Abstract
In Escherichia coli, RelE toxin participates in growth arrest and cell death by inducing mRNA degradation at the ribosomal A-site under stress conditions. The NMR structures of a mutant of E. coli RelE toxin, RelER81A/R83A, with reduced toxicity and its complex with an inhibitory peptide from RelB antitoxin, RelBC (Lys47-Leu79), have been determined. In the free RelER81A/R83A structure, helix α4 at the C terminus adopts a closed conformation contacting with the β-sheet core and adjacent loops. In the RelER81A/R83A-RelBC complex, helix α3* of RelBC displaces α4 of RelER81A/R83A from the binding site on the β-sheet core. This helix replacement results in neutralization of a conserved positively charged cluster of RelE by acidic residues from α3* of RelB. The released helix α4 becomes unfolded, adopting an open conformation with increased mobility. The displacement of α4 disrupts the geometry of critical residues, including Arg81 and Tyr87, in a putative active site of RelE toxin. Our structures indicate that RelB counteracts the toxic activity of RelE by displacing α4 helix from the catalytically competent position found in the free RelE structure.
Toxin-antitoxin (TA)2 systems originally known as suicide or addiction modules, controlling plasmid inheritance through “post-segregational killing” (1, 2), have been documented as an environmental adaptation used by most bacteria (3). Overexpression of certain toxins induces cellular dormancy, also called a quasi-dormant state, which enables cell survival for prolonged times during environmental stresses (4, 5). Recently, TA systems have been linked to medically important phenomena such as biofilm formation and antibiotic resistance (6).
To date, TA toxins are known to perturb one or more vital processes, such as DNA replication, RNA transcription, and protein translation, by targeting DNA gyrase (7), messenger RNA (8, 9), and/or ribosomes (5, 10). A subgroup of toxins, including MazF, RelE, and YoeB, is named as mRNA interferase (11), because they perturb the stability of mRNA by sequence-specific cleavage. Among these toxins, MazF is well established as an ACA sequence-specific endoribonuclease, which cleaves free single-stranded mRNA in the absence of ribosome (12). In contrast, RelE cannot cleave free mRNA transcripts. It cleaves translating mRNA associated with the ribosome at the ribosomal A-site (10). In this manner, RelE is a ribosome-dependent mRNA interferase, and preferential cleavage occurs at the second position of stop codons (UAG, UAA, and UGA) and some sense codons (CAG and UCG), with the UAG (amber) stop codon and the CAG (glutamine) codon being cleaved most efficiently (13). YoeB was initially recognized as a purine-specific endoribonuclease with preference to AG-rich regions, albeit with low efficiency (14). However, it was recently found that YoeB binds to the 50 S subunit in 70 S ribosomes and leads to efficient mRNA cleavage at the ribosomal A-site (15). Therefore, both RelE and YoeB toxins trigger mRNA cleavage in a ribosome-dependent mode, which is distinct from the ribosome-independent mechanism of MazF.
Even though the functionality of Escherichia coli RelE has been extensively characterized, the structural mechanism is still elusive. Here we determined the NMR structures of a low toxicity mutant of RelE, RelER81A/R83A, and its complex with the C-terminal region of RelB, RelBC (Lys47-Leu79). Comparison of the free and RelBC-bound RelER81A/R83A reveals a large conformational change at the putative active site of the RelE toxin. The present structural studies indicate a direct inhibition mechanism for the RelE-RelB addiction module.
EXPERIMENTAL PROCEDURES
Protein Sample Preparation—Recombinant expression and purification were carried out as described previously for RelBC (Lys47-Leu79) and wild-type RelE (16). RelER81A/R83A mutant was obtained using QuikChange site-directed mutagenesis kit (Stratagene). Unlabeled or isotope-enriched (e.g. 15N or 15N,13C) protein was purified from crude lysate using nickel-nitrilotriacetic acid resin (Qiagen) and further purified by size exclusion chromatography. For NMR spectroscopy, all samples were prepared in 25 mm sodium phosphate (pH 6.5) containing 500 mm NaCl and 1 mm dithiothreitol in 90% H2O, 10% D2O, or in 99% D2O.
Protein Synthesis Inhibition Assay on a Prokaryotic Cell-free System—Prokaryotic cell-free protein synthesis was carried out with an E. coli T7 S30 extract system (Promega). The reaction mixture consisted of 10 μl of S30 premix, 7.5 μl of S30 extract, and 2.5 μl of an amino acid mixture (1 mm each of all amino acids except methionine), 1 μl of [35S]methionine, and different amounts of RelE in a final volume of 29 μl. The different amounts of RelE and RelBC were preincubated for 10 min at 25 °C before the assay started by adding 1 μl of pET-11a-MazG plasmid-DNA (0.16 μg/μl). The reaction was performed for 1.5 h at 37 °C, and proteins were then precipitated with acetone and analyzed by SDS-PAGE followed by autoradiography.
Preparation of E. coli 70 S Ribosomes—70 S ribosomes were prepared from E. coli MRE 600 as described previously (17) with minor modifications. Bacterial cells (2 g) were suspended in buffer A (10 mm Tris-HCl (pH 7.8) containing 10 mm MgCl2, 60 mm NH4Cl, and 6 mm 2-mercaptoethanol). The cells were lysed by French press. After incubation with RNase-free DNase (30 min at 0 °C), cell debris was removed by centrifugation two times at 30,000 rpm for 30 min at 4 °C with a Beckman 50Ti rotor. The supernatant (three-fourth volume from the top) was then layered over an equal volume of 1.1 m sucrose in buffer B (buffer A containing 0.5 m NH4Cl) and centrifuged at 45,000 rpm for 15 h at 4 °C with a Beckman 50Ti rotor. After washing with buffer A, the ribosome pellets were resuspended in buffer A and applied to a linear 5-40% (w/v) sucrose gradient prepared in buffer A and centrifuged at 35,000 rpm for 3 h at 4 °C with a Beckman SW41Ti rotor. Gradients were fractionated, and the 70 S ribosome fractions were pooled and pelleted at 45,000 rpm for 20 h at 4 °C with a Beckman 50Ti rotor. The 70 S ribosome pellets were resuspended in buffer A before they were stored at -80 °C.
Toeprinting Assays—Toeprinting was carried out as described previously (18) with a minor modification. The mixture for primer-template annealing containing mRNA and 32P-end-labeled DNA primer was incubated at 70 °C for 5 min and then cooled slowly to room temperature. The ribosome-binding mixture contained 2 μl of 10× buffer (100 mm Tris-HCl (pH 7.8) containing 100 mm MgCl2, 600 mm NH4Cl, and 60 mm 2-mercaptoethanol), different amounts of RelE, 0.375 mm dNTP, 0.05 μm 70 S ribosomal subunits, 1 μm tRNAfMet, and 2 μl of the annealing mixture in a final volume of 20 μl. The final mRNA concentration was 0.035 μm. This ribosome-binding mixture was incubated at 37 °C for 10 min, and then reverse transcriptase (2 units) was added. The cDNA synthesis was carried out at 37 °C for 15 min. The reaction was stopped by adding 12 μl of the sequencing loading buffer (95% formamide, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol EF). The sample was incubated at 90 °C for 5 min prior to electrophoresis on a 6% polyacrylamide sequencing gel. The ompA mRNA was synthesized in vitro from a DNA fragment containing a T7 promoter and a part of the opening reading frame using T7 RNA polymerase. The DNA fragment for ompA (248 bp), which had the initiation codon at the center, was amplified by PCR using appropriate primers and chromosome DNA as the template. The 5′-end primers for ompA contained the T7 promoter sequence.
Nuclear Magnetic Resonance Spectroscopy—NMR spectra were recorded on Inova 500 MHz (Varian) and Avance 600 and 800 MHz (Bruker) spectrometers. All data were collected at 23.5 °C. Backbone and side chain resonance assignments for both free and bound states of RelBC and RelER81A/R83A were accomplished with the standard triple resonance experiments described previously (19) (i.e. HNCACB, CBCACONH, CCCTOCSYNH, HCCTOCSYNH, and HNCO with samples in 90% H2O; HCCHCOSY and HCCHTOCSY with samples in 99% D2O). Both 15N- and 13C-edited nuclear Overhauser effect spectroscopy-HSQC spectra were acquired on 15N,13C-RelER81A/R83A, 15N,13C-RelER81A/R83A-RelBC and 15N,13C-RelBC-RelER81A/R83A samples for the final structural calculations. The intermolecular nuclear Overhauser effects (NOE) were distinguished from the intramolecular NOEs by 13C/15N-filtered (F1) 13C-edited (F2) nuclear Overhauser effect spectroscopy-HSQC spectra (20). Compound chemical shift changes of both RelER81A/R83A and RelBC upon their interactions were calculated from the chemical shift of HN, N, CA, and CB nuclei with the weighted formula as described previously (21). All data were processed by using NMRPIPE (22) and analyzed with XEASY (23) and NMRVIEW (24) software packages. Chemical shift data have been deposited in the Biological Magnetic Resonance Bank with accession codes 16065, 16066, and 16067 for the RelER81A/R83A-RelBC complex, free RelER81A/R83A, and free RelBC, respectively.
Structure Calculation and Refinement—The three-dimensional structures of free RelER81A/R83A and the RelER81A/R83A-RelBC complex were calculated using CYANA (25) with standard protocols. NOE-based distance constraints were obtained from a combination of manual and CYANA-based automated NOE assignment procedures (26). Dihedral angle (ϕ/ψ) constraints were estimated from chemical shifts using TALOS (27). Hydrogen bond constraints were generated based on the locations of predicted secondary structure for the protected NH groups in H2O/D2O solvent exchange experiments. The final structures were refined using CNS with water as the explicit solvent (28). The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 2KC8 and 2KC9 for the RelER81A/R83A-RelBC complex and free RelER81A/R83A, respectively.
RESULTS
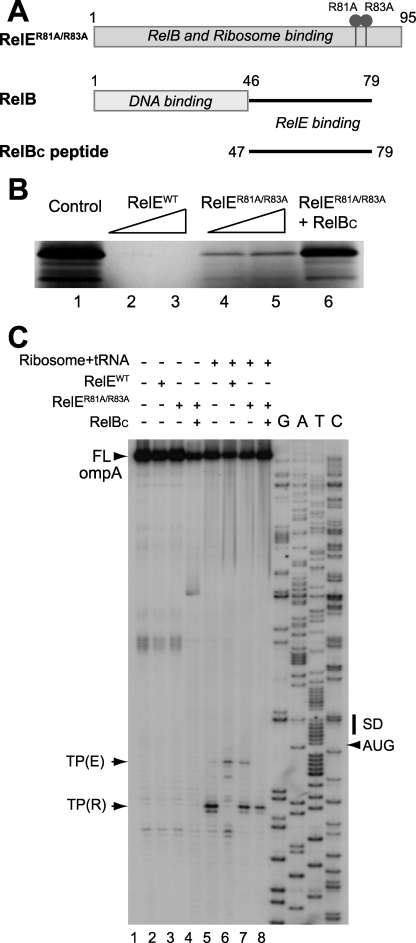
RelBC Abolishes the Residual Catalytic Activity of RelER81A/R83A—Overexpression of wild-type RelE alone in E. coli markedly hindered cell growth because of its cytotoxicity. RelB antitoxin neutralizes RelE toxicity by forming a nontoxic complex. Therefore, RelE can be coexpressed with RelB and isolated from the RelB-RelE complex through a denaturation and refolding procedure (16, 29). However, the refolded RelE protein is unstable in solution at elevated concentrations (i.e. 0.1-0.2 mm). The resonance assignments of refolded wild-type RelE were hampered by a low signal-to-noise ratio and poor magnetization transfer in three-dimensional NMR experiments. We overcame this obstacle by using a low toxicity mutant, RelER81A/R83A (Fig. 1A), which allowed us to purify enough protein under native conditions for structural studies.
FIGURE 1.
Functional characterization of RelE R81A/R83A and RelER81A/R83A-RelBC. A, domain architects of RelE and RelB. The constructs of RelBC and RelER81A/R83A were used in this structural study. B, analysis of translation inhibition activity for wild-type RelE, RelER81A/R83A, and RelER81A/R83A-RelBC complex on MazG synthesis in a prokaryotic cell-free protein synthesis. The control experiment without protein (lane 1) and inhibition experiments with 0.15 μg/μl (lane 2) and 0.35 μg/μl (lane 3) wild type RelE, respectively, are shown; and 0.15 μg/μl (lane 4) and 0.35 μg/μl RelER81A/R83A (lane 5), respectively, and 0.35 μg/μl RelER81A/R83A-RelBC (lane 7) are shown. C, analysis of the ribosome-dependent mRNA cleavage activity for RelE, RelER81A/R83A, and RelER81A/R83A-RelBC complex on ompA mRNA in a toeprinting analysis. The ompA mRNAs were synthesized in vitro from a 248-bp DNA fragment containing a T7 promoter using T7 RNA polymerase. The sequence ladder shown at the right-hand side was obtained using the same primer used for toeprinting with pCR®2.1-TOPO®-ompA as template. Control experiments are shown without protein and ribosome (lane 1), with 0.1125 μg/μl wild-type RelE (lane 2), with 0.1125 μg/μl RelER81A/R83A (lane 3), and with 0.1125 μg/μl RelER81A/R83A-RelBC (lane 4). Toeprinting experiments are shown with 0.05 μm 70 S ribosomes and 1 μm tRNAfMet (lane 5), 0.05 μm 70 S ribosomes and 1 μm tRNAfMet with 0.1125 μg/μl wild-type RelE (lane 6), 0.05 μm 70 S ribosomes and 1 μm tRNAfMet with 0.1125 μg/μl RelER81A/R83A (lane 7), and 0.05 μm 70 S ribosomes and 1 μm tRNAfMet with 0.1125 μg/μl RelER81A/R83A-RelBC (lane 8). The initiation codon, AUG, is indicated with an arrow. The full-length product of primer extension is denoted as FL. Positions of ribosome toeprinting and RelE toeprinting bands are indicated as TP(R) and TP(E), respectively.
Comparison of the mRNA interferase activity of wild-type RelE with RelER81A/R83A indicates that the mutation significantly reduces but does not abolish its activity in both cell-free protein synthesis (Fig. 1B) and toeprinting assays (Fig. 1C). The residual activity of RelER81A/R83A is completely abolished by the addition of the C-terminal domain of RelB antitoxin, RelBC (residues Lys47 to Leu79) (Fig. 1, A-C). These observations suggest that RelER81A/R83A represents a structural model of wild type in an active conformation, whereas the complex of RelER81A/R83A and RelBC represents a model of an inactive conformation.
The structural properties of RelER81A/R83A mutant and wild-type RelE were compared using NMR spectroscopy. The 1H-15N HSQC spectra of the mutant in both free and RelBC-bound states show high similarity to those of the wild type, indicating the structural integrity and the ability for RelB binding were not significantly affected by the mutagenesis (Fig. 2A and supplemental Fig. S1). The affinities of RelBC binding to wild-type RelE and RelER81A/R83A mutant were then measured using an intrinsic tryptophan fluorescence method, by the virtue of only one tryptophan residue (Trp15) existing in RelE toxin. The dissociation constant (KD) of wild type is 154 ± 15 nm and that of RelER81A/R83A is 200 ± 24 nm, indicating that the mutational effect on the affinity is marginal (supplemental Fig. S2).
FIGURE 2.
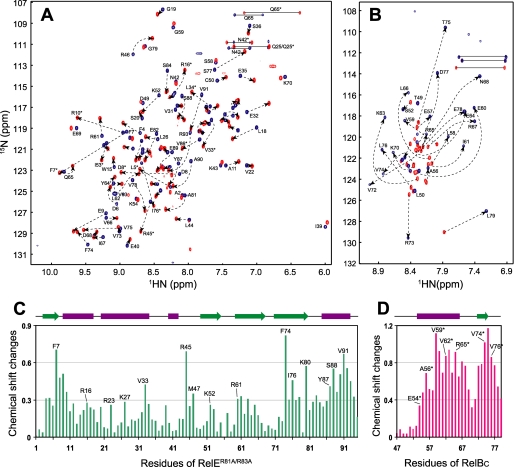
Interaction of RelER81A/R83A and RelBC characterized by NMR. A, superimposed 1H-15N HSQC spectra of free (blue) and RelBC-bound (red) RelER81A/R83A. B, superimposed 1H-15N HSQC spectra of free (red) and RelER81A/R83A-bound RelBC (blue). C, averaged chemical shift changes of individual residues in RelER81A/R83A perturbed by RelBC binding. D, averaged chemical shift changes of individual residues in RelBC upon binding to RelER81A/R83A. The averaged chemical shift changes are calculated by a weighted combination of chemical shifts of HN, N, CA, and CB nuclei (21).
RelER81A/R83A and RelBC Interaction Characterized by NMR Spectroscopy—A substantial improvement in line width and magnetization transfer was observed for spectra recorded on RelER81A/R83A compared with the refolded wild-type RelE (Fig. 2A and supplemental Fig. S1). Titration of 15N,13C-labeled RelER81A/R83A with unlabeled RelBC showed significant chemical shift perturbation in the 1H-15N HSQC spectrum (Fig. 2, A and C). A pair of NH resonances corresponding to each residue in both free and RelBC-bound states was observed during the titration, indicative of a slow exchange regime on the NMR time scale. The slow exchange spectral change is consistent with a high affinity in the range of 10-7 m. A 1H-15N HSQC spectrum of 15N,13C-labeled RelBC alone displays poor dispersion of NH resonance (7.9-8.5 ppm), indicating that this C-terminal region of RelB is largely unstructured in its free state (Fig. 2B). Titration of labeled RelBC with unlabeled RelER81A/R83A shows dramatic chemical shift changes in a similar slow exchange regime (Fig. 2, B and D). The well dispersed spectrum of RelBC in the bound state suggests that RelER81A/R83A binding induces the folding of RelBC.
Resonance assignments of RelER81A/R83A and RelBC in both free and bound states were accomplished using a standard set of triple resonance procedures (19). The chemical shift index analysis (30) of RelBC peptide in both free and bound states revealed a disordered-to-ordered conformation change upon the complex formation (supplemental Fig. S3, A and B). On the other hand, the chemical shift index analysis of RelER81A/R83A revealed an unfolding of a C-terminal helix (α4) coupled with RelBC binding (supplemental Fig. S3, C and D). The three-dimensional structures of RelER81A/R83A alone and in complex with RelBC were determined by using a combination of manual and automated NOE assignment procedures (supplemental Table 1).
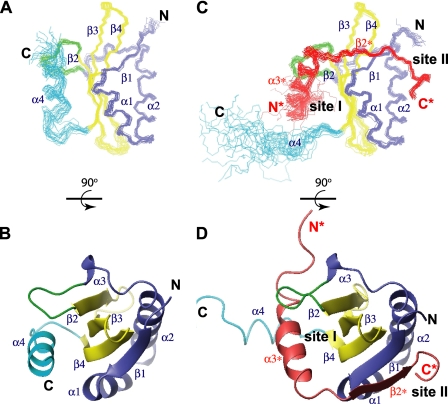
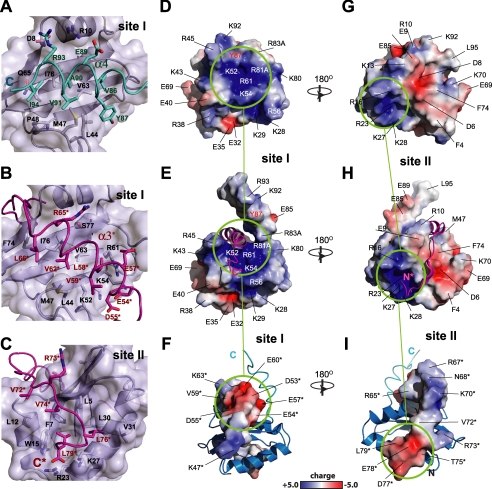
Structures of RelER81A/R83A—The ensemble of the 20 lowest energy structures of RelER81A/R83A shows a well defined α/β sandwich fold with approximate dimensions of 36 × 30 × 30 Å (Fig. 3A). The overall topology is βαααβββα, in which four strands form a β-sheet core surrounded by four α-helices (Fig. 3B). Three consecutive anti-parallel β-strands (β2-β4) pack into a classic meander motif (31), providing a scaffold for folding of the protein. The N-terminal strand (β1) and the helix hairpin, formed by two long helices (α1 and α2), are tightly associated to one side of the β-meander core through numerous hydrophobic contacts. In particular, the hydrophobic residues (Val22, Leu26, Leu30, Val31, Val33, and Leu34) from the amphipathic α2 form extensive contacts with one side of the twisted β-sheet, which is composed entirely of hydrophobic residues (Leu5 and Phe7 from β1; Ile53 and Leu55 from β2; Tyr60, Leu62, Tyr64, and Val66 from β3; and, Val73, Val75, and Val78 from β4). A long linking region that connects α2 and β2 wraps around the same side of the β-sheet, providing additional stabilizing interactions to the main hydrophobic core. A short 3-10 helix (α3) is situated in the middle of the long loop region. In contrast, the other side of the central β-sheet is more exposed to solvent. The C terminus of RelER81A/R83A forms a helix (α4), which associates with the protruded surface of the central β-meander motif. This interaction is supported by α3-β2 loop and β1-α1 junction regions through both hydrophobic and electrostatic contacts (Fig. 3, A and B and Fig. 4A), burying a total surface area of ∼1100 Å2. A positively charged and highly conserved cluster (including Arg45, Lys52, Lys54, Arg56, Arg61, Lys80, Arg81, and Arg83) is located adjacent to the interface of the β-sheet and helix α4 (Fig. 4D). Structural comparison with known RNases and mutagenesis in the previous (10, 29, 32) and present studies suggest that the positively charged cluster represents a putative mRNA substrate-binding site. Residues from both main domains of RelE and the C-terminal α4 helix form an active site for the toxicity of RelE. Hence the closed conformation of α4 is in a catalytically competent position (details under “Discussion”).
FIGURE 3.
Structures of RelER81A/R83A and RelER81A/R83A-RelBC. A, ensemble of the 20 lowest energy structures of RelER81A/R83A. B, ribbon representation of the lowest energy structure of RelER81A/R83A rotated 90° showing the β-sheet core surrounded by four helices. C, ensemble of the 20 structures of the RelER81A/R83A-RelBC complex. D, ribbon representation showing the helix replacement and formation of the inter-molecular β-sheet. Interface site I and II are indicated in the complex structure. The N-terminal isolated strandβ1 and helix hairpin (α1-α2) region of RelER81A/R83A (Met1-Asn42) is colored in blue, loop α3-β2 in green, the central β-meander motif (β2-β4) in yellow, and the C-terminal tail (Ala81-Leu95) in cyan. The antitoxin RelBC peptide is colored in red.
FIGURE 4.
The interface and electrostatic properties of RelER81A/R83A and RelBC. A, helix α4 occupies the surface of the central β-meander motif and interacts with the α3-β2 loop and β1-α1 junction region. Helix α4 is colored in cyan, and the remaining core structure of RelE is colored in gray. B, interface site I. The helix α3* of RelB (magenta) occupies the surface of β-sheet core of RelE (gray). C, interface site II. The C-terminal extended region of RelB (magenta) anchors on the surface of the RelE β1, α1, and α2(gray). The electrostatic surface analysis of free RelER81A/R83A (D), RelBC-bound RelER81A/R83A (E), and RelER81A/R83A-bound RelBC (F). G-I, opposite views of D--F with a rotation of 180°. Two positively charged clusters on the RelER81A/R83A surface are complemented by negatively charged clusters from RelBC, which are denoted by light green circles. The main positive cluster of the RelER81A/R83A protein shown in D-F is the putative mRNA-binding site.
Structure of RelER81A/R83A-RelBC Complex—The RelBC peptide folds into a helix (α3*, Glu54*-Leu66*, number of residues and secondary structure elements refer to full-length RelB, asterisk is used to denote RelB throughout) and a short β-strand (β2*, Val72*-Val74*) upon binding to RelER81A/R83A (Fig. 3, C and D). Two prolines (Pro69 and Pro71) mediate the formation of a turn between helix α3* and strand β2*, bending the polypeptide chain by ∼90°. This perpendicular orientation of α3* and β2* provides a concave surface that interacts with a convex interface on RelE. The α3* contacts with the surface of the central β-meander motif, termed interface site I, and the β2* contacts with the surface formed by β1-α1-α2 elements of RelE, termed interface site II. At site I, helix α3* of RelBC displaces α4 of RelER81A/R83A, through hydrophobic interactions with β3 (Val63), β4 (Ile76), and loop α3-β2 (Leu44 and Met47) (Fig. 4, A and B). However, the orientation of α3* along the β-sheet surface is tilted by 36° compared with α4. An acidic patch (Asp53*, Glu54*, Asp55*, and Glu57*) at the N terminus of helix α3* of RelBC (Fig. 4, B and F) directly neutralizes the positively charged cluster at the putative RNA-binding site of RelER81A/R83A (Arg45, Lys52, Lys54, and Arg61) (Fig. 4E). At site II, the β2* of RelBC forms an inter-molecular anti-parallel β-sheet with adjacent β1 of RelER81A/R83A. The remaining C-terminal residues of RelBC form a rigid turn conformation and anchor to the surface of the α1-α2 hairpin (Fig. 4C). Hydrophobic side chains of Val74*, Leu76*, and Leu79* from RelBC interact with a hydrophobic patch of RelER81A/R83A formed by residues from β1 (Leu5 and Phe7), α1 (Leu12 and Trp15), and α2 (Leu30 and Val31). The unique chemical shifts of δ1 (0.09 ppm) and δ2(-0.61 ppm) protons of Leu79* are consistent with the determined structure, in which methyl groups of Leu79* point to the center of the hydrophobic pocket and pack against the indole ring of Trp15. Solvent-exposed basic residues (Arg16, Arg23, Lys27, and Lys28) surround the hydrophobic patch, forming another positively charged surface (Fig. 4H), which is neutralized by Asp77* and Glu78* and the C-terminal carboxylate group of Leu79* (Fig. 4I). Overall, both hydrophobic and electrostatic forces stabilize the complex formation of RelER81A/R83A and RelBC. In total, RelBC peptide buries a large solvent-accessible surface area of about 2700 Å2, which encompasses both site I and site II.
RelB Perturbs the Integrity of the Active Site of RelE by Inducing Conformational Changes—Comparison of RelBC-bound RelER81A/R83A to the unbound structure reveals a pronounced conformational change of α4 and adjacent loops (loop α3-β2 and β4-α4). The α4 swings out from the surface of the central β-sheet as it is displaced by the amphipathic helix α3* from RelBC in the complex (Figs. 3 and 4). The large chemical shift perturbation in the C-terminal tail region of RelER81A/R83A by RelBC is because of a conformational change rather than direct interaction (Fig. 2, A and C, and supplemental Fig. S4, A and B). The released α4 becomes unfolded, as evidenced by the chemical shift index analysis results (supplemental Fig. S3, C and D).
To further probe the structure and dynamic nature of the RelE toxin, we examined the internal motion of RelER81A/R83A by measuring 1H-15N heteronuclear NOE (hetNOE) relaxation data (supplemental Fig. S5, A and B). With the exception of the N- and C-terminal residues, the hetNOE values for the main domain of RelER81A/R83A in both free and RelBC-bound states are relatively uniform. The relatively high magnitude of the NOE values (>0.8) for residues within the structured core domain (residues 4-79) is characteristic of a well folded globular structure. By contrast, the C-terminal tail region (residues 80-95) displays lower hetNOE values (0.5-0.7) in the RelBC-free state than that seen for the core domain. These data indicate that the C-terminal helix α4 possesses an increased internal mobility despite the fact that the tail is found as part of the folded structure in the RelBC-free state of RelER81A/R83A. The transverse relaxation rate (R2) values of residues in this tail region as well as in the adjacent loops are significantly higher than the average value of the whole protein in the free state (supplemental Fig. S5C). It is likely that this region undergoes a conformational exchange between the associated (closed) state found in the NMR-driven structure and an isolated (open) state that could not be seen in the structure. Upon binding of RelBC, RelER81A/R83A displays large changes in both the hetNOE and R2 values, especially around the C-terminal helix α4 region. A dramatic reduction in hetNOE and R2 values within the α4 region is observed, indicating an increase in the mobility of this region (supplemental Fig. S5, B and D). This observation is fully consistent with the release of this helix from the core structure upon RelBC binding. Concomitant to this structure and dynamic change associated with α4, the dynamic property of the loop α3-β2 region is also significantly altered by RelBC binding; residues Leu44, Gly46, and Asp49 become more dynamic in the bound state than in the free state (supplemental Fig. S5). This region is in close proximity to the C-terminal helix α4 in the free state and the α3* in the bound state, and is presumably affected by the conformational change associated with the toxin-antitoxin interaction.
DISCUSSION
Among the toxin-antitoxin systems, RelE family toxins have the widest phylogenetic distribution in the prokaryotic genomes (8), being found in diverse bacterial and archaeal lineages (33). The E. coli RelE is one of the best characterized TA toxins in terms of both in vivo and in vitro functional studies (13, 29); however, the structure of E. coli RelE has never been reported. In the previous studies (13, 29), substitution of the last six residues (AVKRIL) into a VTVTVT amino acid sequence resulted in a nontoxic version of RelE, RelECS6, thereby indicating the C-terminal region is functionally important for the toxicity of RelE. Our structural studies of RelER81A/R83A revealed a large conformational rearrangement in the C terminus upon interaction with an inhibitory antitoxin peptide RelBC. In the absence of the antitoxin peptide, the last 10 C-terminal residues form an amphipathic helix α4 folded on the surface of the central β-sheet. The α4 residues (Val86, Tyr87, Ala90, and Arg94) involved in the intra-molecular interaction with the core domain (Fig. 4A) are highly conserved within the RelE family of toxins (8), indicating that the position of the C-terminal helix in a closed conformation may be a common feature in the family of proteins.
In a structural similarity search using DALI (34), YoeB toxin (14) from E. coli (Z score = 10, r.m.s.d. = 1.9 Å) and archaeal RelE (32) (called aRelE) from Pyrococcus horikoshii (Z score = 10, r.m.s.d. = 2.5 Å) were identified as the best matches with the present structure of E. coli RelE. Among these structures, the helix hairpin and the β-meander motif are conserved. However, there are several distinct differences between them (supplemental Fig. S6). First, the elongated C-terminal extension of α4 in E. coli RelE is not present in aRelE and YoeB toxins. Second, there is a short strand inserted at the N-terminal side of the conserved three-strand β-meander motif in both aRelE and YoeB; however, this insertion is too short to form a secondary structural element in the structure of E. coli RelE. Instead, it forms a relatively rigid loop α3-β2 (Fig. 3, B and D). In addition, the lengths of α1 and the strands in the central β-meander motif differ in YoeB and RelE/aRelE. The α1 is much shorter in RelE and aRelE (8 residues) than in YoeB (17 residues), whereas the meander strands are longer in RelE and aRelE than in YoeB (supplemental Fig. S6). RelE also shows structural similarities, with low Z scores and small r.m.s.d. values, to other microbial ribonucleases such as the C-terminal ribonuclease domain of colicin-Glu5 (35), a tRNase from E. coli (Z score = 3.7, r.m.s.d. = 3.0 Å) and RNase SA (36), a guanyl-specific ribonuclease from Streptomyces aureofaciens (Z score = 3.2, r.m.s.d. = 3.0 Å). The structural architecture consisting of a two-layer α/β sandwich and an RNA recognition site on the surface of the central β-sheet are highly conserved among these RNA-binding proteins (supplemental Fig. S6).
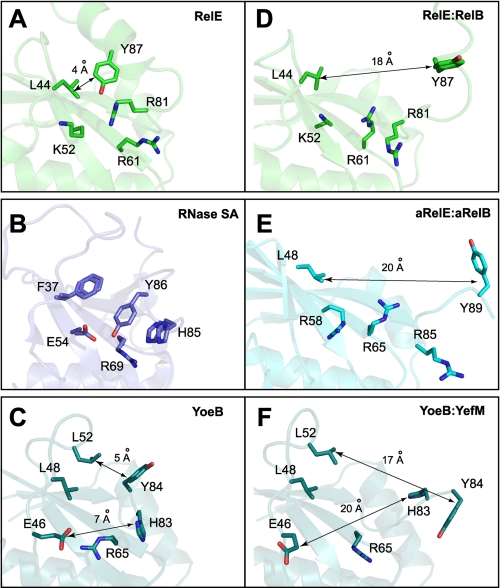
Comparison of RelE with the well characterized RNase SA revealed that RNA substrate-binding residues are conserved in the two proteins (36). In E. coli RelE, Leu44 and Tyr87 are proposed to provide a site for base packing, and Arg61 is proposed to promote backbone phosphate recognition (Fig. 5A). In contrast, residues compared with the canonical catalytic triad of RNase SA are not present in RelE. The catalytic His85 and Glu54 of RNase SA (Fig. 5B) and His83 and Glu46 of YoeB (14) (Fig. 5C) are replaced with Arg81 and Lys52 in E. coli RelE, respectively (Fig. 5A). Despite the nonconservation, RelE and YoeB toxins share the similar microbial RNase fold. The lack of catalytic residues in RelE renders RelE alone nonfunctional in cleaving free mRNA by itself. The enzymatic activity is only achieved upon association with the ribosome. Although YoeB shows weak intrinsic endoribonuclease activity, our recent data (15) demonstrate that YoeB is a potent protein synthesis inhibitor by cleaving mRNA at the ribosomal A-site. It is intriguing to propose that RelE and YoeB toxins share similarity in their mRNA interferase activity in the ribosome-dependent mode, which is distinct from the mechanisms of the canonical microbial RNases and the ribosome-independent mRNA interferase MazF.
FIGURE 5.
RelB induced conformation change in the active site of RelE. A, active conformation of the E. coli RelE mRNA-binding site in the absence of RelB antitoxin. The side chain of Arg81 was modeled based on the orientation of Ala81 in the structure of RelER81A/R83A. B, active site of RNase SA shows the catalytic triad (Glu54, Arg69, and His85) and the hydrophobic site (Phe37 and Tyr86) for base packing. C, catalytic site of YoeB in the YefM-free conformation shows the catalytic triad (Glu46, Arg65, and His83) and base anchor residues (Leu48, Leu52, and Tyr84). D, putative mRNA-binding site of E. coli RelE in the presence of RelBC shows a large conformation disruption to the active site. The side chain of Arg81 is modeled based on the orientation of Ala81 in the structure of RelER81A/R83A-RelBC. E, active site of archaeal aRelE in the aRelB-aRelE complex shows an inactive conformation similar to the RelER81A/R83A-RelBC complex. The C-terminal residues (Tyr89 and Lys90) of aRelE are missing in the crystal structure; they were arbitrarily rebuilt to estimate the position of Tyr89. F, catalytic active site of YoeB in the YoeB-YefM complex shows the conformational change altered by YefM binding.
Structural comparison of RelE in free (active) and antitoxin-bound (inactive) states revealed a large conformational change induced by RelB binding. In the active state of RelE, the conserved Tyr87 in α4 is in close proximity with Arg81 in the β4-α4 loop and Leu44 in the α3-β2 loop (Fig. 5A). This side chain arrangement, apparently required for the cleavage of mRNA at the ribosomal A-site, resembles the active site structure of RNase SA, supporting the hypothesis that this site is an active site of RelE. In the inactive state of RelE complexed with RelB, Tyr87 moves away from Arg81 and Leu44, as α4 is released from the core domain by the binding of RelB. This situation rather resembles the orientation of corresponding side chains found in the crystal structure of the archaean aRelE-aRelB complex (32), known to be an inactive conformation. These results are consistent with the previous study of inactive RelECS6 (13), in which the substitution of the C-terminal six residues alters the conformation of α4 by disrupting the interaction between the C-terminal tail and the core domain. A similar conformational rearrangement in the active site is seen in the YoeB-YefM TA system (14), which also involves a large movement of residues in the C terminus of the protein (His83 and Tyr84 in Fig. 5, C and F), although YoeB toxin has a much shorter C-terminal tail. The perturbation of the proper arrangement of critical residues at an active site seems to be a common theme for the inactivation mechanism of RelE/ParE superfamily TA systems.
At present, the positioning of the catalytically active RelE in the ribosome is unknown. However, mRNA cleavage by RelE requires a vacant A-site and substrate mRNA anchored on the 30 S subunit, suggesting that RelE binds in a region that overlaps with the “decoding center” within the ribosomal A-site (13). It is known that the ribosome is a catalytically active ribozyme that can cleave mRNA even in the absence of RelE. RelE may modulate its substrate specificity by altering the conformation of the ribosome and/or the associated mRNA at the A-site decoding region. It is most likely that RelE functions as a stimulatory factor, which stabilizes the catalytically active conformation of ribosomal RNA. However, in view of the fact that RelE has the side chain arrangement similar to that of RNase SA (36), it is highly possible that a complete catalytic active center may be formed only when RelE and ribosome associate to a holoenzyme.
Although the molecular detail of mRNA and ribosome binding remains to be addressed by further structural studies, previous studies using site-directed mutagenesis in P. horikoshii aRelE (32) and E. coli RelE toxins (29) illuminated that the arginine (Arg85 in aRelE and Arg81 in RelE) at the conserved histidine position in canonical RNases is crucial for the function of RelE toxins. This residue could play a role in the ribosome recognition or be involved in the formation of a catalytic center together with ribosome. Our structural studies provide evidence that RelB antitoxin directly inhibits RelE toxin through binding to the active site, although the formation of a RelE2-RelB2 complex mediated through the N-terminal dimerization domain of RelB (16) may also spatially block RelE from entering the ribosomal A-site where the RNA cleavage takes place, a mechanism proposed by Takagi et al. (32). Nevertheless, further studies are required to elucidate exactly how RelE collaborates with the ribosome for enzymatic activity.
Supplementary Material
Acknowledgments
We thank Christopher B. Marshall, Peter B. Stathopulos, and Emma Gooding for critical reading of the manuscript. CFI funding was received for Bruker 600 and Bruker 800 MHz NMR spectrometers.
The atomic coordinates and structure factors (codes 2KC8 and 2KC9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM081567 (to M. Inouye). This work was also supported by a grant from Canadian Institutes of Health Research (to M. Ikura) and a Canadian Institutes of Health Research operating grant.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1-S6.
Footnotes
The abbreviations used are: TA, toxin-antitoxin; NOE, nuclear Overhauser effect; HSQC, heteronuclear single quantum coherence; hetNOE, heteronuclear NOE; r.m.s.d., room mean square deviation.
References
- 1.Gerdes, K., Jacobsen, J. S., and Franch, T. (1997) Genet. Eng. 19 49-61 [DOI] [PubMed] [Google Scholar]
- 2.Engelberg-Kulka, H., and Glaser, G. (1999) Annu. Rev. Microbiol. 53 43-70 [DOI] [PubMed] [Google Scholar]
- 3.Gerdes, K., Christensen, S. K., and Lobner-Olesen, A. (2005) Nat. Rev. Microbiol. 3 371-382 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki, M., Zhang, J., Liu, M., Woychik, N. A., and Inouye, M. (2005) Mol. Cell 18 253-261 [DOI] [PubMed] [Google Scholar]
- 5.Liu, M., Zhang, Y., Inouye, M., and Woychik, N. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5885-5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis, K. (2008) Curr. Top. Microbiol. Immunol. 322 107-131 [DOI] [PubMed] [Google Scholar]
- 7.Bahassi, E. M., O'Dea, M. H., Allali, N., Messens, J., Gellert, M., and Couturier, M. (1999) J. Biol. Chem. 274 10936-10944 [DOI] [PubMed] [Google Scholar]
- 8.Anantharaman, V., and Aravind, L. (2003) Genome Biol. 4 R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condon, C. (2006) Mol. Microbiol. 61 573-583 [DOI] [PubMed] [Google Scholar]
- 10.Christensen, S. K., and Gerdes, K. (2003) Mol. Microbiol. 48 1389-1400 [DOI] [PubMed] [Google Scholar]
- 11.Yanaguchi, Y., and Inouye, M. (2009) Prog. Mol. Biol. Transl. Sci. 85 467-500 [DOI] [PubMed] [Google Scholar]
- 12.Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003) Mol. Cell 12 913-923 [DOI] [PubMed] [Google Scholar]
- 13.Pedersen, K., Zavialov, A. V., Pavlov, M. Y., Elf, J., Gerdes, K., and Ehrenberg, M. (2003) Cell 112 131-140 [DOI] [PubMed] [Google Scholar]
- 14.Kamada, K., and Hanaoka, F. (2005) Mol. Cell 19 497-509 [DOI] [PubMed] [Google Scholar]
- 15.Zhang, Y., and Inouye, M. (2009) J. Biol. Chem. 284 6627-6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, G. Y., Zhang, Y., Inouye, M., and Ikura, M. (2008) J. Mol. Biol. 380 107-119 [DOI] [PubMed] [Google Scholar]
- 17.Aoki, H., Ke, L., Poppe, S. M., Poel, T. J., Weaver, E. A., Gadwood, R. C., Thomas, R. C., Shinabarger, D. L., and Ganoza, M. C. (2002) Antimicrob. Agents Chemother. 46 1080-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll, I., and Blasi, U. (2002) Biochem. Biophys. Res. Commun. 297 1021-1026 [DOI] [PubMed] [Google Scholar]
- 19.Kanelis, V., Forman-Kay, J. D., and Kay, L. E. (2001) IUBMB Life 52 291-302 [DOI] [PubMed] [Google Scholar]
- 20.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R., and Kay, L. E. (1997) J. Am. Chem. Soc. 119 6711-6721 [Google Scholar]
- 21.Mal, T. K., Masutomi, Y., Zheng, L., Nakata, Y., Ohta, H., Nakatani, Y., Kokubo, T., and Ikura, M. (2004) J. Mol. Biol. 339 681-693 [DOI] [PubMed] [Google Scholar]
- 22.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 23.Bartels, C., Xia, T., Billeter, M., Guntert, P., and Wuthrich, K. (1995) J. Biomol. NMR 6 1-10 [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. A. (2004) Methods Mol. Biol. 278 313-352 [DOI] [PubMed] [Google Scholar]
- 25.Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997) J. Mol. Biol. 273 283-298 [DOI] [PubMed] [Google Scholar]
- 26.Herrmann, T., Guntert, P., and Wuthrich, K. (2002) J. Mol. Biol. 319 209-227 [DOI] [PubMed] [Google Scholar]
- 27.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 28.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 29.Pedersen, K., Christensen, S. K., and Gerdes, K. (2002) Mol. Microbiol. 45 501-510 [DOI] [PubMed] [Google Scholar]
- 30.Wishart, D. S., and Sykes, B. D. (1994) J. Biomol. NMR 4 171-180 [DOI] [PubMed] [Google Scholar]
- 31.Orengo, C. A., and Thornton, J. M. (1993) Structure (Lond.) 1 105-120 [DOI] [PubMed] [Google Scholar]
- 32.Takagi, H., Kakuta, Y., Okada, T., Yao, M., Tanaka, I., and Kimura, M. (2005) Nat. Struct. Mol. Biol. 12 327-331 [DOI] [PubMed] [Google Scholar]
- 33.Gerdes, K., Moller-Jensen, J., and Bugge Jensen, R. (2000) Mol. Microbiol. 37 455-466 [DOI] [PubMed] [Google Scholar]
- 34.Holm, L., and Park, J. (2000) Bioinformatics (Oxf.) 16 566-567 [DOI] [PubMed] [Google Scholar]
- 35.Yajima, S., Inoue, S., Ogawa, T., Nonaka, T., Ohsawa, K., and Masaki, H. (2006) Nucleic Acids Res. 34 6074-6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sevcik, J., Lamzin, V. S., Dauter, Z., and Wilson, K. S. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1307-1313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.