Abstract
We have previously shown that SV40 small t antigen (st) cooperates with deregulated cyclin E to activate CDK2 and bypass quiescence in normal human fibroblasts (NHF). Here we show that st expression in serum-starved and density-arrested NHF specifically induces up-regulation and loading of CDC6 onto chromatin. Coexpression of cyclin E results in further accumulation of CDC6 onto chromatin concomitantly with phosphorylation of CDK2 on Thr-160 and CDC6 on Ser-54. Investigation of the mechanism leading to CDC6 accumulation and chromatin loading indicates that st is a potent inducer of cdc6 mRNA expression and increases CDC6 protein stability. We also show that CDC6 expression in quiescent NHF efficiently promotes cyclin E loading onto chromatin, but it is not sufficient to activate CDK2. Moreover, we show that CDC6 expression is linked to phosphorylation of the activating T loop of CDK2 in serum-starved NHF stimulated with mitogens or ectopically expressing cyclin E and st. Our data suggest a model where the combination of st and deregulated cyclin E result in cooperative and coordinated activation of both an essential origin licensing factor, CDC6, and an activity required for origin firing, CDK2, resulting in progression from quiescence to S phase.
Upon mitogenic stimulation mammalian G1 CDKs4 trigger passage through the restriction point and the transition into DNA replication. In particular, cyclin E/CDK2 is activated in mid to late G1 and phosphorylates a variety of substrates that play critical roles in these processes. CDK2 cooperates with D-type cyclin/CDKs to inactivate E2F/pocket protein repressor complexes inducing the expression of DNA synthesis factors and other cell cycle regulators (reviewed in Refs. 1 and 2). CDK2 also phosphorylates DNA replication factors facilitating prereplication complex assembly and origin firing and plays additional roles in centrosome duplication and histone synthesis (reviewed in Ref. 1). In particular, it has been proposed that CDK2 phosphorylates the essential origin licensing factor CDC6 promoting its stabilization prior to inactivation of the APCCdh1 ubiquitin ligase (3). This is thought to ensure that CDC6 accumulation precedes accumulation of other APC substrates that inhibit origin licensing. Moreover, CDK2-independent cyclin E functions have also been reported to be important for prereplication complex assembly in cells in transit from G0 into G1 (4, 5). In keeping with its role as positive regulator of major G1 transitions, deregulation of the cyclin E via gene amplification or defective protein turnover is commonly seen in primary tumors and is associated with poor prognosis (6–8). In normal fibroblasts, ectopic expression of cyclin E has been associated with shortening of the G1 phase of the cell cycle (9, 10), and with induction of DNA damage (reviewed in Ref. 8). Cyclin E deregulation in certain human tumor cell lines and immortalized rat fibroblasts is associated with mitogen-independent cell cycle entry and progression through the cell cycle (11). However, when cyclin E is ectopically expressed in quiescent normal human fibroblasts (NHF), cells remain in G0 (12).
We have recently reported that coexpression of SV40 small t antigen (st) in quiescent NHF with deregulated cyclin E expression is sufficient to trigger mitogen-independent cell cycle progression, proliferation beyond cell confluence, and foci formation. The bypass of quiescence induced by the expression of st and cyclin E is dependent on CDK2 activation (12). Thus, contrary to what is seen in normal murine cells (13), CDK2 activity appears essential for cell cycle progression when it is oncogenically driven by cyclin E and st expression (12). Because st is known to target pathways uniquely required for the transformation of human cells (14, 15), tumor cells with altered pathways that mimic st/cyclin E expression could predictably be sensitive to selective inhibition of CDK2 activity.
Given the critical role of CDK2 activity in cyclin E and st cooperation in inducing cell proliferation and transformation of NHF, we sought to determine the factors and mechanisms by which st modulates CDK2 activation. In this report we have identified the CDC6 replication licensing factor as a cellular target of st. We also uncover CDC6 as a participant in the events leading to chromatin association of cyclin E and CDK2 and in phosphorylation of CDK2 on its activating T loop both in response to mitogenic stimulation, as well as expression of cyclin E and st in NHF.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Cycle Synchronization—BJ-hTERT immortalized fibroblasts were a gift from William C. Hahn (Harvard Medical School, Dana-Farber Cancer Institute). All cell lines were maintained in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Sigma) at 37 °C in a humidified atmosphere with 5% CO2. Serum starvation was performed as previously described (11). Density arrest was achieved by growing cells to high density until no dividing cells were visible under the microscope and maintained for an additional 48 h prior to any treatment. Progression of cells through the cell cycle was measured by propidium iodide staining followed by flow cytometric analysis (PI/FACS) using a BD FACSCalibur flow cytometer and quantified with Cell Quest software (BD) as described earlier (11).
Recombinant Virus Production and Transduction—Recombinant adenoviruses encoding cyclin E were provided by Jeffrey H. Albrecht (Hennepin County Medical Center). Adenoviruses encoding p16 were provided by Juan Fueyo (The University of Texas MD Anderson Cancer Center). Adenoviruses encoding p21 were provided by Wafik El-Deiry (University of Pennsylvania). β-Galactosidase adenoviruses were from Clontech. Adenoviruses encoding EGFP were provided by Pilar Ruiz-Lozano (Burnham Institute for Medical Research). Adenoviruses encoding st were provided by Kathleen Rundell (Northwestern University). Adenoviruses encoding CDC6 were previously described (16). A mutant derivative of cdc6 (CDC6-S3D) in which all three CDK target residues were converted to aspartic acid was constructed by standard molecular biology methods and packaged identically to normal Cdc6. Adenoviral stocks were amplified using Ad-293 cells and purified by CsCl density gradient centrifugation as described previously (11, 17). Viral titers were determined with the Adeno-X™ Rapid Titer Kit (BD Bioscience). NHF were transduced at a multiplicity of infection (MOI) of 150 plaque-forming units/cell when synchronized by serum starvation, or with an MOI of 40 when synchronized by growth to high density. pBabe-puro-cyclin E was obtained from Bruce Clurman (Fred Hutchinson Cancer Research Center). pBabe-puro-Myc-CDC6 was generated by cloning a BamHI/XhoI fragment encoding Myc-tagged CDC6 from pcDNA3-Myc5-CDC6 into the BamHI/XhoI site of pBabe-puro. Retroviral vector expressing dominant negative E2F1-pRB-A/B was kindly provided by Erik S. Knudsen (Kimmel Cancer Center, Thomas Jefferson University). 10 μg of retroviral plasmids were cotransfected with 5 μg of pCL-Ampho packaging vector (Imgenex), into 293T cells following the calcium phosphate precipitation method (18). Retroviral particles were harvested at 24- and 48-h post-transfection and used to super-infect exponentially growing BJ-hTERT cells. When indicated, clones were selected in the presence of 1 μg/ml puromycin.
Western Blot Analysis—Whole cell lysates were obtained in buffer containing 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 250 mm NaCl, 50 mm NaF, 0.1% Triton X-100, 0.1 mm Na3VO4, 2 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml leupeptin, 4 μg/ml aprotinin, and 4 μg/ml pepstatin as previously described (11). Whole cell lysates were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes for Western blot analysis as previously described (11). The following antibodies were used for Western blot analysis: anti-cyclin A (C19), anti-cyclin E (HE12), anti-CDK2 (M20), anti-CDC6 (180.2), anti-CDC6 phospho-Ser54, anti-PP1 (FL18), and anti-PCNA (PC10) antibodies were from Santa Cruz Biotechnology, anti-MCM2 (BM28) antibody was from BD. Anti-pRB phosphospecific and anti-phospho-CDK2-T160 antibodies were from Cell Signaling. Anti-st/LT 419 monoclonal antibodies were a gift from Elizabeth Moran (Temple University).
Preparation of Chromatin Fractions—Separation of chromatin-bound from soluble proteins was performed as previously described (5) using cytoskeletal (CSK) buffer: 10 mm PIPES-KOH (pH 6.8), 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 0.5 mm PMSF, 0.1 mm glycerolphosphate, 50 mm NaF, 1 mm Na3VO4, containing 0.1% Nonidet P-40 and protease inhibitors 2 mm PMSF, 10 μg/ml leupeptin, 4 μg/ml aprotinin, and 4 μg/ml pepstatin. Briefly, cell pellets were lysed for 10 min on ice followed by 4000 rpm centrifugation for 5 min. The soluble fraction was collected and cleared by high-speed centrifugation, 13,000 rpm for 5 min. Protein concentration was determined by Bradford assays. Pellets containing chromatin-bound proteins were washed with CSK buffer and centrifuged at 4000 rpm for 5 min. Chromatin-bound proteins were solubilized in 1× Laemmli Sample Buffer and boiled for 5 min. Equal amounts of chromatin and soluble fraction were loaded for each sample.
RNA Extraction and Northern Blots Analysis—RNA was extracted using RNeasy kit (Qiagen) and 5–10 μg of RNA were resolved in 1% agarose/0.6 m formaldehyde/1× MOPS gels. RNA was transferred and UV-crosslinked onto Hybond-N+. Prehybridization and hybridization were performed at 45 °C in UltraHyb solution (Ambion). 25 ng of denatured cDNA were end-labeled at room temperature for 1 h with [α-32P]ATP. Radioactive signals were quantified using Image J software (NIH) and normalized using ribosomal RNA bands. When required, membranes were stripped by boiling in water for 5 min. siRNA—Exponentially growing BJ-hTERT cells were plated at 5,000 cells/cm density in 10-cm plates in complete DMEM. Sixteen hours later, cells were serum-starved and transfected with a mix of three different siRNA targeting CDC6 sequences (1) 5′-tctagccaatgtgcttgcaagtgta-3′, (2) 5′-caccatgctcagccattaaggtat-3′, and (3) 5′-aagaatctgcatgtgtgagac-3′, (siRNAs targeting sequence 3 have been previously described in Ref. 19), or control siRNA targeting EGFP or Renilla luciferase at a final concentration of 75 nm siRNA using Dharmafect Reagent 4 (Dharmacon). A second siRNA transfection was performed 24 h later, and after 48 h cells were stimulated with FBS or transduced with adenoviruses expressing cyclin E and st.
Cycloheximide (CHX) Assay—Density-arrested BJ-hTERT cells expressing Myc-tagged CDC6 from a retroviral promoter were transduced with adenovirus expressing st or control adenoviruses. Forty hours after transfection, cells were treated with 10 μm CHX, harvested at the indicated time points and processed for Western blot analysis as previously described (17, 20). Quantitation was done using Image J software (NIH) and normalized by loading. To measure endogenous CDC6 stability, BJ-hTERT cells were serum-starved for 48 h and transduced with st or control adenoviruses and 16 h later stimulated with FBS. Treatment with CHX started 20 h after serum stimulation.
RESULTS
We have previously shown that deregulation of cyclin E in certain quiescent tumor/immortal cell lines leads to mitogen-independent cell cycle entry and proliferation (11). In contrast, expression of cyclin E in NHF at levels found in cancer cells is not sufficient to induce mitogen-independent exit from quiescence (12). However, when cyclin E is deregulated in quiescent NHF, co-expression of SV40 st induces mitogen-independent cell cycle entry. Similarly, coexpression of st and cyclin E in NHF bypasses quiescence induced by growth to high density. In these scenarios, cyclin E/st driven cell cycle entry is characterized by a marked phosphorylation of CDK2 on its activating T-loop and CDK2 activation. Importantly, CDK2 activation is essential for these processes.
Our recent work shows that st induces activation of a fraction of cellular CDK2 complexes devoid from CKIs when coexpressed with cyclin E in quiescent NHF. However, we have not observed overt disruption of cyclin E/CDK2/CKI complexes, and cyclin E and st coexpression can induce cell cycle entry when CKIs are expressed at very high levels (12). This suggests that st creates a pool of cyclin E/CDK2 complexes that is protected from CKI inhibition. Thus, we hypothesized that a factor and/or a subcellular location may mediate this process. Because cyclin E is recruited to replication complexes and cyclin E and/or CDK2 play multiple roles in the modulation of these complexes and the initiation of DNA replication (reviewed in Ref. 1), we sought to determine if st modulates loading of cyclin E/CDK2 complexes onto chromatin.
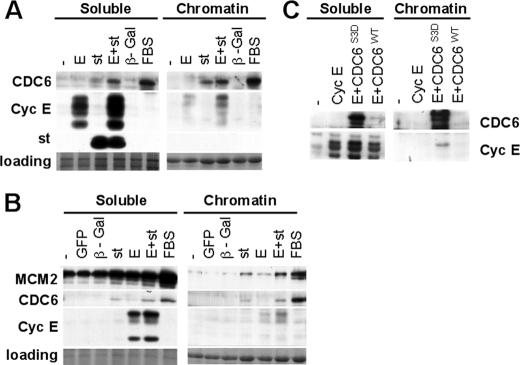
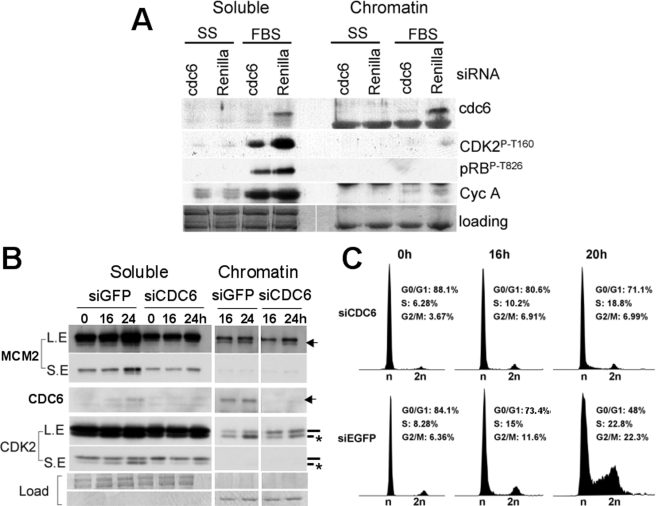
Expression of st in Quiescent Cells Induces Expression and Loading of CDC6 onto Chromatin—hTERT immortalized NHF were serum-starved for 72 h and then stimulated with serum or transduced with the indicated adenoviruses (Fig. 1A). The levels of exogenous cyclin E expressed under these conditions are comparable to the levels of cyclin E expressed in tumor cell lines with deregulated cyclin E (12). Also, st expression is comparable to that observed in NHF immortalized by SV40 (see supplemental Fig. S1). Cells were collected and lysed in CSK buffer to separate soluble proteins from chromatin-bound proteins. Strikingly, we found that st, but not cyclin E or control β-galactosidase, induces expression and loading onto chromatin of the replication factor CDC6. CDC6, together with CDT1 associate with origins of replication through interaction with origin recognition complex proteins (ORC) in G1 (reviewed in Ref. 21). In cooperation with ORC and CDT1, CDC6 loads the MCM DNA helicase complex at origins to prepare them for replication initiation during S phase.
FIGURE 1.
st induces expression and loading of CDC6 onto chromatin in quiescent NHF, and CDC6 facilitates loading of cyclin E. BJ-hTERT fibroblasts were serum-starved for 72 h (A) or grown to high density as described under “Experimental Procedures” (B and C) and then transduced with the indicated adenoviruses or re-stimulated with FBS. Forty-eight hours following transduction, cells were harvested, and chromatin-bound and soluble fractions were obtained as described under “Experimental Procedures.” 15 μg of the soluble protein fractions and the corresponding equivalent chromatin-bound fractions were analyzed by Western blot using the indicated antibodies. Same exposure times are shown for both fractions. Coomassie Blue staining of the gel is shown as a loading control. Relevant proteins are indicated. Note that as reported earlier exogenously expressed cyclin E migrates as a series of bands that migrate faster that endogenous cyclin E (12).
CDC6 mRNA expression is negatively regulated via E2F-dependent transcriptional repression, while the CDC6 protein is actively targeted by the APCCdh1 ubiquitin-ligase for proteasomal-dependent degradation in quiescent cells (22). Thus, CDC6 expression and stabilization is thought to be required for its loading onto chromatin and further recruitment of other members of the replication machinery to replication origins. Recent work has implicated cyclin E/CDK2 in CDC6 protein stabilization via phosphorylation of specific residues that protect CDC6 from ubiquitination and subsequent degradation (3). Consistent with this finding plus our previous results that coexpression of cyclin E and st stimulates CDK2 activity (12), CDC6 expression and loading onto chromatin is further up-regulated when both proteins are coexpressed (Fig. 1A). We also observed that ectopically expressed cyclin E loaded onto chromatin in the absence of st, but st enhanced cyclin E loading. It is important to note that st has not been detected on the chromatin-bound fraction (Fig. 1A), nor was phosphorylated pRB (data not shown), demonstrating the quality of our chromatin preparations. Similarly, in density arrested BJ-hTERT fibroblasts, st but not cyclin E or control adenovirus induced CDC6 expression and loading onto chromatin, and CDC6 accumulation on chromatin correlated with increased cyclin E loading (Fig. 1B). Moreover, the increase in CDC6 was accompanied by enhanced loading of the MCM subunit, MCM2, onto chromatin (Fig. 1B).
To determine if CDC6 expression and loading onto chromatin facilitates cyclin E recruitment to chromatin, we expressed cyclin E and wild-type CDC6 or a CDC6 mutant (CDC6-S3D) resistant to degradation in density arrested BJ-hTERT fibroblasts. Resistance to degradation is due to substitution of three primary CDK sites by Asp to mimic phosphorylation (3). This mutant was selected, as exogenous wild type CDC6 is very unstable in quiescent cells, even when its expression is controlled by a CMV promoter (Fig. 1C). As expected, expression of CDC6-S3D was efficient resulting in its own loading onto chromatin and clearly enhanced cyclin E loading, as compared with cyclin E alone or cyclin E coexpressed with wild type CDC6 (Fig. 1C). This mutant did not substitute for st, as serum-starved fibroblasts co-expressing CDC6-S3D and cyclin E did not exit quiescence (data not shown). This could indicate that st has other targets in addition to CDC6 potentially important for CDK2 activation. For instance, we have previously shown st increases CAK activity in serum-starved NHF (12).
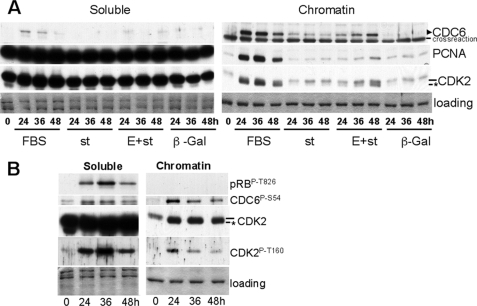
A time course experiment was performed to correlate the effects of st on CDC6 with the chromatin loading of CDK2 and other replication factors. Fig. 2A shows that st, but not β-galactosidase, induced CDC6 loading, even at the earliest time point analyzed. Co-expression of cyclin E and st resulted in time-dependent increase in the amount of CDC6 over that observed with st alone, and this increase correlates with CDK2 accumulation and phosphorylation on Thr-160, and subsequent PCNA loading (Fig. 2A). Of note, we did not observe recruitment of CKIs with CDK2 onto chromatin (data not shown). FBS induced earlier and higher accumulation and loading of CDC6, which correlates with Thr-160 phosphorylation and PCNA loading (Fig. 2A). As CDK2 has been implicated in phosphorylation and stabilization of CDC6, we determined the effects of stimulating serum-starved NHF with FBS on Thr-160 phosphorylation and the phosphorylation of CDK2 substrates on both the soluble and chromatin-bound fraction. Interestingly, we found that CDK2 Thr-160 phosphorylation correlates with pRB phosphorylation (Thr-826) in soluble fraction and with CDC6 phosphorylation on Ser-54 in the chromatin-bound fraction (Fig. 2B). Thus, st expression suffices to induce expression and chromatin loading of CDC6, a component of the prereplication complex, with little effect on CDK2 Thr-160 phosphorylation, and coexpression of cyclin E triggers additional CDC6 accumulation along with coordinated Thr-160 phosphorylation.
FIGURE 2.
st expression in serum-starved NHF induces CDC6 expression and loading, which is enhanced by coexpression of cyclin E. The kinetics of phosphorylation of CDK2 on Thr-160 and CDK2 substrates is distinct in the soluble and chromatin-bound fraction. BJ-hTERT fibroblasts were serum-starved for 72 h and transduced with indicated adenoviruses (A) or re-stimulated with serum (A and B) and harvested at indicated time points. Soluble and chromatin-bound proteins were obtained and simultaneously analyzed by Western blot. Same exposure times are shown for both fractions. Coomassie Blue staining is shown as a loading control. Relevant proteins are indicated. The CDK2 band corresponding to phosphorylated Thr-160 is indicated with an asterisk.
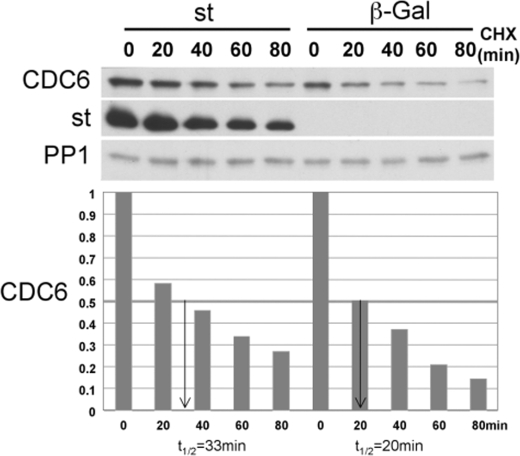
st Induces Modest Stabilization of CDC6—We next determined the effects of st on CDC6 protein stability using protein half-life CHX assays. Because the levels of CDC6 in quiescent cells are extremely low and CDC6 becomes undetectable shortly after addition of CHX, we generated BJ-hTERT fibroblasts stably expressing wild type Myc-tagged CDC6 via retroviral transduction and short puromycin selection. Exogenous Myc-CDC6 levels were comparable to endogenous CDC6 levels in the presence of serum (data not shown). Serum-starved BJ-hTERT-CDC6 cells were transduced with st or β-galactosidase adenoviruses for 24 h and then treated with CHX for the indicated times (Fig. 3). Expression of st in serum-starved BJ-hTERT-CDC6 fibroblasts increased the stability of CDC6 by 65% (from 20 to 33 min, Fig. 3). This experiment was also performed using CDC6 transiently expressed from an adenoviral vector with similar results (data not shown).
FIGURE 3.
st increases CDC6 protein stability. Density-arrested BJ-hTERT fibroblasts stably expressing Myc-tagged CDC6 from a retroviral plasmid were transduced with adenovirus expressing st or control β-galactosidase. Forty hours after transduction, cells were treated with 10 μm CHX and harvested at the indicated time points. Whole cell lysates were analyzed by Western blot. CDC6 signal was quantified relative to time 0 and normalized to the PP1 signal, which is stable through the time course.
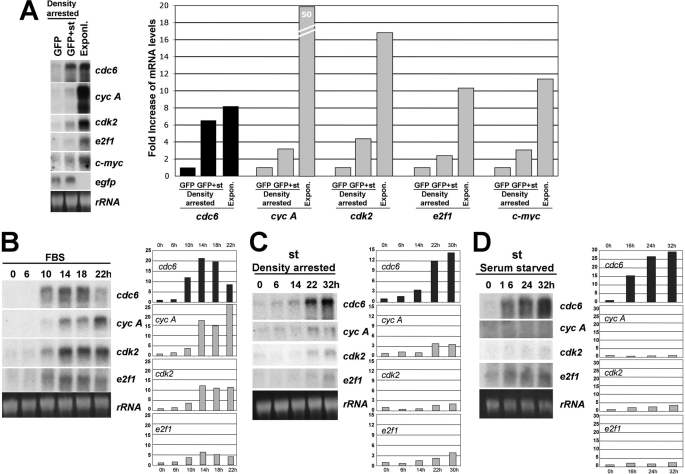
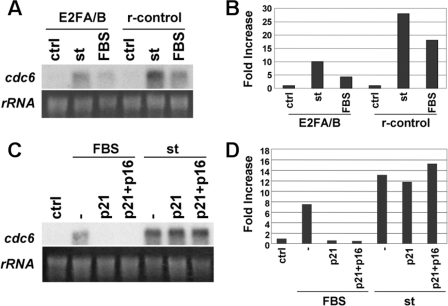
cdc6 mRNA Expression Is Potently Up-regulated by st in Quiescent BJ-hTERT Cells to Levels Similar to Those Induced by Mitogens—As mentioned above, CDC6 expression is not only regulated via proteasomal-dependent degradation, but also by transcriptional, E2F-dependent, and independent mechanisms. Therefore, we determined whether cdc6 and other inducible mRNAs are regulated by st expression in density arrested NHF as compared with proliferating cells. Interestingly, we found that st expression increased cdc6 mRNA to levels comparable to those seen in exponentially growing cells (Fig. 4A). In contrast, the mRNAs corresponding to other E2F-dependent genes and c-Myc, were only slightly up-regulated by st expression but were robustly expressed in exponentially growing cells (Fig. 4A, right panel). Next we determined the kinetics of expression of these mRNAs in response to serum stimulation (Fig. 4B) or st expression in density-arrested (Fig. 4C) or serum-starved (Fig. 4D) BJ-hTERT fibroblasts. In these cells, cdc6 mRNA reaches its maximum 14 h after serum stimulation, closely followed by other E2F targets, such as cdk2 and cyclin A, and then is quickly down-regulated, as cells progress through S and G2/M (Fig. 4B). Strikingly, transduction with st adenovirus induced robust cdc6 mRNA accumulation within a relatively short period of time after transduction in both density-arrested (Fig. 4C) and serum-starved cells (Fig. 4D). Moreover, these experiments clearly show that st expression preferentially modulates cdc6 as compared with three other E2F targets, cyclin A, cdk2, and e2f1. In particular, st induces cdc6 expression ∼14- and ∼29-fold in serum-starved and density-arrested NHF, respectively, while the other mRNAs analyzed increased only by 2–4-fold. This 2–4-fold increase may be due to a modest general effect on gene expression, as we have seen similar effects on the expression of viral promoters (data not shown). Because st induces both cdc6 mRNA and protein accumulation, we wanted to determine if both events occur simultaneously, or if one clearly preceded the other. Hence, density-arrested BJ-hTERT cells were transduced with st adenoviruses and collected at indicated time points (supplemental Fig. S2). Each cell pellet was divided in two for RNA and protein extractions. Northern and Western blot analyses revealed that the kinetics or up-regulation at mRNA and protein levels are similar at the time points analyzed suggesting that these processes are coordinated (supplemental Fig. S2).
FIGURE 4.
cdc6 mRNA expression is potently up-regulated by st in serum-starved and density-arrested BJ-hTERT cells. A, BJ-hTERT cells were grown to high density and transduced with 40 MOI of adenovirus expressing st and/or control EGFP and harvested 30 h later. Exponentially growing cells were used as controls. Levels of expression of indicated mRNAs were analyzed by Northern blot as described under “Experimental Procedures.” Radioactive signals were measured by densitometry, quantified with NIH Image J, and represented as fold increase. B, BJ-hTERT fibroblasts were synchronized by serum starvation for 72 h, re-stimulated with 10% FBS and harvested at indicated time points for Northern blot analysis as in A. C, BJ-hTERT cells were grown to high density, transduced with adenovirus expressing st and harvested at indicated time points after transduction and treated as in A. D, BJ-hTERT cells were serum-starved for 72 h, transduced with adenovirus expressing st and harvested at indicated time points after transduction and treated as in A.
st Induces cdc6 mRNA Expression in an E2F-dependent, but CDK-independent Manner—cdc6 expression is controlled at the transcriptional level via E2F-dependent and independent mechanisms (23–26). As cells enter G1, cdc6 mRNA expression is thought to result from mitogenic activation of G1 CDKs that in turn phosphorylate pocket proteins disrupting E2F/pocket protein repressor complexes located at E2F promoter elements. However, st-mediated up-regulation of cdc6 mRNA occurs in the absence of CDK2 activity or pocket protein phosphorylation (12). Therefore, we sought to determine whether st mediated up-regulation of cdc6 mRNA was dependent or independent of E2F and CDK activity. BJ-hTERT cells were transduced with a retroviral vector expressing a dominant negative version of E2F1 (E2F1A/B) consisting of the E2F DNA-binding and dimerization domains fused to the A/B pocket of RB (27), or with retrovirus control (r-control). After antibiotic selection, cells were serum-starved for 72 h and subsequently re-stimulated with serum or transduced with st or control adenoviruses. Northern blot analyses confirmed that cdc6 mRNA expression induced by FBS was greatly inhibited (∼4-fold) in the presence of the dominant-negative E2F (Fig. 5, A and B). Upon st expression, dominant negative E2F also caused prominent inhibition of cdc6 mRNA expression (3-fold) (Fig. 5, A and B), indicating that transcription of cdc6 induced by st in quiescent cells depends, at least partially, on E2F.
FIGURE 5.
st induces CDC6 mRNA expression in a CDK-independent, E2F-dependent manner. A, BJ-hTERT cells stably expressing a dominant negative E2F/pRB mutant chimera or the empty expression vector (r-control) (see text) were serum-starved for 72 h immediately after antibiotic selection and re-stimulated with FBS or transduced with control or st adenovirus. Cells were harvested for RNA analysis 40 h later. B, radioactive signals were quantified, normalized by ribosomal RNA loading, and represented as fold increase in mRNA as in Fig. 4. C, BJ-hTERT cells were serum-starved for 48 h and transduced with adenovirus expressing p21 and/or p16. Twenty-four hours later, cells were re-stimulated with FBS or transduced with st adenovirus. Forty hours later, cells were harvested for Northern blot analysis. D, radioactive signal was quantified as in B.
To assess the importance of CDK activity on st-induced cdc6 mRNA, BJ-hTERT cells were serum-starved for 48 h and transduced with p16 and p21 adenoviruses for 24 h. Subsequently, cells were stimulated with serum or transduced with st or control adenoviruses. Northern blot analysis confirmed that serum-dependent expression of cdc6 requires CDK activity (Fig. 5, C and D). In contrast, st-mediated induction of cdc6 mRNA was not affected by overexpression of CKIs. Considering that co-overexpression of p21 and p16 potently blocks the kinase activity of all cell cycle CDKs, it appears that st-mediated up-regulation of cdc6 mRNA is independent of CDK-mediated phosphorylation of pocket proteins despite being dependent on E2F promoter elements. This striking result suggests that st regulates CDC6 mRNA expression by mechanisms upstream of pocket proteins that do not require pocket protein inactivation via canonical CDK inactivation pathways. These and other potential alternative mechanisms are considered in the discussion.
CDC6 Expression and Loading onto Chromatin Are Coupled to Phosphorylation of CDK2 on Thr-160 during Mitogen-dependent Cell Cycle Entry—Taking into account the close correlation that we have observed between CDC6 expression with its loading onto chromatin and the phosphorylation of CDK2 on Thr-160 (Fig. 2, A and B), we next determined the effect that CDC6 depletion in cells has on CDK2 phosphorylation under physiological conditions. BJ-hTERT cells were transfected with chemically synthesized siRNA directed to CDC6 (a mixture of two distinct siRNAs 1 and 2) or Renilla firefly (control) and subsequently serum-starved for 48 additional hours in Dulbecco's modified Eagle's medium. Next, cells were stimulated with FBS or left untreated. Fig. 6A shows that the CDC6 but not the Renilla siRNA effectively knocked-down CDC6. Most importantly, preventing CDC6 expression reduces phosphorylation of CDK2 on Thr-160 dramatically, both in the soluble and chromatin-bound fraction (Fig. 6A), suggesting a close link between CDC6 expression and CDK2 activation. This decrease in CDK2 Thr-160 phosphorylation was accompanied by a noticeable but much less prominent decrease on pRB phosphorylation and cyclin A expression. Near normal pRB inactivation and E2F-dependent gene expression is consistent with the effects of the double cyclin E1;E2 knock-out in mice, where these events are only slightly reduced, as D-type cyclin/CDK complexes are sufficient to inactivate pocket proteins and induce E2F gene expression (4). To confirm that knockdown of the essential factor CDC6 prevents cell cycle entry and progression through S phase a time course experiment was performed. Exponentially growing BJ-hTERT cells were transfected with a mixture of three siRNAs targeting CDC6 in serum-free medium. Cells were re-transfected with the same siRNAs 24 h later and maintained without serum for additional 48 h prior to re-stimulation with FBS. Cells were collected at indicated time points, and the pellets were divided in two for PI/FACS analysis and preparation of soluble and chromatin-bound proteins. Fig. 6B shows that the CDC6, but not EGFP, siRNAs effectively knocked-down CDC6. As in Fig. 6A, preventing CDC6 expression reduced phosphorylation of CDK2-Thr-160 in both the soluble and chromatin fractions (Fig. 6B). Inhibition of CDC6 expression also inhibited loading of MCM2 onto chromatin (Fig. 6B), as well as cell cycle entry induced by serum (Fig. 6C). Thus, CDC6 expression appears important for effective phosphorylation and activation of CDK2.
FIGURE 6.
CDC6 expression is coupled to phosphorylation of CDK2 on Thr-160 and exit from quiescence induced by serum. A, BJ-hTERT cells were transfected with siRNAs targeting CDC6 or renilla luciferase in conditions of serum starvation. Forty-eight hours later, cells were restimulated with 10% FBS, and 48 h after restimulation, cells were harvested and lysed for soluble/chromatin-bound protein fractionation and Western blot analysis. B and C, BJ-hTERT cells were transfected with siRNAs targeting CDC6 or EGFP in conditions of serum starvation. Seventy-two hours later, cells were re-stimulated with FBS and harvested at the indicated time points. Cell pellets were divided in two for soluble/chromatin-bound protein fractionation and Western blot analysis (B) and PI/FACS analysis (C). A and B, same exposure times are shown for both fractions. B, CDK2 band corresponding to phosphorylated Thr-160 is indicated with an asterisk. LE indicates long exposure. SE indicates short exposure. C, percent of cells in each cell cycle phase is indicated.
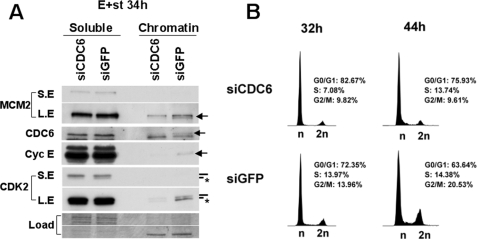
CDC6 Expression and Loading onto Chromatin Are Coupled to Phosphorylation of CDK2 on Thr-160 and Cell Cycle Entry Induced by Deregulated Cyclin E and st Co-expression—We next analyzed if CDC6 was also necessary for CDK2 phosphorylation and cell cycle entry of quiescent fibroblasts expressing deregulated cyclin E and st. BJ-hTERT cells were transfected with siRNA targeting CDC6 as described above. Forty-eight hours after the second round of transfection, cells were transduced with adenoviruses expressing cyclin E and st. As shown in Fig. 7A, knock-down of CDC6 inhibits CDK2 phosphorylation in both the soluble and chromatin fractions. The most striking effect of CDC6 knockdown on cyclin E/CDK2 was observed in the chromatin fractions. Reducing CDC6 by siRNA not only reduced the normal G1 loading of MCM2 onto chromatin as expected, but also greatly impaired the amount of both cyclin E and CDK2 in the chromatin fractions, suggesting that Cdc6 is required for robust association of cyclin E/CDK2 with chromatin. Concomitantly, cells transfected with CDC6 siRNA showed a clear delay in cell cycle entry (Fig. 7B). This result indicates that CDC6 is critical for the cooperative effects that deregulated cyclin E and st have on NHF, including the phosphorylation and chromatin association of cyclin E/CDK2 and entry in to S phase from quiescence.
FIGURE 7.
CDC6 expression is required for phosphorylation of CDK2 on Thr-160 and exit from quiescence induced by deregulated cyclin E and st co-expression. BJ-hTERT cells were transfected with the indicated siRNA and serum-starved as in Fig. 6. Subsequently the cells were transduced with adenovirus expressing cyclin E and st and harvested 34 h later. Cell pellets were divided in two for soluble/chromatin-bound protein fractionation and Western blot analysis (A) and PI/FACS analysis (B). A, same exposure times are shown for both fractions. Relevant proteins are indicated. The CDK2 band corresponding to phosphorylated Thr-160 is indicated with an asterisk. LE indicates long exposure. SE indicates short exposure. B, percent of cells in each cell cycle phase is indicated.
DISCUSSION
Cyclin E deregulation is frequently observed in breast carcinomas, leukemias and lymphomas, cervical carcinomas, gastrointestinal cancers, as well as early bladder lesions and colon adenomas among others and is often linked to poor prognosis (Refs. 6 and 7, reviewed in Ref. 8). We have previously shown that deregulated cyclin E expression cooperates with SV40 st to induce exit from quiescence in the absence of mitogens, as well as continuous proliferation of density-arrested NHF and foci formation. In this oncogenic context, CDK2 activation is required for cell cycle progression (12). Here we have identified the replication licensing factor CDC6 as a cellular target strongly and specifically induced by st expression. When cyclin E is concomitantly deregulated, subsequent accumulation of CDC6 protein stimulates origin licensing and is linked to CDK2 phosphorylation on its activating T loop leading to CDK2 activation and driving quiescent cells into S phase.
st Induces Unscheduled CDC6 Expression, Stabilization, and Loading onto Chromatin—CDC6 is an essential component of the pre-replication complex that is not expressed in quiescent mammalian cells and is tightly regulated throughout the cell cycle to prevent unscheduled or multiple firing of replication origins. Efficient firing of an origin of replication requires the binding of CDT1 and CDC6 to the origin of replication complex (ORC), as well as subsequent loading of the MCM helicase (reviewed in Refs. 21, 28–30). It has been shown that loading of the MCM helicase requires cyclin E protein (but not necessarily CDK2 kinase activity) as cells are stimulated to enter G1 from quiescence (5). Also, cyclin E/CDK2 complexes phosphorylate and stabilize CDC6 in G1 by preventing its ubiquitination by the APCCdh1 ligase, ensuring accumulation of CDC6 and formation of prereplication complexes (3). Here we report that expression of st in quiescent NHF induces accumulation of both cdc6 mRNA and protein, as well as loading onto chromatin, but st-mediated CDC6 accumulation is not sufficient to induce mitogen independent DNA synthesis in untransformed cells. However, when cyclin E is co-expressed with st in quiescent cells, CDC6 expression, and loading onto chromatin are significantly increased and this coincides with phosphorylation of CDK2 on Thr-160 and DNA synthesis.
Investigation of the mechanisms leading to increased cdc6 mRNA expression by st showed sensitivity to E2F-mediated repression. Because we have not observed st-mediated stabilization of cdc6 mRNA species (data not shown), our data suggest E2F-dependent transcriptional regulation. The sensitivity to E2F could indicate a mechanism of regulation similar to that induced by mitogens in quiescent cells, which is mediated via derepression of E2F-pocket protein complexes and subsequent transactivation by activating E2Fs (31). However, our data show that st alone induces cdc6 mRNA in quiescent NHF overexpressing CKIs, which presumably inactivate any potential trace of G1/S CDK activity. In this regard, we have not detected CDK2 activity in st expressing NHF (12). It has been suggested that st-mediated regulation of the cyclin A promoter may be dependent on inactivation of p27 (32). In contrast, cdc6 mRNA induction by st expression is just as efficient in the presence of ectopically expressed CKIs as it is in their absence. Thus, st-dependent up-regulation of cdc6 expression does not occur strictly via canonical downstream stimulation of mitogenic pathways that activate CDKs.
Instead, st may modulate E2F repressor complexes by a distinct and novel mechanism. One possibility is that since E2F/pocket protein complexes recruit histone deacetylases (HDAC), chromatin remodeling complexes (SWI/SNF), and histone methyltransferases at E2F elements (31), st expression may modulate these complexes independently of CDK-mediated phosphorylation. An alternative option may be that st affects the activity of YY1 or YY1 interaction with E2F, since that E2F cofactor is more specific for cdc6 than for other E2F targets (33). Yet another possibility is based on a recent study showing that IκB kinase activation inhibits cell growth and E2F-dependent transcription in NHF in a pocket protein-independent manner. NF-κB/p65 disrupts the physical interaction between activator E2Fs and the HAT cofactor TRRAP, resulting in reduction in E2F-responsive gene expression (34). Furthermore, previous studies have shown that st induces cyclin A mRNA expression in quiescent monkey CV-1 cells and foreskin human diploid fibroblasts (35). Although we have also detected very modest up-regulation of other E2F-regulated genes such as cyclin A in st-expressing density-arrested BJ-TERT cells, induction of cdc6 mRNA expression by st is much more potent and comparable in magnitude to that induced via mitogenic stimulation. Moreover, the induction of cdc6 mRNA results in a significant increase in CDC6 protein and its loading onto chromatin, but cyclin A protein and the products of other E2F-dependent genes are not substantially up-regulated. Thus, it appears that cdc6 is a particularly specific target of st. Clearly, the regulation of cdc6 mRNA accumulation is more complex than simple CDK-mediated pocket protein inactivation, and our findings demonstrate that the effects of st on cdc6 mRNA accumulation cannot be fully explained by global changes in E2F-dependent gene expression.
In addition to increased cdc6 mRNA expression, we have found that the CDC6 protein is induced by st in serum-starved and density-arrested cells as result of increased half-life. The reported CDC6 half-life is about 15–30′ in G1 and increases to >2 h in S phase (22, 36). The increase in protein stability induced by st is modest in the absence of serum (from 20′ to 33′), but appears consistent with the magnitude of increase in protein expression. Also, the increase in CDC6 expression correlates with its phosphorylation on a CDK site, which has been previously shown to stabilize CDC6 in G1 (3). st may promote CDC6 phosphorylation by targeting PP2A holoenzymes containing the PR70 B subunit, which has been recently implicated in stabilization of CDC6 by dephosphorylation of CDK sites (37). The implication of PR70 and other B subunits in this process will require further studies.
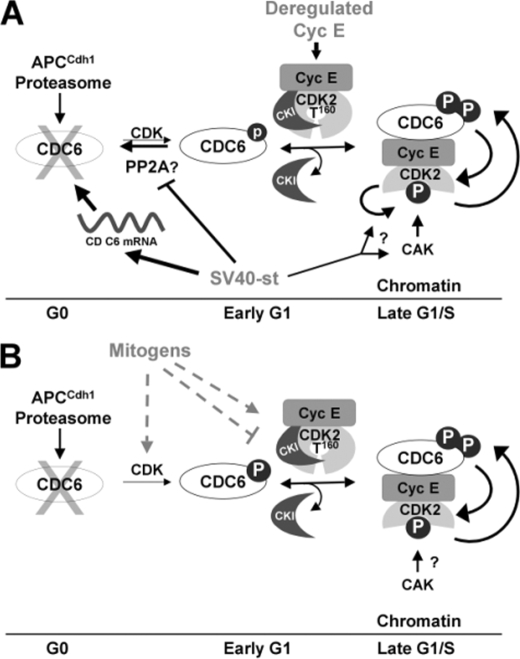
CDC6 May Play a Role as an Upstream Regulator of CDK2—Our data show that phosphorylation of CDK2 on its activating T loop and, hence, its activation is coupled to CDC6 expression in both serum-starved NHF coexpressing cyclin E and st and mitogen-stimulated NHF. We have previously shown that expression of st in quiescent NHF with deregulated cyclin E does not result in activation of CDK2 by inducing the abrupt dissociation of CKIs from CDK2 (12). In this report, we have shown that CDC6 knockdown diminishes CDK2 loading onto chromatin and conversely ectopic expression of CDC6 increases cyclin E loading onto chromatin. Therefore, as has been suggested by earlier studies in vitro (38), CDC6 may play a role in recruitment of CDK2 complexes to origins of replication. The mechanism of this recruitment could be by competing with CKIs for binding to the cyclin E/CDK2 complex (Fig. 8). Supporting this possibility, it has recently been shown that during recovery from a chemical DNA damage checkpoint, p21 dissociation from CDK2 complexes is accelerated by overexpression of CDC6, which binds CDK2 coinciding with p21 dissociation (39). More importantly, CDC6 activates p21-bound CDK2 in an ATP-dependent manner in vitro and mutation of CDC6 cyclin binding or ATPase domains precludes recovery from the checkpoint arrest in cells (39). Thus, considering that CDC6 can displace p21 in vitro, it is conceivable that CDC6 may compete with CKIs for binding to CDK2, thereby recruiting CDK2 complexes to origins of replication to phosphorylate specific substrates. Moreover, it has been previously proposed based on structural data that CKI binding to CDK2 interferes with the docking of CAK (40, 41). Thus, the absence of CKI corecruitment with CDK2 onto chromatin, the correlation of CDC6 expression and Thr-160 phosphorylation, and the dependence of this phosphorylation event on CDC6 expression may indicate that recruitment of CKI-free cyclin E/CDK2 complexes by CDC6 to chromatin facilitates CAK access to the T loop (Fig. 8). Conversely, the ability of monomeric CDK2 to autophosphorylate on Thr-160 has been recently proposed (42). Hence, st may alternatively stimulate CDK2 autophosphorylation in a manner that is dependent on deregulated cyclin E expression and independent from CAK. This st activity could precede a subsequent CDC6 role in displacing CKIs from cyclin E/CDK2 complexes prephosphorylated on Thr-160 leading to CDK2 activation. This scenario is compatible with our previous finding that the fractions of CKI/cyclin E/CDK2 and cyclin E/CDK2 complexes are phosphorylated on Thr-160 at similar levels (12). Further studies are needed to distinguish among these possibilities.
FIGURE 8.
Model of events linking CDC6 and CDK2 activation during exit from quiescence induced by cyclin E and st coexpression or mitogens. A, expression of st and deregulated cyclin E expression in quiescent NHF result in cooperative activation of the essential origin licensing factor CDC6 and coordinated CDK2 activation. st potently induces CDC6 mRNA expression and to a lower extent CDC6 phosphorylation on a CDK site(s) and stabilization. st also facilitates loading of CKI free cyclin E/CDK2 onto chromatin, which correlates with CDK2 phosphorylation on the activating T loop. CDC6 may facilitate CAK access to the CDK2 T loop, which could be prevented by CKIs. Alternatively st may induce CDK2 autophosphorylation on Thr-160 (arrow with question mark; see text). Coexpression of cyclin E and st also enhances CDC6 phosphorylation and loading onto chromatin suggesting positive feedback loops leading to activation of CDK2 and CDC6 accumulation (indicated by reciprocal arrows). B, mitogens may use similar mechanisms to activate CDK2 on chromatin.
Supplementary Material
Acknowledgments
We thank P. Kaldis, J. Albrecht, E. Moran, K. Rundell, J. Fueyo, W. El-Deiry, P. Ruiz, F. Graham, J. B. Clurman, E. Knudsen, and W. Hahn for reagents used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant CA095569 (to X. G. and J. G.) and a Career Development Award (K02 AI01823) (to X. G.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: CDK, cyclin-dependent kinase; st, small t antigen; FBS, fetal bovine serum; MOI, multiplicity of infection; PIPES, 1,4-piperazinediethanesulfonic acid; MOPS, 4-morpholinepropanesulfonic acid; CHX, cycloheximide; NHF, normal human fibroblasts; PI/FACS, propidium iodide/fluorescent-activated cell sorting; EGFP, enhanced green fluorescent protein.
References
- 1.Malumbres, M., and Barbacid, M. (2005) Trends Biochem. Sci. 30 630–641 [DOI] [PubMed] [Google Scholar]
- 2.Graña, X., Garriga, J., and Mayol, X. (1998) Oncogene 17 3365–3383 [DOI] [PubMed] [Google Scholar]
- 3.Mailand, N., and Diffley, J. F. (2005) Cell 122 915–926 [DOI] [PubMed] [Google Scholar]
- 4.Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J. E., Bhattacharya, S., Rideout, W. M., Bronson, R. T., Gardner, H., and Sicinski, P. (2003) Cell 114 431–443 [DOI] [PubMed] [Google Scholar]
- 5.Geng, Y., Lee, Y. M., Welcker, M., Swanger, J., Zagozdzon, A., Winer, J. D., Roberts, J. M., Kaldis, P., Clurman, B. E., and Sicinski, P. (2007) Mol. Cell 25 127–139 [DOI] [PubMed] [Google Scholar]
- 6.Moroy, T., and Geisen, C. (2004) Int. J. Biochem. Cell Biol. 36 1424–1439 [DOI] [PubMed] [Google Scholar]
- 7.Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K., Guldberg, P., Sehested, M., Nesland, J. M., Lukas, C., Orntoft, T., Lukas, J., and Bartek, J. (2005) Nature 434 864–870 [DOI] [PubMed] [Google Scholar]
- 8.Hwang, H. C., and Clurman, B. E. (2005) Oncogene 24 2776–2786 [DOI] [PubMed] [Google Scholar]
- 9.Resnitzky, D., Gossen, M., Bujard, H., and Reed, S. I. (1994) Mol. Cell. Biol. 14 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtsubo, M., and Roberts, J. M. (1993) Science 259 1908–1912 [DOI] [PubMed] [Google Scholar]
- 11.Calbó, J., Parreño, M., Sotillo, E., Yong, T., Mazo, A., Garriga, J., and Graña, X. (2002) J. Biol. Chem. 277 50263–50274 [DOI] [PubMed] [Google Scholar]
- 12.Sotillo, E., Garriga, J., Kurimchak, A., and Graña, X. (2008) J. Biol. Chem. 283 11280–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortega, S., Prieto, I., Odajima, J., Martin, A., Dubus, P., Sotillo, R., Barbero, J. L., Malumbres, M., and Barbacid, M. (2003) Nat. Genet. 35 25–31 [DOI] [PubMed] [Google Scholar]
- 14.Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., DeCaprio, J. A., and Weinberg, R. A. (2002) Mol. Cell. Biol. 22 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeb, K. K., Michalowska, A. M., Yoon, C. Y., Krummey, S. M., Hoenerhoff, M. J., Kavanaugh, C., Li, M. C., Demayo, F. J., Linnoila, I., Deng, C. X., Lee, E. Y., Medina, D., Shih, J. H., and Green, J. E. (2007) Cancer Res. 67 8065–8080 [DOI] [PubMed] [Google Scholar]
- 16.Cook, J. G., Park, C. H., Burke, T. W., Leone, G., DeGregori, J., Engel, A., and Nevins, J. R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1347–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garriga, J., Bhattacharya, S., Calbo, J., Marshall, R. M., Truongcao, M., Haines, D. S., and Graña, X. (2003) Mol. Cell. Biol. 23 5165–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel, F. M., Brent, R., Kington, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. E. (1988) Current Protocols in Molecular Biology, Greene Publishing Associates and Wiley-Interscience, NY
- 19.Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004) J. Cell Biol. 165 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya, S., Garriga, J., Calbo, J., Yong, T., Haines, D. S., and Graña, X. (2003) Oncogene 22 2443–2451 [DOI] [PubMed] [Google Scholar]
- 21.Diffley, J. F. (2004) Curr. Biol. 14 R778–786 [DOI] [PubMed] [Google Scholar]
- 22.Petersen, B. O., Wagener, C., Marinoni, F., Kramer, E. R., Melixetian, M., Lazzerini Denchi, E., Gieffers, C., Matteucci, C., Peters, J. M., and Helin, K. (2000) Genes Dev. 14 2330–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hateboer, G., Wobst, A., Petersen, B. O., Le Cam, L., Vigo, E., Sardet, C., and Helin, K. (1998) Mol. Cell. Biol. 18 6679–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtani, K., Tsujimoto, A., Ikeda, M., and Nakamura, M. (1998) Oncogene 17 1777–1785 [DOI] [PubMed] [Google Scholar]
- 25.Vilaboa, N., Bermejo, R., Martinez, P., Bornstein, R., and Cales, C. (2004) Nucleic Acids Res. 32 6454–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, Z., DeGregori, J., Shohet, R., Leone, G., Stillman, B., Nevins, J. R., and Williams, R. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellers, W. R., Rodgers, J. W., and Kaelin, W. G., Jr. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11544–11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005) Cell 123 13–24 [DOI] [PubMed] [Google Scholar]
- 29.Machida, Y. J., and Dutta, A. (2005) J. Biol. Chem. 280 6253–6256 [DOI] [PubMed] [Google Scholar]
- 30.Sclafani, R. A., and Holzen, T. M. (2007) Annu. Rev. Genet. 41 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowland, B. D., and Bernards, R. (2006) Cell 127 871–874 [DOI] [PubMed] [Google Scholar]
- 32.Skoczylas, C., Henglein, B., and Rundell, K. (2005) Virology 332 596–601 [DOI] [PubMed] [Google Scholar]
- 33.Schlisio, S., Halperin, T., Vidal, M., and Nevins, J. R. (2002) EMBO J. 21 5775–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki, K., Kawauchi, K., and Tanaka, N. (2008) Oncogene 27 5696–5705 [DOI] [PubMed] [Google Scholar]
- 35.Porras, A., Bennett, J., Howe, A., Tokos, K., Bouck, N., Henglein, B., Sathyamangalam, S., Thimmapaya, B., and Rundell, K. (1996) J. Virol. 70 6902–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duursma, A. M., and Agami, R. (2005) Cell Cycle 4 1725–1728 [DOI] [PubMed] [Google Scholar]
- 37.Davis, A. J., Yan, Z., Martinez, B., and Mumby, M. C. (2008) J. Biol. Chem. 283 16104–16114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furstenthal, L., Kaiser, B. K., Swanson, C., and Jackson, P. K. (2001) J. Cell Biol. 152 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan, Q., Jinno, S., Yamamoto, H., Kobayashi, K., and Okayama, H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4757–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lolli, G., and Johnson, L. N. (2005) Cell Cycle 4 572–577 [PubMed] [Google Scholar]
- 41.Lolli, G., Lowe, E. D., Brown, N. R., and Johnson, L. N. (2004) Structure 12 2067–2079 [DOI] [PubMed] [Google Scholar]
- 42.Abbas, T., Jha, S., Sherman, N. E., and Dutta, A. (2007) Cell Cycle 6 843–852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.