Abstract
Almost every protein-coding gene undergoes pre-mRNA splicing, and the majority of these pre-mRNAs are alternatively spliced. Alternative exon usage is regulated by the transient formation of protein complexes on the pre-mRNA that typically contain heterogeneous nuclear ribonucleoproteins (hnRNPs). Here we characterize hnRNP G, a member of the hnRNP class of proteins. We show that hnRNP G is a nuclear protein that is expressed in different concentrations in various tissues and that interacts with other splicing regulatory proteins. hnRNP G is part of the supraspliceosome, where it regulates alternative splice site selection in a concentration-dependent manner. Its action on alternative exons can occur without a functional RNA-recognition motif by binding to other splicing regulatory proteins. The RNA-recognition motif of hnRNP G binds to a loose consensus sequence containing a CC(A/C) motif, and hnRNP G preferentially regulates alternative exons where this motif is clustered in close proximity. The X-chromosomally encoded hnRNP G regulates different RNAs than its Y-chromosomal paralogue RNA-binding motif protein, Y-linked (RBMY), suggesting that differences in alternative splicing, evoked by the sex-specific expression of hnRNP G and RBMY, could contribute to molecular sex differences in mammals.
All protein-coding genes undergo pre-mRNA processing, and the large majority of these genes are alternatively spliced (1). Alternative exons can change many functional aspects of mRNAs and their encoded proteins. The best understood functions are stop codons or frameshifts that are introduced by 20–35% of alternative exons, which often destine the altered mRNA to nonsense-mediated decay. Examples described in the literature show that alternative splicing regulates the binding properties, intracellular localization, enzymatic activity, protein stability, and post-translational modifications of a large number of proteins (reviewed in Ref. 2). Thus, it appears that alternative pre-mRNA processing is a key mechanism regulating the gene expression of complex organisms by generating multiple mRNA isoforms, which encode functionally diverse proteins. Despite its importance, the exact mechanisms governing splice site selection are still poorly understood. In vertebrate systems, protein complexes assemble transiently on exons, and their interaction with the splicing machinery as well as RNA-RNA interactions between spliceosomal proteins and pre-mRNA determine whether an exon is included or skipped (reviewed in Refs. 3 and 4).
When isolated from nuclei of mammalian cells, RNA polymerase II transcripts are found assembled in large ribonucleoprotein 21-MDa complexes, the supraspliceosome, composed of all five spliceosomal small nuclear ribonucleoproteins as well as additional proteins. The entire repertoire of nuclear pre-mRNAs, independent of their length or number of introns, is individually found assembled in supraspliceosomes (reviewed in Ref. 5). Structural studies revealed that the supraspliceosome is composed of four substructures considered to be native spliceosomes that are connected by the pre-mRNA (6–11). Supraspliceosomes are active in splicing (7) and contain regulatory splicing factors such as all phosphorylated SR proteins, which are essential for splicing and splicing regulation (12). Supraspliceosomes also harbor other components of pre-mRNA processing, such as the editing enzymes ADAR1 and ADAR2 (13), cap-binding proteins, and components of the 3′-end processing activity (14). Taken together, these results support the view that the supraspliceosome is a nuclear pre-mRNA processing machine.
hnRNP3 G was discovered some time ago as an autoantigen that is glycosylated, binds to RNA, and is present in lampbrush chromosomes (15). The protein has an N-terminal RNA recognition motif, followed by a C-terminal domain rich in RG and SR repeats, as well as tyrosine residues. hnRNP G suppresses growth of squamous carcinoma cells, whereas mutated hnRNP G fails to stop cancerous cell growth, indicating that hnRNP G could be a tumor suppressor (16). In humans, hnRNP G is encoded by the RBMX gene, located on the X chromosome. Its paralogue on the Y chromosome is the testis-specific RBMY, which is necessary for normal sperm development in humans (17). In addition, humans express a testis-specific gene, hnRNP G-T, which is autosomally encoded. RBMY and hnRNP G are highly similar and share 88% identity in their RNA binding domain. Both RBMY and hnRNP G were shown to interact with the SR-like protein Tra2-β and the STAR proteins SAM68, SLM-1, and SLM-2 (18). hnRNP G has been shown to regulate alternative splicing in several model systems, including SMN2 and tau and α-tropomyosin, but its mechanism of action is not clear (19–21). RBMY binds RNA stem structures containing a C(A/U)CAA loop sequence. RBMY recognizes this loop in a sequence-specific manner and also binds to the stem in a structure-specific manner. This represents a new way of RNA binding by an RRM (22).
Here we show that hnRNP G is a component of the supraspliceosome that regulates alternative splice site selection. hnRNP G influences splice site selection in two ways. The first way is independent of RNA binding and depends on the C terminus of hnRNP G binding to splicing regulatory proteins. The second way depends on the RRM of hnRNP G that binds to sequences containing CC(A/C) motifs. In contrast to RBMY, these sequences do not contain stable stems. Although the hnRNP G and RBMY sequences are similar, both proteins recognize different sets of pre-mRNA, suggesting that the sex-specific expression of hnRNP G and RBMY contributes to mammalian molecular sex differences.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen—Molecular cloning was performed using standard protocols (23), and RT-PCR was performed as described (24). A matchmaker two-hybrid rat brain postnatal day 5 and an embryonic day 16 library (Stratagene) were screened using hTra2-β pGBT9 (25) as bait. The yeast Gal4 two-hybrid screen was performed according to Fields and Song (26) using the strain HF7c. For each screen, ∼6 × 106 transformants were screened with 100 μg of bait DNA and 50 μg of prey DNA. To test the interaction of hnRNP G with other proteins, 1 μg of hnRNP G fused to the Gal4 activation domain and 1 μg of interacting proteins fused to the Gal4 binding domain were cotransformed and plated onto triple dropout plates lacking leucine, tryptophan, and histidine. Surviving colonies were restreaked on triple dropout plates supplemented with 10 mm 3-aminotriazole and isolated as described (27).
Immunoprecipitation and Western blot were performed as described (13, 27) using the anti-green fluorescent protein (Roche Applied Science) antibody. Western blot was performed with anti-Tra2 (pan Tra2-β) (28) 1:1000 and anti-hnRNP G (generated against peptide RDDGYSTKD) 1:2000; horseradish peroxidase-conjugated secondary antibodies were from either AP-Biotech Rockland Immunochemicals for Research or Santa Cruz Biotechnology and used in 1:10,000 dilution. In all experiments the protein in the “load control” represents about 10% of the input protein for the immunoprecipitation.
Gel shift experiments were performed as described (29). RNA was end-labeled, and 2 pmol of 32P-labeled RNA were incubated with protein for 15 min at 30 °C. The mobility was tested in 5% native polyacrylamide gels.
In Vivo Splicing—Splicing assays and cotransfection were performed as described (24), employing the SMN2 minigene (30), containing the alternative exon 7 flanked by the constitutive exons 6 and 8 and the original introns (31). The tau MG contains the alternative exon 10, flanked by exons 9 and 11 (32). Transfection of the minigenes was done in HEK293 cells. 1 μg of indicated minigenes was used together with increasing amounts of the indicated pEGFP constructs and with the appropriate amount of empty pEGFP-C2 vector to ensure that equal amounts of DNA were transfected. The calcium phosphate method was used for transfection. RNA was isolated 16–18 h post-transfection by using the total RNA kit (Qiagen, Germany). Reverse transcription was performed in a total volume of 8.5 μl by using 2 μl of RNA, 1 μl of oligo(dT) (0.5 mg/ml), 1 μl of first-strand buffer (Invitrogen), 10 mm dithiothreitol, 1 mm dNTP, 5 units of SuperScript II RT (Invitrogen) at 42 °C for 60 min. To ensure that only plasmid-derived minigene transcript was detected, subsequent amplification was performed with a vector-specific forward primer (pCI-forward, 5′-GGTGTCCACTCCCAGTTCAA) and the SMN-specific reverse primer SMNex8-rev (SMNex8-reverse, 5′-GCCTCACCACCGTGCTGG) for SMN2 MG (31); and antisense INS3 (cac ctc cag tgc caa ggt ctg aag gtc acc) for SV9/10L/11 tau MG. PCR products resolved on ethidium bromide-stained agarose gels resulted in similar ratios. The ratio of exon inclusion to exon skipping was determined by using the ImageJ program.
Supraspliceosome Isolation—Nuclear supernatants enriched for supraspliceosomes were prepared according to a procedure originally developed by Sperling et al. (33) with minor changes (34). Briefly, nuclear supernatants enriched for supraspliceosomal particles were prepared from purified nuclei of HeLa cells (CILBIOTECH, Mons, Belgium) or from HEK293 cells by microsonication of the nuclei and precipitation of the chromatin in the presence of tRNA. The nuclear supernatants were fractionated in 10–45% glycerol gradients. Supraspliceosomes sediment at the 200 S region of the glycerol gradient (using tobacco mosaic virus as a sedimentation coefficient marker).
Supraspliceosome Fractionation on Native Gels—For fractionation of supraspliceosomes in native composite gels (1.5% polyacrylamide, 0.6% agarose), we used a protocol adapted from Ref. 35 with modifications. Supraspliceosomes prepared from cells overexpressing EGFP-hnRNP G were concentrated at 4 °C, using ultrafiltration columns (Vivaspin 15R, Sartorius), and electrophoresed at 10 V/cm for 4 h at 4 °C in 40 mm Tris base, 25 mm acetic acid, and 2 mm MgCl2. Samples were extracted from the EGFP fluorescent band and visualized by EM using negative staining (1% uranyl acetate). The gel was also Western-blotted with antibodies against hnRNP G.
In Vitro SELEX—An initial PCR to amplify the DNA pool was performed using primers T7 pro (5′-TAATACGACTCACTATAGGGATCCGAATTCCCGACT-3′) and RT (5′-GCGTCTCGAGAAGCTTCCC-3′). The PCR product was cut from an 8% polyacrylamide gel, crushed, and 600 μl of extraction solution (4 ml of ammonium acetate, 0.8 ml of EDTA 0.5 m, pH 8.3, 5.2 ml of H2O) added. The mixture was incubated at 37 °C overnight on a rotating wheel. After spinning down at full speed for 5 min, the supernatant was taken to a fresh tube, mixed with 1 ml of isopropyl alcohol, and placed at -80 °C for 3 h. After centrifuging the pellet was washed with 70% ethanol and dissolved in 10 μl of H2O.
After performing in vitro transcription with 1 μg of the amplified DNA pool in a total of 250 μl for 3 h at 37 °C, DNase treatment and phenol/chloroform extraction were carried out, and the transcribed RNA was precipitated overnight at -80 °C. The mixture was centrifuged and washed with 70% ethanol and resuspended in 70 μl of binding buffer (10 mm Tris, pH 7.5, 100 mm KCl, 2.5 mm MgCl2, 0.1% Triton X-100 + 0.1 mg/ml tRNA).
25 μl of nickel-nitrilotriacetic acid-agarose was washed in binding buffer without tRNA, and the RNA was preincubated with the resin for 15 min at 4 °C. The resin was removed, and 20 μl of purified His-hnRNP G was added and incubated at room temperature for 30 min. 25 μl of resin, washed with binding buffer, was added and incubated for 30 min at 4 °C. After washing the agarose resin twice with binding buffer, 50 ml of 2× proteinase K buffer (200 mm Tris, pH 7.5, + 25 mm EDTA, 300 mm NaCl + 2% SDS) and 5 μg of proteinase K were added and incubated at 37 °C for 30 min. The RNA was isolated by phenol/chloroform extraction and ethanol precipitation containing glycogen.
For performing reverse transcription reaction, the RNA was resuspended in 1× RT buffer (10 μl of first strand buffer, 4 μl of RT primer, 5 μl of 10 mm dNTPs, 31 μl of H2O), incubated for 10 min at 65 °C, spun down, and put on ice for 2 min. 0.5 μl of 0.1 m DTT, 1.0 μl of RNasin and 2.0 ml of Superscript II were added and incubated at 42 °C for 2 h. The reaction was heated for 15 min at 72 °C, and PCR was performed, using primers T7 pro and RT.
The amplified DNA pool was purified from an 8% polyacrylamide gel as described above and another round of SELEX performed. After five rounds, the PCR products of the last SELEX round were cloned into TOPO vector and sequenced.
Antiserum Generation—After coupling to keyhole limpet hemocyanin, the peptide RDDGYSTKD was used to immunize rabbits. After 121 days, serum was purified by affinity chromatography, using the peptide. Dilution for Western blot was 1:1000 and for immunohistochemistry was 1:200.
Northern Blot—A rat tissue Northern blot (Clontech) was first hybridized with a radioactively labeled hnRNP G cDNA probe lacking the first 320 nucleotides that encode RRM. Hybridization was according to the manufacturer's instructions. The probe was removed, and the membrane was reprobed with a radioactively labeled β-actin probe to verify loading in each lane. The membrane was then analyzed using a phosphorimager.
Bioinformatics Analysis—To determine the single strandedness of a motif, we computed the probability that the motif is completely unpaired (PU value), as described previously (36). Briefly, we computed the PU value of the motif for 20 different context lengths (symmetric contexts ranging from 11 to 30 nucleotides up- and downstream of the given motif) using RNAfold from the Vienna RNA package (37). To obtain a single PU value for each motif, we averaged these 20 values. Fisher's exact test and Wilcoxon rank-sum tests were performed using R.
To analyze the distance between the CCM (M = A or C) motifs in exons, we first determined the average distance in the hnRNP G regulated and control exon set. As the control exons contain much less CCM motifs, it is expected that the distance between CCM in the controls is much larger. Consequently, we cannot directly compare the average distances between both groups. Therefore, we determined the expected distribution of the average values separately for both exon sets as follows. For each exon sequence, we generated 10,000 sequences of the same length containing the same number of CCM motifs at randomly chosen positions. This yields 10,000 random sets of exons. For each set, we determined the average distance of the randomly placed CCM motifs and derived the empirical expected distribution from these 10,000 values. This procedure was done separately for hnRNP G regulated and control exons. It should be noted that computing the median distance (instead of the average distance) leads to the same conclusion.
RESULTS
Domain Structure, Expression, and Intranuclear Localization of hnRNP G—We identified rat hnRNP G in two-hybrid screens as an interacting partner of hTra2-β1, YT521-B, and scaffold attachment factor 1 (SAF-B1). Similar to human hnRNP G, the rat protein consists of an N-terminal RRM, followed by a proline-rich region, a region with RGG repeats, and an area rich in SR dipeptides (Fig. 1A and supplemental Fig. 1). The nucleic acid and protein sequences show 90 and 93% identity to human hnRNP G (supplemental Fig. 2). Unless otherwise indicated, all experiments were made with this rat hnRNP G cDNA.
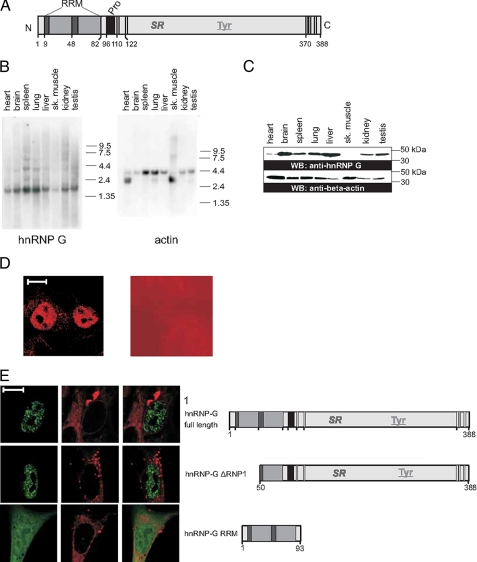
FIGURE 1.
Sequence and expression of rat hnRNP G. A, domain structure of hnRNP G. The RRM is indicated in yellow with the two ribonucleoprotein boxes in dark gray. Pro, proline-rich region, marked in pink;RG sequences are indicated as vertical lines. Numbers indicate the amino acids position shown in supplemental Fig. 1. B, Northern blot with mRNA from different rat tissues was probed with the cDNA of hnRNP G lacking the RRM sequences (left), and the same blot was reprobed with β-actin to demonstrate loading in each lane (right). C, Western blot with lysates from different tissues analyzed with an antiserum against hnRNP G (top) and an antibody against actin (bottom). WB, Western blot. D, immunohistochemistry of hnRNP G. HEK293 cells were stained with the hnRNP G antiserum that stains the nucleoplasma excluding the nucleolus (left). Preimmune serum gave no specific staining (right). The size bar represents 10 μm. E, localization of EGFP-tagged hnRNP G variants in 293 cells. Representative transfected cells analyzed by confocal microscopy are shown. Cells were transfected with N-terminal tagged EGFP variants shown on the right. The EGFP signal is shown in green on the left, and cells were counterstained with the membrane tracer DiI (red), and the two pictures were overlaid on the right.
To get the first insight into the biological function, we next determined the expression of hnRNP G by Northern blot analysis. We probed a rat multiple tissue Northern blot with radiolabeled hnRNP G cDNA and subsequently tested for loading by probing the same membrane with β-actin cDNA. As shown in Fig. 1B, we observed hnRNP G expression in all tested tissues. Because there is often a discrepancy between RNA and protein expression, we determined hnRNP G expression using a peptide antiserum raised against residues 207–215, encoded by a constitutive exon of hnRNP G. As shown in Fig. 1C, hnRNP G was expressed in all tested tissues, although the amount detected was lower in heart and muscle when compared with other tissues. The expression in muscle becomes evident at longer exposure times (supplemental Fig. 4). Although there are numerous splice variants annotated in the data bases, we observed a single major band using this peptide encoded from a constitutive exon, indicating that the other splicing products of hnRNP G are either minor forms or not translated into protein. There is less hnRNP G protein in muscle and heart tissue, which is in accordance with the mRNA expression detected by Northern blot analysis.
The hnRNP G sequence shows no predicted cellular localization signal. We therefore determined the intracellular localization of endogenous hnRNP G and performed immunohistochemistry with our antiserum. As shown in Fig. 1D, left, the immunoreactivity is localized in the nuclei of cells. The signal for hnRNP G depends on the peptide used for immunization, as preimmune serum shows no specific signal (Fig. 1D, right). Furthermore, the antiserum detects only a single band in Western blots after long exposure times (supplemental Fig. 4). hnRNP G staining excludes the nucleoli and is dispersed throughout the nucleoplasma. The nucleoplasmic staining is not homogeneous, as the protein is concentrated in numerous small granules. We next determined whether overexpressed protein has a similar localization and analyzed the expression of EGFP-tagged protein. As shown in Fig. 1E, EGFP-tagged full-length hnRNP G shows a nuclear localization very similar to the endogenous protein. It excludes the nucleoli, and the staining is concentrated in numerous granules. Numerous costaining experiments showed that these granules do not coincide with other known nuclear structures, such as SC35 speckles, coiled bodies, promyelocytic leukemia bodies, or YT bodies (38) (data not shown).
We next determined which protein parts are involved in the intracellular localization of hnRNP G and performed a deletion analysis using the EGFP-tagged mutants. Destruction of the RNA recognition motif by deleting its first half did not change this nuclear localization, indicating that the RRM and RNA binding are not required for nuclear localization. This was confirmed by determining the localization of the RRM of hnRNP G, which is localized in both the nucleus and cytosol (Fig. 1E).
Together, these data show that hnRNP G shows ubiquitous expression in various rat tissues. The expression levels are variable among tissues. The protein is generated from the major splicing variant, and we could not detect expression from other predicted isoforms generated by alternative splicing. The protein is nuclear, and its cellular localization is not determined by its RRM.
Proteins Interacting with hnRNP G—We next used hnRNP G in a yeast two-hybrid screen against a rat embryonic brain library as described previously (27). We identified the proteins hnRNP G, SAF-B, YT521-B, Tra2-β1, SAM68, its related proteins SLM-1 and SLM-2 and mCLK2 as interacting partners for hnRNP G, as the presence of their cDNA allowed yeast to grow under high stringency conditions achieved by adding 10 mm 3-aminotriazole to the selection plates (Fig. 2A). To determine the protein parts involved in the interaction, we tested deletion clones of hnRNP G for its binding to these interaction partners and found that the interactions do not require a functional RRM (Fig. 2A). The interactions detected in yeast were observed when hnRNP G and its interacting partners were fused either to the Gal4 activation or binding domain (data not shown).
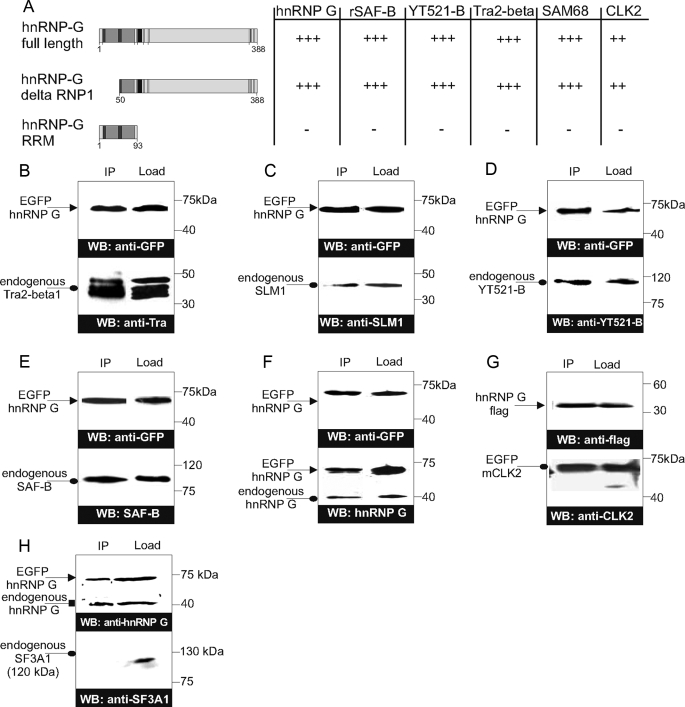
FIGURE 2.
Interaction of hnRNP G with several endogenous splicing factors. A, summary of the two-hybrid interactions. Growth on 10 mm 3-aminotriazole indicator plates is indicated by +++. The structure of hnRNP G variants tested are indicated on the left. B–F, coimmunoprecipitation of hnRNP G and its interacting proteins. EGFP-hnRNP G (wt) was expressed in HEK293 cells and precipitated with anti-GFP antibody. Precipitates were analyzed for the presence of proteins found to interact in yeast. The presence of the interacting proteins in immunoprecipitates was compared with their expression in cellular lysates, which represent ∼1/10 of the immunoprecipitates. The EGFP-fused hnRNP was detected by anti-GFP in the coimmunoprecipitates and the lysates (upper panels). Coimmunoprecipitated (IP) endogenous Tra2-β1 (B), SLM1 (C), YT521-B (D), SAF-B (E), and hnRNP G (F) and the lysates (load) were detected by Western blot (WB) using their specific antibodies (lower panels). Tra2-β1 runs as three different bands because of reversible phosphorylation (41). For mClk2, both interacting partners were overexpressed, and then tagged mClk2 and its endogenous form could be detected in the immunoprecipitates (G). hnRNP G does not immunoprecipitate with endogenous SF3A1, which serves as a negative control (H).
Because yeast two-hybrid interactions are prone to experimental artifacts, we verified the interactions of these proteins by immunoprecipitation. We expressed full-length EGFP-tagged hnRNP G in HEK293 cells and precipitated it with anti-GFP antiserum. The immunoprecipitates contained benzonase, to ensure that we detect protein-protein interactions and not nucleic acid-mediated interactions. The immunoprecipitates were analyzed by Western blot by probing for the endogenous interacting proteins. As shown in Fig. 2, B–G, this analysis could verify the protein-protein interactions against Tra2-β1, rSLM-1, YT521-B, and SAF-B and with endogenous hnRNP G. In addition, we detected an interaction between hnRNP G and mClk2, when both proteins were expressed from tagged cDNAs. Together, these data show that hnRNP G interacts with proteins involved in RNA metabolism and pre-mRNA splicing. These interactions are dependent on protein-protein interaction and do not require the RRM of hnRNP G.
hnRNP G Is a Component of the Supraspliceosome—The protein interaction data indicated that hnRNP G acts in pre-mRNA processing, as all interacting proteins have been implicated in this process (39). We therefore determined whether hnRNP G is part of the supraspliceosome, a macromolecular machine, active in splicing, in which the entire repertoire of nuclear pre-mRNAs is individually packaged (7, 9, 10, 12). As shown in Fig. 3A, when nuclear supernatants enriched for supraspliceosomes are fractionated in glycerol gradients, endogenous hnRNP G is localized in the 21-MDa supraspliceosome fractions that peak around 200 S. Its distribution in the supraspliceosomal fractions is similar to that of the phosphorylated SR proteins (Fig. 3B), which have been previously shown to be associated with supraspliceosomes in these fractions (8, 11, 13).
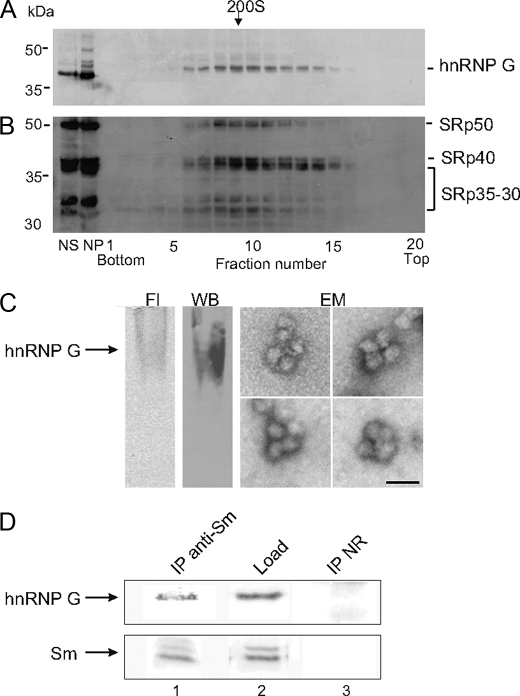
FIGURE 3.
Endogenous hnRNP G is found in supraspliceosomes. Nuclear supernatants enriched in supraspliceosomes prepared from HEK293 cells and fractionated in a 10–45% glycerol gradient were collected (bottom to top) in 20 fractions. A, aliquots from each fraction were analyzed by Western blot using anti-hnRNP G antibodies. B, same blot was probed with monoclonal antibody 104, which is directed against the phosphorylated epitopes of the SR proteins. NS, nuclear supernatant; NP, nuclear pellet. The gradient was calibrated with 200S tobacco mosaic virus run in a parallel gradient. C, Fl, supraspliceosomes, prepared from cells transiently transfected with an EGFP-hnRNP G construct, were fractionated in native gel. The migration of EGFP-hnRNP G was visualized by fluorescence. WB, hnRNP G was detected by Western blot analysis of the native gel, using anti-hnRNP G antiserum (arrow), showing that both endogenous and exogenous hnRNP G comigrate. EM, electron microscopy visualization of aliquots extracted from the gel at the hnRNP G band revealed typical images of supraspliceosomes. Bar indicates 50 nm. D, aliquots from the supraspliceosome peak fractions (fractions 9 and 10) were immunoprecipitated by anti-Sm Y12 monoclonal antibodies bound to protein A-agarose beads. The precipitate (IP, lane 1), the proteins loaded (Load, lane 2, one-fifth of the total load), and the control-IP with nonrelevant IgG (NR, lane 3) were analyzed by Western blotting with anti-hnRNP G antibodies (upper panel). The same blot was reprobed with Y12 monoclonal antibodies. Similar results were obtained in at least three different experiments.
Previous EM structural studies revealed that the supraspliceosome is composed of four substructures (native spliceosomes) interconnected by the pre-mRNA (5–7). To substantiate the association of hnRNP G with supraspliceosomes, we expressed full-length EGFP-tagged hnRNP G in HEK293 cells and prepared supraspliceosomes from these cells. Both endogenous and exogenous hnRNP G are localized in supraspliceosomes, as determined by fractionation on glycerol gradients (data not shown). When supraspliceosomes were fractionated in native gels, the localization of EGFP-hnRNP G was visualized by fluorescence (Fig. 3C, Fl). Both endogenous and exogenous hnRNP G were localized to this band as shown here by Western blots of the native gel, using the anti-hnRNP G antiserum (Fig. 3C, WB). Aliquots extracted from the hnRNP G fluorescent band in the native gel were visualized by EM and revealed the typical tetrameric structure of supraspliceosomes (Fig. 3C, EM). To further substantiate the association of hnRNP G with supraspliceosomes, we performed co-IP experiments on supraspliceosomes using anti-Sm Y12 monoclonal antibodies. Fig. 3D shows that hnRNP G is specifically associated with supraspliceosomes, together with Sm proteins. The control shows that IgG did not precipitate hnRNP G. Together, these data show that hnRNP G is localized with other splicing factors in the supraspliceosome.
hnRNP G Influences Splicing Patterns by Sequestration—The association of hnRNP G with the supraspliceosome and its interaction with proteins implicated in splice site selection is consistent with the previously shown role of hnRNP G in splice site selection (20). It was shown for Tra2-β1 that a splicing factor can influence exon recognition either directly by binding to it or indirectly by sequestering other splicing regulatory proteins (28). To test whether hnRNP G influences alternative splicing patterns of these genes directly or through sequestration, we compared wild-type hnRNP G with a mutant lacking the RRM on their action of reporter minigenes. We cotransfected an increasing amount of expression constructs encoding hnRNP G wild-type and Δ RRM with reporter genes in HEK293 cells. As can be seen in Fig. 4, A and B, both proteins have an influence on the splicing patterns of these minigenes, with the wild-type protein having a stronger effect. At lower concentrations of hnRNP G, the differences between hnRNP G wild-type and Δ RRM variants are more pronounced. In contrast, at higher concentrations, the differences are smaller and are not detectable for the SMN2 reporter. Western blot analysis reveals that both wild-type and mutant hnRNP G were expressed at similar levels (Fig. 4C).
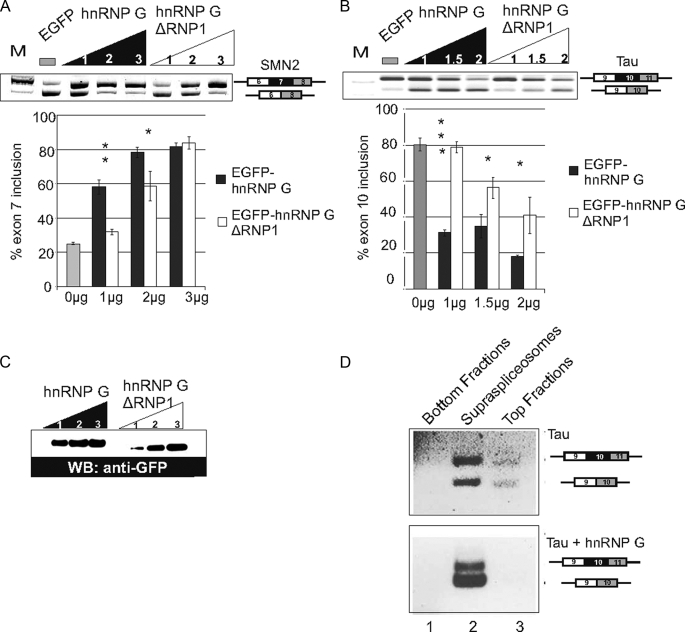
FIGURE 4.
hnRNP G influences splice site selection of reporter minigenes. An increasing amount of EGFP-hnRNP G and EGFP-hnRNP G-ΔRNP1 were cotransfected with the SMN2 (A) or tau minigene (B) in HEK293 cells. Alternative splicing of the minigene was determined by RT-PCR using primers specific for the minigenes. A representative ethidium bromide-stained gel of each experiment is shown. The statistical evaluation of at least three independent experiments is shown below the gels. Asterisks indicate p values from Student's t test comparing wild type and mutant hnRNP G: SMN2 minigene, 1 μg = 0.008; 2 μg = 0.03; tau minigene, 1 μg = 0.0002; 1.5 μg = 0.04; 2 μg = 0.03. C, representative Western blot (WB) showing the expression of the transfected EGFP-tagged proteins. D, changes in splice site selection of tau minigene by hnRNP G occur in supraspliceosomes. Nuclear supernatants enriched for supraspliceosomes were prepared from HEK293 cells transiently transfected with either tau minigene construct, or tau and EGFP-hnRNP G minigene constructs, and fractionated on glycerol gradients. Fractions 1–4 (bottom fractions), 8–11 (Supraspliceosomes), and 17–20 (top fractions) were pooled and were analyzed by RT-PCR using tau-specific primers. Upper panel, transfection with tau; lower panel, transfection with tau and EGFP-hnRNP G. The identity of the splice variants is schematically shown on the right.
hnRNP G influences the alternative splicing pattern of tau reporter minigene and is found in the supraspliceosome. We therefore asked whether alternatively spliced tau transcripts, expressed in the presence and absence of exogenous hnRNP G, are packaged in supraspliceosomes. We transiently transfected HEK293 cells with the tau minigene in the presence and absence of EGFP-hnRNP G. We next prepared supraspliceosomes from both, and we analyzed the splicing pattern of exon 10 of tau by semi-quantitative RT-PCR analyses. For the RT-PCR experiments the bottom fractions 1–4, the supraspliceosomal fractions 8–11, and the top fractions 17–20 were pooled and analyzed together. In supraspliceosomes prepared from cells that are transfected with the tau reporter minigene in the absence of exogenous hnRNP G, predominantly the exon 10 inclusion form can be detected. In contrast, when hnRNP G is cotransfected with the reporter into the cells, exon 10 skipping is the predominant product (Fig. 4D). The skipping of exon 10 has also been observed in our transfection studies (Fig. 4B). In both cases the alternatively spliced forms are found packaged in supraspliceosomes. Taken together, these studies indicate that hnRNP G influences splice site selection in a concentration-dependent manner. The protein can influence splice site selection without its RRM, most likely because of sequestration and participates in splice site selection within supraspliceosomes.
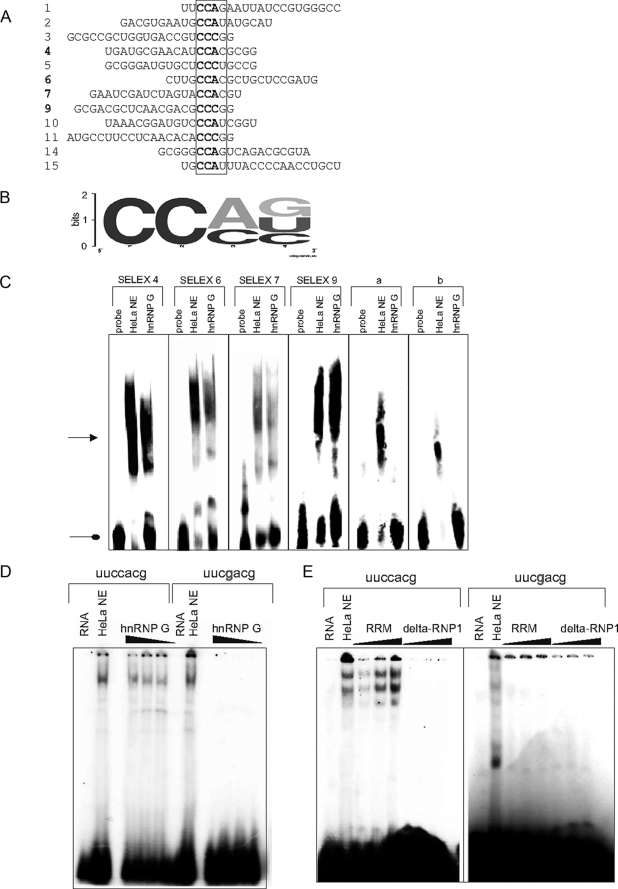
hnRNP G Binds to CCA/CCC-rich Sequences— hnRNP G contains a canonical RRM, and we therefore hypothesize that it influences pre-mRNA processing by directly binding to RNA. The RRM of hnRNP G has not been characterized yet. To determine whether it binds to specific sequences, we performed in vitro SELEX (systematic evolution of ligands by exponential enrichment) experiments. A pool of DNA was transcribed in vitro to generate an RNA pool. The initial pool contained random 20-mers flanked by a T7 promoter and a reverse primer. Recombinant His-tagged hnRNP G protein, generated from baculovirus, was incubated with this RNA pool. After binding, the protein was digested with proteinase K, and the bound RNA was purified by phenol/chloroform precipitation and amplified by RT-PCR. The resulting cDNA was gel-purified and in vitro transcribed, and another round of SELEX was performed. After six rounds of SELEX, the DNA was subcloned and sequenced (Fig. 5A). The majority of SELEX sequences contained a CCA or CCC motif and can collectively be described by the matrix shown in Fig. 5B. We found no evidence that these CCA/CCC motifs are located in the loop of a hairpin structure, as described previously for RBMY (22). To analyze whether hnRNP G binds to these sequences, we performed RNA gel retardation experiments. The final SELEX sequences fell into four related classes, and we transcribed representatives of each class in vitro and performed gel retardation assays with recombinant hnRNP G. We tested three sequences with a CCA motif (SELEX 4, 6, and 7), one with a CCC motif (SELEX 9) and two without CCA/CCC motifs that were identified in earlier SELEX rounds (Fig. 5C, sequences a and b). As shown in Fig. 5C, we observed a shift with SELEX sequences 4, 6, 7, and 9 but not with sequences a and b. To confirm these findings, we next analyzed the binding of hnRNP G to 7-mer RNA oligonucleotides that contain the CCA-binding motif. As shown in Fig. 5D, recombinant hnRNP G causes a shift in an oligonucleotide that contains the CCA repeat, but this retardation in mobility is not observed when the CCA is changed to CGA. It was surprising that an hnRNP G variant without the RRM is able to change splice site selection (Fig. 4, A and B). To investigate whether the C terminus is able to bind to the same RNA, we performed gel retardation assays using the 7-mer probe containing the hnRNP G-binding motif. We observed a mobility shift when using protein corresponding to the RRM but not when using the C terminus (Fig. 5E, left). As with the full-length protein, we did not observe a change in mobility when CCA motif was mutated in the 7-mer (Fig. 5E, right). Together, these data indicate that hnRNP G binds in vitro to CCA/CCC motifs via its RRM.
FIGURE 5.
hnRNP G binds to CCA-rich region. A, SELEX motifs for hnRNP G. The sequences are flanked by the SELEX primers used for amplification of the DNA pool, which are not indicated in the figures. Numbers marked in boldface are the sequences used for gel retardation assay. B, sequence logo describing the common sequence motif in the SELEX winner sequences. The logo was created using WebLogo. C, binding to SELEX sequences to recombinant hnRNP G in gel retardation assay, and 1 μg of nuclear extract (NE) and recombinant hnRNP G were incubated with a number of probes, whose sequences were found by SELEX and analyzed by native gel electrophoresis. The pointed arrow indicates the RNA-protein complexes, the round arrow the free probes. Sequences a, AAGGCATGAGGAAGCTGCC; b, AAGCCTGCAGCGGACGCTGT. D, binding of 7-mer RNA oligonucleotides to recombinant hnRNP G. On the left side a UUCCACG 7-mer containing the hnRNP G-binding site was used in a gel retardation assay. On the right side, a 7-mer with a mutated hnRNP G-binding site (A → g) was used. The protein concentration was 15, 30, and 45 pmol, and the RNA concentration was 2 pm. E, binding of the RRM and C terminus of hnRNP G to 7-mer oligonucleotides. The left side shows the a gel mobility shift assay with a 7-mer containing the hnRNP G-binding site. On the right side an oligonucleotide with a mutated binding site was employed.
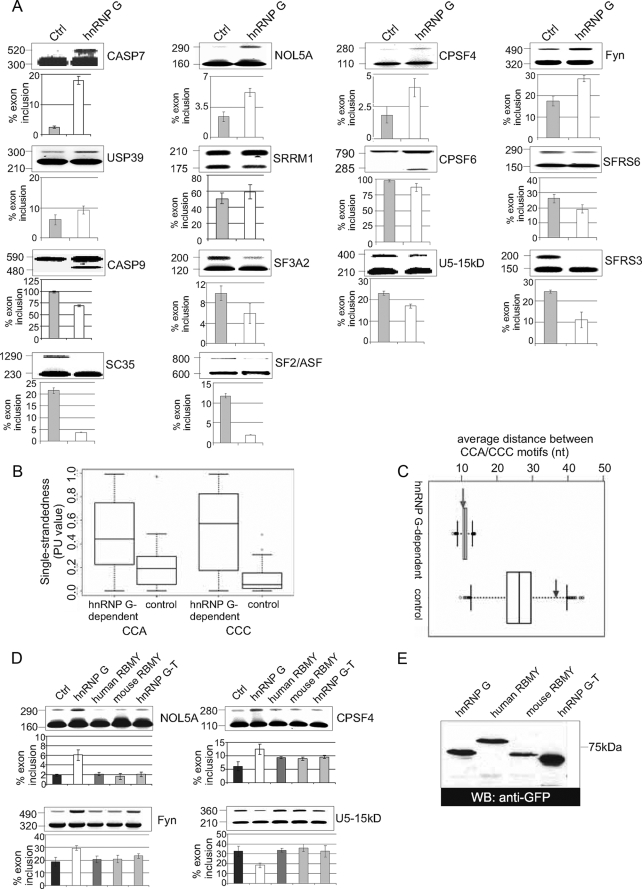
hnRNP G Influences Splice Site Selection of Exons Containing CCA-rich Regions—To test whether hnRNP G directly influences alternative splice site selection, we performed analysis using splice site-sensitive DNA arrays. These arrays detect splicing events using a combination of exon junction and exon body probes and detect 942 splicing events, as described previously (40, 41). We compared HEK293 cells where EGFP-hnRNP G is overexpressed with cells expressing just EGFP. For the array analysis, we did not choose a small interfering RNA knockdown approach because there are several hnRNP G isogenes with possibly redundant functions. RNA was isolated 18 h after transfection. The alternative splicing events were validated by semiquantitative RT-PCR using specific primers in the flanking regions. As shown in Fig. 6A, overexpression of hnRNP G can cause both exon inclusion and skipping.
FIGURE 6.
hnRNP G overexpression changes splice site selection in human genes. A, RNA from HEK293 cells transfected with pEGFP-C2 as control (Ctrl), HEK293 cells overexpressing EGFP-hnRNP G were amplified by RT-PCR using specific primers. Below each gel is the quantification from at least three experiments that expresses the ratio of exon inclusion to exon skipping. The p values of the Student's test were as follows: CASP7, 0.006; NOL5A, 0.004; CPSF4, 0.0169; Fyn, 0.002; USP39, 0.009; SRRM1, 0.082; CPSF6, 0.058; SFRS6, 0.047; CASP9, 0.006; SF3A2, 0.0581; U5-15kD, 0.027; SFRS3, 0.0621; SC35, 0.0132; SF2/ASF, 0.004. B, box plots showing the distribution of the single-strandedness of CCA and CCC motifs in hnRNP G-regulated and control exons. The sequences of the exons are shown in supplemental Fig. 3. PU values (29) were used to measure the single-strandedness. Each box represents 50% of the distribution (upper and lower end of the box are the first and third quartile, and the horizontal line is the median). Whiskers indicate the location of the smallest (largest) value that is located at most 1.5 times the interquartile range (third to first quartile) below (above) the box. C, box plots showing the distribution of the distances between CCA/CCC motifs under random expectation in hnRNP G-regulated and control exons. The expected distribution controls for differences in the overall motif count. The left and right arrows indicate the observed average distance for hnRNP G-regulated and control exons, respectively. The figure clearly shows that the distance in hnRNP G regulated exons is biased toward the lower end of the random distribution (∼75% of the random distribution has larger average distances), whereas the distance in controls is strongly biased toward the upper end (98% of the random distribution has smaller distances). Note that a direct comparison of the distances between hnRNP G-regulated and control exons is not valid, as this strongly depends on the total number of CCA/CCC motifs (the higher the motif number, the smaller the expected distance). D, comparison between hnRNP G, RBMY, and hnRNP G-T. 1 μg of cDNA constructs expressing hnRNP G, RBMY, or hnRNP G-T were transfected into HEK293 cells, and RNA was analyzed by RT-PCR. E, expression levels of the hnRNP G expression constructs used. 1 μg of a construct expression EGFP-tagged hnRNP G, hRBMY, mRBMY, and hnRNP G-T was transfected into HEK293 cells. Lysates representing 1/50 of the total cells were analyzed by Western blot (WB) using an antiserum against GFP.
We next asked whether there are sequential features in the regulated exons that reflect the in vitro binding of hnRNP G to RNA. We used the alternative exon sequences from Casp7, NOL5A, CPSF4, Fyn, USP39, SRRM1, SFRS6, Casp9, SFRS3, and SF2/ASF as examples for exons that are regulated by hnRNP G. As control data, we used the exons that did not show an effect of splicing by hnRNP G, as determined by RT-PCR. The sequences of the 23 exons used are shown in supplemental Fig. 3.
We first analyzed the frequency of CCA and CCC, the sequences that bind hnRNP G in vitro (Fig. 5). There are a total of 1631 triplets in the hnRNP G-dependent sequences, from which 61 (3.7%) are CCA and 50 (3.1%) are CCC. In the control set, of the 932 triplets only 15 (1.6%) are CCA and only 16 (1.7%) are CCC. Thus, hnRNP G-dependent sequences have ∼2-fold enrichment in hnRNP G-binding motifs, which is statistically significant (Fisher's exact test, p = 0.02 for CCA and p = 0.04 for CCC). To further test whether CCA/CCC triplets are enriched in hnRNP G-dependent exons, we compared the frequency of all permutations of CCA (ACC and CAC) in these exon sets and found no significant enrichment. Interestingly, CAA motifs, which are binding sites for RBMY, as well as its permutations (AAC and ACA) are more frequent in the control exon set, thus showing that the hnRNP G-dependent exons are probably not regulated by RBMY and that hnRNP G and RBMY have different target exons.
Although hnRNP G-dependent exons are significantly enriched in CCA/CCC motifs, there are still 31 occurrences of these triplets in the control set. We recently showed that splicing regulatory elements are more frequently found in single-stranded regions in the pre-mRNA (36). We investigated the single strandedness of the CCA/CCC motifs by computing the probability that these motifs are completely unpaired (denoted as PU value), as described previously (36).
As shown in Fig. 6B, CCA/CCC motifs have a significantly higher single strandedness in the hnRNP G-dependent exons compared with the controls (CCA, median PU value 0.5 versus 0.25; Wilcoxon rank sum test, p = 0.038; CCC, median PU value 0.57 versus 0.05, p = 0.001). As highly double-stranded motifs are unlikely to be bound by hnRNP G, this would explain why the control exons are not regulated by hnRNP G despite having few CCA/CCC motifs.
As hnRNP G homomultimerizes (Fig. 2F), it can potentially bind RNA as a homodimer. Therefore, we compared the distance between the CCA/CCC motifs in the hnRNP G-regulated and control exons. Comparing the observed distance with the one expected from random CCA/CCC spacing, we found a strong bias for CCA/CCC motifs to be located close to each other in the hnRNP G-regulated exons. In contrast, in the controls, the CCA/CCC motifs that occur in these exons are strongly biased toward being located farther away from each other (Fig. 6C). This suggests that hnRNP G frequently regulates exon inclusion by binding as a homodimer to RNA. In summary, exons that are regulated by hnRNP G contain significantly more CCA/CCC motifs; these motifs are significantly more single-stranded and are located close to each other.
hnRNP G Regulates Different Exons than the Related Proteins RBMY and hnRNP G-T— hnRNP G is encoded in the X chromosome, and humans express a related Y-chromosome-encoded protein, RBMY, as well as the related testes-specific hnRNP G-T. Whereas RBMY binds RNA stem structures containing a C(A/U)CAA loop sequence, we found that hnRNP G binds to CCA/CCC motifs localized in a single-stranded conformation, suggesting that both proteins regulate different RNAs.
We therefore compared the ability of these proteins to change alternative exon usage of hnRNP G-regulated genes. As shown in Fig. 6D, we found several cases where the related hnRNP G proteins have no effect on exons responding to hnRNP G. The constructs expressed RBMY at a level that is comparable with hnRNP G. hnRNP G-T was expressed at a higher level. Even at this increased level, hnRNP G-T had no effect on these hnRNP G-responsive exons. Together, these data indicate the existence of sex-specific differences in pre-mRNA processing that depend on the ratios of hnRNP G and RBMY.
DISCUSSION
hnRNP G Is a Nuclear Protein That Functions in Splice Site Selection—Although hnRNP G has been identified about 15 years ago (15, 42), its molecular functions remained elusive. It was shown to be an autoantigen, glycosylated, associated with lampbrush chromosomes, and being able to bind RNA (15). Subsequent studies showed that hnRNP G binds in yeast and in pulldown experiments to the splicing factor Tra2-β1, and yeast-two-hybrid interactions suggested binding to RBMY, hnRNP G-T, and SRp30c (18). The binding properties suggested a role in splice site selection, and it was subsequently shown that hnRNP G modulates alternative splicing of tau exon 10, SMN2 exon 7, and the SK exon of α-tropomyosin(s) (19–21, 43). However, the mechanistic details of hnRNP G action were not clear.
To obtain better insight into its physiological role, we generated an antiserum specific for hnRNP G and demonstrated that the endogenous protein is localized in the nucleus. However, the diffuse staining pattern is different from other splicing factors that most frequently localize in nuclear speckles (44). The nuclear localization of hnRNP G could be caused in part by its binding to the nuclear proteins YT521-B and rSLM-1, -2, or SAM68, none of which are localized exclusively in speckles (38, 45). This nuclear localization suggests that hnRNP G could also function in aspects of pre-mRNA metabolism different from splice site selection, similar to other processing proteins such as hnRNP A1 or SF2/ASF (46, 47). We next used the antiserum to determine the expression levels of hnRNP G in various tissues and found that although it can be detected in all tissues, there are different expression levels. Most striking are the low expression levels in heart and muscle that are reflected by a lower mRNA expression detected by Northern blot analysis. Although both analysis methods used the same amount of protein and RNA in the individual tissues, they cannot identify strong concentrations of hnRNP G in certain tissue sections. However, by assuming a homogeneous expression within a tissue, the data suggest that the ratio between hnRNP G and other splicing factors varies among tissues, which could contribute to tissue-specific splice site selection. In contrast, in humans, a strong expression of hnRNP G mRNA was seen in heart and muscle, and it remains to be determined whether this difference to the rat system used here reflects species differences or the collection of tissues (21).
To further corroborate the interaction between hnRNP G and various nuclear proteins, we performed coimmunoprecipitations. We confirmed the binding of hnRNP G to itself, to Tra2-β1, scaffold attachment factor B (SAF-B), YT521-B, and Clk2. It was previously shown that hnRNP G coimmunoprecipitates with rSLM-1 (48). We did not see coimmunoprecipitation with SRp30c, which was suggested by yeast two-hybrid interactions (18). As these experiments were performed in the presence of benzonase, these interactions are not nucleic acid-mediated, which is also supported by the finding that in yeast these interactions take place when the RRM of hnRNP G is deleted.
We then tested whether these protein interactions are functional and determined the presence of endogenous hnRNP G in the supraspliceosome (7). hnRNP G was found in the peak fractions of the supraspliceosome. Using the combination of native gel fractionation, Western blotting, extraction from the gel, and visualization by EM, we could show that hnRNP G is physically associated with the supraspliceosomes. This finding was further substantiated by co-IP experiments using anti-Sm antibodies, which showed that hnRNP G is specifically associated with supraspliceosomes. Finally, we found that usage of tau exon 10 in the supraspliceosome is increased when hnRNP G is overexpressed. Together, these data suggest that hnRNP G is directly involved in splice site selection by direct interaction within the supraspliceosome.
hnRNP G Can Change Splice Site Selection without Its RRM— Based on earlier experiments using the SMN2 system, it was proposed that hnRNP G acts by unspecific binding to mRNA while bound to Tra2-β1 (19). Tra2-β1 is specifically bound to the RNA of an exonic enhancer in the regulated exon 7 of the SMN2-pre-mRNA (30). However, the Drosophila TRA example shows that a molecule can influence an RNA-regulatory complex without directly binding to RNA. Tra does not contain an RNA-binding domain and does not bind RNA. Tra binds to Tra2, which contains a canonical RRM. In female flies, only the Tra-Tra2 complex regulates alternative splicing of several target genes, which ultimately determines the sex of the fly (49). We therefore investigated whether the RNA recognition motif is necessary for hnRNP G to exert its influence on splice site selection. We used SMN2 exon 7 and tau exon 10 as model systems, because both exons are regulated by a central Tra2-β1-dependent enhancer (30, 41, 50) and because Tra2-β1 strongly interacts with hnRNP G (Fig. 2B) (18). Using these test systems, we compared the influence of hnRNP G possessing or lacking an RRM on splice site selection. To our surprise, both proteins influence splice site selection. The influence is indistinguishable at higher hnRNP G concentrations, indicating that the action of hnRNP G does not require direct binding to RNA. It is not clear how hnRNP G can influence splice site selection without RNA binding. We tested the hnRNP G protein without an RRM for its ability to bind to RNA in gel shift experiments and could not detect any binding (Fig. 5E). This suggests that hnRNP G binds RNA predominantly with its RRM. It is possible that hnRNP G changes splice site selection independent from RNA binding through sequestration. In this model, hnRNP G would bind to an interacting protein, e.g. Tra2-β1, which would remove Tra2-β1 from its site of action. The predicted effect would be antagonistic, i.e. an inhibition of Tra2-β1 action. Such an antagonistic effect has been observed for Tra2-β1 pre-mRNA system (28) where hnRNP G without an RRM antagonizes exon II inclusion and for the α-tropomyosin system where full-length hnRNP G antagonizes Tra2-β1 in SK exon inclusion. However, this model cannot explain why hnRNP G lacking an RRM promotes exon inclusion in the SMN2 system. The competition with a factor inhibiting Tra2-β1 activity could explain this effect. Both Tra2-β1 and hnRNP G bind to the dual-specificity kinase Clk2 (28) (Fig. 2G). Phosphorylation evoked by Clk2 inhibits Tra2-β1 action, most likely because it disrupts the formation of exonic enhancer complexes on the pre-mRNA (41). In this model, hnRNP G would promote Tra2-β1-dependent exon inclusion by competing with Tra2-β1 for binding to Clk2. Finally, it is possible that binding to hnRNP G would cause a structural change in Tra2-β1, i.e. lock its RS domains in a certain conformation, which could promote exon inclusion. Although the exact mechanism needs to be determined, it is clear that similar to Drosophila TRA, hnRNP G can change splice site selection independent of an RRM.
RNA Binding Properties of hnRNP G—To analyze the role of the RRM for hnRNP G function, we determined its preferred RNA-binding motifs by in vitro SELEX. Recently, for the hnRNP G paralogue RBMY, a CA/UCAA loop was identified as binding motif. RBMY binds to this loop only when it is preceded by an extended stem structure (22). The inspection of our SELEX winner sequences and alternative exons regulated by hnRNP G showed the presence of a CCA/CCC motif that was most likely in single-stranded conformation, but it is clearly not flanked by a stable stem structure. The direct binding to CCA-rich sequences could explain previous data that showed hnRNP G regulating a patient-derived dystrophin pseudo exon (21), because this exon contains several CCA clusters. To test the hypothesis that hnRNP G promotes exons with a CCA signature, we performed array experiments to detect the influence of hnRNP G overexpression on splice site selection. hnRNP G-dependent exons were shown to contain significantly more CCA signatures than control exons, suggesting that hnRNP G can directly regulate such exons. Therefore, hnRNP G can influence alternative splicing also directly by binding CCA-rich sequences in single-stranded conformation via its RRM.
Finally, our work shows that the X chromosome-encoded hnRNP G, also called RBMX, has different RNA binding than the Y chromosome-encoded paralogue RBMY. Although the primary structure of their RNA-binding motifs is quite similar, their RNA binding abilities are different. NMR structures show that Lys-84 in RBMY interacts with both adenines in the CAA motif (22). This amino acid is a Thr in hnRNP G. As all other residues that contact the CAA motif are identical between hnRNP G and RBMY (22), it is possible that this Lys-Thr exchange triggers the different binding properties. The β2-β3 loop of RBMY was shown to be responsible for the shape-specific interaction of the protein with the RNA stem. hnRNP G differs from RBMY in this β2-β3 loop, and because of these changes hnRNP G cannot recognize the stem (22). These findings agree with our SELEX and functional splicing data, as we detected no preference for stem-loop structures.
We then compared directly hnRNP G and RBMY in transfection assays and found that they differ in their ability to regulate certain pre-mRNAs, which can be explained by their different binding properties. It is therefore possible that both proteins are part of a splicing-dependent mechanism that determines the sexual phenotype, which is well understood in Drosophila (49) but has been only a hypothesis in vertebrate systems (51).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM079549 (to R. S. and J. S.), P20 RR020171, and R21HD056195-01 (to S. S.). This work was also supported by the German Research Council, the Families of SMA, EURASNET, the Bundesministerium für Bildung und Forschung, and a grant from the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (to J. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: hnRNP, heterogeneous nuclear ribonucleoprotein; RRM, RNA-recognition motif; GFP, green fluorescent protein; EGFP, enhanced green fluorescent protein; RT, reverse transcription; IP, immunoprecipitated; co-IP, co-immunoprecipitated; SMN2, survival of motoneuron 2.
References
- 1.Moore, M. J. (2005) Science 309 1514-1518 [DOI] [PubMed] [Google Scholar]
- 2.Stamm, S., Ben-Ari, S., Rafalska, I., Tang, Y., Zhang, Z., Toiber, D., Thanaraj, T. A., and Soreq, H. (2005) Gene (Amst.) 344 1-20 [DOI] [PubMed] [Google Scholar]
- 3.Maniatis, T., and Reed, R. (2002) Nature 416 499-506 [DOI] [PubMed] [Google Scholar]
- 4.Stamm, S. (2008) J. Biol. Chem. 283 1223-1227 [DOI] [PubMed] [Google Scholar]
- 5.Sperling, J., Azubel, M., and Sperling, R. (2008) Structure (Lond.) 16 1605-1615 [DOI] [PubMed] [Google Scholar]
- 6.Muller, S., Wolpensinger, B., Angenitzki, M., Engel, A., Sperling, J., and Sperling, R. (1998) J. Mol. Biol. 283 383-394 [DOI] [PubMed] [Google Scholar]
- 7.Azubel, M., Habib, N., Sperling, R., and Sperling, J. (2006) J. Mol. Biol. 356 955-966 [DOI] [PubMed] [Google Scholar]
- 8.Azubel, M., Wolf, S. G., Sperling, J., and Sperling, R. (2004) Mol. Cell 15 833-839 [DOI] [PubMed] [Google Scholar]
- 9.Miriami, E., Angenitzki, M., Sperling, R., and Sperling, J. (1995) J. Mol. Biol. 246 254-263 [DOI] [PubMed] [Google Scholar]
- 10.Spann, P., Feinerman, M., Sperling, J., and Sperling, R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 466-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen-Krausz, S., Sperling, R., and Sperling, J. (2007) J. Mol. Biol. 368 319-327 [DOI] [PubMed] [Google Scholar]
- 12.Yitzhaki, S., Miriami, E., Sperling, R., and Sperling, J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8830-8835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raitskin, O., Cho, D. S., Sperling, J., Nishikura, K., and Sperling, R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6571-6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raitskin, O., Angenitzki, M., Sperling, J., and Sperling, R. (2002) J. Struct. Biol. 140 123-130 [DOI] [PubMed] [Google Scholar]
- 15.Soulard, M., Della Valle, V., Siomi, M. C., Pinol-Roma, S., Codogno, P., Bauvy, C., Bellini, M., Lacroix, J. C., Monod, G., Dreyfuss, G., and Larsen, C.-J. (1993) Nucleic Acids Res. 21 4210-4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin, K. H., Kang, M. K., Kim, R. H., Christensen, R., and Park, N. H. (2006) Clin. Cancer Res. 12 3222-3228 [DOI] [PubMed] [Google Scholar]
- 17.Elliott, D. J. (2004) Int. J. Androl. 27 328-334 [DOI] [PubMed] [Google Scholar]
- 18.Venables, J. P., Elliott, D. J., Makarova, O. V., Makarov, E. M., Cooke, H. J., and Eperon, I. C. (2000) Hum. Mol. Genet. 9 685-694 [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, Y., and Wirth, B. (2002) Hum. Mol. Genet. 11 2037-2049 [DOI] [PubMed] [Google Scholar]
- 20.Wang, J., Gao, Q.-S., Wang, Y., Lafyatis, R., Stamm, S., and Andreadis, A. (2004) J. Neurochem. 88 1078-1090 [DOI] [PubMed] [Google Scholar]
- 21.Nasim, M. T., Chernova, T. K., Chowdhury, H. M., Yue, B. G., and Eperon, I. C. (2003) Hum. Mol. Genet. 12 1337-1348 [DOI] [PubMed] [Google Scholar]
- 22.Skrisovska, L., Bourgeois, C. F., Stefl, R., Grellscheid, S. N., Kister, L., Wenter, P., Elliott, D. J., Stevenin, J., and Allain, F. H. (2007) EMBO Rep. 8 372-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 24.Stoss, O., Stoilov, P., Hartmann, A. M., Nayler, O., and Stamm, S. (1999) Brain Res. Brain Res. Protoc. 4 383-394 [DOI] [PubMed] [Google Scholar]
- 25.Beil, B., Screaton, G., and Stamm, S. (1997) DNA Cell Biol. 16 679-690 [DOI] [PubMed] [Google Scholar]
- 26.Fields, S., and Song, O. (1989) Nature 340 245-247 [DOI] [PubMed] [Google Scholar]
- 27.Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., and Stamm, S. (1999) Mol. Biol. Cell 10 3909-3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoilov, P., Daoud, R., Nayler, O., and Stamm, S. (2004) Hum. Mol. Genet. 13 509-524 [DOI] [PubMed] [Google Scholar]
- 29.Stamm, S., Casper, D., Hanson, V., and Helfman, D. M. (1999) Mol. Brain Res. 64 108-118 [DOI] [PubMed] [Google Scholar]
- 30.Hofmann, Y., Lorson, C. L., Stamm, S., Androphy, E. J., and Wirth, B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9618-9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorson, C. L., Hahnen, E., Androphy, E. J., and Wirth, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6307-6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartmann, A. M., Rujescu, D., Giannakouros, T., Nikolakaki, E., Goedert, M., Mandelkow, E. M., Gao, Q. S., Andreadis, A., and Stamm, S. (2001) Mol. Cell. Neurosci. 18 80-90 [DOI] [PubMed] [Google Scholar]
- 33.Sperling, R., Sperling, J., Levine, A. D., Spann, P., Stark, G. R., and Kornberg, R. D. (1985) Mol. Cell. Biol. 5 569-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miriami, E., Sperling, J., and Sperling, R. (1994) Nucleic Acids Res. 22 3084-3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadano, D., Aoki, C., Yoshinaka, T., Irie, S., and Sato, T. A. (2001) Biochemistry 40 15184-15193 [DOI] [PubMed] [Google Scholar]
- 36.Hiller, M., Zhang, Z., Backofen, R., and Stamm, S. (2007) PLoS Genet. 3 2147-2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofacker, I. L. (2003) Nucleic Acids Res. 31 3429-3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nayler, O., Hartmann, A. M., and Stamm, S. (2000) J. Cell Biol. 150 949-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jurica, M. S., and Moore, M. J. (2003) Mol. Cell 12 5-14 [DOI] [PubMed] [Google Scholar]
- 40.Fehlbaum, P., Guihal, C., Bracco, L., and Cochet, O. (2005) Nucleic Acids Res. 33 e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novoyatleva, T., Heinrich, B., Tang, Y., Benderska, N., Butchbach, M. E., Lorson, C. L., Lorson, M. A., Ben-Dov, C., Fehlbaum, P., Bracco, L., Burghes, A. H., Bollen, M., and Stamm, S. (2008) Hum. Mol. Genet. 17 52-70 [DOI] [PubMed] [Google Scholar]
- 42.Soulard, M., Barque, J. P., Della Valle, V., Hernandez-Verdun, D., Masson, C., Danon, F., and Larsen, C.-J. (1991) Exp. Cell Res. 193 59-71 [DOI] [PubMed] [Google Scholar]
- 43.Sergeant, K. A., Bourgeois, C. F., Dalgliesh, C., Venables, J. P., Stevenin, J., and Elliott, D. J. (2007) J. Cell Sci. 120 309-319 [DOI] [PubMed] [Google Scholar]
- 44.Lamond, A. I., and Spector, D. L. (2003) Nat. Rev. Mol. Cell Biol. 4 605-612 [DOI] [PubMed] [Google Scholar]
- 45.Chen, T., Boisvert, F.-M., Bazett-Jones, D. P., and Richard, S. (1999) Mol. Biol. Cell 10 3015-3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guil, S., Long, J. C., and Caceres, J. F. (2006) Mol. Cell. Biol. 26 5744-5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaustein, M., Pelisch, F., Tanos, T., Munoz, M. J., Wengier, D., Quadrana, L., Sanford, J. R., Muschietti, J. P., Kornblihtt, A. R., Caceres, J. F., Coso, O. A., and Srebrow, A. (2005) Nat. Struct. Mol. Biol. 12 1037-1044 [DOI] [PubMed] [Google Scholar]
- 48.Stoss, O., Novoyatleva, T., Gencheva, M., Olbrich, M., Benderska, N., and Stamm, S. (2004) Mol. Cell. Neurosci. 27 8-21 [DOI] [PubMed] [Google Scholar]
- 49.Baker, B. S. (1989) Nature 340 521-524 [DOI] [PubMed] [Google Scholar]
- 50.Jiang, Z., Tang, H., Havlioglu, N., Zhang, X., Stamm, S., Yan, R., and Wu, J. Y. (2003) J. Biol. Chem. 278 18997-19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lalli, E., Ohe, K., Latorre, E., Bianchi, M. E., and Sassone-Corsi, P. (2003) J. Cell Sci. 116 441-445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.