of a computational prediction model suggested that M1652I and V1833M were likely to be neutral. While the results for the M1652I were a rationalization of a preliminary qualitative functional assay, the V1833M was a prediction by the model. The rule-based decision tree used by the model arrived at a neutral classification by considering solely that the valine to methionine substitution was a likely amino acid change. Indeed, the Grantham score [53] for Val/Met is very low, which is unusual for a deleterious missense mutation. However, the bioinformatics analysis and rationalization of structural impact performed here (Tables 1 and 3) suggests that the Val/Met change, in the context of the tight packing of the BRCT-C domain, may be more disruptive than previously believed.
In conclusion, we analyzed a series of missense, frameshift, and in-frame deletion variants of BRCA1 for which clinical information was not available or not informative. Using a combination of functional analysis and structural predictions we tentatively classify variants V1833M, 5673insC and Δ exons 16/17 as likely to be deleterious and missense variants M1652I, S1613C, and Q1826H as likely to be neutral.
Acknowledgments
This work was supported by National Institutes of Health grants CA116167 and CA92309, and the Florida Breast Cancer Coalition (ANM), the Italian Association and Foundation for Cancer Research (AIRC/FIRC) special project “Tumori Ereditari” (PR), and a Susan G. Komen Foundation grant KG080137 (RK). Additional support for this work was provided by the Department of Pharmaceutical Sciences at St. John’s University (BB) and the Molecular Imaging and the Molecular Biology cores at the H.L. Moffitt Cancer Center (ANM). We thank Dr. Dana Rollison for help with statistical analysis.
Abbreviations
- BRCT
BRCA1 COOH terminal domain
- DBD
DNA-binding domain
- DCIS
ductal carcinoma in situ
- ESE
exonic splicing enhancer
- LOH
loss of heterozygosity
- ONPG
o-Nitrophenyl-β-D-galactopyranoside
- SD
synthetic drop-out medium
- WT
wild type
Footnotes
Competing Interests
The authors declare that there are no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final ci form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 3.Begg CB, Haile RW, Borg A, Malone KE, Concannon P, Thomas DC, Langholz B, Bernstein L, Olsen JH, Lynch CF, nton-Culver H, Capanu M, Liang X, Hummer AJ, Sima C, Bernstein JL. Variation of Breast Cancer Risk Among BRCA1/2 Carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, Tang J, Li S, Zhang S, Shaw PA, Narod SA. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 6.Levy-Lahad E, Plon SE. Cancer. A risky business--assessing breast cancer risk. Science. 2003;302:574–575. doi: 10.1126/science.1091465. [DOI] [PubMed] [Google Scholar]
- 7.Narod SA, Offit K. Prevention and Management of Hereditary Breast Cancer. J Clin Oncol. 2005;23:1656–1663. doi: 10.1200/JCO.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 8.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 9.Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo CI, Worley T, Monteiro AN. Understanding Germ-Line Mutations in BRCA1. Cancer Biol Ther. 2004;3:515–520. doi: 10.4161/cbt.3.6.841. [DOI] [PubMed] [Google Scholar]
- 11.Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, len-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho MA, Couch FJ, Monteiro AN. Functional assays for BRCA1 and BRCA2. Int J Biochem Cell Biol. 2007;39:298–310. doi: 10.1016/j.biocel.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domchek S, Weber BL. Genetic Variants of Uncertain Significance: Flies in the Ointment. J Clin Oncol. 2008;26:16–17. doi: 10.1200/JCO.2007.14.4154. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, Baumbach L, Sutphen R, Pickard-Brzosowicz JL, Nathanson KL, Sali A, Goldgar D, Couch FJ, Radice P, Monteiro ANA. Determination of Cancer Risk Associated with Germ Line BRCA1 Missense Variants by Functional Analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phelan CM, Dapic V, Tice B, Favis R, Kwan E, Barany F, Manoukian S, Radice P, van der Luijt RB, van Nesselrooij BP, Chenevix-Trench G, Caldes T, De La HM, Lindquist S, Tavtigian SV, Goldgar D, Borg A, Narod SA, Monteiro AN. Classification of BRCA1 missense variants of unknown clinical significance. J Med Genet. 2005;42:138–146. doi: 10.1136/jmg.2004.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, Borg A, Monteiro AN. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirkovic N, Marti-Renom MA, Weber BL, Sali A, Monteiro AN. Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Res. 2004;64:3790–3797. doi: 10.1158/0008-5472.CAN-03-3009. [DOI] [PubMed] [Google Scholar]
- 18.Karchin R, Monteiro AN, Tavtigian SV, Carvalho MA, Sali A. Functional Impact of Missense Variants in BRCA1 Predicted by Supervised Learning. PLoS Comput Biol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams RS, Glover JN. Structural consequences of a cancer-causing BRCA1-BRCT missense mutation. J Biol Chem. 2003;278:2630–2635. doi: 10.1074/jbc.M210019200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson DH. PCR: A practical approach. IRL Press; Oxford: 2003. [Google Scholar]
- 21.Golemis EA, Gyuris J, Brent R. Two-hybrid system/interaction traps. In: Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith JA, Struhl K, editors. Current protocols in molecular biology. John Wiley & Sons; New York: 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- 22.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 24.Hayes F, Cayanan C, Barilla D, Monteiro AN. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 2000;60:2411–2418. [PMC free article] [PubMed] [Google Scholar]
- 25.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 26.Karchin R, Kelly L, Sali A. Improving functional annotation of non-synonomous SNPs with information theory. Pac Symp Biocomput. 2005:397–408. doi: 10.1142/9789812702456_0038. [DOI] [PubMed] [Google Scholar]
- 27.Karchin R, Agarwal M, Sali A, Couch F, Beattie M. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Informatics. 2008;6:9–12. doi: 10.4137/cin.s618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karplus K, Katzman S, Shackleford G, Koeva M, Draper J, Barnes B, Soriano M, Hughey R. SAM-T04: what is new in protein-structure prediction for CASP6. Proteins. 2005;61 Suppl 7:135–142. doi: 10.1002/prot.20730. [DOI] [PubMed] [Google Scholar]
- 29.Monteiro AN. BRCA1: exploring the links to transcription. Trends Biochem Sci. 2000;25:469–474. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- 30.Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J Biol Chem. 2003;278:53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- 32.Tischkowitz M, Hamel N, Carvalho MA, Birrane G, Soni A, van Beers EH, Joose SA, Wong N, Novak D, Quenneville LA, Grist S, ConFab k, Nederlof PM, Goldgar DE, Tavtigian SV, Monteiro AN, Ladias JA, Foulkes WD. Pathogenicity of a BRCA1 missense variant is determined by the disruption of the phosphopeptide binding pocket - a multi-modal approach. European Journal of Human Genetics. 2008 doi: 10.1038/ejhg.2008.13. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 34.Varma AK, Brown RS, Birrane G, Ladias JA. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochemistry. 2005;44:10941–10946. doi: 10.1021/bi0509651. [DOI] [PubMed] [Google Scholar]
- 35.Judkins T, Hendrickson BC, Deffenbaugh AM, Eliason K, Leclair B, Norton MJ, Ward BE, Pruss D, Scholl T. Application of embryonic lethal or other obvious phenotypes to characterize the clinical significance of genetic variants found in trans with known deleterious mutations. Cancer Res. 2005;65:10096–10103. doi: 10.1158/0008-5472.CAN-05-1241. [DOI] [PubMed] [Google Scholar]
- 36.Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de SD, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, King MC. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 38.Szabo CI, King MC. Inherited breast and ovarian cancer. Hum Mol Genet. 1995;4 Spec No:1811–1817. doi: 10.1093/hmg/4.suppl_1.1811. [DOI] [PubMed] [Google Scholar]
- 39.Lovelock PK, Spurdle AB, Mok MT, Farrugia DJ, Lakhani SR, Healey S, Arnold S, Buchanan D, Investigators K, Couch FJ, Henderson BR, Goldgar DE, Tavtigian SV, Chenevix-Trench G, Brown MA. Identification of BRCA1 missense substitutions that confer partial functional activity: potential moderate risk variants? Breast Cancer Res. 2007;9:R82. doi: 10.1186/bcr1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:W480–W482. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de IC, Orozco XM. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 44.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci U S A. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 47.Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer- associated protein BRCA1. Nat Struct Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- 49.Monteiro AN, August A, Hanafusa H. Common BRCA1 variants and transcriptional activation. Am J Hum Genet. 1997;61:761–762. doi: 10.1086/515515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deffenbaugh AM, Frank TS, Hoffman M, Cannon-Albright L, Neuhausen SL. Characterization of common BRCA1 and BRCA2 variants. Genet Test. 2002;6:119–121. doi: 10.1089/10906570260199375. [DOI] [PubMed] [Google Scholar]
- 51.Humphrey JS, Salim A, Erdos MR, Collins FS, Brody LC, Klausner RD. Human BRCA1 inhibits growth in yeast: potential use in diagnostic testing. Proc Natl Acad Sci U S A. 1997;94:5820–5825. doi: 10.1073/pnas.94.11.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolopoulos G, Pyrpassopoulos S, Thanassoulas A, Klimentzou P, Zikos C, Vlassi M, Vorgias CE, Yannoukakos D, Nounesis G. Thermal unfolding of human BRCA1 BRCT-domain variants. Biochim Biophys Acta. 2007;1774:772–780. doi: 10.1016/j.bbapap.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng ECTE. Ferrin, UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Shackleford G, Karplus K. Contact prediction using mutual information and neural nets. Proteins. 2007;69 Suppl 8:159–164. doi: 10.1002/prot.21791. [DOI] [PubMed] [Google Scholar]
- 56.Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, et al. Genetic and Histopathologic Evaluation of BRCA1 and BRCA2 DNA Sequence Variants of Unknown Clinical Significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 57.Malacrida S, Agata S, Callegaro M, Casella C, Barana D, Scaini MC, et al. BRCA1 p.Val1688del Is a Deleterious Mutation That Recurs in Breast and Ovarian Cancer Families From Northeast Italy. J Clin Oncol. 2008;26:26–31. doi: 10.1200/JCO.2007.13.2118. [DOI] [PubMed] [Google Scholar]