Abstract
The RAD51 protein is a central player in homologous recombinational repair. The RAD51B protein is one of five RAD51 paralogs that function in the homologous recombinational repair pathway in higher eukaryotes. In the present study, we found that the human EVL (Ena/Vasp-like) protein, which is suggested to be involved in actin-remodeling processes, unexpectedly binds to the RAD51 and RAD51B proteins and stimulates the RAD51-mediated homologous pairing and strand exchange. The EVL knockdown cells impaired RAD51 assembly onto damaged DNA after ionizing radiation or mitomycin C treatment. The EVL protein alone promotes single-stranded DNA annealing, and the recombination activities of the EVL protein are further enhanced by the RAD51B protein. The expression of the EVL protein is not ubiquitous, but it is significantly expressed in breast cancer-derived MCF7 cells. These results suggest that the EVL protein is a novel recombination factor that may be required for repairing specific DNA lesions, and that may cause tumor malignancy by its inappropriate expression.
Chromosomal DNA double strand breaks (DSBs)2 are potential inducers of chromosomal aberrations and tumorigenesis, and they are accurately repaired by the homologous recombinational repair (HRR) pathway, without base substitutions, deletions, and insertions (1–3). In the HRR pathway (4, 5), single-stranded DNA (ssDNA) tails are produced at the DSB sites. The RAD51 protein, a eukaryotic homologue of the bacterial RecA protein, binds to the ssDNA tail and forms a helical nucleoprotein filament. The RAD51-ssDNA filament then binds to the intact double-stranded DNA (dsDNA) to form a three-component complex, containing ssDNA, dsDNA, and the RAD51 protein. In this three-component complex, the RAD51 protein promotes recombination reactions, such as homologous pairing and strand exchange (6–9).
The RAD51 protein requires auxiliary proteins to promote the homologous pairing and strand exchange reactions efficiently in cells (10–12). In humans, the RAD52, RAD54, and RAD54B proteins directly interact with the RAD51 protein (13–17) and stimulate the RAD51-mediated homologous pairing and/or strand exchange reactions in vitro (18–21). The human RAD51AP1 protein, which directly binds to the RAD51 protein (22), was also found to stimulate RAD51-mediated homologous pairing in vitro (23, 24). The BRCA2 protein contains ssDNA-binding, dsDNA-binding, and RAD51-binding motifs (25–33), and the Ustilago maydis BRCA2 ortholog, Brh2, reportedly stimulated RAD51-mediated strand exchange (34, 35). Most of these RAD51-interacting factors are known to be required for efficient RAD51 assembly onto DSB sites in cells treated with ionizing radiation (10–12).
The RAD51B (RAD51L1, Rec2) protein is a member of the RAD51 paralogs, which share about 20–30% amino acid sequence similarity with the RAD51 protein (36–38). RAD51B-deficient cells are hypersensitive to DSB-inducing agents, such as cisplatin, mitomycin C (MMC), and γ-rays, indicating that the RAD51B protein is involved in the HRR pathway (39–44). Genetic experiments revealed that RAD51B-deficient cells exhibited impaired RAD51 assembly onto DSB sites (39, 44), suggesting that the RAD51B protein functions in the early stage of the HRR pathway. Biochemical experiments also suggested that the RAD51B protein participates in the early to late stages of the HRR pathway (45–47).
In the present study, we found that the human EVL (Ena/Vasp-like) protein binds to the RAD51 and RAD51B proteins in a HeLa cell extract. The EVL protein is known to be involved in cytoplasmic actin remodeling (48) and is also overexpressed in breast cancer (49). Like the RAD51B knockdown cells, the EVL knockdown cells partially impaired RAD51 foci formation after DSB induction, suggesting that the EVL protein enhances RAD51 assembly onto DSB sites. The purified EVL protein preferentially bound to ssDNA and stimulated RAD51-mediated homologous pairing and strand exchange. The EVL protein also promoted the annealing of complementary strands. These recombination reactions that were stimulated or promoted by the EVL protein were further enhanced by the RAD51B protein. These results strongly suggested that the EVL protein is a novel factor that activates RAD51-mediated recombination reactions, probably with the RAD51B protein. We anticipate that, in addition to its involvement in cytoplasmic actin dynamics, the EVL protein may be required in homologous recombination for repairing specific DNA lesions, and it may cause tumor malignancy by inappropriate recombination enhanced by EVL overexpression in certain types of tumor cells.
EXPERIMENTAL PROCEDURES
Purification of the Human EVL Protein—The human EVL gene was isolated from a human cDNA pool (purchased from Clontech) by polymerase chain reaction and was cloned in the pET-15b vector (Novagen). In this construct, the hexahistidine tag sequence was fused at the N-terminal end of the gene. The EVL protein was expressed in the Escherichia coli JM109(DE3) strain, which also carried an expression vector for the minor tRNAs (Codon(+)RIL; Stratagene). The cells producing the EVL protein were resuspended in buffer containing 20 mm potassium phosphate (pH 8.5), 700 mm NaCl, 5 mm 2-mercaptoethanol, 10 mm imidazole, and 10% glycerol and were disrupted by sonication. The cell debris was removed by centrifugation for 20 min at 30,000 × g, and the lysate was mixed gently by the batch method with Ni2+-NTA-agarose beads (8 ml; Qiagen) at 4 °C for 1 h. The EVL-bound beads were washed with 80 ml of buffer containing 20 mm potassium phosphate (pH 8.5), 700 mm NaCl, 5 mm 2-mercaptoethanol, 30 mm imidazole, and 10% glycerol. The beads were then washed with 80 ml of buffer containing 20 mm potassium phosphate (pH 8.5), 700 mm NaCl, 5 mm 2-mercaptoethanol, 60 mm imidazole, and 10% glycerol and then were washed again with buffer containing 20 mm potassium phosphate (pH 8.5), 700 mm NaCl, 5 mm 2-mercaptoethanol, 30 mm imidazole, and 10% glycerol. The beads were then packed into an Econo-column (Bio-Rad) and were washed with 300 ml of buffer containing 20 mm potassium phosphate (pH 8.5), 100 mm NaCl, 5 mm 2-mercaptoethanol, 30 mm imidazole, and 10% glycerol. The His6-tagged EVL protein was eluted with a 30-column volume linear gradient of 30–300 mm imidazole, in 20 mm potassium phosphate (pH 8.5), 100 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol. The fractions containing the His6-tagged EVL protein were diluted with the same volume of buffer, containing 10 mm potassium phosphate (pH 8.5), 100 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol, and were mixed gently by the batch method with hydroxyapatite resin (10 ml; Bio-Rad) at 4 °C for 1 h. The resin was then washed with 80 ml of buffer, containing 20 mm potassium phosphate (pH 8.5), 100 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol, and was packed into an Econo-column (Bio-Rad). The resin was further washed with 300 ml of buffer, containing 20 mm potassium phosphate (pH 8.5), 225 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol, and then the His6-tagged EVL protein was eluted with a 30-column volume linear gradient of 225–1000 mm NaCl and 10–300 mm potassium phosphate (pH 8.5). The His6 tag was uncoupled from the EVL portion by a digestion with 5.5 units of thrombin protease (GE Healthcare) per mg of the His6-tagged EVL protein, and then the protein was immediately dialyzed against buffer containing 20 mm potassium phosphate (pH 7.5), 200 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol at 4 °C. After uncoupling of the His6 tag, the EVL protein was further purified by Superdex 200 gel filtration column (HiLoad 26/60 preparation grade; GE Healthcare) chromatography, followed by Mono S column chromatography. The peak fractions were diluted with the same volume of buffer, containing 20 mm potassium phosphate (pH 7.5), 5 mm 2-mercaptoethanol, and 10% glycerol, and were subjected to MonoS (GE Healthcare) column chromatography. The column was washed with 20 column volumes of buffer, containing 20 mm potassium phosphate (pH 7.5), 100 mm NaCl, 5 mm 2-mercaptoethanol, and 10% glycerol, and the EVL protein was eluted with a 12-column volume linear gradient of 100–600 mm NaCl. The purified EVL protein was dialyzed against buffer containing 20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 5 mm 2-mercaptoethanol, and 30% glycerol and was stored at -80 °C. The concentration of the purified EVL protein was determined by the Bradford method, using bovine serum albumin (BSA) as the standard.
Purification of the Human RAD51, RAD51B, DMC1, and RPA Proteins—The human RAD51 protein (50), human RAD51B protein (47), human DMC1 protein (51), and human RPA protein (52) were purified by the methods described previously.
DNA Substrates—Single-stranded φX174 viral (+) strand DNA and double-stranded φX174 replicative form I DNA were purchased from New England Biolabs. The linear dsDNA was prepared from the φX174 replicative form I DNA by PstI digestion. In the D-loop formation assay, superhelical dsDNA (pB5Sarray DNA) was prepared by a method to prevent irreversible denaturation of the dsDNA substrate by alkaline treatment of the cells harboring the plasmid DNA. The cells were gently lysed using Sarkosyl, as described previously (15). The pB5Sarray DNA contained 11 repeats of a sea urchin 5 S rRNA gene (207-bp fragment) within the pBlueScript II SK(+) vector. For the ssDNA annealing assay, the following high performance liquid chromatography (HPLC)-purified oligonucleotides were purchased from Nihon Gene Research Laboratory: 5′-GTC CCA GGC CAT TAC AGA TCA ATC CTG AGC ATG TTT ACC AAG CGC ATT G-3′ and 5′-CAA TGC GCT TGG TAA ACA TGC TCA GGA TTG ATC TGT AAT GGC CTG GGA C-3′. For the ssDNA substrate used in the D-loop assay, the following HPLC-purified oligonucleotide was purchased: 50-mer, 5′-GGA ATT CGG TAT TCC CAG GCG GTC TCC CAT CCA AGT ACT AAC CGA GCC CT-3′. The DNA sequences used in the strand exchange assay with the oligonucleotides are as follows: 63-mer, 5′-TCC TTT TGA TAA GAG GTC ATT TTT GCG GAT GGC TTA GAG CTT AAT TGC TGA ATC TGG TGC TGT-3′; 32-mer top strand, 5′-CCA TCC GCA AAA ATG ACC TCT TAT CAA AAG GA-3′; 32-mer bottom strand, 5′-TCC TTT TGA TAA GAG GTC ATT TTT GCG GAT GG-3′. The 5′-ends of the oligonucleotide (32-mer bottom strand) were labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP at 37 °C for 30 min. All DNA concentrations are expressed in moles of nucleotides.
Pulldown Assays with EVL-conjugated Beads—The EVL protein was covalently conjugated to Affi-Gel 10 beads (100 μl; Bio-Rad), according to the manufacturer's instructions. To block the remaining active ester sites, ethanolamine (pH 8.0) was added to a final concentration of 100 mm, and the resin was incubated at 4 °C overnight. The unbound proteins were removed by washing the Affi-Gel 10-EVL beads three times with 500 μl of binding buffer, which contained 20 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 5 mm 2-mercaptoethanol, 30% glycerol, and 0.05% Triton X-100. After washing the resin, the Affi-Gel 10-protein matrices were adjusted to 50% slurries and were stored at 4 °C.
For the RAD51B-binding and RAD51-binding assays, the EVL beads were incubated with a HeLa whole cell extract (2 mg of protein), and the beads were washed three times with 200 μl of washing buffer, containing 50 mm potassium phosphate (pH 7.5), 100 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, and protease inhibitor mixture (Nacalai Tesque). The proteins that copelleted with the EVL beads were separated into two portions for the RAD51 and RAD51B analyses and were fractionated by 12% SDS-PAGE. The RAD51 and RAD51B proteins were detected with the RAD51-specific and RAD51B-specific rabbit polyclonal antibodies, respectively.
Pulldown Assays with the Ni2+-NTA-Agarose Beads—The His6-tagged EVL protein (0.45 μm) was prepared and was mixed with the RAD51 or RAD51B protein (2 μm) in 20 mm HEPES buffer (pH 7.3), containing 95 mm NaCl, 0.1 mm EDTA, 0.045% Triton X-100, 1.8 mm ammonium sulfate, 2 mm 2-mercaptoethanol, 4.5 mm imidazole, and 29% glycerol. After a 30-min incubation at room temperature, 1.5 μl of the Ni2+-NTA-agarose beads were added to the reaction mixture, and the RAD51 or RAD51B protein bound to the His6-tagged EVL protein was captured by the beads. The beads were washed two times with 100 μl of washing buffer, containing 20 mm HEPES-NaOH (pH 7.3), 90 mm NaCl, 0.1 mm EDTA, 0.05% Triton X-100, 2 mm ammonium sulfate, 2 mm 2-mercaptoethanol, 5 mm imidazole, and 30% glycerol. The proteins that copelleted with the Ni2+-NTA beads were eluted with a buffer, containing 14 mm HEPES-NaOH (pH 7.3), 100 mm NaCl, 3.5 mm 2-mercaptoethanol, 300 mm imidazole, and 21% glycerol. The eluted fractions were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining.
Surface Plasmon Resonance Analysis—The surface plasmon resonance (SPR) signals were measured with a Biacore J instrument (GE Healthcare). Flow cells were maintained at 25 °C during the measurement, and the instrument was operated at the mid-flow rate (∼30 μl/min). The EVL protein was conjugated to the activated surface of CM5 sensor chips (GE Healthcare), using the standard amine coupling conditions recommended by the manufacturer. The level of the conjugated EVL protein was 3,860 resonance units. Reference SPR signals of the flow cell containing a sensor chip without the EVL protein were subtracted from those of the SPR signals of the flow cell containing the EVL-conjugated sensor chip. The running buffer was 20 mm HEPES-NaOH (pH 7.3), 150 mm NaCl, 2 mm ammonium sulfate, 5% glycerol, and 0.05% Triton X-100. Before each binding assay, the sensor chip was regenerated by a 1-min wash with 10 mm NaOH and two 1-min washes with 20 mm citrate buffer (pH 2.65), followed by a 25-min wash with the running buffer. For each binding assay, 1 μm analyte solution (i.e. the RAD51 protein, the RAD51B protein, the DMC1 protein, or the BSA solution) was injected for 5 min.
Assays for DNA Binding—The φX174 circular ssDNA (20 μm) or the linearized φX174 dsDNA (20 μm) was mixed with the EVL protein in 10 μl of a standard reaction solution, containing 36 mm HEPES-NaOH buffer (pH 7.5) with 1 mm dithiothreitol, 4 mm 2-mercaptoethanol, 80 mm NaCl, 1 mm MgCl2, 24% glycerol, and 0.1 mg/ml BSA. The reaction mixtures were incubated at 37 °C for 15 min and were then analyzed by 0.8% agarose gel electrophoresis in 1× TAE buffer (40 mm Tris acetate and 1 mm EDTA) at 3.3 V/cm for 2 h. The bands were visualized by ethidium bromide staining.
The D-loop Formation Assay—The indicated amount of the EVL protein was incubated in the presence or absence of the RAD51 protein (0.1 μm) at 37 °C for 5 min in the reaction buffer, containing 26 mm HEPES-NaOH (pH 7.5), 40 mm NaCl, 0.02 mm EDTA, 0.9 mm 2-mercaptoethanol, 5% glycerol, 1 mm MgCl2, 1 mm DTT, 2 mm AMPPNP, and 0.1 mg/ml BSA. After this incubation, the 32P-labeled 50-mer oligonucleotide (1 μm) was added, and the samples were further incubated at 37 °C for 5 min. The reactions were then initiated by the addition of the pB5Sarray superhelical dsDNA (60 μm) along with 9 mm MgCl2 and were continued at 37 °C for 30 min. The reactions were stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K and were further incubated at 37 °C for 15 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1× TAE buffer at 3.3 V/cm for 2.5 h. The gels were dried, exposed to an imaging plate, and visualized using an FLA-7000 imaging analyzer (Fujifilm).
Assays for Homologous Pairing with Oligonucleotides—For the homologous pairing assay with oligonucleotides, the RAD51 protein and the indicated amounts of the EVL protein were incubated with a 63-mer ssDNA (15 μm) in 10 μl of standard reaction buffer, containing 28 mm HEPES-NaOH (pH 7.5), 50 mm NaCl, 1 mm ATP, 1 mm DTT, 0.1 mg/ml BSA, 1 mm MgCl2, 1 mm CaCl2, 0.02 mm EDTA, 1.4 mm 2-mercaptoethanol, 20 mm creatine phosphate, 75 μg/ml creatine kinase, and 8% glycerol at 37 °C for 10 min. The strand exchange reaction was initiated by the addition of the 32-mer dsDNA (1.5 μm), which shared sequence homology with the 63-mer ssDNA. After a 30-min incubation at 37 °C, the reaction was stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K. The reaction mixtures were further incubated for 10 min at 37 °C. After 6-fold loading dye was added, the reaction mixtures were subjected to 15% polyacrylamide gel electrophoresis in 0.5× TBE buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA) at 10 V/cm for 5 h. The gels were dried, exposed to an imaging plate, and visualized using an FLA-7000 imaging analyzer.
Assay for Strand Exchange—The RAD51 protein (1 μm) and RPA (1 μm) were incubated with the indicated amounts of the EVL protein and/or the RAD51B protein at 30 °C for 10 min, in 8 μl of reaction buffer, containing 28 mm HEPES-NaOH (pH 7.5), 2.5 mm Tris-HCl (pH 7.5), 49 mm NaCl, 5 mm KCl, 0.03 mm EDTA, 1.1 mm 2-mercaptoethanol, 9% glycerol, 0.2 mm ammonium sulfate, 0.005% Triton X-100, 1 mm MgCl2, 1 mm CaCl2, 1.1 mm DTT, 1 mm ATP, 0.1 mg/ml BSA, 20 mm creatine phosphate, and 75 μg/ml creatine kinase. After this incubation, φX174 circular ssDNA (20 μm) was added, and the samples were further incubated at 30 °C for 10 min. The reactions were then initiated by the addition of φX174 linear dsDNA (20 μm) and were continued at 30 °C for 60 min. The reactions were stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K (Roche Applied Science) and were further incubated for 20 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1 × TAE buffer at 3.3 V/cm for 4 h. The products were visualized by SYBR Gold (Invitrogen) staining.
ssDNA Annealing Assay—The ssDNA oligonucleotide 49-mer (0.2 μm) was incubated with the indicated amounts of the EVL protein at 30 °C for 5 min in 9 μl of reaction buffer, containing 28 mm HEPES-NaOH (pH 7.5), 50 mm NaCl, 2 mm 2-mercaptoethanol, 12% glycerol, 0.1 mm MgCl2, 1 mm DTT, and 0.1 mg/ml BSA. For the experiment with the RAD51B protein, the EVL protein was preincubated with the indicated amounts of the RAD51B protein at 30 °C for 10 min, in 8 μl of reaction buffer, containing 28 mm HEPES-NaOH (pH 7.5), 38 mm NaCl, 1.4 mm 2-mercaptoethanol, 12% glycerol, 0.4 mm ammonium sulfate, 0.01% Triton X-100, 1 mm MgCl2, 1 mm DTT, 1 mm ATP, and 0.1 mg/ml BSA, before the addition of the ssDNA oligonucleotide. The reactions were initiated by the addition of 0.2 μm antisense 32P-labeled 49-mer oligonucleotide. At the times indicated, the reactions were quenched with an excess of the unlabeled 49-mer oligonucleotide. The DNA substrates and products were deproteinized by 0.2% SDS and 1.5 mg/ml proteinase K at 30 °C for 10 min. The products were fractionated by 10% PAGE in 0.5× TBE. The gels were dried, exposed to an imaging plate, and visualized using an FLA-7000 imaging analyzer.
EVL and RAD51B Knockdown Experiments—Rabbit polyclonal antibodies for the EVL and RAD51B proteins were obtained with the purified recombinant EVL and RAD51B proteins as antigens. The RAD51 antibody was purchased from Calbiochem (anti-RAD51 Ab-1 rabbit polyclonal antibody), and the Cdk2 antibody was purchased from Santa Cruz Biotechnology (Anti-Cdk2 M2 rabbit polyclonal antibody). The siRNA duplexes (Stealth™ RNAi) were designed by the BLOCK-iT™ RNAi Designer (Invitrogen) and were purchased from Invitrogen. The siRNA sequences of EVL and RAD51B are as follows: 5′-CCAGUAAGAAAUGGGUACCAAUCAA-3′ (for EVL) and 5′-GAGGUGUCCAUGAACUUCUAUGUAU-3′ (for RAD51B). The sequence of the control siRNA is 5′-CCAAGAAGUAAAUGGAACCUUGCAA-3′, in which the EVL siRNA sequence was shuffled. The cells were seeded in a 6-well plate. A 15-μl aliquot of siRNA (20 μm) was diluted with 185 μl of serum-free Dulbecco's modified Eagle's medium. In a separate tube, 2 μl of DharmaFECT 4 Transfection Reagent (Dharmacon) were diluted with 198 μl of serum-free Dulbecco's modified Eagle's medium, and the mixture was incubated at room temperature for 5 min. The diluted DharmaFECT 4 was combined with the diluted siRNA duplex, and the mixture was incubated at room temperature for 20 min. A 2-ml aliquot of Dulbecco's modified Eagle's medium was then added to the DharmaFECT 4-siRNA complex. This mixture was applied to the cells, which were then incubated at 37 °C for 48 h.
Western Blotting—The cells were harvested and were resuspended in lysis buffer, containing 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 1.7 μg/ml aprotinin, 50 nm cantharidin, 1 mm phenylmethylsulfonyl fluoride, and 10 nm microcystine. After a 30-min incubation on ice, the cell lysates were sonicated and were centrifuged at 20,000 × g for 5 min at 4 °C. The lysates (30 μg of protein/lane) were separated by 12% SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. The membrane was probed with anti-EVL rabbit polyclonal antibody, anti-RAD51B rabbit polyclonal antibody, or anti-Cdk2 rabbit polyclonal antibody and was developed using a horseradish peroxidase-conjugated anti-rabbit IgG antibody and the ECL Western blotting detection reagent (GE Healthcare).
Immunostaining—MCF7 cells were grown on glass coverslips and were either nontreated or irradiated with 8 or 20 grays, using the 137Cs source in the Facilities for Biological Research, Department of Nuclear Engineering and Management, Graduate School of Engineering, University of Tokyo. At 2 h after irradiation, the cells were fixed in 4% paraformaldehyde for 10 min, washed with PBS, and permeabilized with 0.5% Triton X-100 and 0.1% SDS in PBS for 5 min. The coverslips were incubated for 1 h at 37 °C with an anti-RAD51 antibody diluted with blocking buffer (1% BSA in PBS) and were then washed again. The RAD51 foci were visualized by an incubation with a fluorescein isothiocyanate-conjugated secondary antibody diluted in blocking buffer for 30 min at 37 °C. The nuclei of the cells were stained with 4′,6′-diamidino-2-phenylindole.
For MMC treatment, the cells were incubated in medium containing 0.8 μg/ml MMC (KYOWA) for 1 h at 37 °C. The cells were then washed with PBS, incubated for 24 h at 37 °C, and immunostained as described above.
Sensitivity to MMC—MCF7 cells were treated with MMC in suspension for 1 h and were washed three times with PBS. The cells were then plated at a density of 2 × 103 cells/60-mm dish. After 12 days of culture, colonies were counted.
RESULTS
The EVL Protein Is Involved in RAD51 Assembly—To identify novel HRR factors, we screened a human cDNA library by the two-hybrid method, with the human RAD51 protein and/or the RAD51 paralogs as bait. We obtained 73 positive clones as putative RAD51B-interacting proteins and found that 30 clones among them contained EVL gene fragments. Six clones encoding the RAD51C protein, which is a known RAD51B-interacting protein, were also found among these positive clones, suggesting that the screening was properly performed.
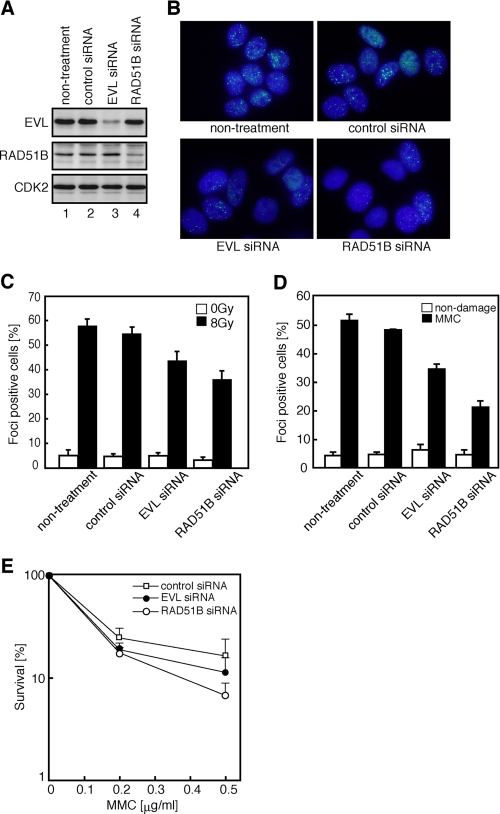
Since RAD51B is known to be required for the RAD51 assembly onto DSB sites after exposure to ionizing radiation, we then tested whether the EVL protein functions in RAD51 assembly in vivo. To do so, we performed RNA interference experiments. A treatment with a short interfering RNA (siRNA) designed to target the EVL mRNA caused about a 90% reduction in the EVL level in MCF7 cells, as compared with the cells treated with a control siRNA (Fig. 1A). The siRNA designed to target the RAD51B mRNA caused about an 80% reduction in the RAD51B level, without influencing the EVL level (Fig. 1A).
FIGURE 1.
Analyses of the EVL and RAD51B knockdown cells. A, the EVL, RAD51B, and CDK2 proteins in the siRNA-treated MCF7 cells. The top, middle, and bottom panels indicate the expression levels of the EVL, RAD51B, and CDK2 proteins, respectively. The proteins were detected by Western blotting. Lanes 1 and 2, control experiments without siRNA and with a control siRNA, respectively. Lanes 3 and 4, experiments with EVL siRNA and RAD51B siRNA, respectively. B, RAD51 foci formation. The MCF7 cells were treated with γ-ray irradiation (8 grays). The cells without treatment, with a control siRNA treatment, with an EVL siRNA treatment, or with a RAD51B siRNA treatment are presented. C, graphic representation of the RAD51 foci formation after γ-ray irradiation (8 grays). The cells containing more than 10 RAD51 foci were scored as positive and were plotted. The averages of three independent experiments are plotted with S.D. values. D, graphic representation of the RAD51 foci formation after MMC treatment. The cells containing more than 10 RAD51 foci were scored as positive and were plotted. The averages of three independent experiments are plotted with S.D. values. E, sensitivity to MMC. Open squares, closed circles, and open circles, experiments with a control siRNA, EVL siRNA, and RAD51B siRNA, respectively. The averages of three independent experiments are plotted with S.D. values.
The cells containing more than ten RAD51 foci after ionizing radiation treatment were scored as positive, because many cells containing fewer than 10 RAD51 foci were observed, even in the absence of DNA damage. The EVL knockdown MCF7 cells exhibited a clear reduction in RAD51 foci formation (Fig. 1, B and C, and supplemental Fig. 1). Consistent with previous studies (39, 44), the RAD51B knockdown MCF7 cells exhibited a comparable reduction in RAD51 foci formation on the DSB sites after ionizing radiation (Fig. 1, B and C). Reduced RAD51 foci formation was also observed in the EVL knockdown cells after treatment with an interstrand cross-linking agent, MMC (Fig. 1D). These results suggested that, like the RAD51B protein, the EVL protein functions in RAD51 assembly on DSB sites in the HRR pathway.
On the other hand, the EVL knockdown cells did not exhibit significant increase in sensitivity to MMC (Fig. 1E). The weak MMC sensitivity in the EVL knockdown cells, as compared with that in the RAD51B knockdown cells, may be explained by the presence of EVL paralogs, such as the MENA and VASP proteins, which may complement the functions of the EVL protein.
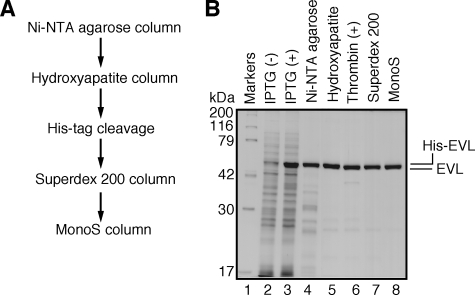
Interaction of the EVL Protein with the RAD51 and RAD51B Proteins—The human EVL protein was purified as a recombinant protein by a five-step procedure (Fig. 2A). In this procedure, the His6 tag was uncoupled with thrombin protease from the EVL portion, which then migrated slightly faster than the His6-tagged EVL protein upon SDS-polyacrylamide gel electrophoresis (Fig. 2B, lane 6).
FIGURE 2.
Purification of the human EVL protein. A, schematic representation of the purification steps for the Evl protein. B, proteins from each purification step were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1, molecular mass markers; lanes 2 and 3, whole cell lysates before and after induction with isopropyl-1-thio-β-d-galactopyranoside (IPTG), respectively. Lanes 4–8, samples from the peak Ni2+-NTA-agarose (Invitrogen) fraction, the peak hydroxyapatite (Bio-Rad) fraction, the fraction after the removal of the hexahistidine tag, the peak Superdex 200 fraction (GE Healthcare), and the peak MonoS fraction (GE Healthcare), respectively.
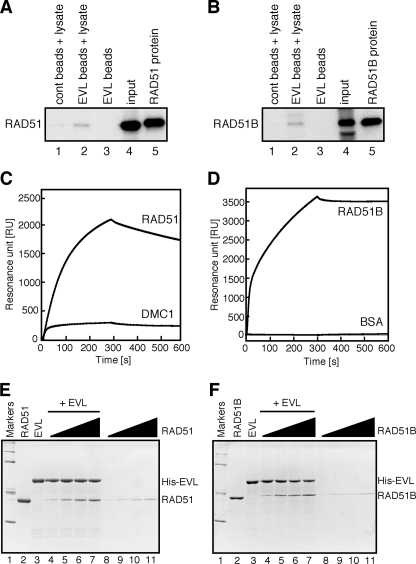
We then tested whether the EVL protein binds to the RAD51 protein, because the EVL knockdown cells exhibited a reduction in RAD51 foci formation (Fig. 1, B and C). To do so, we prepared Affi-Gel 10 beads chemically conjugated with the EVL protein and performed pulldown assays with HeLa cell extracts. As shown in Fig. 3A (lane 2), the endogenous RAD51 protein in the HeLa cell extract was detected in the EVL-bound fraction over the background level. Like the RAD51 protein, the endogenous RAD51B protein in the HeLa cell extract was also detected in the EVL-bound fraction (Fig. 3B, lane 2). These results suggested that the EVL protein binds to the RAD51 and RAD51B proteins.
FIGURE 3.
The EVL protein binds to the RAD51 and RAD51B proteins. A and B, pulldown assay. Affi-Gel 10 beads chemically conjugated with the EVL protein were incubated with a HeLa whole cell extract. Proteins bound to the EVL beads were separated by SDS-PAGE and were analyzed by Western blotting. A, the endogenous RAD51 protein was probed with an anti-RAD51 polyclonal antibody. Lanes 1 and 2, experiments with the control Affi-Gel 10 beads and the EVL beads, respectively, in the presence of the HeLa cell lysate. Lane 3, a control experiment with the EVL beads in the absence of the HeLa cell lysate. The input HeLa cell lysate (10 μg of protein) and the purified RAD51 (2 ng) protein were applied in lanes 4 and 5, respectively. B, the endogenous RAD51B protein was probed with an anti-RAD51B polyclonal antibody. Lanes 1 and 2, experiments with the control Affi-Gel 10 beads and the EVL beads, respectively, in the presence of the HeLa cell lysate. Lane 3, a control experiment with the EVL beads in the absence of the HeLa cell lysate. The input HeLa cell lysate (10 μg) and the purified RAD51B protein (2 ng) were applied in lanes 4 and 5, respectively. C, surface plasmon resonance analyses of the EVL-RAD51 and EVL-DMC1 interactions. Sensorgrams for RAD51 and DMC1 binding to the immobilized EVL protein are presented. The RAD51 and DMC1 concentrations were 1 μm. D, surface plasmon resonance analysis of the EVL-RAD51B interaction. Sensorgrams for RAD51B and BSA binding to the immobilized EVL protein are presented. The RAD51B and BSA concentrations were 1 μm. E and F, the Ni2+-NTA-agarose pulldown assay with the His6-tagged EVL and RAD51 (E) or RAD51B (F) proteins. Lane 1, molecular mass markers (200, 116, 79, 42, and 30 kDa). Lanes 2 and 3, RAD51 (E) or RAD51B (F) and the His6-tagged EVL proteins. Lanes 4–7 and lanes 8–11, experiments in the presence and absence of the His6-tagged EVL, respectively. The concentration of the His6-tagged EVL protein was 0.45 μm. The RAD51 or RAD51B concentrations were 0.5 μm (lanes 4 and 8), 1 μm (lanes 5 and 9), 1.5 μm (lanes 6 and 10), and 2 μm (lanes 7 and 11).
To verify the EVL binding to the RAD51 and RAD51B proteins, we performed SPR analyses. The RAD51 protein interacted with the EVL-conjugated sensor chip (Fig. 3C). The RAD51B protein also significantly interacted with the EVL protein (Fig. 3D). In contrast, the DMC1 protein, which shares about 50% amino acid identity with the RAD51 protein, exhibited little binding to the EVL protein (Fig. 3C). These results indicated that the EVL protein specifically binds to the RAD51 and RAD51B proteins.
We finally confirmed the EVL binding to the RAD51 and RAD51B proteins by the Ni2+-NTA-agarose pulldown assay. In this assay, the His6-tagged EVL protein was used as the bait protein, since the EVL protein covalently conjugated to the Affi-gel beads did not enter the polyacrylamide gel. Consistently, the purified RAD51 and RAD51B proteins bound to the His6-tagged EVL protein were captured by the Ni2+-NTA beads in a concentration-dependent manner (Fig. 3, E and F). In the presence of excess amounts of the RAD51 and RAD51B proteins, the EVL:RAD51 and EVL:RAD51B ratios detected in the gel were 2.4:1 and 1.4:1, respectively (Fig. 3, C and D, lane 7). Therefore, we conclude that the EVL protein directly interacts with the RAD51 and RAD51B proteins.
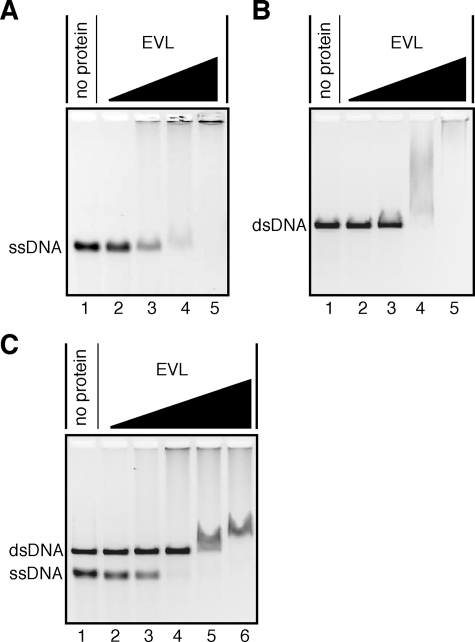
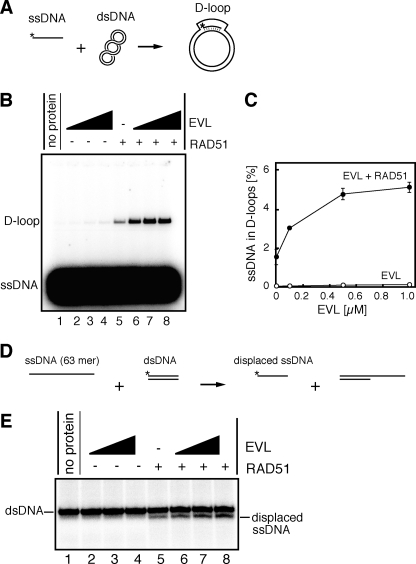
The EVL Protein Stimulates RAD51-mediated Homologous Pairing—As shown in Fig. 4, A and B, the EVL protein bound to both ssDNA and dsDNA. A competitive DNA-binding assay revealed that the EVL protein preferentially bound to ssDNA rather than dsDNA (Fig. 4C). To study the EVL activity in the recombination reaction, we tested whether the EVL protein affects the RAD51-mediated homologous pairing. To do so, we performed the D-loop formation assay (Fig. 5A). The superhelical dsDNA used in this assay was prepared by a method without alkali treatment to avoid denaturation of the double helix of the dsDNA (15). As shown in Fig. 5, B (lanes 2–4) and C, the EVL protein alone did not promote D-loop formation. However, intriguingly, the amount of D-loops formed by the RAD51 protein synergistically increased, in an EVL concentration-dependent manner (Fig. 5, B (lanes 5–8) and C). Therefore, the EVL protein may have an activator function in homologous pairing by the RAD51 protein.
FIGURE 4.
DNA binding activities of the EVL protein. φX174 ssDNA (20 μm) and/orφX174 linear dsDNA (20 μm) were each incubated with the EVL protein at 37 °C for 15 min. The samples were then separated by 0.8% agarose gel electrophoresis in TAE buffer and were visualized by ethidium bromide staining. A, ssDNA binding. Lane 1, a negative control experiment without the EVL protein. The EVL concentrations were 0.1 μm (lane 2), 0.2 μm (lane 3), 0.4 μm (lane 4), and 0.8 μm (lane 5). B, dsDNA binding. Lane 1, a negative control experiment without the EVL protein. The EVL concentrations were 0.1 μm (lane 2), 0.2 μm (lane 3), 0.4 μm (lane 4), and 0.8 μm (lane 5). C, competitive binding to ssDNA and dsDNA. Lane 1, a negative control experiment without the EVL protein. The EVL concentrations were 0.1 μm (lane 2), 0.2 μm (lane 3), 0.4 μm (lane 4), 0.8 μm (lane 5), and 1.2 μm (lane 6).
FIGURE 5.
The EVL protein stimulates the RAD51-mediated homologous pairing. A, a schematic representation of the D-loop formation assay. Asterisks indicate the 32P-labeled end of the 50-mer ssDNA. B, the D-loop formation assay. The reactions were conducted without the RAD51 protein (lanes 1–4) or with the RAD51 protein (0.1 μm) (lanes 5–8) in the presence of increasing amounts of the EVL protein. The EVL concentrations were 0 μm (lanes 1 and 5), 0.1 μm (lanes 2 and 6), 0.5 μm (lanes 3 and 7), and 1 μm (lanes 4 and 8). C, graphic representation of the experiments shown in B. Closed and open circles, experiments with and without the RAD51 protein, respectively. D, a schematic representation of the homologous pairing assay with oligonucleotides. The asterisks indicate the 32P-labeled end of the 32-mer DNA strand. E, the homologous pairing assay. The reactions were conducted without the RAD51 protein (lanes 1–4) or with the RAD51 protein (1 μm) (lanes 5–8) in the presence of increasing amounts of the EVL protein. The EVL concentrations were 0 μm (lanes 1 and 5), 0.5 μm (lanes 2 and 6), 1 μm (lanes 3 and 7), and 2 μm (lanes 4 and 8).
To confirm this EVL-mediated activation of homologous pairing, we performed the homologous pairing assay with short oligonucleotides (Fig. 5D). In this assay, a 63-mer ssDNA and a homologous 32-mer dsDNA were used as substrates. In the reaction, the 32P-labeled 32-mer strand, which contains a sequence identical to that of the 63-mer ssDNA, is displaced from the dsDNA, as a consequence of the homologous pairing and subsequent strand exchange reactions by RAD51. As shown in Fig. 5E (lanes 5–8), the EVL protein enhanced the RAD51-mediated homologous pairing between the 63-mer ssDNA and the homologous 32-mer dsDNA. The displaced 32P-labeled 32-mer product was not detected when the RAD51 protein was omitted from the reaction mixture (Fig. 5E, lanes 2–4), suggesting that the EVL protein alone did not promote homologous pairing. These results obtained from two independent homologous pairing assays are perfectly consistent; therefore, we conclude that the EVL protein stimulates the RAD51-mediated homologous pairing in vitro.
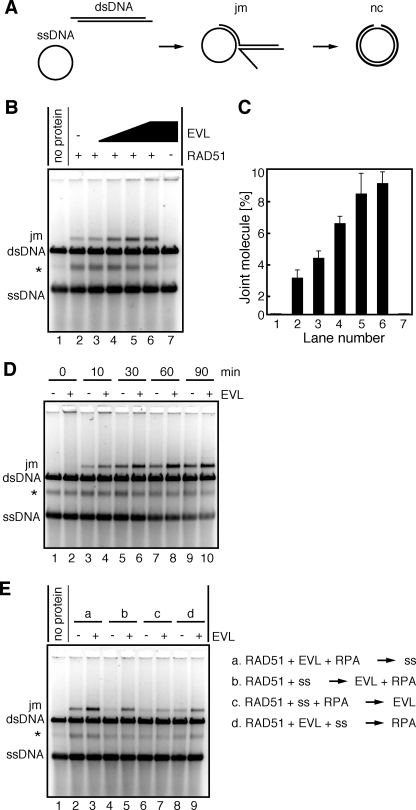
The EVL Protein Stimulates RAD51-mediated Strand Exchange—After homologous pairing, the RAD51 protein promotes a long tract of strand exchange, probably for the fine matching of homologous sequences. We next tested whether the EVL protein affects the RAD51-mediated strand exchange with long DNA substrates. In this assay, φX174 phage circular ssDNA (5,386 bases) and linearized φX174 dsDNA (5,386 base pairs) were used as DNA substrates (Fig. 6A). RPA is required for the efficient promotion of the strand exchange reaction by the RAD51 protein, but this RPA-dependent stimulation of strand exchange was not significant when the ssDNA was pre-incubated with RPA before the RAD51-ssDNA binding. This suppressive effect of RPA is considered to be overcome by the RAD51-binding proteins (11).
FIGURE 6.
The EVL protein stimulates the RAD51-mediated strand exchange. A, a schematic representation of the strand exchange reaction. The two reaction products, joint molecule and nicked circular DNAs, are denoted as jm and nc, respectively. B, the RAD51 protein (1 μm) and RPA (1 μm) were incubated with the indicated amounts of the EVL protein at 30 °C for 10 min. After this incubation, φX174 circular ssDNA (20 μm) was added to the reaction mixture, which was incubated at 30 °C for 10 min. The reactions were then initiated by the addition of φX174 linear dsDNA (20 μm) and were continued at 30 °C for 60 min. The deproteinized products were separated by 1% agarose gel electrophoresis and were visualized by SYBR Gold (Invitrogen) staining. The asterisk indicates the self-annealing products of the ssDNA. The EVL concentrations were 0 μm (lanes 1 and 2), 0.25 μm (lane 3), 0.5 μm (lane 4), 1 μm (lane 5), and 2 μm (lanes 6 and 7). Lane 7, a control experiment without the RAD51 protein. C, graphic representation of the experiments shown in B. The band intensities of the joint molecule DNA products were quantified, and the average values of three independent experiments are shown with the S.D. values. D, time course experiment. The RAD51 protein (1 μm) and RPA (1 μm) were incubated with the EVL protein (1 μm) at 30 °C for 10 min. After this incubation, φX174 circular ssDNA (20 μm) was added to the reaction mixture, which was further incubated at 30 °C for 10 min. The reactions were then initiated by the addition of φX174 linear dsDNA (20 μm) and were continued for the indicated times. The deproteinized products were separated by 1% agarose gel electrophoresis and were visualized by SYBR Gold staining. E, the strand exchange assay with various reaction orders. The RAD51 protein (1 μm), RPA (1 μm), the EVL protein (1 μm), and ssDNA were incubated in the indicated reaction orders at 30 °C for 10 min. The reactions were then initiated by the addition of φX174 linear dsDNA (20 μm) and were continued at 30 °C for 60 min. The deproteinized products were separated by 1% agarose gel electrophoresis and were visualized by SYBR Gold staining. The asterisk indicates the self-annealing products of the ssDNA.
We then conducted the strand exchange reaction under the suppressive conditions with RPA. Interestingly, the EVL protein clearly enhanced the RAD51-mediated strand exchange, in a concentration-dependent manner (Fig. 6, B and C). No products were observed when the reactions were conducted without the RAD51 protein (Fig. 6B, lane 7), suggesting that the EVL protein itself did not possess the strand exchange activity. A time course experiment also revealed significant enhancement of the RAD51-mediated strand exchange by the EVL protein (Fig. 6D). Therefore, the EVL protein may be a novel stimulation factor for RAD51-mediated strand exchange as well as homologous pairing.
We next tested the effects of the reaction orders on strand exchange. As shown in Fig. 6E (lanes 2 and 3), the EVL-mediated stimulation of strand exchange was clearly observed when all three proteins, RAD51, RPA, and EVL, were mixed before the addition of ssDNA. Similarly, substantial enhancement was observed when the RAD51 protein was incubated with ssDNA followed by the addition of the RPA and EVL proteins (Fig. 6E, lanes 4 and 5). A small but clear increase in the products was detected when the RAD51 protein, RPA, and ssDNA were mixed before the addition of the EVL protein (Fig. 6E, lanes 6 and 7). Preincubation of the RAD51 and EVL proteins with ssDNA before the addition of RPA also enhanced the strand exchange (Fig. 6E, lanes 8 and 9). Therefore, the EVL protein actually stimulates the RAD51-mediated strand exchange in various reaction orders, although the stimulation efficiencies significantly depend on the reaction order.
It should be noted that the partial ssDNA annealing products (indicated by asterisks in Fig. 6) generated by the RAD51 protein were detected under the reaction conditions used in the present study (30 °C). This RAD51-dependent ssDNA annealing product was resolved by heating and was not significantly formed when the reactions were performed at 37 °C (supplemental Fig. 2).
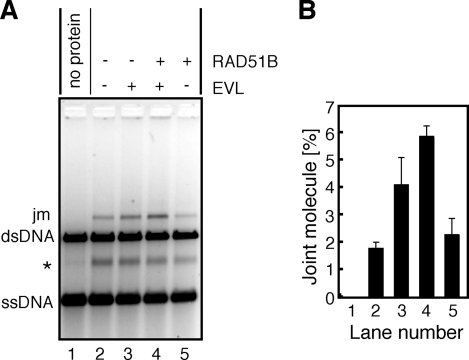
The RAD51B Protein Enhances RAD51-mediated Strand Exchange with the EVL Protein—We next tested whether the RAD51B protein affects RAD51-mediated strand exchange in the presence of the EVL protein, because the RAD51B protein interacts with the EVL protein (Fig. 3, B and D). The RAD51B protein was added to the reaction mixture in the presence or absence of the EVL protein. We found that the RAD51B protein stimulated RAD51-mediated strand exchange in the presence of the EVL protein (Fig. 7, A (lane 4) and B). In contrast, the RAD51B-dependent stimulation was not observed in the absence of the EVL protein (Fig. 7, A (lane 5) and B). Therefore, the RAD51B protein enhances the RAD51-mediated recombination reaction only in the presence of the EVL protein.
FIGURE 7.
The EVL protein further stimulates the RAD51-mediated strand exchange in the presence of the RAD51B protein. A, the RAD51 protein (1 μm) and RPA (1 μm) were incubated with the EVL protein (0.5 μm) and/or the RAD51B protein (0.5 μm) at 30 °C for 10 min. After this incubation, φX174 circular ssDNA (20 μm) was added to the reaction mixture, which was further incubated at 30 °C for 10 min. The reactions were then initiated by the addition of φX174 linear dsDNA (20 μm) and were continued for 60 min. The deproteinized products were separated by 1% agarose gel electrophoresis and were visualized by SYBR Gold staining. The asterisk indicates the self-annealing products of the ssDNA. The EVL concentrations were 0 μm (lanes 1, 2, and 5) and 0.5 μm (lanes 3 and 4). The RAD51B protein concentrations were 0 μm (lanes 1–3) and 0.5 μm (lanes 4 and 5). B, graphic representation of the experiments shown in A. The band intensities of the joint molecule DNA products (jm) were quantified, and the average values of three independent experiments are shown with the S.D. values.
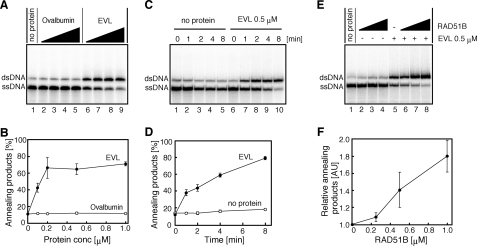
The EVL Protein Promotes Annealing between ssDNA Molecules—We found that the EVL protein possesses robust ssDNA annealing activity. In this assay, complementary ssDNA 49-mers were used as the substrates. As shown in Fig. 8, A–D, the EVL protein alone promoted annealing between complementary strands. In contrast, the RAD51B protein alone did not exhibit annealing activity (Fig. 8, E (lanes 2–4) and F). Interestingly, the EVL-mediated annealing was significantly enhanced by the RAD51B protein (Fig. 8, E (lanes 5–8) and F). These results indicated that the EVL protein may be a novel factor that functions in the HRR pathway and that the RAD51B protein stimulates the EVL-mediated recombination reactions.
FIGURE 8.
ssDNA annealing activity of the EVL protein. A, protein titration. The EVL protein was first complexed with 2 μm ssDNA, followed by the addition of a complementary ssDNA. The reactions were conducted at 30 °C for 8 min. Lane 1, a control experiment without protein; lanes 6–9, experiments with the EVL protein. The EVL concentrations were 0 μm (lane 1), 0.1 μm (lane 6), 0.2 μm (lane 7), 0.5 μm (lane 8), and 1 μm (lane 9). Lanes 2–5, negative control experiments with ovalbumin; the amounts of protein were the same as those of the EVL protein, in terms of weight. B, graphical representation of the experiments shown in A. The band intensities of the annealing products were quantified, and the average values of three independent experiments are shown with the S.D. values. Closed circles and open circles, experiments with the EVL protein and with ovalbumin, respectively. C, time course. Lanes 1–5, negative control experiments without the EVL protein. Lanes 6–10, experiments with the EVL protein (0.5 μm). Reaction times were 0 min (lanes 1 and 6), 1 min (lanes 2 and 7), 2 min (lanes 3 and 8), 4 min (lanes 4 and 9), and 8 min (lanes 5 and 10). D, graphical representation of the experiments shown in C. The band intensities of the annealing products were quantified, and the average values of three independent experiments are shown with the S.D. values. Closed circles and open circles, experiments with and without the EVL protein, respectively. E, effect of the RAD51B protein. The EVL protein was preincubated with or without the RAD51B protein, followed by an incubation with 2 μm ssDNA. The reactions were initiated by the addition of a complementary ssDNA and were conducted at 30 °C for 8 min. Lanes 1–4, experiments without the EVL protein; lanes 5–8, experiments with the EVL protein. The RAD51B concentrations were 0 μm (lanes 1 and 5), 0.25 μm (lanes 2 and 6), 0.5 μm (lanes 3 and 7), and 1 μm (lanes 4 and 8). F, graphical representation of the experiments shown in E. The band intensities of the annealing products, relative to the experiment in the absence of the RAD51B protein, were quantified, and the average values of three independent experiments are shown with the S.D. values.
DISCUSSION
The human EVL protein is a member of the Ena/Vasp family, which reportedly functions in cytoplasmic actin dynamics (48). In the present study, we discovered an unexpected function of the EVL protein in the HRR pathway. Specifically, we found that the EVL protein (i) directly binds to the RAD51 and RAD51B proteins, (ii) functions in DSB repair, probably in the RAD51 assembly step on DSB sites, (iii) stimulates RAD51-mediated homologous pairing and strand exchange, and (iv) preferentially binds to ssDNA and promotes ssDNA annealing. We also found that (v) the RAD51B protein enhances the EVL-dependent strand exchange stimulation and ssDNA annealing reactions. These EVL activities discovered in the present study strongly suggest that the EVL protein is a novel activator for the RAD51-mediated homologous recombination reaction.
The biochemical properties of the EVL protein presented here are surprisingly similar to those of the RAD52 protein, which is a prominent RAD51 activator in eukaryotes. The RAD52 protein directly binds to the RAD51 protein (13–15) and stimulates RAD51-mediated homologous pairing (18) and strand exchange (53–55). The RAD52 protein itself possesses robust ssDNA annealing activity (56–59). These functional similarities between the EVL and RAD52 proteins imply that they may have overlapping functions in cells.
In the present study, we found that MMC sensitivity in the EVL knockdown cells was not as remarkable as the MMC sensitivity of the RAD51B knockdown cells. This fact implies that a factor(s) that complements the EVL function may exist. The RAD52 protein, which shares significant biochemical similarity with the EVL protein, may be a factor. In addition, two more paralogs of the EVL protein, MENA and VASP, exist in humans (48) and may also contribute to the functional redundancy of the EVL protein.
The RAD52 knock-out chicken DT40 cells reportedly did not exhibit defects in DSB repair (60). The cells with the double knock-out of the RAD52 and XRCC2 proteins were significantly defective in the HRR pathway (61), indicating that the RAD52 protein has an overlapping function with the XRCC2 protein. The XRCC2 protein forms the BCDX2 complex, which is composed of the RAD51B, RAD51C, RAD51D, and XRCC2 proteins (62, 63), and like the EVL and RAD52 proteins, the BCDX2 complex possesses robust ssDNA annealing activity (64). Therefore, redundant annealing activities may be required in the HRR pathway in higher eukaryotes. The RAD52-mediated annealing is suggested to be important in the second end capture step, just after homologous pairing and strand exchange (65). Therefore, the EVL protein may also function in the second end capture step, in addition to the RAD51 assembly and homologous pairing/strand exchange steps. It is intriguing to study the functional interactions between these ssDNA annealing proteins, such as RAD52, BCDX2, and EVL-RAD51B.
We found that the EVL protein was highly expressed in MCF7 cells. However, we noticed that EVL expression was very low in other cells, such as RPE, HT1080, T47D, HepG2, and HCT116 (data not shown). The rat EVL homologue is reportedly expressed in brain, lung, spleen, thymus, and testis but not in liver, muscle, and prostate (66). Given that the EVL protein is a novel recombination activator for the RAD51-dependent HRR pathway, it may not constitutively function like the RAD51 and RAD51B proteins. The EVL protein may be required for repairing specific DNA lesions in certain tissues or cells, and it may cause tumor malignancy due to inappropriate recombination activation by its overexpression in certain types of tumor cells. The MCF7 cell line, which robustly expresses the EVL protein, was derived from a breast cancer tumor. Intriguingly, the abnormal expression of the EVL mRNA was also found in another breast cancer tumor (49). Given that EVL overexpression is a source for the malignancy of tumor cells, such as breast cancer, the EVL protein may be a potential target of anti-cancer treatments.
Supplementary Material
Acknowledgments
We thank Dr. W. Kagawa (RIKEN) for critical reading of the manuscript and also thank the Cs-137 γ-ray Irradiation Facilities for Biological Research, Department of Nuclear Engineering and Management, Graduate School of Engineering, University of Tokyo.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: DSB, double strand break; HRR, homologous recombinational repair; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; MMC, mitomycin C; NTA, nitrilotriacetic acid; HPLC, high performance liquid chromatography; SPR, surface plasmon resonance; DTT, dithiothreitol; AMPPNP, 5′-adenylyl-β,γ-imidodiphosphate; BSA, bovine serum albumin; siRNA, small interfering RNA.
References
- 1.Weinstock, D. M., Richardson, C. A., Elliott, B., and Jasin, M. (2006) DNA Repair 5 1065-1074 [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., Tafel, A. A., and Kanaar, R. (2006) DNA Repair 5 1075-1081 [DOI] [PubMed] [Google Scholar]
- 3.Wyman, C., and Kanaar, R. (2006) Annu. Rev. Genet. 40 363-383 [DOI] [PubMed] [Google Scholar]
- 4.Sung, P., and Klein, H. (2006) Nat. Rev. Mol. Cell Biol. 7 739-750 [DOI] [PubMed] [Google Scholar]
- 5.West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4 435-445 [DOI] [PubMed] [Google Scholar]
- 6.Sung, P. (1994) Science 265 1241-1243 [DOI] [PubMed] [Google Scholar]
- 7.Baumann, P., Benson, F. E., and West, S. C. (1996) Cell 87 757-766 [DOI] [PubMed] [Google Scholar]
- 8.Maeshima, K., Morimatsu, K., and Horii, T. (1996) Genes Cells 1 1057-1068 [DOI] [PubMed] [Google Scholar]
- 9.Gupta, R. C., Bazemore, L. R., Golub, E. I., and Radding, C. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 463-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66 630-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung, P., Krejci, L., Van Komen, S., and Sehorn, M. G. (2003) J. Biol. Chem. 278 42729-42732 [DOI] [PubMed] [Google Scholar]
- 12.San Filippo, J., Sung, P., and Klein, H. (2008) Annu. Rev. Biochem. 77 229-257 [DOI] [PubMed] [Google Scholar]
- 13.Shen, Z., Cloud, K. G., Chen, D. J., and Park, M. S. (1996) J. Biol. Chem. 271 148-152 [DOI] [PubMed] [Google Scholar]
- 14.Kurumizaka, H., Aihara, H., Kagawa, W., Shibata, T., and Yokoyama, S. (1999) J. Mol. Biol. 291 537-548 [DOI] [PubMed] [Google Scholar]
- 15.Kagawa, W., Kurumizaka, H., Ikawa, S., Yokoyama, S., and Shibata, T. (2001) J. Biol. Chem. 276 35201-35208 [DOI] [PubMed] [Google Scholar]
- 16.Golub, E. I., Kovalenko, O. V., Gupta, R. C., Ward, D. C., and Radding, C. M. (1997) Nucleic Acids Res. 25 4106-4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka, K., Hiramoto, T., Fukuda, T., and Miyagawa, K. (2000) J. Biol. Chem. 275 26316-26321 [DOI] [PubMed] [Google Scholar]
- 18.Benson, F. E., Baumann, P., and West, S. C. (1998) Nature 391 401-404 [DOI] [PubMed] [Google Scholar]
- 19.Sigurdsson, S., Van Komen, S., Petukhova, G., and Sung, P. (2002) J. Biol. Chem. 277 42790-42794 [DOI] [PubMed] [Google Scholar]
- 20.Mazina, O. M., and Mazin, A. V. (2004) J. Biol. Chem. 279 52042-52051 [DOI] [PubMed] [Google Scholar]
- 21.Wesoly, J., Agarwal, S., Sigurdsson, S., Bussen, W., Van Komen, S., Qin, J., van Steeg, H., van Benthem, J., Wassenaar, E., Baarends, W. M., Ghazvini, M., Tafel, A. A., Heath, H., Galjart, N., Essers, J., Grootegoed, J. A., Arnheim, N., Bezzubova, O., Buerstedde, J. M., Sung, P., and Kanaar, R. (2006) Mol. Cell. Biol. 26 976-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovalenko, O. V., Golub, E. I., Bray-Ward, P., Ward, D. C., and Radding, C. M. (1997) Nucleic Acids Res. 25 4946-4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modesti, M., Budzowska, M., Baldeyron, C., Demmers, J. A., Ghirlando, R., and Kanaar, R. (2007) Mol. Cell 28 468-481 [DOI] [PubMed] [Google Scholar]
- 24.Wiese, C., Dray, E., Groesser, T., San Filippo, J., Shi, I., Collins, D. W., Tsai, M. S., Williams, G. J., Rydberg, B., Sung, P., and Schild, D. (2007) Mol. Cell 28 482-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies, A. A., Masson, J. Y., McIlwraith, M. J., Stasiak, A. Z., Stasiak, A., Venkitaraman, A. R., and West, S. C. (2001) Mol. Cell 7 273-282 [DOI] [PubMed] [Google Scholar]
- 26.Pellegrini, L., Yu, D. S., Lo, T., Anand, S., Lee, M., Blundell, T. L., and Venkitaraman, A. R. (2002) Nature 420 287-293 [DOI] [PubMed] [Google Scholar]
- 27.Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., Zheng, N., Chen, P. L., Lee, W. H., and Pavletich, N. P. (2002) Science 297 1837-1848 [DOI] [PubMed] [Google Scholar]
- 28.Esashi, F., Christ, N., Gannon, J., Liu, Y., Hunt, T., Jasin, M., and West, S. C. (2005) Nature 434 598-604 [DOI] [PubMed] [Google Scholar]
- 29.Galkin, V. E., Esashi, F., Yu, X., Yang, S., West, S. C., and Egelman, E. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8537-8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shivji, M. K., Davies, O. R., Savill, J. M., Bates, D. L., Pellegrini, L., and Venkitaraman, A. R. (2996) Nucleic Acids Res. 34 4000-4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies, O. R., and Pellegrini, L. (2007) Nat. Struct. Mol. Biol. 14 475-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esashi, F., Galkin, V. E., Yu, X., Egelman, E. H., and West, S. C. (2007) Nat. Struct. Mol. Biol. 14 468-474 [DOI] [PubMed] [Google Scholar]
- 33.Petalcorin, M. I., Galkin, V. E., Yu, X., Egelman, E. H., and Boulton, S. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8299-8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazloum, N., Zhou, Q., and Holloman, W. K. (2007) Biochemistry 46 7163-7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazloum, N., Zhou, Q., and Holloman, W. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 105 524-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albala, J. S., Thelen, M. P., Prange, C., Fan, W., Christensen, M., Thompson, L. H., and Lennon, G. G. (1997) Genomics 46 476-479 [DOI] [PubMed] [Google Scholar]
- 37.Rice, M. C., Smith, S. T., Bullrich, F., Havre, P., and Kmiec, E. B. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7417-7422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cartwright, R., Dunn, A. M., Simpson, P. J., Tambini, C. E., and Thacker, J. (1998) Nucleic Acids Res. 26 1653-1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takata, M., Sasaki, M. S., Sonoda, E., Fukushima, T., Morrison, C., Albala, J. S., Swagemakers, S. M. A., Kanaar, R., Thompson, L. H., and Takeda, S. (2000) Mol. Cell. Biol. 20 6476-6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bleuyard, J. Y., Gallego, M. E., Savigny, F., and White, C. I. (2005) Plant J. 41 533-545 [DOI] [PubMed] [Google Scholar]
- 41.Hatanaka, A., Yamazoe, M., Sale, J. E., Takata, M., Yamamoto, K., Kitao, H., Sonoda, E., Kikuchi, K., Yonetani, Y., and Takeda, S. (2005) Mol. Cell. Biol. 25 1124-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osakabe, K., Abe, K., Yamanouchi, H., Takyuu, T., Yoshioka, T., Ito, Y., Kato, T., Tabata, S., Kurei, S., Yoshioka, Y., Machida, Y., Seki, M., Kobayashi, M., Shinozaki, K., Ichikawa, H., and Toki, S. (2005) Plant Mol. Biol. 57 819-833 [DOI] [PubMed] [Google Scholar]
- 43.Yonetani, Y., Hochegger, H., Sonoda, E., Shinya, S., Yoshikawa, H., Takeda, S., and Yamazoe, M. (2005) Nucleic Acids Res. 33 4544-4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Date, O., Katsura, M., Ishida, M., Yoshihara, T., Kinomura, A., Sueda, T., and Miyagawa, K. (2006) Cancer Res. 66 6018-6024 [DOI] [PubMed] [Google Scholar]
- 45.Sigurdsson, S., Van, Komen, S., Bussen, W., Schild, D., Albala, J. S., and Sung, P. (2001) Genes Dev. 15 3308-3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lio, Y. C., Mazin, A. V., Kowalczykowski, S. C., and Chen, D. J. (2003) J. Biol. Chem. 278 2469-2478 [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama, H., Kurumizaka, H., Ikawa, S., Yokoyama, S., and Shibata, S. (2003) J. Biol. Chem. 278 2767-2772 [DOI] [PubMed] [Google Scholar]
- 48.Kwiatkowski, A. V., Gertler, F. B., and Loureiro, J. J. (2003) Trends Cell Biol. 13 386-392 [DOI] [PubMed] [Google Scholar]
- 49.Hu, L. D., Zou, H. F., Zhan, S. X., and Cao, K. M. (2008) Oncol. Rep. 19 1015-1020 [PubMed] [Google Scholar]
- 50.Ishida, T., Takizawa, Y., Sakane, I., and Kurumizaka, H. (2008) Genes Cells 13 91-103 [DOI] [PubMed] [Google Scholar]
- 51.Kinebuchi, T., Kagawa, W., Enomoto, R., Tanaka, K., Miyagawa, K., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2004) Mol. Cell 14 363-374 [DOI] [PubMed] [Google Scholar]
- 52.Henricksen, L. A., Umbricht, C. B., and Wold, M. S. (1994) J. Biol. Chem. 269 11121-11132 [PubMed] [Google Scholar]
- 53.Sung, P. (1997) J. Biol. Chem. 272 28194-28197 [DOI] [PubMed] [Google Scholar]
- 54.New, J. H., Sugiyama, T., Zaitseva, E., and Kowalczykowski, S. C. (1998) Nature 391 407-410 [DOI] [PubMed] [Google Scholar]
- 55.Shinohara, A., and Ogawa, T. (1998) Nature 391 404-407 [DOI] [PubMed] [Google Scholar]
- 56.Mortensen, U. H., Bendixen, C., Sunjevaric, I., and Rothstein, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10729-10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy, G., Golub, E. I., and Radding, C. M. (1997) Mutat. Res. 377 53-59 [DOI] [PubMed] [Google Scholar]
- 58.Sugiyama, T., New, J. H., and Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6049-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagawa, W., Kagawa, A., Saito, K., Ikawa, S., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2008) J. Biol. Chem. 283 24264-24273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi-Iwai, Y., Sonoda, E., Buerstedde, J. M., Bezzubova, O., Morrison, C., Takata, M., Shinohara, A., and Takeda, S. (1998) Mol. Cell. Biol. 18 6430-6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimori, A., Tachiiri, S., Sonoda, E., Thompson, L. H., Dhar, P. K., Hiraoka, M., Takeda, S., Zhang, Y., Reth, M., and Takata, M. (2001) EMBO J. 20 5513-5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masson, J. Y., Tarsounas, M. C., Stasiak, A. Z., Stasiak, A., Shah, R., McIlwraith, M. J., Benson, F. E., and West, S. C. (2001) Genes Dev. 15 3296-3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schild, D., Lio, Y. C., Collins, D. W., Tsomondo, T., and Chen, D. (2000) J. Biol. Chem. 275 16443-16449 [DOI] [PubMed] [Google Scholar]
- 64.Yokoyama, H., Sarai, N., Kagawa, W., Enomoto, R., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2004) Nucleic Acids Res. 32 2556-2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McIlwraith, M. J., and West, S. C. (2008) Mol. Cell 29 510-516 [DOI] [PubMed] [Google Scholar]
- 66.Ohta, S., Mineta, T., Kimoto, M., and Tabuchi, K. (1997) Biochem. Biophys. Res. Commun. 237 307-312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.