Abstract
OBJECTIVE
The pathogenesis of type 1 diabetes has a strong genetic component. Genome-wide association scans recently identified novel susceptibility genes including the phosphatases PTPN22 and PTPN2. We hypothesized that PTPN2 plays a direct role in β-cell demise and assessed PTPN2 expression in human islets and rat primary and clonal β-cells, besides evaluating its role in cytokine-induced signaling and β-cell apoptosis.
RESEARCH DESIGN AND METHODS
PTPN2 mRNA and protein expression was evaluated by real-time PCR and Western blot. Small interfering (si)RNAs were used to inhibit the expression of PTPN2 and downstream STAT1 in β-cells, allowing the assessment of cell death after cytokine treatment.
RESULTS
PTPN2 mRNA and protein are expressed in human islets and rat β-cells and upregulated by cytokines. Transfection with PTPN2 siRNAs inhibited basal- and cytokine-induced PTPN2 expression in rat β-cells and dispersed human islets cells. Decreased PTPN2 expression exacerbated interleukin (IL)-1β + interferon (IFN)-γ–induced β-cell apoptosis and turned IFN-γ alone into a proapoptotic signal. Inhibition of PTPN2 amplified IFN-γ–induced STAT1 phosphorylation, whereas double knockdown of both PTPN2 and STAT1 protected β-cells against cytokine-induced apoptosis, suggesting that STAT1 hyperactivation is responsible for the aggravation of cytokine-induced β-cell death in PTPN2-deficient cells.
CONCLUSIONS
We identified a functional role for the type 1 diabetes candidate gene PTPN2 in modulating IFN-γ signal transduction at the β-cell level. PTPN2 regulates cytokine-induced apoptosis and may thereby contribute to the pathogenesis of type 1 diabetes.
Type 1 diabetes is a chronic autoimmune disease with a strong genetic etiology. Genetic predisposition to type 1 diabetes depends on a small number of genes having large effects and a larger number of genes having small effects (1). These genes interact with putative environmental factors, which may include viral infections, triggering insulitis and eventually diabetes (2). Recent genome-wide association studies have shown association between type 1 diabetes and four chromosome regions, pointing to several new candidate genes for the disease (3). Most of the newly and previously identified genes are assumed to regulate immune function. This contrasts with type 2 diabetes, where similar studies indicate a major role for genes regulating β-cell function (4).
We have presently evaluated whether the recently identified candidate genes for type 1 diabetes (3,5) are expressed in pancreatic β-cells and whether their expression is modulated by proinflammatory cytokines. This was done by examining our previous array analyses of cytokine-treated or virus-infected rodent and human β-cells/pancreatic islets (6–10; complete information on these arrays is available at the Beta Cell Gene Expression Bank [(11)]) and new array analyses performed in our laboratory using the new Affymetrix rat array Genechip 230.2.0 (unpublished data). This analysis identified β-cell expression of the candidate gene protein tyrosine phosphatase (PTP)N2.
PTPN2 (also known as TC-PTP or PTP-S2) is a member of the first nontransmembrane (NT1) subfamily of PTPs. PTPs are a superfamiliy of enzymes with opposite roles compared with protein tyrosine kinases (12). PTPN2 is expressed in immune cells, and its expression is modulated by cell cycle, mitogenic agents, and cytokines (13). PTPN2 exists as two isoforms generated from alternative splicing: a major TC45 isoform (45 kDa) containing a nuclear localization sequence and that shuttles between the nucleus and the cytoplasm and a less abundant TC48 isoform (48 kDa) that is anchored to the endoplasmic reticulum (13). Many targets have already been identified for TC45, including Janus kinases (JAKs) and signal transducer and activator of transcription (STATs), p42/44 mitogen-activated protein kinase (MAPK) (extracellular signal–related kinase [ERK]), epidermal growth factor receptor (EGFR), and insulin receptor β (IRβ) (14–17). Several of these pathways have been implicated in the control of β-cell physiology, survival, and expansion (18–20).
We have previously suggested that islet inflammation and subsequent β-cell death develops in the context of a “dialogue” between the immune system and β-cells (21). Thus, β-cells exposed to viral agents (6,22), or to endogenous Toll-like receptor ligands (23), release cytokines and chemokines that attract and activate macrophages, T-cells, and B-cells. These immune cells will then trigger β-cell apoptosis via contact mediators such as Fas-FasL and/or via secretion of proinflammatory cytokines such as interferon (IFN)-γ, interleukin (IL)-1β, and tumor necrosis factor (TNF)-α (21,24). IFN-γ has a key role in this process. Thus, neither IL-1β nor TNF-α alone induce human or rodent β-cell death, but combinations of these cytokines with IFN-γ lead to 50% β-cell death after 6–9 days (21,24). IFN-γ signal transduction involves activation of the tyrosine kinases JAK1 and JAK2 that phosphorylate STAT1, which then dimerizes and translocates to the nucleus where it binds γ-activated sites of diverse genes (21). Excessive activation of JAK/STAT signaling may lead to cell death, and STAT1 transcriptional activity is regulated by multiple negative feedback mechanisms, including inhibition of JAKs by specific phosphatases and by the suppressors of cytokine signaling and inhibition of STAT1 activity by protein inhibitor of activated STAT (PIAS) proteins (25). β-Cells from STAT1-deficient mice (STAT1−/−) are protected against IL-1β + IFN-γ–induced apoptosis, and STAT1−/−mice are more resistant to diabetes induced by multiple low doses of streptozotocin or after backcross into NOD mice (18,26). This protective effect takes place at the β-cell level, since islets from STAT1−/−, but not from wild-type mice, prevent diabetes when transplanted into wild-type mice subsequently treated with multiple low doses of streptozotocin (27). However, it still remains unclear whether modulation of phosphatases modifies IFN-γ–induced β-cell apoptosis.
We presently report that PTPN2 is regulated by the cytokines IL-1β, IFN-γ, and TNF-α in rodent and human pancreatic islet cells. Importantly, small interfering (si)RNA-mediated PTPN2 knockdown increases STAT1 activation and aggravates cytokine-induced β-cell apoptosis in a STAT1-dependent manner. These observations indicate that one of the new candidate genes for type 1 diabetes may act at the immune and β-cell level, exacerbating cytokine-induced β-cell apoptosis under inflammatory conditions.
RESEARCH DESIGN AND METHODS
Culture of primary fluorescence-activated cell–sorted rat β-cells, human islets, and INS-1E cells.
Male Wistar rats (Charles River Laboratories, Brussels, Belgium) were housed and used according to the guidelines of the Belgian Regulations for Animal Care. Islets were isolated by collagenase digestion and handpicked under a stereomicroscope. β-Cells were purified by autofluorescence-activated cell sorting (FACSAria; BD Bioscience, San Jose, CA) (28,29). The preparations contained 90.4 ± 3.2% β-cells (n = 4). β-Cells were cultured for 2 days in Ham's F-10 medium containing 10 mmol/l glucose, 2 mmol/l glutaMAX, 50 μmol/l 3-isobutyl-1-methylxanthine, 5% fetal bovine serum (FBS), 0.5% charcoal-absorbed BSA (Boehringer, Indianapolis, IN), 50 units/ml penicillin, and 50 μg/ml streptomycin (29,30). During cytokine exposure, cells were cultured in the same medium but without serum.
Human islets were isolated from 11 nondiabetic organ donors (age 63 ± 5 years; BMI 25.1 ± 0.8 kg/m2) in Pisa, Italy, with the approval of the local ethics committee. Islets were isolated by enzymatic digestion and density-gradient purification (31) and cultured in M199 medium containing 5.5 mmol/l glucose. Their functional status was determined using glucose-stimulated insulin release and was 2.8 ± 0.4 (expressed as stimulation index). The human islets were shipped to Brussels within 1–5 days of isolation. After overnight recovery in Ham's F-10 containing 6.1 mmol/l glucose, 10% FBS, 2 mmol/l GlutaMAX, 50 μmol/l 3-isobutyl-1-methylxanthine, 1% BSA, 50 units/ml penicillin, and 50 μg/ml streptomycin, islets were exposed to cytokines in the same medium without FBS for 2 days. The percentage of β-cells, examined in the 11 dispersed islet preparations by staining with anti-insulin antibody (1:1,000; Sigma, Bornem, Belgium) and donkey anti-mouse IgG rhodamine (1:200; Lucron Bioproducts, De Pinte, Belgium), was 49 ± 5%.
The rat insulin-producing INS-1E cell line (a gift from Dr. C. Wollheim, Centre Medical Universitaire, Geneva, Switzerland) was cultured as previously described (32,33).
RNA interference.
The siRNAs used in this study are listed in Supplementary Table A1 (available in an online-only appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db08-1510/DC1). The best settings for the transfection of siRNAs in INS-1E cells, primary fluorescence-activated cell–sorted rat β-cells, and dispersed human islets were established by using a fluorescein isothiocyanate (FITC)-conjugated siRNA (siGLO Green Transfection Indicator, Thermo Scientific). Different transfection methods were tested, including electroporation, and several lipid reagents. Among those, DharmaFECT 1 (Thermo Scientific) was selected for its high transfection efficiency, namely 95 ± 2.7% in INS-1E cells and 80.7 ± 5.6% in primary rat β-cells. No cell toxicity was observed with this transfection method (% cell death: 9.4 ± 1.7% after transfection vs. 8.4 ± 2.2% in control for INS-1E cells and 9.8 ± 1.3% after transfection vs. 7.4 ± 1.7% in control for primary rat β-cells). Cells were cultured in antibiotic-free medium for at least 24 h before transfection. For transfection, the siRNA and the DharmaFECT were diluted separately in OptiMEM medium and incubated at room temperature for 5 min. Lipid-siRNA complexes were then formed at room temperature for 20 min in a proportion of 0.75, 1.25, and 1.75 μl DharmaFECT to 150 nmol/l of siRNA for INS-1E cells, primary β-cells, and dispersed human islets, respectively. The complexes were diluted five times in antibiotic-free medium and added to the cells at a final concentration of 30 nmol/l siRNA (except when indicated otherwise) for overnight transfection. The concentration of 30 nmol/l was selected after dose-response studies (data not shown). Afterward, cells were cultured for a 48-h recovery period and subsequently exposed to cytokines.
Cell treatment and nitric oxide measurement.
The following cytokine concentrations were used, based on previous dose-response experiments (10,24,33): recombinant human IL-1β (specific activity 1.8 × 107units/mg; a gift from C.W. Reinolds, National Cancer Institute, Bethesda, MD) at 10, 50, or 100 units/ml as indicated; recombinant murine TNF-α (specific activity: 2 × 108units/mg; Innogenetics, Gent, Belgium) at 1,000 units/ml; and recombinant rat and human IFN-γ (specific activity: 2 × 107units/mg; R&D Systems, Abingdon, U.K.) at 100 and 1,000 units/ml for rat cells and human islets, respectively. When cells were treated with cytokines, culture supernatants were collected for nitrite determination (nitrite is a stable product of nitric oxide [NO] oxidation) at OD540 nm using the Griess method (34). Palmitate (sodium salt, Sigma) was dissolved in 90% ethanol, heated to 60°C, and used at a final concentration of 0.5 mmol/l in medium containing 1% BSA (35). The SERCA blocker CPA (Sigma) was dissolved in DMSO and used at a concentration of 25 μmol/l.
Assessment of cell viability.
The percentage of viable, apoptotic, and necrotic cells was determined after a 15-min incubation with the DNA-binding dyes propidium iodide (PI, 5 μg/ml; Sigma) and Hoechst 33342 (HO, 5 μg/ml; Sigma) (10,29,36). A minimum of 500 cells was counted in each experimental condition. Viability was evaluated by two independent observers, one of them being unaware of sample identity. The agreement between findings obtained by the two observers was >90%. Results are expressed as percent apoptosis, calculated as number of apoptotic cells/total number of cells × 100. In some experiments, apoptosis was confirmed using the Cell Death Detection ELISAplus kit (Roche Diagnostics, Vilvoorde, Belgium), which detects cytoplasmic fragmented DNA.
mRNA extraction and real-time PCR.
Poly(A)+mRNA was isolated from INS-1E cells, rat primary β-cells, and human islets using the Dynabeads mRNA DIRECT kit (Invitrogen) and reverse transcribed as previously described (10,29,37). The real-time PCR amplification reaction was done as described (29,37), using SYBR Green and compared with a standard curve (38). Expression values were corrected for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). IL-1β alone does not modify GAPDH expression, whereas exposure to IL-1β + IFN-γ reduces its expression according to cell death in INS-1E cells (Supplementary Fig. A1 and (7,10)). The primers used in this study are detailed in Supplementary Table A2 (online appendix).
Western blot analysis.
Cells were washed with cold PBS and lysed with either Laemmli buffer or phospho lysis buffer (the compositions of the buffers are provided in Supplementary Table A3). Lysates were then resolved by 8–10% SDS-PAGE and transferred to a nitrocellulose membrane. The antibodies used in this study are listed in Supplementary Table A4. Immunoreactive bands were revealed using the SuperSignal West Femto chemiluminescent substrate (Thermo Scientific), detected using a LAS-3000 charge-coupled device camera and quantified with the Aida Analysis software (Fujifilm).
Immunofluorescence.
INS-1E cells were seeded onto glass coverslips and treated as indicated. Cells were washed with cold PBS, fixed for 10 min in 100% methanol at −20°C, washed three times with PBS, quenched 5 min in PBS/0.1% sodium borohydride, washed twice with PBS, and incubated for 5 min in PBS containing 0.2% Triton X-100 (PBST). After a 1-h blocking with 5% normal goat serum in PBST, cells were incubated overnight with PTPN2 antibodies (0.7 μg/ml) in PBST/0.5% normal goat serum (Supplementary Table A4). Cells were washed three times with PBST, and FITC-conjugated goat anti-mouse antibodies (Lucron Bioproducts) were applied for 2 h at 1:200 in PBST. After two washes with PBST, nuclei were counterstained with Hoechst 33342 (HO) for 5 min and washed three times with PBS. Coverslips were mounted in mounting medium (Dakocytomation), and immunofluorescence was visualized on a Zeiss microscope.
Statistical analysis.
Data are presented as means ± SE. Comparisons were performed by two-tailed paired Student's t test or by ANOVA followed by Student's t test with Bonferroni correction. A P value <0.05 was considered statistically significant.
RESULTS
Cytokines upregulate PTPN2 mRNA and protein expression in primary rat β-cells, human islets, and INS-1E cells.
We first evaluated the expression of PTPN2 mRNA (both TC45 and TC48 isoforms) in primary fluorescence-activated cell–sorted rat β-cells, human islets, and rat insulin–producing INS-1E cells and tested whether cytokine treatment would affect its expression. PTPN2 mRNA was expressed in untreated primary rat β-cells and human islets cells, and IL-1β + IFN-γ exposure upregulated its expression by 9.6-fold (rat β-cells) and 2.2-fold (human islets) after 24 and 48 h, respectively (Fig. 1A and B). A time course of IL-1β + IFN-γ treatment in INS-1E cells indicated that PTPN2 mRNA was already induced after 2 h, increasing progressively up to 24 h (Fig. 1C). We then confirmed by Western blot that PTPN2 was upregulated by two- and threefold, respectively, in human islets and INS-1E cells after IL-1β + IFN-γ treatment (Fig. 1D and E andFig. 3B and C). In accordance with previous reports (13), we observed TC45 (arrow,Figs. 1D and 3 B) to be the major PTPN2 isoform expressed in β-cells, with TC48 being poorly expressed (Fig. 1D and 3 B). In contrast, the expression of PTPN22 (another phosphatase associated with type 1 diabetes risk; (39)) was only present in samples from rat spleen and lymph nodes (used as positive controls), whereas no or marginal PTPN22 expression was observed in primary β-cells, human islets, and INS-1E cells. The expression of PTPN22 was not modified by cytokines (Supplementary Fig. A2 in the online appendix).
FIG. 1.
Cytokines upregulate PTPN2 expression in primary fluorescence-activated cell–sorted rat β-cells, human islets, and INS-1E cells. A: Rat β-cells were cultured for 2 days and subsequently left untreated or treated with the combination of IL-1β (10 units/ml) + IFN-γ (100 units/ml) for 24 h. B, D, and E: Hand-picked human islets were cultured overnight and then left untreated or exposed to IL-1β (50 units/ml) + IFN-γ (1,000 units/ml) for 48 h. C: INS-1E cells were left untreated or treated with IL-1β (10 units/ml) + IFN-γ (100 units/ml) for 2, 6, and 24 h as indicated. A– C: PTPN2 mRNA expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. D: PTPN2 and α-tubulin expression in human islets were evaluated by Western blot. E: Mean optical density measurements of PTPN2 Western blots corrected for protein loading by α-tubulin (representative figure in D). Results are means ± SE of three to five independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. untreated cells by Student's t test. NS, nonstimulated (untreated).
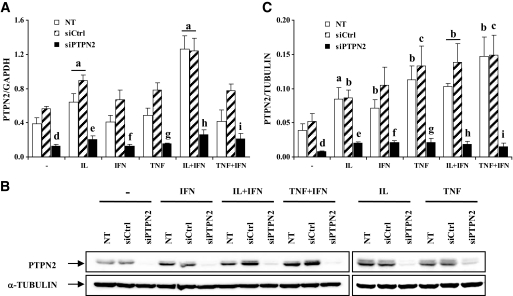
FIG. 3.
A siRNA targeting PTPN2 inhibits basal and cytokine-induced PTPN2 expression in INS-1E cells. INS-1E cells were left untransfected (NT □) or transfected with 30 nmol/l of either a control siRNA (siCtrl, ▨) or a pool of siRNAs targeting PTPN2 (siPTPN2, ■). After 2 days of recovery, cells were left untreated, or treated for 24 h with IL-1β (100 units/ml), IFN-γ (100 units/ml), TNF-α (1,000 units/ml), IL-1β (10 units/ml) + IFN-γ (100 units/ml), or TNF-α (1,000 units/ml) + IFN-γ (100 units/ml). A: PTPN2 mRNA expression was assayed by RT-PCR and normalized for the housekeeping gene GAPDH. Results are means ± SE of four independent experiments. B: PTPN2 and α-tubulin protein expression were evaluated by Western blot. The results are representative of five independent experiments. C: Mean optical density measurements of PTPN2 Western blots, corrected for protein loading by α-tubulin. Results are means ± SE of five independent experiments; a: P < 0.001, b: P < 0.01, and c: P < 0.05 vs. untreated NT or untreated transfected with the same siRNA; d: P < 0.01 vs. untreated NT and siCtrl; e: P < 0.01 vs. IL-1β–treated NT and siCtrl; f: P < 0.01 vs. IFN-γ–treated NT and siCtrl; g: P < 0.01 vs. TNF-α–treated NT and siCtrl; h: P < 0.001 vs. IL-1β + INF-γ–treated NT and siCtrl, i: P < 0.01 vs. TNF-α + INF-γ–treated NT and siCtrl; ANOVA followed by Student's t test with Bonferroni correction.
PTPN2 subcellular distribution.
PTPN2 was mainly located in the nucleus under unstimulated conditions (Fig. 2, left panels). However, a 15-min treatment with IFN-γ alone or in combination with IL-1β or TNF-α resulted in redistribution of the protein between the cytoplasm and the nucleus (Fig. 2, middle and right panels).
FIG. 2.
Effects of cytokine exposure on PTPN2 subcellular distribution. INS-1E cells were plated onto glass coverslips and left untreated or treated for 15 min with IFN-γ (100 units/ml), IL-1β (10 units/ml) + IFN-γ (100 units/ml), or TNF-α (1,000 units/ml) + IFN-γ (100 units/ml). Cells were then fixed and processed for immunofluorescence as described in research design and methods. The figure is representative of five independent experiments. (A high-quality digital representation of this figure is available in the online issue.)
siRNA targeting PTPN2 inhibits basal and cytokine-induced PTPN2 expression and exacerbates cytokine-induced apoptosis in INS-1E cells, primary rat β-cells, and dispersed human islets.
We next examined the role of PTPN2 in β-cells. To this end, INS-1E cells were left untransfected, transfected with an irrelevant control siRNA (siCtrl), or with an siRNA targeting both PTPN2 isoforms (siPTPN2). Cells were then left untreated, or treated for 24 h with IL-1β, IFN-γ, TNF-α, or with the combination of IL-1β + IFN-γ or TNF-α + IFN-γ. PTPN2 mRNA expression was significantly upregulated after IL-1β and IL-1β + IFN-γ treatment in untransfected and siCtrl-transfected INS-1E cells (Fig. 3A). The expression of PTPN2 proteins was increased by all cytokines tested, alone or in combination, in both untransfected and siCtrl-transfected cells (Fig. 3B and C), suggesting that PTPN2 expression is regulated at both transcriptional and post-transcriptional levels. Transfection with PTPN2 siRNA potently inhibited basal and cytokine-induced PTPN2 expression at the mRNA and protein level (Fig. 3A–C). We then evaluated whether siRNA-mediated PTPN2 inhibition affects cytokine-induced apoptosis in INS-1E cells. As expected, a 24-h treatment with IL-1β + IFN-γ or TNF-α + IFN-γ induced apoptosis in untransfected and siCtrl-transfected cells, whereas each individual cytokine did not (Fig. 4A; of note, the necrotic component was <2% under all experimental conditions tested; data not shown). The inhibition of PTPN2 exacerbated β-cell apoptosis after exposure to both IL-1β + IFN-γ and TNF-α + IFN-γ treatments. Importantly, PTPN2 inhibition also rendered IFN-γ treatment toxic to the cells, whereas this cytokine alone failed to induce apoptosis in both control counterparts (Fig. 4A). These results were confirmed using a second method to evaluate apoptosis, namely quantification of cytoplasmic fragmented DNA (Fig. 4B), and also with a second siRNA targeting PTPN2 (see below). The increased apoptosis in cytokine-treated PTPN2-deficient INS-1E was not accompanied by a more severe decrease in Ins1 and Ins2 mRNA contents in these cells (Supplementary Fig. A3) and could not be explained by a higher release of NO, since nitrite was similarly produced by untransfected, siCtrl-, and siPTPN2-transfected cells after IL-1β, IL-1β + IFN-γ, and TNF-α + IFN-γ treatments (Fig. 4C). Blocking PTPN2 using siRNA also augmented IFN-γ–, IL-1β + IFN-γ–, and TNF-α + IFN-γ–induced apoptosis in primary rat β-cells (Fig. 4D). Similar observations were made in dispersed human islet cells, in which siRNA-mediated PTPN2 inhibition exacerbated IL-1β + IFN-γ–induced cell death by 30–38% (Fig. 4E and F). This was again independent of NO production (data not shown). This effect seems to be specific for cytokines, since PTPN2 inhibition did not increase β-cell apoptosis in response to palmitate, high glucose, or the SERCA blocker CPA (Supplementary Fig. A4).
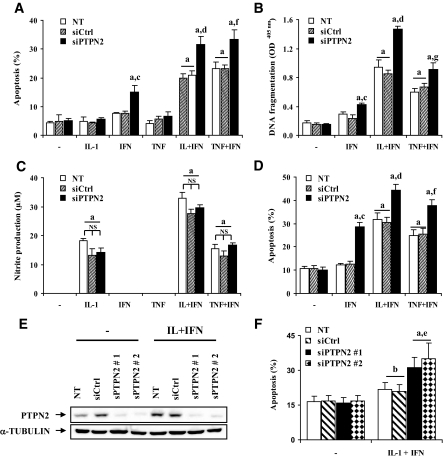
FIG. 4.
siRNA-mediated PTPN2 inhibition exacerbates cytokine-induced apoptosis in INS-1E cells, primary rat β-cells, and dispersed human islets, independently of NO production. INS-1E cells were transfected and treated as described in Fig. 3. Apoptosis was evaluated after 24 h using HO/PI staining (A) and a Cell Death Detection ELISAplus kit (B). C: INS-1E cells were transfected and treated as described in Fig. 3, and nitrite concentrations in supernatants were measured as described in research design and methods (D). Primary fluorescence-activated cell–sorted rat β-cells were cultured for 2 days and then transfected as described in Fig. 3. After 2 days of recovery, cells were left untreated (NT) or treated for 72 h with IFN-γ (100 units/ml), IL-1β (10 units/ml) + IFN-γ (100 units/ml), or TNF-α (1,000 units/ml) + IFN-γ (100 units/ml). E and F: Dispersed human islets were left untransfected, or transfected with 30 nmol/l of siCtrl or human siPTPN2 #1 or #2 and cultured for a 48-h recovery period. Cells were then treated with IL-1β (50 units/ml) + IFN-γ (1,000 units/ml) for 48 h when PTPN2 and α-tubulin were evaluated by Western blot (E) and 96 h when apoptosis was evaluated by HO/PI staining (F). Results are means ± SE of four experiments; a: P < 0.001 and b: P < 0.05 vs. untreated NT or untreated transfected with the same siRNA; c: P < 0.001 vs. IFN-γ–treated NT and siCtrl, d: P < 0.001 and e: P < 0.05 vs. IL-1β + INF-γ–treated NT and siCtrl, f: P < 0.001 and g: P < 0.05 vs. TNF-α + INF-γ–treated NT and siCtrl; ANOVA followed by Student's t test with Bonferroni correction.
PTPN2 inhibition increases IFN-γ–induced STAT1 and STAT3 phosphorylation.
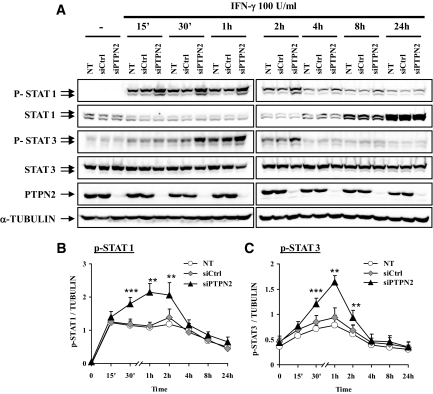
Taking into account that STAT1 is a substrate of PTPN2 in other cell types (13) and that it is an important mediator of cytokine-induced β-cell apoptosis (18), we next examined the effect of PTPN2 inhibition on the kinetics and magnitude of IFN-γ–induced STAT1 phosphorylation. STAT1 phosphorylation was highly induced after 15 min of IFN-γ treatment in both untransfected and siCtrl-transfected controls, slowly decreasing between 2 and 24 h (Fig. 5A and B). However, IFN-γ–induced STAT1 phosphorylation was markedly enhanced in cells lacking PTPN2, reaching a peak at 1–2 h and slowly decreasing afterward (Fig. 5A and B). We confirmed that the increased STAT1 phosphorylation in PTPN2-deficient cells was not due to an augmentation of total STAT1 content in these cells (Fig. 5A). Comparable results were observed for STAT3, another target of PTPN2 phosphatase activity, with clearly increased STAT3 phosphorylation in PTPN2-deficient cells (Fig. 5A and C). These data demonstrate that the phosphatase PTPN2 is a major modulator of IFN-γ–induced STAT1 and STAT3 activity in β-cells. We also evaluated the p42/44 MAPK (ERK), EGFR, and IRβ activation pathways, previously described as PTPN2 targets in other cell types (14,16,40). Neither ERK nor EGFR signaling pathways were affected after PTPN2 inhibition in INS-1E cells (Supplementary Fig. A5). On the other hand, there was an increase in IRβ phosphorylation after 30 min of cytokine treatment in PTPN2-inhibited INS-1E cells that lasted until 14 h (Supplementary Fig. A5).
FIG. 5.
PTPN2 inhibition increases IFN-γ–induced STAT1 and STAT3 phosphorylation. INS-1E cells were left untransfected (NT) or transfected with 30 nmol/l of either a control siRNA (siCtrl) or with a pool of siRNAs targeting PTPN2 (siPTPN2). After 2 days of recovery, cells were left untreated or treated with IFN-γ (100 units/ml) for 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 24 h. A: phospho-STAT1, total STAT1, phospho-STAT3, total STAT3, PTPN2, and α-tubulin proteins were evaluated by Western blot. These results are representative of five independent experiments. B and C: Mean optical density measurements of phospho-STAT1 (B) and phospho-STAT3 (C) Western blots corrected for protein loading by α-tubulin. Results are means ± SE of five independent experiments; **P < 0.01 and ***P < 0.001 vs. NT and siCtrl at the same time point, ANOVA followed by Student's t test with Bonferroni correction.
Double knockdown of PTPN2 and STAT1 protects INS-1E cells against cytokine-induced apoptosis.
To evaluate the role of STAT1 in the exacerbation of cytokine-induced β-cell apoptosis observed after PTPN2 inhibition, we additionally interfered with STAT1 in a double knockdown approach. We first confirmed by Western blot that both PTPN2 and STAT1 siRNAs adequately inhibited their respective targets without affecting the expression of the other protein and also that the double transfection of PTPN2 and STAT1 siRNAs potently inhibited both target proteins (Fig. 6A). As previously shown (Fig. 4), IL-1β + IFN-γ or TNF-α + IFN-γ induced apoptosis to a similar degree in both control conditions, and PTPN2 inhibition exacerbated cytokine-induced apoptosis, also rendering treatment with IFN-γ alone toxic to the cells (Fig. 6B). Inhibition of STAT1 protected the cells against IL-1β + IFN-γ- and TNF-α + IFN-γ–induced apoptosis (by 56 and 67%, respectively). STAT1 knockdown abrogated the proapoptotic effect of PTPN2 inhibition in cells exposed to IFN-γ or to combinations of cytokines (Fig. 6B). These results were confirmed using a second siRNA targeting PTPN2 expression (Supplementary Fig. A6) and suggest that increased STAT1 activity contributes to the aggravation of cytokine-induced apoptosis in PTPN2-deficient β-cells.
FIG. 6.
Double knockdown of PTPN2 and STAT1 protects INS-1E cells from cytokine-induced apoptosis. INS-1E cells were left untransfected (NT) or were transfected with 60 nmol/l of a control siRNA (siCtrl), or with 30 nmol/l of either a pool of siRNAs targeting PTPN2 (siPTPN2), or a siRNA targeting STAT1 (siSTAT1), or double transfected with 30 nmol/l of both siPTPN2 and siSTAT1. After 2 days of recovery, cells were left untreated, or treated for 24 h with IFN-γ (100 units/ml), IL-1β (10 units/ml) + IFN-γ (100 units/ml), or TNF-α (1,000 units/ml) + IFN-γ (100 units/ml) as indicated. A: Expression of STAT1, PTPN2, and α-tubulin proteins were evaluated by Western blot. The results are representative of three independent experiments. B: Apoptosis was evaluated using HO/PI staining. Results are means ± SE of four independent experiments; a: P < 0.001 vs. untreated NT or untreated transfected with the same siRNA; b: P < 0.001 vs. IFN-γ–treated NT and siCtrl; c: P < 0.001 vs. IFN-γ–treated siSTAT1 and siPTPN2 + siSTAT1; d: P < 0.001 vs. IL-1β + INF-γ–treated NT and siCtrl; e: P < 0.001 vs. IL-1β + INF-γ–treated siSTAT1 and siPTPN2 + siSTAT1; f: P < 0.001 vs. TNF-α + INF-γ–treated NT and siCtrl; g: P < 0.001 vs. TNF-α + INF-γ–treated siSTAT1 and siPTPN2 + siSTAT1; ANOVA followed by Student's t test with Bonferroni correction.
DISCUSSION
We presently show that the phosphatase PTPN2 is expressed in human islets and rat primary and immortal β-cells and that its expression is regulated by the pro-inflammatory cytokines IL-1β, IFN-γ, and TNF-α in these cells. PTPN2 was first identified in humans as a T-cell PTP (41) with two spliced variants, namely TC45 and TC48 (13,42). In rat β-cells and human islets, the TC45 isoform is by far the most prevalent (Fig. 1D and 3 B), and it is able to shuttle between the nucleus and the cytoplasm after cytokine exposure (Fig. 2). This change in subcellular location of PTPN2 has been shown previously in other cell types (14,16) and allows the phosphatase to target numerous cellular substrates, including cytoplasmic JAK tyrosine kinases, EGFR, the IRβ and nuclear STAT1, STAT3, STAT5, and STAT6 (43,44). Because of its modulatory role in a wide variety of signaling pathways, perturbations in PTPN2 expression are associated with diverse pathogenic processes, including ABC-like diffuse large B-cell lymphomas (45), inflammation-associated tumorigenesis (44), and several autoimmune disorders (see below).
PTPN2 inhibition in β-cells results in an early and more intense STAT1 and STAT3 activation after IFN-γ treatment, suggesting that this phosphatase plays an important role in the dephosphorylation and consequent inactivation of these transcription factors in β-cells. This early regulation of IFN-γ–induced STAT1 activation seems to be critical for the subsequent triggering of β-cell apoptosis, since PTPN2-deficient β-cells show an aggravation of cytokine-induced apoptosis, which is reverted by the combined inhibition of PTPN2 and STAT1. Importantly, PTPN2 inhibition unmasked the proapoptotic effect of IFN-γ alone in both INS-1E and primary β-cells, while it failed to augment palmitate- or CPA-induced β-cell death. Taken together, these observations indicate two interesting aspects that require additional investigation:1) The crucial negative feedback of PTPN2 on the STAT1 signaling pathway suggests that PTPN2 is part of the “defense mechanisms” triggered by β-cells in response to cytokines. It remains to be clarified why this and other defense mechanisms (24) are not sufficient to protect β-cells against apoptosis. 2) IFN-γ alone induces STAT1 activation, but this only leads to apoptosis when PTPN2 activation is prevented. This suggests that STAT1 must cross a “nuclear activation threshold” before becoming proapoptotic to β-cells. It remains to be clarified whether this threshold depends on the intensity/length and/or periodicity of STAT1 activation, as previously suggested for nuclear factor κB (33).
The exacerbation of apoptosis in β-cells with decreased PTPN2 expression is independent of augmented NO production. This contrasts with observations reporting that PTPN2-deficient macrophages produce higher amounts of nitric oxide under inflammatory conditions (40,46), highlighting the differential regulation of inflammation-associated genes in cells from different backgrounds.
As discussed above, the transcription factor STAT1 has a major role in cytokine-induced β-cell apoptosis in vitro (18) and in vivo (27). It is thus conceivable that a genetically determined modification in the expression and/or the function of PTPN2 may sensitize β-cells to IFN-γ/STAT1–driven proapoptotic signaling, amplifying β-cell loss under inflammatory conditions, and in some cases, contributing to diabetes. The potential effect of the transient hyper-phosphorylation of STAT3 or IRβ in PTPN2-inhibited cells remains to be clarified. STAT3 has been reported to be dispensable for β-cell development and function (47), but it is generally associated with increased survival of tumoral cells (48), whereas the insulin receptor may exert positive feedback on β-cell function and survival (20).
Over the past few years, several new candidate genes have been identified in human type 1 diabetes. Most of these genes are expressed in the immune system, including major histocompatibility complex (e.g., HLA-DRB1), CTLA4, IL-2Rα (CD25), IFIH1 (MDA5), and several protein tyrosine phosphatases, namely PTPN22, PTPN11, and the recently reported PTPN2 (3,49). Modified function or regulation of these genes could contribute to the development of an autoimmune response. Thus, nonsynonymous single-nucleotide polymorphisms (SNPs) in PTPN22 (human LYP) have been associated with type 1 diabetes and other autoimmune processes, probably secondary to defective deletion of autoreactive T-cells during thymic selection (39). The fact that PTPN22 is not expressed in β-cells (present data) reinforces the likelihood that this is solely due to effects on the immune system. PTPN2-null mice present severe abnormalities in the immune system, resulting in a systemic inflammatory disease and widespread tissue infiltration by mononuclear cells (13,46,50). Moreover, the PTPN2 gene is associated with several other autoimmune disorders besides type 1 diabetes, including Crohn's disease, ulcerative colitis, and rheumatoid arthritis (49,51,52). It is currently unknown whether the SNPs identified in the PTPN2 gene on chromosome 18p11 will lead to a gain or a loss of function of the protein. PTPN2 overexpression in other cell types has been shown to induce a p53- and caspase 1–dependent apoptosis (53), whereas suppression of PTPN2 sensitize β-cells to cytokine-induced apoptosis (present data). In experiments on INS-1E cells, we observed that DNA vector–based overexpression of PTPN2 induced 30–50% of apoptosis 36 h after transfection, whereas there was <8% apoptotic cells in control-transfected cells (data not shown). These preliminary results require experimental confirmation in primary rat and human β-cells using adenoviral vectors to overexpress PTPN2, but they suggest that PTPN2 must be tightly regulated to avoid deleterious effects.
Pancreatic β-cells themselves may play an active role in the pathogenesis of type 1 diabetes (21), and at least three of the type 1 diabetes–associated genes are expressed in β-cells, namely insulin, IFIH1 (MDA5) (6,54), and PTPN2 (present study). β-Cells have been shown to participate actively in the recruitment and activation of the immune system by secreting various chemokines and cytokines under inflammatory conditions (18,21,55) and by providing “danger signals” to the immune system during apoptosis, especially in the context of local inflammation (56). We presently demonstrate that changes in PTPN2 function in β-cells sensitize these cells to surrounding proapoptotic inflammatory signals (e.g., cytokines), potentially amplifying β-cell loss and insulitis. This suggests that PTPN2, a type 1 diabetes–associated gene, may modulate β-cell apoptosis in early type 1 diabetes independently of its potential effects on the immune system.
Supplementary Material
Acknowledgments
This work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS-FRSM) Belgium, the Communauté Française de Belgique–Actions de Recherche Concertées (ARC), the European Union (STREP Savebeta, contract number 036903; in the Framework Programme 6 of the European Community), and the Belgium Program on Interuniversity Poles of Attraction initiated by the Belgium State (IUAP P6/40). F.M. is the recipient of a post-doctoral fellowship from FNRS, Belgium. M.L.C. is the recipient of a scholarship from CAPES (Brazilian Coordination for the Improvement of Higher Education Personnel).
No other potential conflicts of interest relevant to this article were reported.
We thank John A. Todd and Dr. Kate Downes, Department of Medical Genetics, University of Cambridge, Cambridge, U.K., for helpful discussions, and Hindrik Mulder for helpful suggestions on siRNA transfection methods. We thank M.A. Neef, G. Vandenbroeck, M. Urbain, J. Schoonheydt, R. Leeman, A.M. Musuaya, and S. Mertens from the Laboratory of Experimental Medicine, ULB, for excellent technical support.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Wang WY, Barratt BJ, Clayton DG, Todd JA: Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet 2005; 6: 109– 118 [DOI] [PubMed] [Google Scholar]
- 2.Drescher KM, Tracy SM: The CVB and etiology of type 1 diabetes. Curr Top Microbiol Immunol 2008; 323: 259– 274 [DOI] [PubMed] [Google Scholar]
- 3.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG: Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007; 39: 857– 864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Florez JC: Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia 2008; 51: 1100– 1110 [DOI] [PubMed] [Google Scholar]
- 5.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, Heap GA, Dubois PC, Clayton DG, Hunt KA, van Heel DA, Todd JA: Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008; 359: 2767– 2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ylipaasto P, Kutlu B, Rasilainen S, Rasschaert J, Salmela K, Teerijoki H, Korsgren O, Lahesmaa R, Hovi T, Eizirik DL, Otonkoski T, Roivainen M: Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia 2005; 48: 1510– 1522 [DOI] [PubMed] [Google Scholar]
- 7.Cardozo AK, Kruhoffer M, Leeman R, Orntoft T, Eizirik DL: Identification of novel cytokine-induced genes in pancreatic β-cells by high-density oligonucleotide arrays. Diabetes 2001; 50: 909– 920 [DOI] [PubMed] [Google Scholar]
- 8.Cardozo AK, Heimberg H, Heremans Y, Leeman R, Kutlu B, Kruhoffer M, Orntoft T, Eizirik DL: A comprehensive analysis of cytokine-induced and nuclear factor-κ B-dependent genes in primary rat pancreatic β-cells. J Biol Chem 2001; 276: 48879– 48886 [DOI] [PubMed] [Google Scholar]
- 9.Rasschaert J, Liu D, Kutlu B, Cardozo AK, Kruhoffer M, ORntoft TF, Eizirik DL: Global profiling of double stranded RNA- and IFN-γ-induced genes in rat pancreatic beta cells. Diabetologia 2003; 46: 1641– 1657 [DOI] [PubMed] [Google Scholar]
- 10.Kutlu B, Cardozo AK, Darville MI, Kruhoffer M, Magnusson N, Orntoft T, Eizirik DL: Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 2003; 52: 2701– 2719 [DOI] [PubMed] [Google Scholar]
- 11.Hulbert EM, Smink LJ, Adlem EC, Allen JE, Burdick DB, Burren OS, Cassen VM, Cavnor CC, Dolman GE, Flamez D, Friery KF, Healy BC, Killcoyne SA, Kutlu B, Schuilenburg H, Walker NM, Mychaleckyj J, Eizirik DL, Wicker LS, Todd JA, Goodman N: T1DBase: integration and presentation of complex data for type 1 diabetes research. Nucleic Acid Res 2007; 35: D742– D746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP: Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol 2001; 21: 7117– 7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simoncic PD, McGlade CJ, Tremblay ML: PTP1B and TC-PTP: novel roles in immune-cell signaling. Can J Physiol Pharmacol 2006; 84: 667– 675 [DOI] [PubMed] [Google Scholar]
- 14.Galic S, Klingler-Hoffmann M, Fodero-Tavoletti MT, Puryer MA, Meng TC, Tonks NK, Tiganis T: Regulation of insulin receptor signaling by the protein tyrosine phosphatase TCPTP. Mol Cell Biol 2003; 23: 2096– 2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K: Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol 2002; 22: 5662– 5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiganis T, Bennett AM, Ravichandran KS, Tonks NK: Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol Cell Biol 1998; 18: 1622– 1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walchli S, Curchod ML, Gobert RP, Arkinstall S, Hooft vH: Identification of tyrosine phosphatases that dephosphorylate the insulin receptor: a brute force approach based on “substrate-trapping” mutants. J Biol Chem 2000; 275: 9792– 9796 [DOI] [PubMed] [Google Scholar]
- 18.Gysemans CA, Ladriere L, Callewaert H, Rasschaert J, Flamez D, Levy DE, Matthys P, Eizirik DL, Mathieu C: Disruption of the γ-interferon signaling pathway at the level of signal transducer and activator of transcription-1 prevents immune destruction of β-cells. Diabetes 2005; 54: 2396– 2403 [DOI] [PubMed] [Google Scholar]
- 19.Miettinen P, Ormio P, Hakonen E, Banerjee M, Otonkoski T: EGF receptor in pancreatic beta-cell mass regulation. Biochem Soc Trans 2008; 36: 280– 285 [DOI] [PubMed] [Google Scholar]
- 20.Xu GG, Rothenberg PL: Insulin receptor signaling in the beta-cell influences insulin gene expression and insulin content: evidence for autocrine beta-cell regulation. Diabetes 1998; 47: 1243– 1252 [DOI] [PubMed] [Google Scholar]
- 21.Eizirik DL, Moore F, Flamez D, Ortis F: Use of a systems biology approach to understand pancreatic beta-cell death in type 1 diabetes. Biochem Soc Trans 2008; 36: 321– 327 [DOI] [PubMed] [Google Scholar]
- 22.Liu D, Cardozo AK, Darville MI, Eizirik DL: Double-stranded RNA cooperates with interferon-γ and IL-1 β to induce both chemokine expression and nuclear factor-κ B-dependent apoptosis in pancreatic β-cells: potential mechanisms for viral-induced insulitis and β-cell death in type 1 diabetes mellitus. Endocrinology 2002; 143: 1225– 1234 [DOI] [PubMed] [Google Scholar]
- 23.Goldberg A, Parolini M, Chin BY, Czismadia E, Otterbein LE, Bach FH, Wang H: Toll-like receptor 4 suppression leads to islet allograft survival. FASEB J 2007; 21: 2840– 2848 [DOI] [PubMed] [Google Scholar]
- 24.Eizirik DL, Mandrup-Poulsen T: A choice of death: the signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia 2001; 44: 2115– 2133 [DOI] [PubMed] [Google Scholar]
- 25.Shuai K, Liu B: Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol 2005; 5: 593– 605 [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Kim HS, Chung KW, Oh SH, Yun JW, Im SH, Lee MK, Kim KW, Lee MS: Essential role for signal transducer and activator of transcription-1 in pancreatic β-cell death and autoimmune type 1 diabetes of nonobese diabetic mice. Diabetes 2007; 56: 2561– 2568 [DOI] [PubMed] [Google Scholar]
- 27.Callewaert HI, Gysemans CA, Ladriere L, D'Hertog W, Hagenbrock J, Overbergh L, Eizirik DL, Mathieu C: Deletion of STAT1 pancreatic islets protects against streptozotocin-induced diabetes and early graft failure but not against late rejection. Diabetes 2007; 56: 2169– 2173 [DOI] [PubMed] [Google Scholar]
- 28.Pipeleers DG, in't Veld PA, Van de Winkel M, Maes E, Schuit FC, Gepts W: A new in vitro model for the study of pancreatic A and B cells. Endocrinology 1985; 117: 806– 816 [DOI] [PubMed] [Google Scholar]
- 29.Rasschaert J, Ladriere L, Urbain M, Dogusan Z, Katabua B, Sato S, Akira S, Gysemans C, Mathieu C, Eizirik DL: Toll-like receptor 3 and STAT1 contribute to double-stranded RNA + interferon-γ-induced apoptosis in primary pancreatic β-cells. J Biol Chem 2005; 280: 33984– 33991 [DOI] [PubMed] [Google Scholar]
- 30.Ling Z, Hannaert JC, Pipeleers D: Effect of nutrients, hormones and serum on survival of rat islet beta cells in culture. Diabetologia 1994; 37: 15– 21 [DOI] [PubMed] [Google Scholar]
- 31.Lupi R, Dotta F, Marselli L, Del Guerra S, Masini M, Santangelo C, Patane G, Boggi U, Piro S, Anello M, Bergamini E, Mosca F, Di Mario U, Del Prato S, Marchetti P: Prolonged exposure to free fatty acids has cytostatic and proapoptotic effects on human pancreatic islets: evidence that β-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes 2002; 51: 1437– 1442 [DOI] [PubMed] [Google Scholar]
- 32.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB: Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 1992; 130: 167– 178 [DOI] [PubMed] [Google Scholar]
- 33.Ortis F, Cardozo AK, Crispim D, Storling J, Mandrup-Poulsen T, Eizirik DL: Cytokine-induced proapoptotic gene expression in insulin-producing cells is related to rapid, sustained, and nonoscillatory nuclear factor-κB activation. Mol Endocrinol 2006; 20: 1867– 1879 [DOI] [PubMed] [Google Scholar]
- 34.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR: Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem 1982; 126: 131– 138 [DOI] [PubMed] [Google Scholar]
- 35.Cnop M, Ladriere L, Hekerman P, Ortis F, Cardozo AK, Dogusan Z, Flamez D, Boyce M, Yuan J, Eizirik DL: Selective inhibition of eukaryotic translation initiation factor 2α dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic β-cell dysfunction and apoptosis. J Biol Chem 2007; 282: 3989– 3997 [DOI] [PubMed] [Google Scholar]
- 36.Hoorens A, Van de CM, Kloppel G, Pipeleers D: Glucose promotes survival of rat pancreatic beta cells by activating synthesis of proteins which suppress a constitutive apoptotic program. J Clin Invest 1996; 98: 1568– 1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen MC, Proost P, Gysemans C, Mathieu C, Eizirik DL: Monocyte chemoattractant protein-1 is expressed in pancreatic islets from prediabetic NOD mice and in interleukin-1β-exposed human and rat islet cells. Diabetologia 2001; 44: 325– 332 [DOI] [PubMed] [Google Scholar]
- 38.Overbergh L, Valckx D, Waer M, Mathieu C: Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 1999; 11: 305– 312 [DOI] [PubMed] [Google Scholar]
- 39.Bottini N, Vang T, Cucca F, Mustelin T: Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol 2006; 18: 207– 213 [DOI] [PubMed] [Google Scholar]
- 40.Simoncic PD, Bourdeau A, Lee-Loy A, Rohrschneider LR, Tremblay ML, Stanley ER, McGlade CJ: T-cell protein tyrosine phosphatase (Tcptp) is a negative regulator of colony-stimulating factor 1 signaling and macrophage differentiation. Mol Cell Biol 2006; 26: 4149– 4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cool DE, Tonks NK, Charbonneau H, Walsh KA, Fischer EH, Krebs EG: cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci U S A 1989; 86: 5257– 5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosinger B, Jr, Tillmann U, Westphal H, Tremblay ML: Cloning and characterization of a mouse cDNA encoding a cytoplasmic protein-tyrosine-phosphatase. Proc Natl Acad Sci U S A 1992; 89: 499– 503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourdeau A, Dube N, Tremblay ML: Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol 2005; 17: 203– 209 [DOI] [PubMed] [Google Scholar]
- 44.Stuible M, Doody KM, Tremblay ML: PTP1B and TC-PTP: regulators of transformation and tumorigenesis. Cancer Metastasis Rev 2008; 27: 215– 230 [DOI] [PubMed] [Google Scholar]
- 45.Lu X, Chen J, Sasmono RT, Hsi ED, Sarosiek KA, Tiganis T, Lossos IS: T-cell protein tyrosine phosphatase, distinctively expressed in activated-B-cell-like diffuse large B-cell lymphomas, is the nuclear phosphatase of STAT6. Mol Cell Biol 2007; 27: 2166– 2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinonen KM, Nestel FP, Newell EW, Charette G, Seemayer TA, Tremblay ML, Lapp WS: T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood 2004; 103: 3457– 3464 [DOI] [PubMed] [Google Scholar]
- 47.Lee JY, Hennighausen L: The transcription factor Stat3 is dispensable for pancreatic beta-cell development and function. Biochem Biophys Res Commun 2005; 334: 764– 768 [DOI] [PubMed] [Google Scholar]
- 48.Al Zaid SK, Turkson J: STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res 2008; 18: 254– 267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourdeau A, Dube N, Heinonen KM, Theberge JF, Doody KM, Tremblay ML: TC-PTP-deficient bone marrow stromal cells fail to support normal B lymphopoiesis due to abnormal secretion of interferon-γ. Blood 2007; 109: 4220– 4228 [DOI] [PubMed] [Google Scholar]
- 51.Cope AP, Schulze-Koops H, Aringer M: The central role of T cells in rheumatoid arthritis. Clin Exp Rheumatol 2007; 25: S4– S11 [PubMed] [Google Scholar]
- 52.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M, Schreiber S: Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet 2008; 40: 713– 715 [DOI] [PubMed] [Google Scholar]
- 53.Gupta S, Radha V, Sudhakar C, Swarup G: A nuclear protein tyrosine phosphatase activates p53 and induces caspase-1-dependent apoptosis. FEBS Lett 2002; 532: 61– 66 [DOI] [PubMed] [Google Scholar]
- 54.Flodstrom M, Tsai D, Fine C, Maday A, Sarvetnick N: Diabetogenic potential of human pathogens uncovered in experimentally permissive β-cells. Diabetes 2003; 52: 2025– 2034 [DOI] [PubMed] [Google Scholar]
- 55.Cardozo AK, Proost P, Gysemans C, Chen MC, Mathieu C, Eizirik DL: IL-1β and IFN-γ induce the expression of diverse chemokines and IL-15 in human and rat pancreatic islet cells, and in islets from pre-diabetic NOD mice. Diabetologia 2003; 46: 255– 266 [DOI] [PubMed] [Google Scholar]
- 56.Filippi CM, von Herrath MG: Islet beta-cell death: fuel to sustain autoimmunity? Immunity 2007; 27: 183– 185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.