Abstract
The spindle assembly checkpoint (SAC) is an evolutionarily conserved surveillance mechanism that delays anaphase onset and mitotic exit in response to the lack of kinetochore attachment. The target of the SAC is the E3 ubiquitin ligase anaphase-promoting complex (APC) bound to its Cdc20 activator. The Cdc20/APC complex is in turn required for sister chromatid separation and mitotic exit through ubiquitin-mediated proteolysis of securin, thus relieving inhibition of separase that unties sister chromatids. Separase is also involved in the Cdc-fourteen early anaphase release (FEAR) pathway of nucleolar release and activation of the Cdc14 phosphatase, which regulates several microtubule-linked processes at the metaphase/anaphase transition and also drives mitotic exit. Here, we report that the SAC prevents separation of microtubule-organizing centers (spindle pole bodies [SPBs]) when spindle assembly is defective. Under these circumstances, failure of SAC activation causes unscheduled SPB separation, which requires Cdc20/APC, the FEAR pathway, cytoplasmic dynein, and the actin cytoskeleton. We propose that, besides inhibiting sister chromatid separation, the SAC preserves the accurate transmission of chromosomes also by preventing SPBs to migrate far apart until the conditions to assemble a bipolar spindle are satisfied.
INTRODUCTION
Chromosome segregation during anaphase relies on the assembly of a bipolar spindle followed by the amphitelic attachment of sister kinetochores to opposite spindle poles. Microtubule-organizing centers, namely, spindle pole bodies (SPBs) in yeast and centrosomes in higher eukaryotic systems, are essential in many organisms for bipolar spindle formation (Doxsey et al., 2005). The budding yeast SPB duplicates at the G1/S transition, and the two SPBs initially remain side by side connected by a bridge. During S phase, the bridge is severed by an ill-defined mechanism, and SPBs migrate away from each other to form the two poles of a short bipolar spindle, constituted by an array of interdigitated microtubules (Jaspersen and Winey, 2004). SPB separation requires the activity of two partially redundant plus-end–directed kinesins of the BimC family, Cin8 and Kip1 (Hoyt et al., 1992; Roof et al., 1992), as well as mitotic cyclin-dependent kinases (CDKs) (Fitch et al., 1992; Lim et al., 1996). Lack of Cin8 and Kip1 or ablation of mitotic cyclins prevents spindle formation, causing cells to arrest with duplicated but unseparated chromatids, large buds, and SPBs arranged side by side (Hoyt et al., 1992; Roof et al., 1992; Fitch et al., 1992; Lim et al., 1996). The function of Cin8 and Kip1 is counteracted by the minus-end–directed kinesin Kar3, which binds to the cytoplasmic side of SPBs and is required for spindle positioning (Hildebrandt and Hoyt, 2000). Stu1, the yeast member of the CLASP family of microtubule-associated proteins, is also needed for SPB separation and bipolar spindle assembly (Pasqualone and Huffaker, 1994; Yin et al., 2002). Mitotically active CDKs (dephosphorylated on Y19) are required to stabilize Cin8, Kip1, and the Ase1 microtubule-binding protein, and allow SPB separation (Crasta et al., 2006).
Unlike vertebrate cells, budding yeast cells undergo “closed mitosis” without nuclear envelope breakdown. Because SPBs are embedded in the nuclear envelope, different sets of microtubules, cytoplasmic and nuclear, are physically separated. Whereas nuclear microtubules are directly involved in kinetochore attachment and chromosome segregation, cytoplasmic microtubules are required for spindle positioning. Two sequential processes, the Kar9 pathway and the dynein pathway, contribute to spindle positioning. Either pathway is dispensable for cell viability, whereas inactivation of both is lethal (Miller and Rose, 1998). Kar9 is localized at the SPB, and it is translocated to microtubule plus ends through interaction with the plus-end–directed motor Kip2 and the microtubule-associated Bim1 protein (Lee et al., 2000; Maekawa et al., 2003). The Kar9–Bim1 complex guides microtubules along polarized actin cables into the bud by interacting with the type V myosin Myo2 (Yin et al., 2000). Through these interactions, the Kar9 pathway promotes capture of cytoplasmic microtubules with the bud cortex primarily before anaphase. The second pathway of spindle positioning requires the minus-end–directed motor dynein (Yeh et al., 1995) that associates with the cortical anchor Num1 (Heil-Chapdelaine et al., 2000; Farkasovsky and Kuntzel, 2001). Targeting of dynein to microtubule plus ends requires the plus end tracking protein Bik1 (Sheeman et al., 2003), which in turn binds microtubule ends through Kip2 (Carvalho et al., 2004). The dynein pathway acts predominantly during anaphase and might contribute to the cytoplasmic microtubule capture by the bud tip through sliding plus ends of cytoplasmic microtubules along the cortex (reviewed in Pearson and Bloom, 2004).
Once all replicated chromatids are attached and bioriented, the Scc1/Mcd1 subunit of the cohesin complex, which holds sister chromatids together, is cut by an endoprotease called separase (Esp1 in yeast), leading to sister separation. Esp1 is kept inactive by the association with its inhibitor securin (Pds1 in budding yeast; Yamamoto et al., 1996) that is targeted for degradation at anaphase onset by the ubiquitin-ligase anaphase-promoting complex (APC) bound to its activator Cdc20 (Uhlmann, 2001; Nasmyth, 2002). Cdc20/APC activity is therefore required to activate separase and to promote anaphase onset. Among other substrates of the APC, B-type cyclins are also targeted for degradation by the Cdc20–APC complex at the onset of anaphase and by Cdh1/APC in telophase and G1 (Peters, 2006). Inactivation of mitotic CDKs through cyclin degradation is in turn important for spindle disassembly and mitotic exit.
The activity of cyclin B–CDKs in yeast is counteracted by the activity of the Cdc14 phosphatase, which is also essential for mitotic exit (Stegmeier and Amon, 2004). Cdc14 is sequestered in the nucleolus during a large window of the cell cycle, which restrains its activity by preventing the accessibility to substrates. Two temporally distinct pathways mediate Cdc14 release from the nucleolus and its subsequent activation: the Cdc fourteen early anaphase release (FEAR) network (D'Amours and Amon, 2004; Stegmeier and Amon, 2004) and the mitotic exit network (MEN) (Bardin and Amon, 2001; Simanis, 2003). Whereas the MEN is absolutely necessary for mitotic exit and cytokinesis, as is Cdc14, the FEAR pathway is dispensable for cell cycle progression. However, an increasing number of processes that ensure the fidelity of chromosome segregation, such as stabilization of the spindle midzone, segregation of the rDNA, regulation of microtubule dynamics, and spindle positioning, have been reported to depend on the FEAR pathway (Pereira and Schiebel, 2003; D'Amours et al., 2004; Ross and Cohen-Fix, 2004; Sullivan et al., 2004; Higuchi and Uhlmann, 2005; Woodbury and Morgan, 2007). To date, five proteins have been involved in the FEAR pathway: the Esp1 separase, the kinetochore/spindle protein Slk19, Spo12 and its paralogue Bns1, and the Polo kinase Cdc5. Securin and the nucleolar protein Fob1 negatively regulate the cascade (Stegmeier et al., 2002, 2004).
When the attachment of kinetochores to spindle microtubules is defective, the spindle assembly checkpoint (SAC) delays anaphase onset and mitotic exit by inhibiting Cdc20/APC. The SAC involves the Mad1, Mad2, Mad3/BubR1, Bub1, Bub3, and Mps1 proteins. The AuroraB/Ipl1 protein kinase also participates to the SAC by correcting faulty kinetochore attachments (Musacchio and Salmon, 2007).
Although the role of the SAC in regulating anaphase progression and mitotic exit is well established, its possible involvement in controlling other mitotic events, such as SPB separation and mitotic spindle organization/dynamics, has not been investigated. In this article, we characterize a new function of the SAC in restraining SPBs separation and aberrant chromosome segregation when spindle assembly is impaired. Failure to activate the SAC under these conditions leads to unscheduled SPB separation that involves activation of Cdc20/APC, the FEAR pathway of Cdc14 nucleolar release, dynein, and the actin cytoskeleton, suggesting that the SAC probably modulates forces acting on cytoplasmic microtubules. We propose that, in case of SPB or spindle defects, the SAC delays progression into anaphase and SPB separation, thus providing the time necessary for the assembly of a functional bipolar spindle and ultimately increasing the fidelity of chromosome segregation.
MATERIALS AND METHODS
Strains, Media, and Reagents
All yeast strains (Supplemental Table 1) were derivatives of or were backcrossed at least three times to W303 (ade2-1, trp1-1, leu2-3,112, his3-11,15, ura3, ssd1). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, and 50 mg/l adenine) supplemented with 2% glucose (YEPD) or 2% raffinose (YEPR) or 2% raffinose and 2% galactose (YEPRG). Sectoring YEPD medium plates were prepared without adenine supplement. Unless differently stated, α-factor was used at 2 μg/ml and nocodazole at 15 μg/ml. Cdc14 overexpression was achieved by 2% galactose addition 90 min after G1 release. CLB2-Δdb induction was performed in G1 cells arrested for 150 min with 3 μg/ml α-factor 45 min before the G1 release. Latrunculin-B (Lat-B) was used at 0.2 mM and added 90 min after G1 release to avoid interference with budding.
Measure of Distances between SPBs
For imaging, cells were mounted on a thin layer of 2% agarose between coverslip and slide; digital images were acquired in a single plane at room temperature with MetaMorph imaging system software (Molecular Devices, Sunnyvale, CA) and wide-field fluorescence microscope (Eclipse 90i, Nikon, Tokyo, Japan; 100 × 0.5–1.3 PlanFluor objective) equipped with a charge-coupled device camera (CoolSNAP; Photometrics, Tucson, AZ). Distances between SPBs were measured with MetaMorph analysis tool and logged to a text file of Excel (Microsoft, Redmond, WA). The significance of the differences between distance distributions was statistically tested by means of a two-tailed t test, assuming unequal variances (*p < 10−3, **p < 10−6, ***p < 10−9). For all presented experiments, >100 cells were scored for each strain. The distance data were binned in groups spanning 1 μm apart and then plotted using the histogram tool in the Data Analysis plug-in of Excel.
In each experiment, kinetics of SPB separation (i.e., two distinguishable Spc42-green fluorescent protein [GFP] dots) was scored during the cell cycle, and SPB distances were measured at the time point at which the percentage of SPB separation was highest. Kinetics of SPB separation varied depending on the temperature, medium, and growth kinetics of the different mutants.
Screen for Mutations Synthetically Lethal with the Bub3-WDd1 Allele
To screen for mutations synthetically lethal with bub3-WDd1, the ade2/ade3 red-white colony sectoring system was used, according to Cvrckova and Nasmyth (1993). In brief, a bub3-WDd1 ade2 ade3 ura3 mutant carried the wild-type BUB3 gene on an ADE3 URA3 plasmid with an unstable centromere and was therefore undergoing frequent plasmid loss forming red-white–sectored colonies able to grow on 5-fluoroorotic acid (5-FoAR Sect+). This strain was treated with 188 mM ethylmethansulfonate to obtain 40–50% viability, plated on sectoring YEPD medium, and then incubated at 25°C. Of >300,000 surviving clones, we selected mutants giving rise to red colonies (Sect−) unable to grow on 5-FoA plates (5-FoAS) because of their inability to lose the ADE3 URA3 BUB3 plasmid. The selected mutants were then transformed with a LEU2 BUB3-bearing plasmid or vector alone to discard all mutants where the sectoring phenotype was not rescued by the presence of the additional copy of BUB3. In two mutants, synthetic lethal with bub3 (slb)1 and slb2, the sectoring phenotype was rescued, and genetic analysis showed that slb1 and slb2 belonged to the same complementation group, were allelic to each other, and exhibited a temperature-sensitive phenotype that was tightly associated to synthetic lethality with the bub3-WDd1 mutation.
Cloning and Sequencing of slb1 and slb2
To clone the SLB1/SLB2 gene, the bub3-WDd1 slb1 and bub3-WDd1 slb2 mutants bearing the BUB3 URA3 ADE3 plasmid were transformed with a budding yeast genomic DNA library constructed in a LEU2 centromeric plasmid (Jansen et al., 1993). Of 75,000 transformants for each mutant, 1000 Sect+ clones for slb1 and 3000 for slb2 were selected and tested for the recovery of the temperature sensitivity of slb mutations. One slb2 Sect+ temperature-resistant clone was isolated and the plasmid recovered. Because slb1 and slb2 belonged to the same complementation group, this plasmid was also able to restore the Sect+ of bub3-WDd1 slb1 mutant and to recover slb1 thermosensitivity. Sequencing of the insert and comparison with the whole budding yeast genome sequence through WU-BLAST analysis (http://blast.wustl.edu) revealed that the plasmid contained the CIN8 gene. Subsequent genetic analysis showed that the slb mutations belong to the same complementation group as a CIN8 deletion and are linked to the CIN8 locus. In addition, direct sequencing of the CIN8 coding region revealed that slb1 carries a missense mutation changing the 428th codon from CCT into CTT that causes a proline-to-leucine substitution in the Cin8 motor domain, whereas slb2 carries a nonsense mutation changing the 24th codon from CAG into TAG.
Other Techniques
Flow cytometric DNA quantitation and in situ immunofluorescence were performed according to Fraschini et al. (1999). Nuclear division was scored with the fluorescence microscope described above on cells stained with propidium iodide.
To detect spindle formation and elongation, α-tubulin immunostaining was performed on spheroplasts with the YOL34 monoclonal antibody (Serotec, Oxford, United Kingdom) followed by indirect immunofluorescence using rhodamine-conjugated anti-rat antibody (1:100; Pierce Chemical, Rockford, IL). Detection of Spc42-GFP was carried out on ethanol-fixed cells, upon wash with 10 mM Tris, pH 8.5, and sonication.
RESULTS
Mutations in CIN8 Are Synthetically Lethal with the Hypomorphic Bub3-WDd1 Allele
To gain insight into the possible involvement of the SAC in processes other than the control of sister chromatid separation, we performed a genetic screen for mutations causing synthetic lethality in the presence of the Bub3-WDd1 variant, in which Trp31 of one of the WD40 domains of the protein is replaced by glycine. The bub3-WDd1 mutant is checkpoint-defective at 37°C (data not shown) and partially checkpoint proficient at 25°C (Fraschini et al., 2001). We carried out the genetic screen at 25°C (hence with partially active SAC) to increase its stringency. Of >300,000 clones that survived to the mutagenic treatment (see Materials and Methods), we isolated only two independent recessive mutations that were synthetically lethal with the bub3-WDd1 allele and were called slb1 and slb2 (synthetic lethal with bub3). Cloning of SLB1 and SLB2 genes by complementation with a genomic library revealed that they were allelic to CIN8, as confirmed by subsequent linkage analysis and sequencing (see Materials and Methods). Consistent with a role of Cin8 in SPB separation during bipolar spindle formation and elongation (Hoyt et al., 1992), slb1 and slb2 mutants showed a marked delay in spindle formation and elongation compared with wild-type cells (Supplemental Figure 1). Furthermore, the slb1 and slb2 mutations were synthetically lethal with the deletion of any SAC genes (data not shown). A possible explanation for this synthetic lethality is that the SAC becomes essential when spindle dynamics are perturbed by Cin8 inactivation (Geiser et al., 1997). However, that only CIN8 mutations were recovered in the screen raised the possibility that SAC proteins might also regulate spindle formation and/or stabilization. This prompted us to investigate the effects of SAC inactivation in cin8 mutants.
SAC Activation Restrains Unscheduled SPB Separation
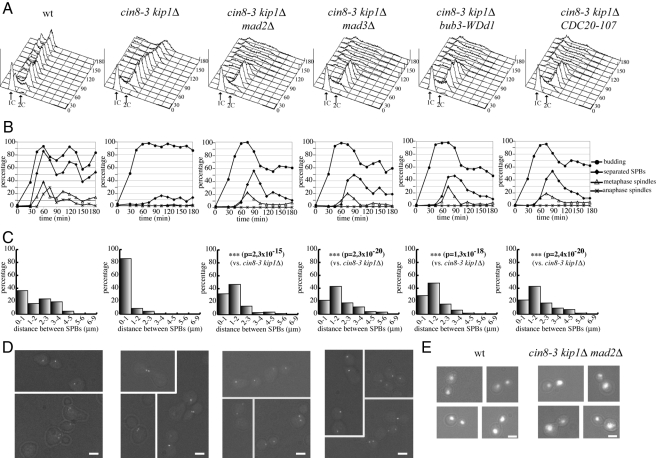
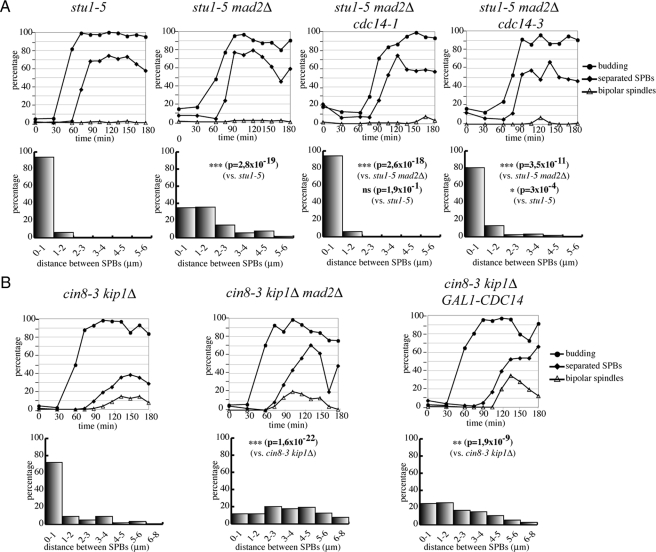
To characterize the phenotype of cin8 mutants in the absence of a functional SAC, we introduced SAC mutations into a cin8-3 temperature-sensitive mutant where the Cin8 paralogue Kip1 was deleted (cin8-3 kip1Δ; Hoyt et al., 1992). SPB separation using the Spc42-GFP marker (Adams and Kilmartin, 1999), as well as spindle formation and elongation, was analyzed during the cell cycle. Cell cultures were arrested in G1 by α-factor and released at the restrictive temperature of 34°C. In these conditions, the ability of cin8-3 kip1Δ cells to assemble mitotic spindle was considerably impaired compared with the wild-type and mutant cells arrested with large buds and replicated DNA (Figure 1, A and B). In agreement with previous data (Hoyt et al., 1992), most cin8-3 kip1Δ cells were unable to separate SPBs. Moreover, in the small fraction of mutant cells in which they separated, the two SPBs remained very close to each other, at a distance that in most cases was ≤1 μm at 90′ after release, whereas wild-type cells displayed a broad distribution of SPB distances at the same time point (Figure 1, C and D). The presence of the bub3-WDd1 allele allowed cin8-3 kip1Δ cells to escape the cell cycle arrest, accumulating aberrant DNA contents and massive chromosome missegregation (Figure 1A), consistent with loss of checkpoint function. Similar effects were observed also in the presence of MAD2 or MAD3 deletion or of the CDC20-107 allele that renders Cdc20 refractory to checkpoint activation (Hwang et al., 1998) (Figure 1, A and E). Remarkably, SAC-defective cin8-3 kip1Δ cells separated SPBs more efficiently and at higher distance than cin8-3 kip1Δ cells (Figure 1, B and C): at 90 min, only a minority of cells (20–30%) separated SPBs to ≤1 μm, whereas a high fraction of cells separated them by 2 μm or more. Moreover, SAC inactivation in cin8-3 kip1Δ cells enabled bipolar spindle assembly, albeit with a delay compared with wild-type cells and without subsequent spindle elongation (Figure 1, C and D). These data suggest that the SAC restrains SPB separation in the cin8-3 kip1Δ mutant through Cdc20 inhibition.
Figure 1.
SAC activation restrains SPBs separation in cin8-3 kip1Δ mutants. Wild-type (ySP4935), cin8-3 kip1Δ (ySP5361), cin8-3 kip1Δ mad2Δ (ySP5364), cin8-3 kip1Δ mad3Δ (ySP5363), cin8-3 kip1Δ bub3-WDd1 (ySP5360), and cin8-3 kip1Δ CDC20-107 (ySP5366) cells were arrested in G1 with α-factor at 25°C and released at 34°C (time 0). (A and B) Samples were collected at the indicated times for fluorescence-activated cell sorting (FACS) analysis of DNA contents and to follow kinetics of budding, SPB separation and mitotic spindle formation and elongation. (C) Distribution of distances between SPBs. (D) Micrographs of SPBs were taken 90′ after release. Bar, 2 μm. (E) Examples of anaphase cells at 90′ after release after nuclear staining with propidium iodide. Bar, 2 μm.
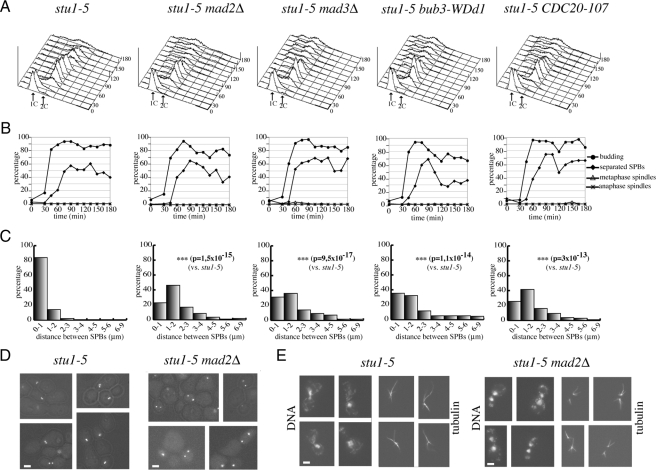
We then asked whether SAC inactivation could allow SPB separation in other mutants impaired in spindle assembly, such as the temperature-sensitive stu1-5 (Yin et al., 2002). We released at the restrictive temperature of 37°C G1-arrested stu1-5, stu1-5 mad2Δ, stu1-5 mad3Δ, stu1-5 bub3-WDd1, and stu1-5 CDC20-107 mutant cells, all expressing the Spc42-GFP fusion protein. As expected, most stu1-5 cells arrested with large buds and replicated DNA (Figure 2, A and B). Cells were unable to assemble bipolar spindles but could partially undergo SPB separation, although SPBs remained next to each other, with a distance of ≤1 μm in >80% of the cells 90 min after the G1 release (Figure 2, B–E). Impairment of SAC components drove stu1-5 cells out of mitosis and allowed them to undergo cytokinesis (Figure 2, A and B). Strikingly, in these double mutants the distance between separated SPBs at 90 min was considerably higher than in stu1-5 cells (Figure 2, C and D), indicating that SAC inactivation promotes SPBs separation also upon loss of Stu1 function. In spite of the higher distance between SPBs, mitotic spindles were undetectable in SAC-defective stu1-5 cells (Figure 2, B–E), indicating that the essential role of Stu1 in bipolar spindle assembly cannot be bypassed by SAC impairment. In contrast, cytoplasmic microtubules were readily apparent (Figure 2E). Thus, it is likely that SPB separation in these conditions is driven by forces that act on cytoplasmic, rather than spindle, microtubules. Altogether, our data suggest that the SAC controls the extent of SPB separation when spindle assembly is impaired.
Figure 2.
SAC inactivation allows SPB separation upon loss of Stu1 function. stu1-5 (ySP1804), stu1-5 mad2Δ (ySP1856), stu1-5 mad3Δ (ySP4595), stu1-5 bub3-WDd1 (ySP6202), and stu1-5 CDC20-107 (ySP6194) mutant cells were arrested in G1 with α-factor at 25°C and released at 37°C (time 0). (A and B) Samples were collected at the indicated times for FACS analysis of DNA contents and to follow kinetics of budding, SPB separation and mitotic spindle formation and elongation. (C) Distribution of distances between SPBs. (D) Micrographs of SPBs were taken 90′ after release. Bar, 2 μm. (E) Examples of microtubules stained by immunofluorescence with anti-tubulin antibodies (time 90′ after release). Bar, 2 μm.
Cdc20/APC Activity Is Required to Separate SPBs upon SAC Inactivation
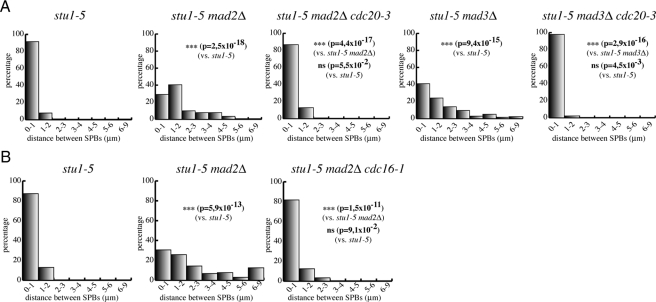
The observation that a version of Cdc20 refractory to checkpoint activation was able to increase SPB separation in cin8-3 kip1Δ and stu1-5 mutants suggested that the SAC might restrain SPB separation through inhibition of Cdc20/APC. To test this hypothesis, we asked whether SAC inactivation in stu1-5 mutants was still sufficient to allow SPB separation when Cdc20 or APC activity was impaired. Therefore, we first inactivated Cdc20 by using the temperature-sensitive cdc20-3 allele in stu1-5 mad2Δ and stu1-5 mad3Δ mutants and measured SPB distances 120 min after release from a G1 arrest at restrictive temperature. Cdc20 inactivation in stu1-5 mad2Δ and stu1-5 mad3Δ mutants completely suppressed the effects of SAC inactivation and led to SPB distances similar to those observed in stu1-5 cells under the same conditions (Figure 3A), confirming that Cdc20 is required to separate SPBs in these cells. Similar results were obtained by inactivating the APC subunit Cdc16 in stu1-5 mad2Δ cells with the cdc16-1 temperature-sensitive allele (Figure 3B). Thus, the SAC is able to restrain SPB separation through inhibition of Cdc20/APC activity in response to spindle defects.
Figure 3.
The SAC restrains SPB separation through inhibition of Cdc20/APC. (A) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625), stu1-5 mad2Δ cdc20-3 (ySP6665), stu1-5 mad3Δ (ySP6891) stu1-5 mad3Δ cdc20-3 (ySP6789) cells and (B) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625), stu1-5 mad2Δ cdc16-1 (ySP7211) mutants were arrested in G1 with α-factor at 25°C and released at 37°C. Distances between SPBs were measured 120′ after release.
Securin Contributes to Restricting SPB Separation in the Presence of Spindle Defects
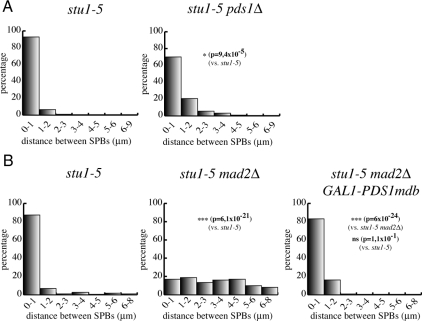
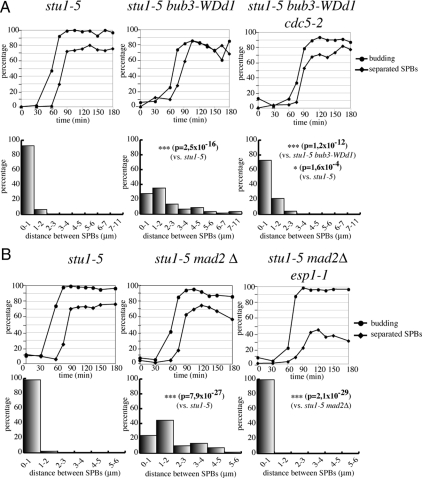
The main target of Cdc20/APC at the metaphase-to-anaphase transition is securin, which prevents sister chromatid separation by inhibiting separase (Peters, 2006). Securin proteolysis had been previously implicated in spindle integrity (Severin et al., 2001) and elongation (Jensen et al., 2001) but never in the control of SPB separation. Because our data indicate that Cdc20/APC activity is required to separate SPBs in stu1-5 mutants upon SAC inactivation, we asked whether knocking out securin could also promote SPB separation in stu1-5 cells. We measured the ability of stu1-5 cells with or without PDS1, encoding for yeast securin, to separate SPBs after a release from a G1 block at the nonpermissive temperature of 37°C. The percentage of stu1-5 pds1Δ cells separating SPBs >1 μm at 150 min increased slightly, but significantly, with respect to the stu1-5 mutant (Figure 4A), although not as high as in SAC-defective stu1-5 cells. These data suggest that securin is partly responsible for the lack of SPB separation in stu1-5 mutants, although other Cdc20/APC targets might be involved. An alternative explanation for the partial effects of lack of securin on SPB separation is that Pds1 not only inhibits separase but also promotes its full activation by regulating its nuclear import and spindle localization (Jensen et al., 2001; Agarwal and Cohen-Fix, 2002). Separase activation, in turn, has been shown to be important for spindle stability and elongation (Jensen et al., 2001; Sullivan et al., 2001; Baskerville et al., 2008), as well as for cytoplasmic microtubule-associated forces (Ross and Cohen-Fix, 2004). Therefore, it is possible that PDS1 deletion allows only partial SPB separation in stu1-5 cells because of improper activation of separase (see below). We therefore asked whether lack of Pds1 degradation, by using a nondegradable version of Pds1 expressed from the GAL1 promoter (Cohen-Fix et al., 1996), was sufficient to reverse the effects of SAC inactivation on SPB separation in stu1-5 cells. Cell cultures of stu1-5, stu1-5 mad2Δ, and stu1-5 mad2Δ GAL1-PDS1mdb strains, the latter of which expressed a nondegradable version of Pds1 from the galactose-inducible GAL1 promoter (Cohen-Fix et al., 1996), were synchronized in G1 by α-factor and released into the cell cycle in the presence of galactose at 37°C. Analysis of SPB distances 135 min after release showed that most stu1-5 mad2Δ GAL1-PDS1mdb cells were not able to separate SPBs >1 μm, similarly to stu1-5 cells (Figure 4B). Thus, Pds1 stabilization prevents SPB separation in stu1 mutants.
Figure 4.
Securin restrains SPB separation in stu1-5 cells. (A) stu1-5 (ySP6892) and stu1-5 pds1Δ (ySP7068) mutants were arrested in G1 with α-factor at 25°C and released at 37°C. Distances between SPBs were measured 150 min after release. (B) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625) and stu1-5 mad2Δ GAL1-PDS1mdb (ySP7572) mutants were grown in YEPR, arrested in G1 with α-factor at 25°C, and then released in YEPRG medium at 37°C. Distances between SPBs were measured 135′ after release.
The FEAR Pathway and Cdc14 Phosphatase Regulate SPB Separation in Cells with Inactivated SAC
Separase is involved in the FEAR pathway for the nucleolar release of the Cdc14 protein phosphatase, which in turn regulates mitotic spindle stability during early anaphase (reviewed in Khmelinskii and Schiebel, 2008). We therefore wondered whether the SAC and securin could restrain SPB separation in stu1-5 cells through Cdc14 inhibition. To this purpose, we tested whether SPBs could still separate in stu1-5 mad2Δ mutants if Cdc14 activity was impaired through the temperature-sensitive cdc14-1 and cdc14-3 alleles. The percentage of cells separating SPBs >1 μm at 120 min after G1 release at restrictive temperature was significantly lower in stu1-5 mad2Δ cdc14-1 and stu1-5 mad2Δ cdc14-3 cells than in stu1-5 mad2Δ cells (Figure 5A), suggesting that Cdc14 is required to separate SPBs under these conditions.
Figure 5.
Cdc14 activity is required for SPB separation induced by SAC inactivation. (A) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625), stu1-5 mad2Δ cdc14-1 (ySP7981), stu1-5 mad2Δ cdc14-3 (ySP6583) mutants were arrested in G1 with α-factor at 25°C and released at 37°C (time 0). Samples were collected at the indicated times to follow kinetics of budding, SPB separation, and mitotic spindle formation. At 120′ after release, distances between SPBs were measured. (B) cin8-3 kip1Δ (ySP5361), cin8-3 kip1Δ mad2Δ (ySP5364), cin8-3 kip1Δ GAL1-CDC14 (ySP7773) cells were grown in YEPR, arrested in G1 with α-factor at 25°C, and released at 34°C. We added 2% galactose 90 min after release. At the indicated times, samples were collected to follow kinetics of budding, SPB separation, and mitotic spindle formation. Distances between SPBs were measured 150′ after release.
We then asked whether CDC14 overexpression from the GAL1 promoter could increase, like SAC inactivation, SPB separation in cells defective in spindle assembly. Because it is difficult to obtain high CDC14 expression levels from the GAL1 promoter at 37°C, we tested the effects of CDC14 overexpression in cin8-3 kip1Δ cells that are defective in SPB separation already at 34°C (Figure 1). As shown in Figure 5B, high levels of Cdc14 allowed cin8-3 kip1Δ cells to separate SPBs more efficiently and to a higher extent with respect to the cin8-3 kip1Δ mutant alone. More specifically, SPBs had separated >1 μm in the majority of cin8-3 kip1Δ GAL1-CDC14 cells 150 min after release from G1, whereas they mostly remained at ≤1-μm distance in similarly treated cin8-3 kip1Δ cells. These data suggest that SPB separation is regulated by Cdc14 activity in presence of spindle defects. In agreement with the notion that Cdc14 dephosphorylates preferentially CDK targets (Stegmeier and Amon, 2004), we found that overexpression of nondegradable CLB2, encoding the main budding yeast mitotic cyclin, partly reversed the effects of SAC inactivation on SPB separation in stu1-5 mad2Δ cells (Supplemental Figure 2).
Cdc14 is released from the nucleolus in early anaphase by the FEAR pathway, which includes separase and polo-like kinase (Stegmeier and Amon, 2004). We therefore asked whether inactivation of the Esp1 separase or the Cdc5 polo-like kinase could prevent SPB separation in SAC-defective stu1-5 cells. Indeed, most stu1-5 bub3-WDd1 cells where the polo-like kinase was inactivated by the temperature-sensitive mutation cdc5-2 (Figure 6A) or stu1-5 mad2Δ cells in which separase was inactivated by the temperature-sensitive esp1-1 mutation (Figure 6B) were mostly unable to separate SPBs >1 μm at restrictive temperature. Altogether, these data indicate that the FEAR pathway and Cdc14 modulate SPB separation in mutants where the SAC is activated by spindle defects.
Figure 6.
Polo-like kinase and separase are required to take apart SPBs in stu1 mutants upon SAC inactivation. (A) stu1-5 (ySP6892), stu1-5 bub3-WDd1 (ySP6204), stu1-5 bub3-WDd1 cdc5-2 (ySP7469) cells were arrested in G1 with α-factor at 25°C and released at 37°C. Distances between SPBs were measured 120′ after release. (B) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625), stu1-5 mad2Δ esp1-1 (ySP7828) mutants were arrested in G1 with α-factor at 25°C and released at 37°C. Distances between SPBs were measured 120′ after release.
Dynein and Actin Cytoskeleton Are Required for SPB Separation Driven by SAC Inactivation
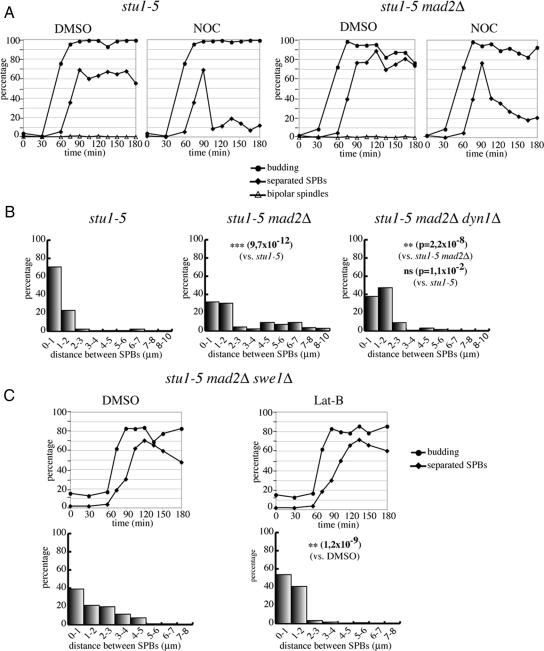
Because our data suggest that SAC inactivation drives SPB separation in stu1-5 cells by acting on cytoplasmic microtubules, we decided to test the effects of microtubule depolymerization on SPB separation under these conditions. We reasoned that treatment with nocodazole would selectively depolymerize cytoplasmic microtubules, because nuclear microtubules are not apparent in stu1-5 and stu1-5 mad2Δ cells. G1-arrested cells were released into fresh medium, and nocodazole was added to half of the cultures after 90 min. As expected, nocodazole treatment caused the already separated SPBs to come close together in both stu1-5 and stu1-5 mad2Δ cells, resulting in most cells showing a single Spc42-GFP signal (Figure 7A). We then knocked out different genes involved in cytoplasmic microtubule dynamics in SAC-defective stu1-5 cells, to test which of them could suppress the effects on SPB separation caused by lack of a functional SAC. Deletion of KAR9, BIK1, KAR3, VIK1 (encoding a regulatory partner of Kar3), and KIP2 had no effect on SPB separation in these conditions (data not shown). In addition, the Ase1 microtubule-binding protein was also dispensable for SPB separation in stu1-5 mad2Δ cells (data not shown). In contrast, deletion of DYN1 (encoding for dynein heavy chain) significantly suppressed SPB separation caused by SAC inactivation in stu1-5 cells, restoring short distances between SPBs (Figure 7B).
Figure 7.
Microtubules, dynein, and actin cytoskeleton are required for SPB separation driven by SAC inactivation in stu1-5 mutants. (A) stu1-5 (ySP6892) and stu1-5 mad2Δ (ySP6625) cells were arrested in G1 with α-factor at 25°C and released at 37°C. At 90 min after release, cultures were split in two, and dimethyl sulfoxide (DMSO) or nocodazole (NOC) was added, respectively, to each half of the culture. At the indicated times, samples were collected to follow kinetics of budding, SPB separation, and mitotic spindle formation. (B) stu1-5 (ySP6892), stu1-5 mad2Δ (ySP6625), stu1-5 mad2Δ dyn1Δ (ySP6605) cells were arrested in G1 with α-factor at 25°C and released at 37°C. Distances between SPBs were measured 150′ after release. (C) stu1-5 mad2Δ swe1Δ (ySP7926) cells were arrested in G1 with α-factor at 25°C and released at 37°C. At 90 min after release, DMSO or Lat-B were added. Distances between SPBs were measured 120′ after release.
Because the actin cytoskeleton is involved in spindle positioning through interaction with cytoplasmic microtubules (Palmer et al., 1992), we tested the effects of actin depolymerization on SPB separation in stu1-5 mad2Δ cells. Because actin perturbations activate the morphogenesis checkpoint, which delays mitotic entry through Swe1-mediated inhibitory phosphorylation of Cdk1 (McMillan et al., 1998), we disrupted the actin cytoskeleton with latrunculin-B in stu1-5 mad2Δ cells that were also deleted for SWE1. Remarkably, actin depolymerization dramatically reduced SPB distances in stu1-5 mad2Δ swe1Δ cells (Figure 7C), suggesting that the actin cytoskeleton plays a role in SPB separation in these conditions. Thus, forces acting on cytoplasmic microtubules seem responsible for SPB separation in stu1-5 cells when the SAC is turned off.
DISCUSSION
A Novel Role for the SAC in Restraining SPB Separation
In this article, we report that SAC inactivation leads to unscheduled SPB separation in cells that are defective in bipolar spindle formation, such as stu1-5 and cin8-3 kip1Δ temperature-sensitive mutants.
The essential role of the Cin8 and Kip1 kinesins in bipolar spindle assembly has been recently linked to their microtubule-bundling activity rather than their motor function (Crasta et al., 2006). Consistently, we find that inactivation of SAC proteins in cin8-3 kip1Δ cells allows also spindle formation. In contrast, loss of SAC activity in stu1-5 cells leads to SPB separation apparently without spindle assembly, suggesting that the SAC controls forces acting on astral, rather than spindle, microtubules. In agreement with this conclusion, we find that microtubules, cytoplasmic dynein and the actin cytoskeleton are required for SPB separation in the absence of the SAC (see below).
The SPB separation that we observe in stu1-5 and cin8-3 kip1Δ cells lacking a functional SAC depends on unscheduled activation of Cdc20/APC and the FEAR pathway of Cdc14 nucleolar release, suggesting that this process occurs at the metaphase-to-anaphase transition, a cell cycle phase where normally SPBs are already separated and have formed bipolar spindles. Although this excludes that the SAC controls SPB separation during the unperturbed cell cycle, we propose that preventing SPB separation, as well as sister chromatid separation, when the conditions to assemble a bipolar spindle have not been satisfied contributes to cell viability by providing a safeguard mechanism that ensures balanced chromosome segregation. Because yeast centromeres are connected to SPBs much earlier than bipolar spindle formation (Tanaka et al., 2002), one can envision that precocious SPB separation in conditions where a functional bipolar spindle cannot be formed would bring duplicated chromosomes too far from the unbound SPB, thus compromising the establishment of bipolar attachments once the conditions become again permissive for spindle assembly. Consistent with this hypothesis, we find that SAC inactivation allows aberrant segregation of chromatin masses in both cin8-3 kip1Δ and stu1-5 cells. Interestingly, premature spindle elongation during S phase has recently been proposed to impair proper kinetochore bipolar attachment (Liu et al., 2008).
Our genetic screen for synthetic lethals with the bub3-WDd1 mutation could also underscore an important role for the SAC in restraining SPB separation when a functional bipolar spindle cannot be formed. Because of the partial checkpoint proficiency of the bub3-WDd1 mutation (Fraschini et al., 2001), this screen has been highly stringent and uncovered only cin8 mutants among a very high number of mutagenized clones. In agreement with a residual SAC activity, the bub3-WDd1 mutation is not lethal when combined with the knockouts of KAR3, CIK1, or BIM1 that, conversely, are synthetically lethal with MAD2 deletion (our unpublished data). Therefore, the viability of cin8 mutants seems to rely on a fully functional SAC, consistent with previous data (Geiser et al., 1997), and providing further evidence that, besides preventing unscheduled sister chromatid separation, this checkpoint preserves genome stability by additional mechanisms. Remarkably, the SAC has been involved in centrosome clustering in cells with extra centrosomes, thus suppressing multipolar mitoses (Basto et al., 2008; Kwon et al., 2008). The analogy with our data raises the possibility that SAC-mediated controls of centrosome/SPB forces are universal and involve similar proteins. Interestingly, Basto et al. (2008) and Kwon et al. (2008) reported that the kinesin 14 motor protein Ncd is required for the coalescence of extra centrosomes in Drosophila. We found that the yeast kinesin 14 Kar3 is also involved in restraining SPB separation in stu1-5 cells (data not shown). However, KAR3 deletion also partially rescues the spindle assembly defects of the stu1-5 mutant (data not shown), thereby possibly accounting for the SPB separation that we observe in stu1-5 kar3Δ cells.
The SAC has also been recently shown to promote symmetric localization of Kar9 at SPBs (Leisner et al., 2008), which in turn would orient both spindle poles toward the bud, thereby disrupting proper spindle orientation (Liakopoulos et al., 2003). Altogether, these data suggest that the SAC controls pulling forces at spindle poles.
The FEAR Pathway and Cdc14 in the Regulation of Spindle Forces
We find that SPB separation driven by SAC inactivation in stu1 cells requires the Cdc14 phosphatase and FEAR pathway for its early anaphase nucleolar release. Consistently, CDC14 overexpression can bring about SPB separation in cin8-3 kip1Δ mutant cells. The FEAR pathway has been implicated in spindle stability during anaphase by recruiting the Aurora B complex to the spindle midzone (Pereira and Schiebel, 2003) and by regulating microtubule turnover at kinetochores (Higuchi and Uhlmann, 2005). In our experimental conditions, Cdc14 seems to regulate SPB separation by controlling the forces at cytoplasmic, rather than nuclear, microtubules. Consistently, the FEAR network and Cdc14 have been shown to regulate nuclear positioning by promoting spindle pulling forces by cytoplasmic microtubules in the mother cell (Ross and Cohen-Fix, 2004). Before anaphase, cytoplasmic microtubules in the mother cell push the nucleus toward the bud neck with the help of pulling forces exerted by cytoplasmic microtubules anchored to the bud (Pearson and Bloom, 2004). The FEAR pathway has been proposed to switch the forces to a pulling mode at the onset of anaphase in the mother cell. This could involve alterations in the interactions between cytoplasmic microtubules and the mother cortex and/or changes in the behavior of motor proteins, such as dynein (Ross and Cohen-Fix, 2004). It is worth noting that Cdc14 localizes at SPBs in anaphase (Pereira et al., 2002), although its targets in this process are unknown. A key candidate is Kar9, which is localized on the bud-directed spindle pole because of Clb4/CDK-dependent phosphorylation that inhibits its recruitment to the mother-bound SPB (Liakopoulos et al., 2003; Maekawa and Schiebel, 2004). However, whether Kar9 is required for the nuclear migration into the bud observed in FEAR mutants has not been investigated. In our experimental conditions, Kar9 seems to be dispensable for SPB separation in stu1-5 mad2Δ cells (data not shown), suggesting that other proteins, such as components of the dynein–dynactin complex, must be dephosphorylated by Cdc14 for this process to take place. Whether components of the dynein/dynactin complex are phosphorylated by CDKs is unknown at the moment and certainly deserves further investigation.
Involvement of Cytoplasmic Microtubules, Dynein, and Actin Cytoskeleton in SPB Separation
We showed that microtubule depolymerization by nocodazole in stu1-5 mad2Δ cells, which seem to lack nuclear microtubules, prevents SPB separation. In addition, cytoplasmic dynein and the actin cytoskeleton, which are involved in spindle positioning, are required for SPB separation when the SAC is inhibited in stu1-5 cells, supporting the notion that this process depends on forces pulling on astral microtubules. The actin cytoskeleton is implicated in spindle positioning by providing a mechanical network for microtubule transport (Palmer et al., 1992; Theesfeld et al., 1999). The class V-myosin Myo2, which interacts through Kar9 with Bim1 at microtubule plus ends, pulls microtubules toward the bud by moving along actin cables (Yin et al., 2000; Hwang et al., 2003). Conversely, we found that Kar9 function is dispensable for SPB separation in stu1-5 mad2Δ cells, whereas the dynein pathway of spindle positioning is required. Dynein promotes spindle positioning predominantly during anaphase through association with the cortical anchor Num1 and might contribute to the capture of cytoplasmic microtubules by the bud tip through sliding plus ends of cytoplasmic microtubules along the cortex (reviewed in Pearson and Bloom, 2004). It is possible that the actin cytoskeleton contributes also to the dynein pathway of spindle positioning by regulating cortical localization of polarity cues, such as Num1. Interestingly, dynein is asymmetrically localized at SPBs in metaphase and this asymmetric distribution is important for proper spindle positioning (Grava et al., 2006). Asymmetric localization of dynein at SPBs depends on the mitotic Clb1/2-dependent CDKs, although their substrate(s) in this process is currently unknown. In any case, Cdc14-dependent dephosphorylation events during anaphase are likely to reverse this asymmetry to generate the pulling forces for spindle elongation and chromosome segregation.
Whether dynein could be involved in SPB separation during a normal cell cycle is still an open question. Cytoplasmic dynein is required for centrosome separation in mammalian cells (Vaisberg et al., 1993). In yeast, DYN1 deletion is lethal for yeast cells lacking Cin8 (Geiser et al., 1997), although lethality seems to be ascribed to defects in spindle elongation during anaphase (Saunders et al., 1995). Dynein and dynactin have also been recently shown to be essential for stu1-5 cells at permissive temperature in some genetic backgrounds, leading to the proposal that dynein/dynactin could provide an SPB-separating activity (Amaro et al., 2008). Our data support the notion that indeed dynein promotes SPB separation at least under certain circumstances.
Interestingly, the actin cytoskeleton (Kwon et al., 2008) and dynein (Quintyne et al., 2005) are required for centrosome clustering in cells with extra centrosomes, thereby preventing spindle multipolarity in cancer cells that are often characterized by centrosome amplification. Thus, by controlling different aspects of centrosome/SPB function in mammalian versus yeast cells actin cytoskeleton and dynein contribute to preventing chromosome missegregation and aneuploidy occurrence.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to N. Pavelka for statistical analysis of data and to R. Primignani for help with the genetic screen for synthetic lethals. We thank J. Kilmartin, A. Murray, A. Hoyt, T. Huffaker, F. Uhlmann, R. Visintin, and K. Nasmyth for strains and plasmids; A. Musacchio, R. Li, S. Jaspersen, E. Schwob, N. Pavelka, and R. Fraschini for critical reading of the manuscript; and all members of our laboratory for useful discussions. E. C. was supported by a Telethon fellowship. This work has been supported by grants from Associazione Italiana Ricerca sul Cancro, Telethon, and Progetti di Ricerca di Interesse Nazionale (to S. P.).
Abbreviations used:
- APC
anaphase-promoting complex
- FEAR
Cdc-fourteen early anaphase release
- SAC
spindle assembly checkpoint
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1150) on April 1, 2009.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R., Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro I. A., Costanzo M., Boone C., Huffaker T. C. The Saccharomyces cerevisiae homolog of p24 is essential for maintaining the association of p150Glued with the dynactin complex. Genetics. 2008;178:703–709. doi: 10.1534/genetics.107.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Amon A. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Baskerville C., Segal M., Reed S. I. The protease activity of yeast separase (esp1) is required for anaphase spindle elongation independently of its role in cleavage of cohesin. Genetics. 2008;178:2361–2372. doi: 10.1534/genetics.107.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J. W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Jr, Hoyt M. A., Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J. M., Kirschner M. W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Crasta K., Huang P., Morgan G., Winey M., Surana U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006;25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F., Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D., Amon A. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- D'Amours D., Stegmeier F., Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Farkasovsky M., Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I., Dahmann C., Surana U., Amon A., Nasmyth K., Goetsch L., Byers B., Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Sironi L., Musacchio A., Lucchini G., Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti E., Lucchini G., Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J. R., Schott E. J., Kingsbury T. J., Cole N. B., Totis L. J., Bhattacharyya G., He L., Hoyt M. A. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine R. A., Oberle J. R., Cooper J. A. The cortical protein num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Hoyt M. A. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K., Saunders W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L. H., Lau L. F., Smith D. L., Mistrot C. A., Hardwick K. G., Hwang E. S., Amon A., Murray A. W. Budding yeast Cdc 20, a target of the spindle checkpoint [correction published in Science (1998). 280, 1331] Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jansen R., Tollervey D., Hurt E. C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jensen S., Segal M., Clarke D. J., Reed S. I. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Schiebel E. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 2008;7:283–286. doi: 10.4161/cc.7.3.5349. [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., Thery M., Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Tirnauer J. S., Li J., Schuyler S. C., Liu J. Y., Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Leisner C., Kammerer D., Denoth A., Britschi M., Barral Y., Liakopoulos D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr. Biol. 2008;18:1249–1255. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lim H. H., Goh P. Y., Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol. Cell. Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liang F., Jin F., Wang Y. The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet. 2008;4:e1000262. doi: 10.1371/journal.pgen.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M., Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. N., Sia R.A.L., Lew D. J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Rose M. D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Sullivan D. S., Huffaker T., Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D., Huffaker T. C. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Pereira G., Manson C., Grindlay J., Schiebel E. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 2002;157:367–379. doi: 10.1083/jcb.200112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Peters J. M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Quintyne N. J., Reing J. E., Hoffelder D. R., Gollin S. M., Saunders W. S. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross K. E., Cohen-Fix O. A role for the FEAR pathway in nuclear positioning during anaphase. Dev. Cell. 2004;6:729–735. doi: 10.1016/s1534-5807(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Saunders W. S., Koshland D., Eshel D., Gibbons I. R., Hoyt M. A. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Hyman A. A., Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J. R., Kho D., Hoyt M. A., Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Simanis V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Huang J., Rahal R., Zmolik J., Moazed D., Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Higuchi T., Katis V. L., Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Lehane C., Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Theesfeld C. L., Irazoqui J. E., Bloom K., Lew D. J. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep. 2001;2:487–492. doi: 10.1093/embo-reports/kve113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg E. A., Koonce M. P., McIntosh J. R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury E. L., Morgan D. O. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J. Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R. V., Cheng J. W., Salmon E. D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T. C., Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Yin H., You L., Pasqualone D., Kopski K. M., Huffaker T. C. Stu1p Is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol. Biol. Cell. 2002;13:1881–1892. doi: 10.1091/mbc.01-09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.