Abstract
When analyzing single-molecule data, a low-dimensional set of system observables typically serve as the observational data. We calibrate stochastic dynamical models from time series that record such observables (our focus throughout is on a molecule’s end-to-end distance). Numerical techniques for quantifying noise from multiple time scales in a single trajectory, including experimental instrument and inherent thermal noise, are demonstrated. The techniques are applied to study time series coming from both simulations and experiments associated with the nonequilibrium mechanical unfolding of titin’s I27 domain. The estimated models can be used for several purposes: (1) detect dynamical signatures of “rare events” by analyzing the effective diffusion and force as a function of the monitored observable; (2) quantify the influence that experimentally unobservable conformational degrees of freedom have on the dynamics of the monitored observable; (3) quantitatively compare the inherent thermal noise to other noise sources, e.g. instrument noise, variation induced by conformational heterogeneity, etc.; (4) simulate random quantities associated with repeated experiments; (5) apply pathwise (i.e. trajectory-wise) hypothesis tests to assess the goodness-of-fit of models and even detect conformational transitions in noisy signals. These items are all illustrated with several examples.
I. INTRODUCTION
Recent advances in single-molecule (SM) experimental techniques have allowed researches to explore small scale systems with high spatial and temporal resolution. This has allowed researchers to gain a better understanding of the kinetics and thermodynamics of various complex biological systems, including SM studies of proteins and nucleic acids [1–10]. Often the dynamics are best described by stochastic models due to the inherent thermal noise which is non-negligible at time/length scales associated with current SM experiments [11].
However, many challenges still exist in SM experiments. The magnitude of the effective noise associated with a low-dimensional system observable is often not known a priori and it tends to be state-dependent in complex systems [3, 12–15]. Another complication encountered in SM experiments is that conformational degrees of freedom correlate heavily with the dynamics of the monitored observable, but are typically experimentally inaccessible; in simulations these relevant degrees of freedom may be nonlinear collective coordinates [16]. Such degrees of freedom can often significantly influence the dynamic and static properties of SM observables. For a concrete example, suppose one is doing an experiment of protein unfolding. The root-mean-square displacement (rmsd) partially characterizes the conformational state of the protein. This type of quantity is not usually accessible in dynamic SM experiments, but the rmsd variability can heavily influence the distribution of an end-to-end coordinate and cause heavily skewed histograms of the latter [9, 15, 17–20]. Furthermore, unobservable conformational transitions can occur on time scales which are fairly slow relative to the experiment. This last type of effect makes the use of a single low-dimensional model to approximate the stochastic dynamics of the entire population of SM experiments questionable, even if the same molecule is studied using the same experimental protocol each time [15, 21]. Fortunately atomistic simulation has also advanced considerably in recent years, and this tool can provide information about the behavior of quantities difficult to physically measure in the laboratory and aid in modeling SM systems. As progress is made in both simulation and SM experimental methods, the time and length-scales accessible to both will have appreciable overlap and will greatly assist in our understanding of the factors influencing the dynamics of complex molecules.
In this article, diffusion models are constructed using time series resulting from nonequilibrium mechanical unfolding of macromolecules and are used to help in addressing the issues mentioned above. Simulation data is generated by constant velocity steered molecular dynamics (SMD) simulations of unfolding of the I27 domain of titin. Experimental unfolding data of the same molecule is obtained using atomic force microscope (AFM) experiments to unfold engineered versions of the I27 domain of human cardiac titin [6]. Instrument noise associated with the experimental apparatus is quantified using time domain techniques developed in statistics [22]. Maximum likelihood type approximations are made throughout to estimate the parameters of the stochastic model; the influence of measurement noise is accounted for explicitly by the estimator.
The estimated time-dependent diffusion models aim at summarizing the wealth of information contained in low-dimensional SM time series. Each time series observed results in the estimation of a new nonlinear time-dependent diffusion model. We refer to the entire collection of estimated models summarizing a batch of time series as the surrogate process approximations (SPAs). We show that the models can be used to:
Approximate quantities associated with the inherent randomness of SM systems, e.g. the effective force and thermal noise.
Indirectly detect subtle dynamical signatures of experimentally unobservable conformational degrees of freedom.
Quantify how such experimentally inaccessible quantities influence the dynamics of observable quantities.
Compare the thermal noise to the noise induced by other noise sources such as conformational heterogeneity [4, 9, 10, 23–26] and instrument noise.
Predict distributions of random quantities, like the nonequilibrium work associated with repeated experiments/simulations.
These points are illustrated with examples from experiments and/or simulations. In addition, we demonstrate methods for quantitatively assessing the goodness-of-fit of the SPA models using time series testing methods [27]; the tests used also check the validity of the assumptions we make on the instrument noise. The article is organized as follows: Section II presents the computational and experimental methods, Section III presents the results and discussion. Section IV contains the conclusion and outlook followed by an appendix summarizing some supplemental statistical results.
II. MATERIALS AND METHODS
A. Local Maximum Likelihood Estimation
Stochastic differential equations (SDEs) are fit to time series coming either from MD output or AFM experiments where an external force is added into the system [28, 29]. It is important to stress that each time series results in the estimation of a new SDE, so a given batch of time series results in a corresponding batch of (estimated) SDEs. The global SPA [15, 21] representing the dynamics of a single time series trajectory is assumed to be a generic nonlinear diffusion of the form:
| (1) |
| (2) |
where ξt represents the system observable value at time t (throughout this the end-to-end extension of the molecule stretched), Bt represents the standard Brownian motion, μ (·, ·) the time dependent drift function and σ2(·) represents the diffusion coefficient 1. The drift term is time dependent because we are adding an external force. The contamination process noise (εti) does not allow us to directly observe ξt, instead we observe yti= ξti + εti where the subscript on the time index is used to stress that our observations are discrete.
In SM systems, the complexity of the atomistic system often cannot be ignored and causes problems in developing physically based, accurate parametric SDE models [13, 30–32] from a priori considerations. Due to this fact, we assume that the global dynamics are completely unknown a priori, so appealing to a standard parametric estimation scheme is problematic. To overcome the difficulty of unknown global drift and diffusion functions, we use local models [15, 21, 32, 33] to fit the coefficients of polynomial SDEs whose functional form is motivated by overdamped Langevin equation [15]. The relevant expressions are:
| (3) |
| (4) |
| (5) |
| (6) |
where kBT represents Boltzmann’s constant times the system temperature, FExt the external force applied to the system, λ(t) is the pulling protocol (common to all experiments/simulations) we desire the observable monitored to follow, kpull is the spring constant associated with the harmonic constraint used to apply the external force, FInt the force due to internal molecular forces, and θ≡(A, B, C, D) is the local parameter vector estimated by approximate maximum likelihood estimation [34]. ξo is a free parameter (used only for estimation purposes). Since a constant velocity protocol is used for λ, we set ξo to the average (temporal) value in local time series windows. The windows were formed by dividing a single global time series into M windows who all represents an equal temporal fraction of the total time series. A can be interpreted as the effective internal system force associated with the value ξo and B the associated linear sensitivity (similarly for C and D). The modeling ideas behind the two-scale realized volatility estimator (TSRV) [22] were used to approximate the variance of the noise process in each local window. We do not assume that the measurement noise magnitude is constant for all state values, however a “white” measurement noise is assumed. Extensions of TSRV can readily accommodate colored noise [35] if the experimental apparatus is believed to produce colored noise. The goodness-of-fit test employed here [27] can detect this type of error 2. TSRV approximation details are summarized in the Appendix. One assumption made is that the measurement noise dominates the diffusive noise of the SDE; simple techniques for correcting the bias introduced when the diffusive noise magnitude is commensurate (but still considerably smaller) than measurement noise are also outlined in the Appendix. Recent studies have applied a Bayesian analysis to work distributions at a fixed time [36]. We would like to note that out interest is in characterizing the noise along the entire trajectory. Our methods could in principle be cast into a Bayesian framework, but we prefer a frequentist approach primarily because it facilitates assessing the goodness-of-fit of the models. The information from a TSRV inspired analysis is used in conjunction with likelihood based methods to estimate quantities describing the ξ dynamics. The fitting criterion we use is motivated by the local linear maximum likelihood type method outlined in [34]. The stage location λ(t) was denoised using the Daubechies (5 vanishing moments) wavelet family and all measurement noise was assumed to be contained in ξ. The validity of the various assumptions (diffusive noise, local linearity, etc.) are tested in an a posteriori fashion using the probability integral transform based Q-test developed in Ref. [27]. It should be noted that if a physics based model is in hand, many of the pathwise testing tools presented here are still applicable and can help in testing theoretical models given nonstationary (or stationary) observations.
To obtain a global model which can be used to predict random quantities like nonequilibrium work distributions, a penalized spline was used to stitch the piecewise polynomial models together [37]. The full numerical details of this spline procedure are outlined in Ref. [38]. Briefly, a sequence of estimated local θs measured along one trajectory are used to construct both μ and σ. Information about the parameter uncertainty is used to determine a regularized global model from the local θs. The procedure is then repeated for each observed time series.
B. Steered Molecular Dynamics Simulations
The NAMD program [39] was used to simulate the unfolding of the I27 domain of titin which was placed in a periodic water box. The water molecules were modeled using the TIP3 model and the CHARMM 27 force field was employed. All simulations were carried out in the NpT ensemble using 20,705 total atoms. The initial atomic coordinates came from the PDB crystal structure (1TIT). This structure was then solvated in water, using the VMD plugin Solvate and then the system was equilibrated. The Cα of the 1st residue was anchored in place using a harmonic constraint kpull 100 kcal=(Å2mol). In the SMD simulations, the Cα of the 89th residue was pulled at constant velocity using a time dependent harmonic potential (spring constant 5 kcal=Å2mol and velocity=25 Å/ns). The time step used for integrating the SMD simulation was 1 fs and data was discretely sampled every 50 fs for estimation purposes.
C. AFM Experiments
Engineered proteins of eight serially linked repeats of the I27 domain of human cardiac titin (Athena ES) were used. 10 μ l of protein solution, 50–100 μg/ml, was incubated on a gold substrate at room temperature for 20 min. A Multimode AFM with Picoforce option (Veeco Instruments) was used for force spectroscopy measurements. Individual protein chains were attached to a silicon nitride cantilever tip with spring constant, kpull = 50 pN/nm. The attached molecule was stretched to unfold several domains, allowed to relax back nearly to the substrate surface, and held at a constant position to allow the molecule to refold before repeating the cycle. The stretch and relax portions of the cycle were performed at a constant velocity of 50 nm/s, followed by a rest time of 30 sec. In this article we focus on analyzing the second peak observed in each cycle, though the results for the other peaks are similar. Discrete time series were recorded at the frequency of 20kHz.
III. RESULTS AND DISCUSSION
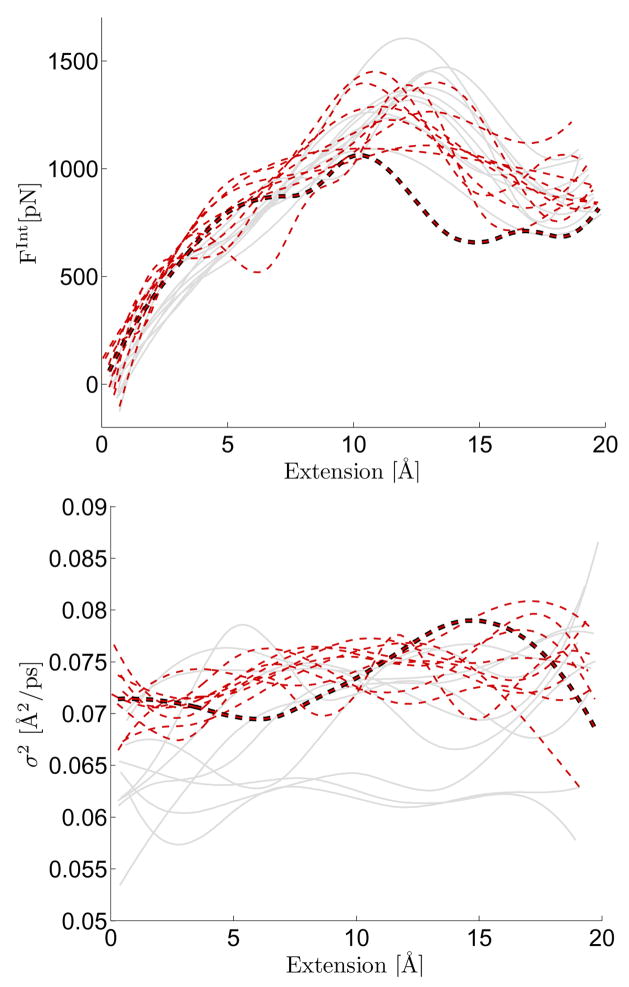
Figure 1 presents results obtained from SMD simulations of unfolding the I27 domain of titin. The effective force and diffusion coefficient estimated from 20 different SMD time series are displayed. The two different curve types, light-solid and red-dashed lines, denote two different batches of SMD data. The batches are distinguished by the initial position coordinates; within each batch the same positional coordinates are used for each simulation. Each trajectory uses different random initial velocities and a different random number stream to simulate the nonequilibrium unfolding of titin in a Langevin heat bath. These batches were analyzed to determine how long the persistence of the initial configurations can be felt and how this manifests itself in the estimated SPA model approximating this system [32, 40–42]. For these trajectories, the diffusion σ(·)2 functions are appreciably different for the two batches of curves. An approximation of the statistical uncertainty associated with estimation is quantified in Supp. Mat. Fig. 1 and this curve indicates that the differences cannot be attributed only to estimation uncertainty associated with a finite sample time series.
FIG. 1.
Simulation results from unfolding the I27 domain of titin. Two batches of 10 trajectories were simulated and each trajectory had the effective SPA force and diffusion coefficient estimated using the procedure outlined here. The first batch (solid grey lines) started one common initial coordinate set and the second batch (dotted red lines) used another initial coordinate set. Different random velocities were assigned at time zero in each case. The curve highlighted by a dark thick line denotes a trajectory where protein denaturation occurred unusually early (discussed further in text).
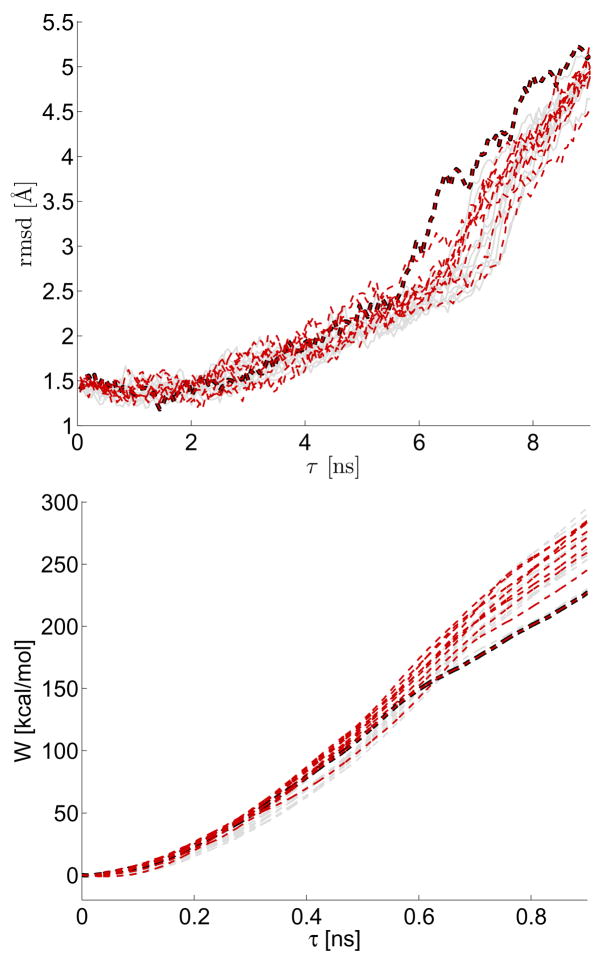
We demonstrate that the variability observed in these batches of measured SPA curves has physical relevance. Different features of the same titin SMD simulation data displayed in the previous figure are shown in Fig. 2. Here, the color-coding for the curves is the same, but the rmsd of the titin molecule from the crystal structure (PDB:1TIT) is plotted as a function of time in the top panel. The bottom panel plots the nonequilibrium work added into the system. In all of the constant velocity simulations and experiments, when we mention “work”, we are referring to the nonequilibrium work definition given in Refs. [30, 43, 44], namely . Close inspection of the rmsd evolution of the two batches reveals that in the initial temporal segment the paths corresponding to two different conformational coordinate initial conditions appear similar, but a distinction between the two batches of curves becomes apparent at later times (≈0.6–0.8 ns). This distinction occurs up until the well-studied I27 domain rupture event occurring around an extension of ≈10–15Å in SMD simulations [2, 45–48]3. In this application these coordinates have appreciable “memory” relative to the time scale of this simulation [40, 49–51]. Recall how the diffusion coefficient, σ2(·) in Fig. 1 depended heavily on the initial coordinate conditions used in the simulation.
FIG. 2.
Two batches of 10 trajectories coming from simulations of unfolding the I27 domain of titin. The same trajectories shown in Fig. 1 are analyzed, but this time the temporal evolution of the root-mean-square-displacement (rmsd) from the crystal structure is plotted as well as that of the nonequilibrium work. Both of the aforementioned quantities were taken directly from the SMD simulation using the program VMD [63]. The curve highlighted by a dark thick line denotes a trajectory where protein denaturation occurred unusually early (discussed further in text).
The differences in dynamical responses can also be attributed in part to “unresolved orthogonal coordinates” [41, 49]. For example, it is known that the number of hydrogen bonds in the molecule correlate heavily with its mechanical strength [45, 48, 52]. Other possible “unresolved orthogonal coordinates” can be related to collective conformational degrees of freedom. For example collective motions associated with allosteric motion are known to modulate the dynamical response of simple low-dimensional models [15, 20]. These types of collective coordinates are typically associated with relatively slow time-scales. Explicitly including a deterministic memory kernel in a scalar model, as in spirtit of generalized Langevin equation [51], may not be able capture the effects of these unresolved collective coordinates.
The variation observed in the functions estimated for the SPA description indirectly reflects the variability introduced by both “long-time” memory and conformational heterogeneity. Comparing the information in a collection/population of SPA models, e.g. comparing the different drift and diffusion coefficients, provides one means for quantify this type of variation. The population of SPA diffusion models, which do not explicitly model “memory”, provides information about the effective dynamics of the underlying complex system. The variability in the SPA models is due to time-scales slow relative to the experiment or simulation [15, 38, 53]. It is usually challenging to numerically carry out various statistical inference procedures for generalized Langevin models, even for stationary signals [51, 54]. The use of collection of SPA models to quantify the effects due to slow time-scale motion is one alternative to using a generalized Langevin description and/or including additional degrees of freedom in the effective model.
Aside from population differences, the SPA model coefficient can also be used to identify “rare events”. A large “outlier” rmsd curve in an unfolding experiment suggests premature mechanically induced denaturation [55]. The dark highlighted curve is used to identify one such “outlier” in the rmsd plot; the curves corresponding to this particular simulation trajectory are also highlighted in Figs. 1–2. The relatively low value of the work path associated with this trajectory, which is measured from the simulation directly, also suggests that significant mechanical denaturation has occurred earlier than usual. Determining the frequency of such rare events has high relevance to free energy computations using nonequilibrium simulation data [30, 56–58]. In complex molecules, extracting the frequency of such events is challenging, but works that use both unfolding and refolding data have demonstrated it might be worth pursuing further [59–62].
Our interest in this article is in extracting as much information as possible from a low-dimensional observable time series. Dynamically monitoring a quantity like the rmsd is a luxury we have in simulations, but analogous structural metrics are not accessible in experiments. In experiments, some other simpler structural metric may be known to be physically important, but again is simply inaccessible in the laboratory. In these situations, the use of a collection of SPA models to summarize system information is also appealing. This is another area where this type of modeling can help in understanding complex SM data.
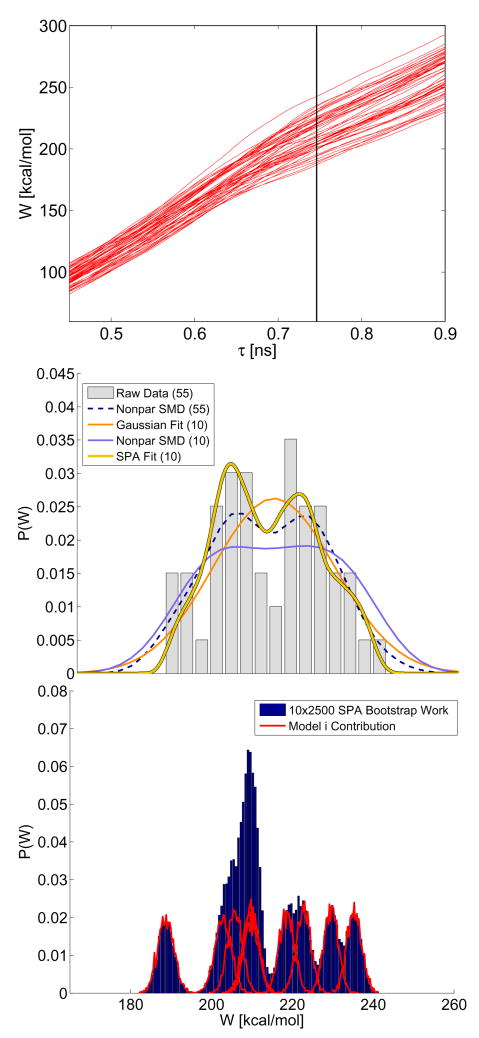
We have shown how information in the SPA functions can be used to detect conformational differences in the underlying molecule by inspecting a collection of SPA models. Now we move on to show how the individual estimated models can be used to make quantitative predictions about variability induced by thermal noise. We demonstrate this first using the titin SMD data where each nonequilibrium simulation was started with a substantially different coordinate initial condition 4. The top panel of Fig. 3 plots the work added into the system as a function of simulation time. The vertical line corresponds to the time where λ ≈18.5Å. Under our simulation conditions, the I27 domain has typically ruptured at this point. We randomly selected 10 curves from the population of 55 to see how well we could approximate features of the work distribution with limited trajectory data. The middle panel plots various estimates (discussed later) of two nonequilibrium work densities obtained by analyzing a subset of the SMD paths.
FIG. 3.
Approximating the nonequilibrium work distribution corresponding to SMD simulation of unfolding the I27 domain of titin. The top panel plots 55 nonequilibrium SMD work paths. The bars in the middle panel plot the normalized histogram obtained by analyzing the SMD data at the time point corresponding the vertical line in the top panel. The solid line curves in the middle panel denote various estimates of the population histogram (see text). The bottom panel displays the simulated SPA work paths used in case labeled “SPA fit” in the middle panel; the bars denote the histogram of all simulated work paths and the solid lines correspond to the contribution of the work density from each individual SPA model. Some SPA models predict effectively disjoint work histograms (all models used passed pathwise goodness-of-fit tests they were subjected to)..
For the 10 curves selected, we noted the work measured directly from the SMD simulation and also calibrated 10 SPA diffusion models using each of the 10 trajectories. The calibrated SPA models were used to generate 2500 realizations using the same initial condition as the corresponding SMD simulation and the random work introduced to the SPA simulation was recorded. The normalized histogram obtained from the 10 × 2500 SPA work is plotted as bars in the bottom panel of Fig. 3. The solid curves in the same figure show the contribution coming from each of the 10 different SPA models (summing these curves would result in the histogram plotted as a bar). Note that many of the work distributions displayed as solid lines in the bottom panel do not appreciably overlap. This means that conformational heterogeneity [4, 9, 10, 23–26] in the simulation data will not allow a single scalar diffusion model to accurately capture the factors making significant contributions to the random work process in this system. Given the complexity of the many-body SMD simulation, this is not surprising, but it should be noted that each of the SPA models do pass pathwise goodness-of-fit tests (see Appendix Fig. 2) indicating that each SPA model adequately approximates the SMD time series used to calibrate it. Also note that a collection of SPA diffusion models, each calibrated from one SMD trajectory, can approximate the the features of the many-body SMD responses (here an ensemble of work paths). Again we stress that we only steer the end-to-end distance of the molecule with a biasing potential. We force this observable to change at a rate much faster than it typically does in the unperturbed case; this rate is very fast relative to the time-scales associated with other slowly changing conformational degrees of freedom which are relevant to the β sandwich structure of this molecule [45, 46, 64, 65]. The conformational degrees of freedom modulate the dynamics of ξ, i.e. although we only steer the ξ coordinate the other degrees of freedom in the system are coupled to this observable. On the time-scale of the SMD simulation, these degrees of freedom are effectively “stuck” in one region of phase space. The variation we observe in the SPA curves (see Fig. 1) can be explained by the factors that cause variability in the SPA models being associated with time-scales slow relative to the simulation or experiment. This type of time-scale separation is not unique to titin simulations, it has been observed in various experimental and simulations shown here and elsewhere [15, 18, 19, 21, 38, 53, 66]. Analyzing the diversity in a population of SPA histograms, i.e. the outlined curves in the middle panel, is one way of indirectly quantifying the variation induced by conformational degrees of freedom in this type of setting.
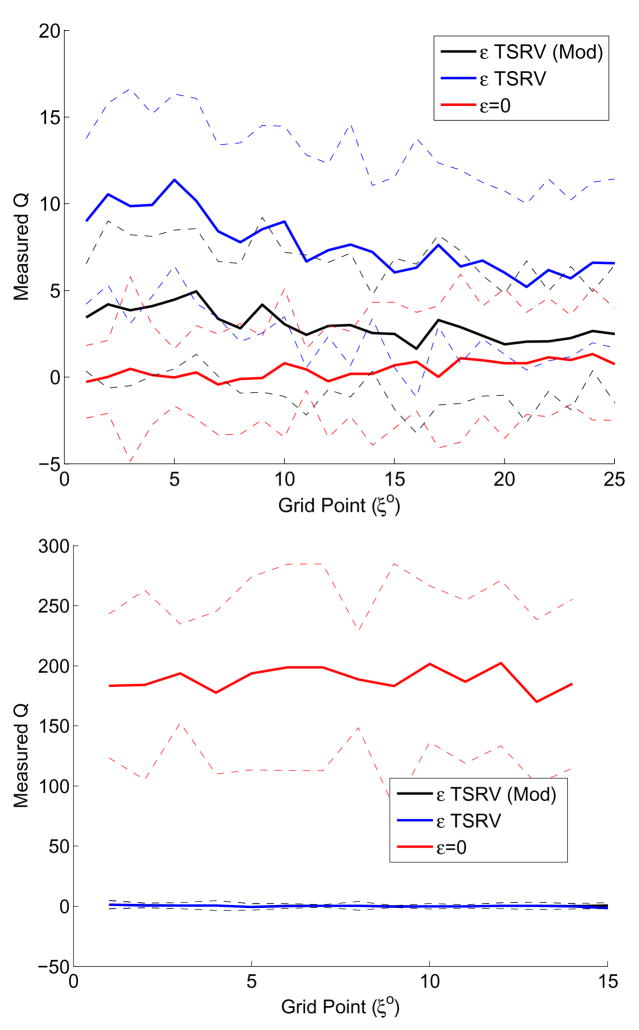
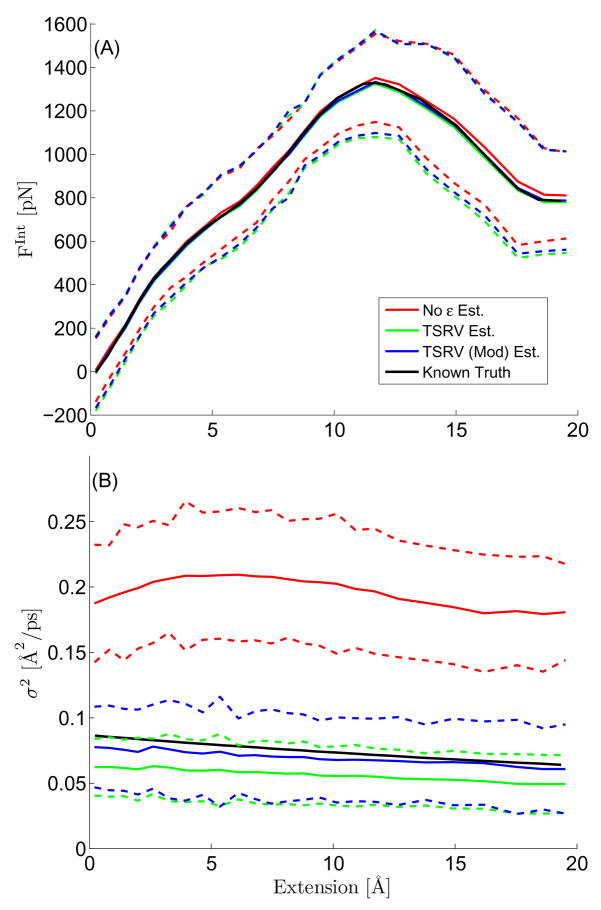
Appendix. Fig. 2.
Top panel: The Q-test statistic average using three noise estimation schemes applied to SMD simulation data. 1) Modified TSRV, 2) TSRV, 3) Ignoring the Measurement noise. Surprisingly the best model using this criterion is the ε = 0 case. The fast-scale motions (the time series were sampled uniformly with 50 fs between observations) cannot be adequately be represented by “white measurement noise” in this system. This tests dictated the model we used for subsequent approximations of the SMD data. Bottom panel: The Q-test statistic average (over the 8 experimental curves) using three noise estimation schemes. 1) Modified TSRV, 2) TSRV, 3) Ignoring the Measurement noise. Note that the test has no problem in rejecting (with virtually no uncertainty) the case where measurement noise is not taken into account. The test statistics under the asymptotic null is a standard normal distribution.
Next, we demonstrate how information in the middle panel of Fig. 3 can be used in a “SPA nonparametric model bootstrapping” type scheme. The details of the scheme used here are provided in the Appendix. The scheme attempts to quantitatively account for variability due to slow time-scale conformational degrees of freedom as well as variability due to thermal noise. In the middle panel of Fig. 3, we plot the normalized work histogram coming from the 55 simulations. We also plot a standard nonparametric density estimate using all 55 work values at the time point studied 5. We then selected 10 curves randomly from the 55 total curves and fit a Gaussian density to the observations. We also compute the non-parametric density estimate using the smaller data set in addition to plotting the results from applying the “SPA nonparametric model bootstrapping” scheme to the corresponding trajectories. Note in the larger population of nonequilibrium work, that two modes are apparent in both the raw data and the nonparametric density estimate. Empirically determining the number of modes in a histogram is extremely difficult if only a small number of random variables from the distribution are available. This would be the situation we faced if only 10 work curves were available to us. Limited data situations are frequently encountered in simulations due to computational cost limitations and can also be relevant to SM experiments [48, 68]. However, by using a small number of trajectories along with the SPA modeling ideas laid out here, we can more easily determine that there are indeed two underlying modes in the data. This is possible because the SPA models have predictive capability. By simulating one SPA model, we can approximate randomness due to inherent thermal noise; the width of the solid densities in the bottom panel reveal this information. In experiments we can also simulate instrument noise. Variation induced by conformational heterogeneity can be determined by comparing the output of multiple SPA models. The two sources of variation can be quantitatively compared using a relatively small number of SMD simulation trajectories. This type of quantitative tool can possibly help in understanding many different complex SM systems. Particularly systems where “multiple conformational states” cause broadened observable histograms and/or multiple modes [4, 9, 10, 23–26].
Next we present results where we approximate the effective force and diffusion from experimental AFM time series in the presence of thermal and instrument noise. The AFM force extension data consists formed the “sawtooth pattern” associated with titin’s I27 domain [6, 64]. Some representative trajectories coming from the experimental apparatus are plotted in Appendix Fig. 3. To minimize variation due to the tip attachment point, we captured a titin molecule on the AFM tip and retained the same molecule for a sequence of force extension cycles. Due to the nature of the non-covalent forces binding the titin molecule to the AFM tip, we could only retain the same molecule for a limited number of repeated force extension cycles. For every distinct “sawtooth peak” observed, we estimated a separate SPA model. That is each global experimental time series used to estimate a SPA model contains only one peak. Results from the second and third peaks observed are plotted in Fig. 4. Results from the first peak observed are not shown because they are more likely to be affected by nonspecific binding artifacts [64, 69].
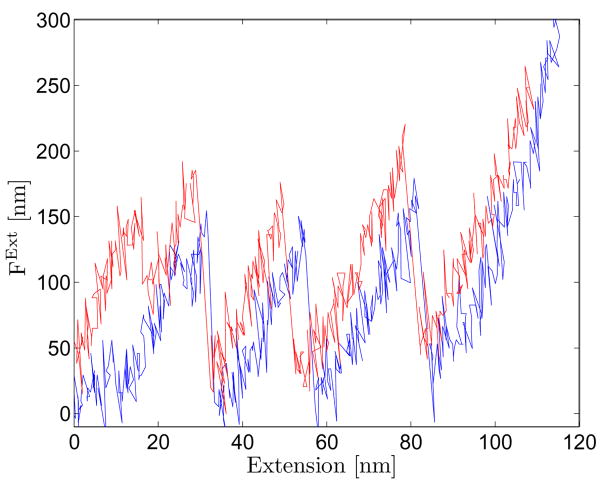
Appendix. Fig. 3.
Sample experimental force extension curve obtained when the AFM is used to unfold the I27 domain of titin.
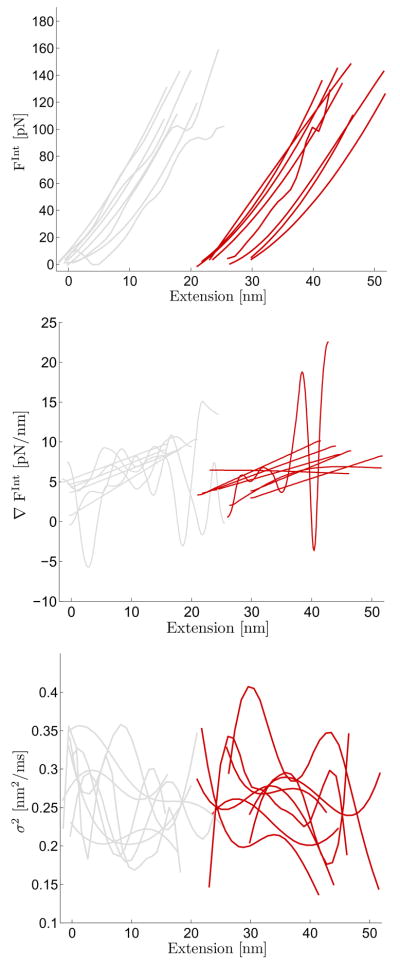
FIG. 4.
Results from calibrating SPA model using experimental AFM data. The force vs extension AFM data consisted of the typical sawtooth pattern [6, 64]. Sample output from the AFM is included in Appendix Fig. 3. We used the second (light grey) and third (dark red) sawtooth in each force extension cycle for estimation purposes. The same I27 molecule remained attached to the tip for a total of 8 force extension cycles. The effective internal force, its gradient with respect to extension, and the effective diffusion coefficient are plotted as a function of extension.
Many of the trends observed in other works involving force-clamp [70] and dynamic force modulation [69] experiments probing the molecular stiffness and internal friction of titin I27 domain appear to also hold in the constant velocity experiments we carried out and analyzed. Namely the gradient of the force shows a generally decreasing magnitude as more domains are unfolded and the internal effective diffusion coefficient demonstrates a roughly decreasing trend as extension increases within one peak. As witnessed in Ref. [69], we also appear to observe that the effective friction, inversely related to the effective diffusion coefficient in our fitted SPA models, also appears to display a slight decrease as the number of unfolded I27 domains increase. However, the effective diffusion coefficient is fairly large and contains noise from multiple sources besides the thermal noise associated with the molecule alone (e.g. cantilever solvent bombardment). Currently we are researching methods for using overlapping windows, variance reduction techniques, and methods which account more explicitly for inherent instrument noise to refine the estimation of the effective diffusion coefficient associated with the molecule given its potential connection/relevance to the internal molecular friction of a single macromolecule [70].
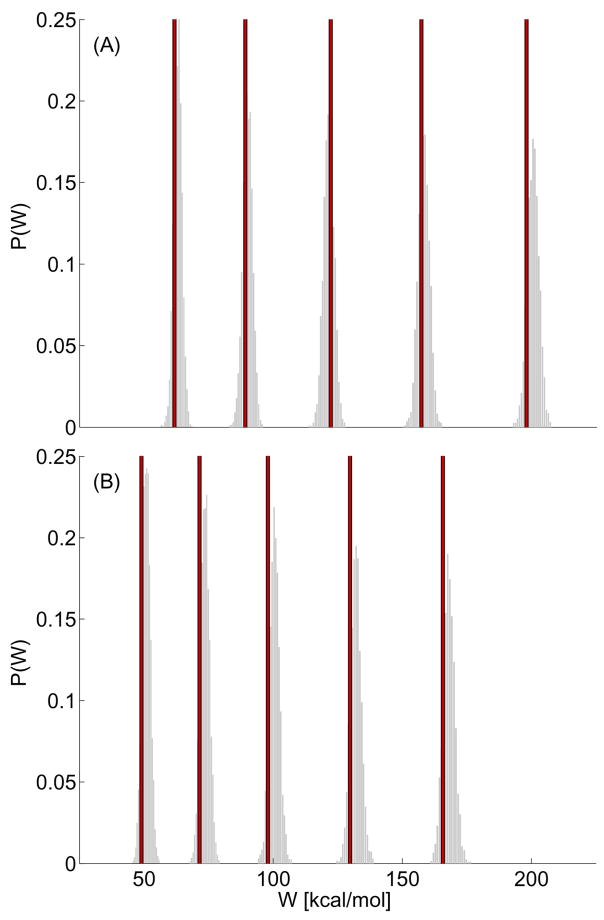
Figure 5 plots the work distribution predicted by the SPA models calibrated from two different trajectories, each panel corresponding to one trajectory, evolving over time as a sequence of histograms. The value measured directly from the experiment at the corresponding point, determined by λ(τ), is also plotted. The observed experimental work paths are consistent with the SPA work densities for all times observed. Results from the other six curves are similar and this demonstrates that we can, on a pathwise basis, reasonably model the uncertainty due to thermal and instrument noise. We can identify that repeated experiments do show variability which might be attributed to conformational heterogeneity; the details of how the titin molecule refolds at “zero extension” may influence the effective molecular force measured. Large, possibly conformationally induced variation, of this sort has also been observed in different experiments probing titin’s dynamics [70]. Note that we make observations on larger time-scales in the experiment, so making direct analogies to simulation results is problematic due to the disparate scales involved [48, 52]. Again advances in simulation techniques may overcome this problem in the near future [68, 71]. The difference between the simulated evolving work distribution are not excessively large, but we believe physically significant structural differences do exist on a trajectory-wise basis due to the fact the fact that observing a “force-hump” [2, 45–48] appears to be a random event.
FIG. 5.
Simulation of SPA work histograms compared to experimental measurements. The solid vertical lines denote the nonequilibrium work value measured from the experimental AFM data corresponding to target extensions of λ, of 1012.51517.520nm; the corresponding work values appear from left to right in each panel. The two panels correspond to two different experimental time series where the same molecule was retained on the AFM tip for multiple force extension cycles. We compare results obtained from two different unfolding cycles where the second sawtooth is used to calibrate two different SPA models. The resulting SPA models were each used to generate 2500 simulated work paths; the time evolving histogram at the λ extensions listed for the experiment are shown. This plot indicates that each individual SPA model can approximate the variation which can be attributed to thermal and instrument noise (see text).
Finally we subject our titin I27 experimental data to goodness-of-fit tests. Our interest is both in determining the validity of the models assumptions and on attempting to detect subtle phase transitions using our models and the observed time series. Regarding the latter item, it is known that a “force-hump” can be observed in stretching the I27 domain of titin [2, 48], but this force hump can be difficult to observe in constant velocity AFM experiments [47]. Recall we are analyzing data coming from the second force peak obtained by pulling the same titin molecule repeatedly. Fig. 5 displays the Q-test results obtained by analyzing 8 force extension curves. Again we remind the reader that each force extension curve was used to calibrate a new SPA diffusion model. The number of local models in each case was M = 15. The estimated SPA models were then used along with the observed data and the Q-test [27] to determine the goodness-of-fit of each SPA model. Out of the 8 AFM trajectories analyzed, 2 were associated with a parameter set that was rejected at the significance level α = 0.01 6. In the rejected models, the local models broke down at a force ≈100 pN where the force-hump “transition” is known to occur in this system [47, 48]. The raw data from the AFM is plotted in the inset for the two rejected cases. In the insets the estimated SPA FInt(·) function is plotted with “o-” lines as well as a wavelet and penalized spline smoothing of the raw AFM data, i.e. FExt(·). In the top right plot, the hump is visible using standard smoothing techniques. Our time series methods confirm that something statistically significant/detetable is changing in the dynamics. In the curve on the left, the time series based methods appear to detect a transition that the other smoothing techniques do not. A sudden change in “thermal noise” magnitude occurs around the 100 pN, which is likely an artifact of the transition. This provides another application of our dynamical models: they can be used to determine when a statistically significant change in the stochastic dynamics occurs. Perhaps more importantly, the testing procedure employed gives us a quantitative metric to test our various model assumptions with.
IV. CONCLUSION AND OUTLOOK
Methods for analyzing single-molecule data in a pathwise fashion were presented. Each trajectory had the measurement noise quantified and this influence of this random noise sources was included in the model estimate. The individual SPA models passed pathwise goodness-of-fit tests. These tests simultaneously tested various model assumptions, e.g. overdamped diffusive dynamics, white measurement noise, etc. The models also demonstrated predictive power, i.e. the probable range of work values predicted by the models was consistent with the simulations/experimental data. More importantly the methods were shown to indirectly quantify the variation induced by conformational degrees of freedom. In experiments this information is typically unobservable. A nonparametric resampling scheme which utilized the work distribution predicted by our surrogate models was demonstrated to qualitatively predict the shape of certain process functionals, e.g. the nonequilibrium work distribution. This is relevant because it allows a researcher studying SM systems to better approximate the basic shape of a nontrivial work distribution using a small number of samples. This can be used for various purposes, e.g. the reliability of a nonequilibrium free energy estimate depending on a non-Gaussian work distribution can more readily be assessed [3, 6, 15, 44, 58]. The collection of SPA models could in principle also be used to predict mean first passage times of complex biolmolecules [74]. In addition, “outlier” curves were shown to correlate with physically relevant structural information which is not typically directly accessible in dynamical experiments [15, 66]. This type of information cannot be inferred from a single SPA model along, but required one to analyze a population of SPA models calibrated from different trajectories. Also certain transitions were detected apparently using the information contained in our estimated models, e.g. transitions known to exist appeared to be detected by pathwise goodness-of-fit tests.
Data-driven numerical tools for analyzing complex systems with a relatively small number of system observables [75, 76] will likely significantly assist researcher in understanding the rich data sets coming from detailed computer simulations and high resolution SM experiments [1–8, 10]. Other systems ranging from double and single stranded DNA [66], ion-channel proteins [38, 53], and small polypeptides [15] have demonstrated that fingerprints of large collective coordinate changes can be detected by analyzing the effective system noise and force as a function of state in both simulations and experiments. Although our focus here was on data-driven methods, many of the estimation and testing procedures could also be applied if a “first principles” physically based model is available [4, 52, 77].
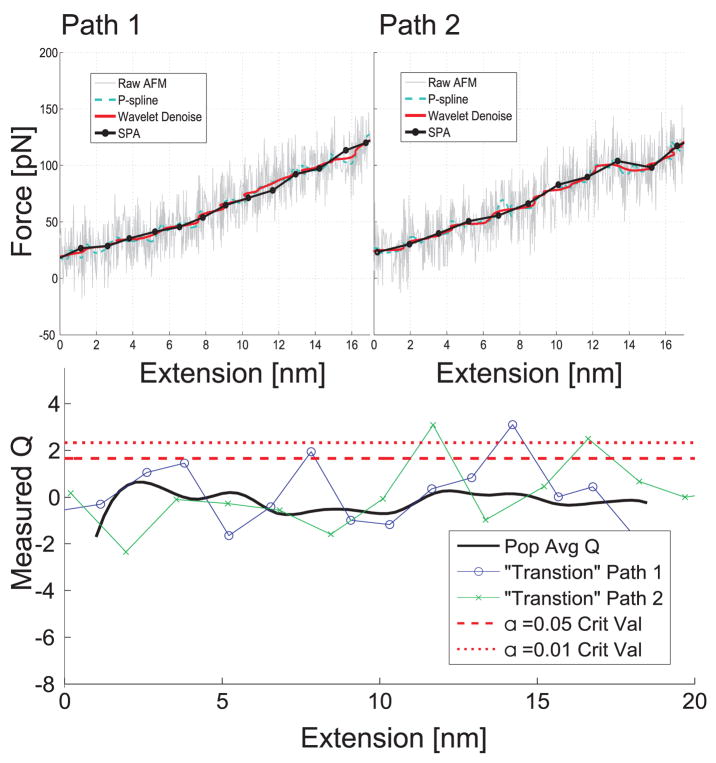
FIG. 6.
Hypothesis tests based on the fitted models and the observed nonstationary time series describing unfolding the I27 domain of titin via AFM. The resulting SPA models were subjected to the Q-test [27] and the population average (over the 8 SPA models) is plotted along with the critical values corresponding to type I rejection rates (α) of 0.01 and 0.05. Only two models were rejected using α = 0.01, those rejected have the corresponding force extension displayed above as an inset. Interestingly both rejection occur near a known “transition” [45–48]. Standard smoothing techniques, penalized spline smoothing [37] and wavelet denoising [72], can readily detect the transition in “Path 2”, but have a difficult time detecting the subtle transition associated with “Path 1” whereas the hypothesis test readily identifies this suspicious point (the rejection is caused mainly to a dramatic change in noise magnitude). The noisy curves in the panel correspond to “FExt” measured directly from the AFM; the standard smoothing techniques were applied to this same quantity. The SPA curves correspond to “FInt”.
Appendix. Fig. 1.
Top panel: Confidence bands for idealized model (genuine SDE). 500 Monte Carlo paths were generated using a single diffusion (with known drift and diffusion). Local parameter estimates were obtained and the average and standard deviation in each local windows were computed. A spline fit to the average represents the mean SPA function obtained. The standard deviation of the 500 paths in each window was also computed and a spline was fit to that data (the dotted curves represent μ(ξ)±2σ(ξ) where the functions represent the spline fits to the corresponding quantities). Normally distributed measurement noise (of known variance) was added and various techniques for estimating the noise were attempted. Under these controlled conditions, it is demonstrated that the estimator we employed [34] does consistently estimate the local parameters. Note how ignoring the noise strongly influences the diffusion coefficient estimate.
Acknowledgments
CPC thanks NSF DMS 0240058, NSF ACI-0325081, and the computer support provided by NSF CNS-0421109 & a partnership between Rice AMD and Cray. The work of NCH and CPC was supported in part by a training fellowship from the Nanobiology Training Program of the W. M. Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NIH T90DK70121). CHK thanks NSF DMR-0505814 and Welch Foundation C-1632 for support. DDC thanks NSF DMS-0505584.
VI. APPENDIX
A. Monte Carlo Simulation Diagnostic Figures
The captions describe information which was alluded to in the main text.
B. SPA Nonparametric Resampling Procedure
The scheme below was carried out in order to generate the nonparametric densities discussed in the main text. An explanation and physical interpretation of each step is provided below the procedure.
Collect N time series from either simulation or experiment. In this paper, these N time series were used to estimate a collection of global drift functions, μi(·, ·), and diffusion functions, σi(·) for i = 1, …, N.
Simulate K sample paths using the SDE dictated by μi(·) and σi(·). For the initial condition, use that of the corresponding time series 7. Do this for all N sets of μi(·, ·) and σi(·).
Draw N′ different random variables U1 which are uniformly distributed over the integers ∈[1, N].
For each N′ draw above, draw another random variable and denote this by U2. A uniform distribution over the integers ∈ [1, K] is the law associated with U2. A realization of U2 is used to resample a work value from the histogram generated in Step 2. For example, if in the step above, we obtain the realizations U1=2 and U2 = 32, we would look up the 32nd work value generated by μ2(·, ·) and σ2(·) and then store this value.
-
Refine the estimate the work density of the simulated process by using the empirical data generated in the step above and a nonparametric density estimate [67]. Each histogram obtained by recording the results above will consist of N′ measurements (for our nonparametric density estimated we use a univariate bandwidth suggested in [67], i.e. , where σ̂ is the empirical variation in the sample). Save the density estimate onto disk.
Repeat steps 3–5 D times, then average the density estimates.
The first step generates the diffusion models based on observational data. Each time series in 1) gives a different model. The differences are due both to statistical uncertainty and also to different conformational details underlying the system (discussed in text).
In Fig. 3, M = 25 local parameter vectors were used to generate 10 drift and diffusion curves from the N = 10 observed time series. The second step sets up a collection of work histograms which we will continually resample from. We set K = 2500 and N′ = N. The third step attempts to include the variability of conformational noise. A scheme motivated by bootstrapping ideas [78] is used to accomplish this. The work resamping is similar in spirit to a traditional bootstrapping scheme, however it should be noted that we are doing two types of resampling. When we draw U1 we are resampling functions (or models), so it should be viewed as a type of functional bootstrapping [79]. N is the same as the observational data in order to approximate finite sampling noise. The fourth step is used to simulate the effects of classic thermal noise assuming a fixed initial positional conformation. The procedure is repeated to average and can also be used to give us an idea of variability caused by finite sample sizes.
C. Two-Scale Realized Volatility
We attempt to model noise coming either from fast-scale motion of the dynamics or experimental apparatus noise as a white noise contamination preventing us from directly monitoring the system observable of interest (ξ). Instead of directly observing ξ, we observe the discrete process yti (see Eq. 2). To estimate the variance of εt from our frequently sampled time series, we appeal to modeling ideas falling under the label Two-Scale Realized Volatiliy (TSRV). The basic idea for estimating the variance of εt within this framework is outlined in Ref. [22], we summarize it here. First compute the following:
| (7) |
in the above, one using all n temporal observations. If the sampling frequency is fairly large, the signal to noise ratio will be such that the signal contained in the εt process dominates that of the ξt process. More formally,
| (8) |
| (9) |
where η denotes a random variable following the standard normal distribution (N(0, 1)) and the symbol
denotes convergence in distribution [22]. The so-called quadratic variation, denoted by 〈ξ, ξ〉T, associated with the dynamics of interest is an 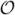 (1) term in the diffusion models we are considering. The second term on the right-hand side is
(1) term in the diffusion models we are considering. The second term on the right-hand side is 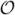 (n) and dominates as n → ∞ 1. In mathematical finance, the interest is usually in the object 〈ξ, ξ〉T. To get at this object in the TSRV frame-work, one subsamples the data on several grids. In other words, one skips every k observations and computes
where the last quantity is an estimator which uses a subsequence Yi, Yi + k, … to get a better estimate of the unobserved quadratic variation. Another idea central to the TSRV estimator is to not waist any time series data, i.e. one creates multiple subsequences from the original time series of length n (e.g. {Y1, Yk, Y2k, …, Yn}, {Y2, Y1+k, Y1+2k, …, Yn−1}, …) and then averages the quadratic variation estimates from the resulting batch of subsequences to obtain a refined estimate denoted by
. Directly approximating
(n) and dominates as n → ∞ 1. In mathematical finance, the interest is usually in the object 〈ξ, ξ〉T. To get at this object in the TSRV frame-work, one subsamples the data on several grids. In other words, one skips every k observations and computes
where the last quantity is an estimator which uses a subsequence Yi, Yi + k, … to get a better estimate of the unobserved quadratic variation. Another idea central to the TSRV estimator is to not waist any time series data, i.e. one creates multiple subsequences from the original time series of length n (e.g. {Y1, Yk, Y2k, …, Yn}, {Y2, Y1+k, Y1+2k, …, Yn−1}, …) and then averages the quadratic variation estimates from the resulting batch of subsequences to obtain a refined estimate denoted by
. Directly approximating 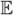 [ε2] by
is often accurate in typical high-frequency financial data sets (and in experimental situations where experimental noise is fairly large and data is sampled frequently), but we will show that some care needs to be taken when these ideas are applied to frequently sampled molecular dynamics data.
[ε2] by
is often accurate in typical high-frequency financial data sets (and in experimental situations where experimental noise is fairly large and data is sampled frequently), but we will show that some care needs to be taken when these ideas are applied to frequently sampled molecular dynamics data.
Before continuing on how we deal with the last issue mentioned, we would like to note that we are trying to estimate a signal which has noise contributions coming from many different time scales. Some noise sources are considered physically interesting (e.g. the magnitude of the thermal noise as a function of ξ) and others are considered uninteresting (e.g. εt). If these sources can be reasonably be quantitatively approximated, there are techniques which can utilize information contained in the entire time series available. Sometimes experimentalists view the thermal noise magnitude as a fundamental limit regarding experimental resolution, we would like to stress that useful physical information can be possibly be extracted even if the signal of interest is “buried” in noise. For example the influence of conformational degrees of freedom on the ξ dynamics are subtle in the titin system we studied in this article, but the pathwise estimation methods used allowed us to quantitatively measure how these sources of (relatively slow time scale) variation influence the observed response and the estimation methods also allowed us to approximate thermal noise which might change as a function of ξ. TSRV type methods can be used in various ways to assist parameter estimation. They can provide an initial guess for a likelihood based method which simultaneously estimates the measurement noise along with the system evolution parameters (resulting in a larger optimization problem). Alternatively they can be used to divide the estimation of the measurement noise and the estimation of parameter’s governing the state into separate problems; i.e., the likelihood conditional on the TSRV type noise estimate ca be estimated. This is the viewpoint we take here (though the results shown throughout this article do not change whichever approach is employed).
Now let us return to how we deal with the contamination noise (2n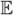 [ε2]) being commensurate in magnitude with 〈ξ, ξ〉T. First we utilize the assumptions behind the TSRV to estimate
[ε2]) being commensurate in magnitude with 〈ξ, ξ〉T. First we utilize the assumptions behind the TSRV to estimate 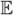 [ε2] for each local time series; with this estimate in hand, we then determine the local parameter θ that maximizes the likelihood of the hidden Markov model in Eqn. 1. The local models used allows us to approximate the thermal noise process, σt, using C +D(ξt − ξo). This quantity allows as to approximate a TSRV subsampling parameter, k*, derived in Ref. [22]:
[ε2] for each local time series; with this estimate in hand, we then determine the local parameter θ that maximizes the likelihood of the hidden Markov model in Eqn. 1. The local models used allows us to approximate the thermal noise process, σt, using C +D(ξt − ξo). This quantity allows as to approximate a TSRV subsampling parameter, k*, derived in Ref. [22]:
| (10) |
| (11) |
It should be mentioned that this rules was determined to balance the standard variance/bias trade-off encountered in nonparametric estimation under a variety of assumptions on the dynamics [22]. Our estimate of σt comes from an likelihood estimate which uses an estimate of 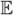 [ε2] coming from
. This will introduce some bias into the estimated parameter θ (and hence our estimate of, σt), and our k*estimate is affected by this. However, the subsampling rule above should really just be viewed as a guide for refining our estimates of
[ε2] coming from
. This will introduce some bias into the estimated parameter θ (and hence our estimate of, σt), and our k*estimate is affected by this. However, the subsampling rule above should really just be viewed as a guide for refining our estimates of 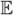 [ε2]. Note that this correction still assumes that the εt process is white (variants of the TSRV accounting for more complex noise structures are possible [80]). Once k* is estimated from the data, one can then compute
. To remove the bias from finite time series lengths, the following adjusted estimator is recommended in Ref. [22]:
[ε2]. Note that this correction still assumes that the εt process is white (variants of the TSRV accounting for more complex noise structures are possible [80]). Once k* is estimated from the data, one can then compute
. To remove the bias from finite time series lengths, the following adjusted estimator is recommended in Ref. [22]:
| (12) |
We subtract the above quantity from
and then divide the result by 2n in order to get a revised estimate of 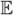 [ε2]. Finally, with this noise estimate we then find a new parameter vector θ that maximizes the likelihood of our hidden Markov model. In simulation studies meant to mimic our MD data sets (in these controlled simulations ε is forced to have the properties assumed by the TSRV), we show that this procedure greatly helps in regards to accuracy (we have a known reference solution). In the empirical MD case studies, we apply this procedure and show that the goodness-of-fit tests are improved. However, surprisingly, in the titin system studied, it turns out a better approximation results when ε is set to zero and a diffusion is estimated (but the hidden Markov model was attempted first). This is likely due to the fact that the fast scale motion has a positive correlation (when spaced by only 50 fs) and this is better approximated by a diffusive noise.
[ε2]. Finally, with this noise estimate we then find a new parameter vector θ that maximizes the likelihood of our hidden Markov model. In simulation studies meant to mimic our MD data sets (in these controlled simulations ε is forced to have the properties assumed by the TSRV), we show that this procedure greatly helps in regards to accuracy (we have a known reference solution). In the empirical MD case studies, we apply this procedure and show that the goodness-of-fit tests are improved. However, surprisingly, in the titin system studied, it turns out a better approximation results when ε is set to zero and a diffusion is estimated (but the hidden Markov model was attempted first). This is likely due to the fact that the fast scale motion has a positive correlation (when spaced by only 50 fs) and this is better approximated by a diffusive noise.
Footnotes
The thermal noise in the “internal system” has contributions coming from internal molecular fluctuations as well as solvent bombardment on the molecule and the cantilever tip. Methods for decoupling these noise sources are currently under investigation.
An analysis of the autocorrelation of the difference of the time series suggested that the assumption of white measurement noise was reasonable (not shown).
In the constant velocity experiments studied, this extension occurs roughly at a time of roughly 0.75 ns. The harmonic constraint used was weak enough to have a readily apparent difference between the target value λ(0.75) ≈ 19Å) and the underlying molecular extension at this time. This discrepancy is of no concern to our dynamical modeling.
We drew configurations from equilibrated samples where the molecular extension was biased by our guiding potential to be at the value observed in the crystal structure.
The bandwidth (h) was determined by , where σ̂ denotes the empirical standard deviation and n the number of observed samples. The Gaussian kernel was employed in the nonparametric estimation [67].
It should be noted that we use carry out a test in each local window and determine our critical value using a single hypothesis test. Alternatively we could aggregate the PIT results from the M windows into a vector for each observed time series and carry out simultaneous multiple comparison, adjusting the significance level accordingly (e.g. use Bonferroni’s method) [73]. In this situation it would make rejecting a model more difficult.
Alternatively, if one believes the initial condition is “equilibrated” at a value, one could set vpull to zero in the SPA model and obtain the stationary distribution.
References
- 1.Hegner M, Smith S, Bustamante C. Proc Natl Acad Sci USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clausen-Schaumann H, Rief M, Gaub HE. Nat Struct Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 3.Collin D, Ritort F, Jarzynski C, Smith S, Tinoco I, Jr, Bustamante C. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther K, Brujic J, Li H, Fernandez J. Biophys J. 2006;90:3806–3812. doi: 10.1529/biophysj.105.076224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans E, Calderwood D. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 6.Harris N, Song Y, Kiang C. Phys Rev Lett. 2007;99:068101. doi: 10.1103/PhysRevLett.99.068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ke C, Humeniuk M, S-Gracz H, Marszalek P. Phys Rev Lett. 2007;99:018302. doi: 10.1103/PhysRevLett.99.018302. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez J, Li H. Science. 2003;303:5664. [Google Scholar]
- 9.Liu S, Bokinsky G, Walter N, Zhuang X. Proc Natl Acad Sci USA. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenleaf W, Frieda K, Foster D, Woodside M, Block S. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffitt J, Chemla Y, Smith S, Bustamante C. Annual Review of Biochemistry. 2008;77:19.1–19.4 . doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 12.Sigg D, Qian H, Bezanilla F. Biophys J. 1999;76:782–803. doi: 10.1016/S0006-3495(99)77243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hummer G. New J Phys. 2005;7:1–14. [Google Scholar]
- 14.Chahine J, Oliveira R, Leite V, Wang J. Proc Natl Acad Sci USA. 2007;104:14646. doi: 10.1073/pnas.0606506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderon C, Chelli R. J Chem Phys. 2008;128:145103. doi: 10.1063/1.2903439. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan J, Runborg O, Kevrekidis I. Comp Chem Eng. 2004;28:557–574. [Google Scholar]
- 17.Procacci PSM, Barducci A, Signorini G, Chelli R. J Chem Phys. 2006;125:164101. doi: 10.1063/1.2360273. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z, Hu H, Yang W, Marszalek P. Biophys J: Biophys Lett. 2006:L57–L59. doi: 10.1529/biophysj.106.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramore S, Ayton G, Voth G. J Chem Phys. 2007;14:105105. doi: 10.1063/1.2764487. [DOI] [PubMed] [Google Scholar]
- 20.Calderon, C. and Arora, K. to be submitted to J. Chemical Theor. and Comp. (2008).
- 21.Calderon C. J Chem Phys. 2007;126:084106. doi: 10.1063/1.2567098. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Mykland P, Ait-Sahalia Y. Journal of the American Statistical Association. 2005;100:1394–1411. [Google Scholar]
- 23.Zhuang X, Kim H, Pereira M, Babcock H, Walter N, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 24.Min W, Gopich I, English B, Kou S, Xie X, Szabo A. J Phys Chem B. 2006;110:20093–20097. doi: 10.1021/jp065187g. [DOI] [PubMed] [Google Scholar]
- 25.Vendruscolo M, Dobson C. Science. 2006;313:1586. doi: 10.1126/science.1132851. [DOI] [PubMed] [Google Scholar]
- 26.Lange O, Lakomek N, Fars C, Schrder G, Walter K, Becker S, Meiler J, Grubmller H, Griesinger C, de Groot B. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 27.Hong Y, Li H. The Review of Financial Studies. 2005;18:37–84. [Google Scholar]
- 28.Balsera M, Stepaniants S, Izrailev S, Oono Y, Schulten K. Biophys J. 1997;73:1281. doi: 10.1016/S0006-3495(97)78161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li P, Makarov D. J Chem Phys. 2003;119:9260–9267. [Google Scholar]
- 30.Park S, Schulten K. J Chem Phys. 2004;120:5946–5961. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 31.Hummer G, Kevrekidis I. J Chem Phys. 2003;118:10762–10773. [Google Scholar]
- 32.Calderon C. Mutliscale Modeling and Simulation. 2007;6:656–687. [Google Scholar]
- 33.Fan J, Fan Y, Jiang J. Journal of American Statistical Association. 2007;102:618–631. [Google Scholar]
- 34.Jimenez J, Ozaki T. J Time Series Analysis. 2005;27:77–97. [Google Scholar]
- 35.Ait-Sahalia, Y., Mykland, P., and Zhang, L. Working Paper 11380, National Bureau of Economic Research, May (2005).
- 36.Maragakis P, Ritort F, Bustamante C, Karplus M, Crooks G. J Chem Phys. 2008 Jul;129:024102. doi: 10.1063/1.2937892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruppert D, Wand M, Carroll R. Semiparametric Regression. Cambridge University Press; New York: 2003. [Google Scholar]
- 38.Calderon, C., Martinez, J., Carroll, R., and Sorensen, D. to be submitted to PRE (2008).
- 39.Phillips J, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R, Kale L, Schulten K. J of Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kupferman R, Stuart A. Physica D. 2004;199:279–316. [Google Scholar]
- 41.Chorin A, Kast A, Kupferman R. Proc Natl Acad Sci USA. 1998;95:4094–4098. doi: 10.1073/pnas.95.8.4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavliotis GA, Stuart AM. J Stat Phys. 2007;127:741–781. [Google Scholar]
- 43.Jarzynski C. Phys Rev E. 1997;56:5018–5035. [Google Scholar]
- 44.Hummer G, Szabo A. Proc Natl Acad Sci USA. 2001;98:3658–3661. doi: 10.1073/pnas.071034098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Biophys J. 1998;75:662671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marszalek P, Lu H, Li H, Carrion-Vazquez M, Oberhauser A, Schulten K, Fernandez J. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 47.Higgins M, Sader J, Jarvis S. Biophys J. 2006;90:640–647. doi: 10.1529/biophysj.105.066571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sotomayor M, Schulten K. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 49.Zwanzig R. Nonequilibrium Statistical Mechanics. Oxford Univeristy Press; New York: 2001. [Google Scholar]
- 50.Kou S, Xie X. Phys Rev Lett. 2004;93:18. doi: 10.1103/PhysRevLett.93.180603. [DOI] [PubMed] [Google Scholar]
- 51.Mamonov A, Kurnikova M, Coalson R. Biophys Chem. 2006;124:268–278. doi: 10.1016/j.bpc.2006.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarov DE, Hansma PK, Metiu H. J Chem Phys. 2001;114:9663. [Google Scholar]
- 53.Janosi CCL, Kosztin I. working paper. 2008 http://www.caam.rice.edu/~cpc1/drafts/cjk_08.pdf.
- 54.Horenko I, Hartmann C, Schütte C. Phys Rev E. 2007;76:016706. doi: 10.1103/PhysRevE.76.016706. [DOI] [PubMed] [Google Scholar]
- 55.Li M, Hu C, Klimov D, Thirumalai D. Proc Natl Acad Sci USA. 2006;103:93–98. doi: 10.1073/pnas.0503758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jarzynski C. Phys Rev Lett. 1997;78:2690–2693. [Google Scholar]
- 57.Crooks GE. J Stat Phys. 1998;90:1481–1487. [Google Scholar]
- 58.Jarzynski C. Phys Rev E. 2006;73:046105. doi: 10.1103/PhysRevE.73.046105. [DOI] [PubMed] [Google Scholar]
- 59.Shirts MEB, Hooker G, Pande V. Physical Review Letters. 2003;91:140601. doi: 10.1103/PhysRevLett.91.140601. [DOI] [PubMed] [Google Scholar]
- 60.Kosztin I, Barz B, Janosi L. J Chem Phys. 2006;124:064106. doi: 10.1063/1.2166379. [DOI] [PubMed] [Google Scholar]
- 61.Chelli R, Marsili S, Procacci P. Phys Rev E. 2008;77:031104. doi: 10.1103/PhysRevE.77.031104. [DOI] [PubMed] [Google Scholar]
- 62.Minh D, Adib A. Phys Rev Lett. 2008:180602. doi: 10.1103/PhysRevLett.100.180602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Carrion-Vazquez M, Oberhauser A, Fisher T, Marszalek P, Li H, Fernandez J. Progress in Biophysics and Molecular Biology. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 65.Becker N, Oroudjev E, Mutz S, Cleveland J, Hansma P, Hayashi C, Makarov D, Hansma H. Nature Materials. 2003;2:282. doi: 10.1038/nmat858. [DOI] [PubMed] [Google Scholar]
- 66.Calderon, C., Chen, W., Harris, N., Lin, K., and Kiang, C. submitted to J. Physics: Condensed Matter (2008).
- 67.Scott D. Multivariate Density Estimation: Theory, Practice, and Visualization. John Wiley & Sons; New York: 1992. [Google Scholar]
- 68.Maragakis P, Lindorff-Larsen K, Eastwood M, Dror R, Klepeis J, Arkin I, Jensen M, Xu H, Trbovic N, Friesner R, Palmer A, Shaw D. J Phys Chem B. 2008;112:6155–6158. doi: 10.1021/jp077018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawakami M, Byrne K, Brockwell D, Radford S, Smith D. Biophys J. 2006;91:L16–L18. doi: 10.1529/biophysj.106.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khatri BS, Byrne K, Kawakami M, Brockwell D, Smith D, Radford S, McLeish T. Faraday Discuss. 2008 doi: 10.1039/b716418c. [DOI] [PubMed] [Google Scholar]
- 71.Simms A, Toofanny R, Kehl C, Benson N, Daggett V. Protein Engineering, Design & Selection. 2008;21:369377. doi: 10.1093/protein/gzn012. [DOI] [PubMed] [Google Scholar]
- 72.Daubechies I. Ten Lectures on Wavelets. SIAM; Philadelphia: 1992. [Google Scholar]
- 73.Lehmann E, Romano J. Testing Statistical Hypotheses. Springer-Verlag; 2008. [Google Scholar]
- 74.Kopelevich D, Panagiotopoulos A, Kevrekidis I. J Chem Phys. 2005;122:044908. doi: 10.1063/1.1839174. [DOI] [PubMed] [Google Scholar]
- 75.Kevrekidis I, Gear C, Hummer G. AIChE Jounral. 2004;50:474–489. [Google Scholar]
- 76.Dudko OK, Mathe J, Szabo A, Meller A, Hummer G. Biophys J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hummer G, Szabo A. Biophys J. 2003;85:5–15. doi: 10.1016/S0006-3495(03)74449-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Efron B, Tibshirani R. An Introduction to the Bootstrap. Chapman & Hall/CRC; Boca Raton, FL: 1994. [Google Scholar]
- 79.Ramsay J, Silverman B. Functional Data Analysis. Springer-Verlag; New York: 2005. [Google Scholar]
- 80.Ait-Sahalia Y, Mancini L. J Econometrics. ((to appear)) [Google Scholar]