Abstract
It is impossible to predict which pathway, direct glutaminylation of tRNAGln or tRNA-dependent transamidation of glutamyl-tRNAGln, generates mitochondrial glutaminyl-tRNAGln for protein synthesis in a given species. The report that yeast mitochondria import both cytosolic glutaminyl-tRNA synthetase and tRNAGln has challenged the widespread use of the transamidation pathway in organelles. Here we demonstrate that yeast mitochondrial glutaminyl-tRNAGln is in fact generated by a transamidation pathway involving a novel type of trimeric tRNA-dependent amidotransferase (AdT). More surprising is the fact that cytosolic glutamyl-tRNA synthetase (cERS) is imported into mitochondria, where it constitutes the mitochondrial nondiscriminating ERS that generates the mitochondrial mischarged glutamyl-tRNAGln substrate for the AdT. We show that dual localization of cERS is controlled by binding to Arc1p, a tRNA nuclear export cofactor that behaves as a cytosolic anchoring platform for cERS. Expression of Arc1p is down-regulated when yeast cells are switched from fermentation to respiratory metabolism, thus allowing increased import of cERS to satisfy a higher demand of mitochondrial glutaminyl-tRNAGln for mitochondrial protein synthesis. This novel strategy that enables a single protein to be localized in both the cytosol and mitochondria provides a new paradigm for regulation of the dynamic subcellular distribution of proteins between membrane-separated compartments.
Keywords: Dual localization, tRNA-dependent amidotransferase, tRNAGln, mitochondria, metabolism, Saccharomyces cerevisiae
In all living organisms, ribosome-mediated protein synthesis requires the supply of a set of at least 20 perfectly paired aminoacyl-transfer RNA (aa-tRNA) species, one for each of the canonical amino acids found in proteins. Although most of the aa-tRNA is made by aminoacyl-tRNA synthetases (aaRS), a family of 20 enzymes that catalyze direct acylation of each tRNA species with its cognate amino acid (Ibba and Söll 2000), it is now clear that the process of aa-tRNA synthesis is far from universally conserved. In different organisms, up to four aa-tRNA species are, or can be, generated by two-step indirect pathways invariably involving tRNA-dependent conversion of a precursor mischarged aa-tRNA (Sheppard et al. 2008). These pathways, which usually compensate for the absence of a given aaRS, are not oddities of the translation machinery, since they sometimes constitute the sole route for synthesis of a given aa-tRNA. Among all aa-tRNA species used for mRNA translation, one, glutaminyl-tRNAGln (Q-tRNAQ), is of particular interest, not only because it displays the unique feature of being generated by kingdom-specific pathways or enzymes (Tumbula et al. 2000), but also because its synthesis is still a matter of debate and remains to be elucidated for the majority of eukaryotic organelles. To date, all eukaryotes studied so far use glutaminyl-tRNA synthetase (QRS) for the formation of cytosolic glutaminyl-tRNAGln (Q-ctRNAQ) (Ibba et al. 2000), whereas the vast majority of prokaryotes use the two-step transamidation pathway in which two enzymes are working in tandem. First, a nondiscriminating glutamyl-tRNA synthetase (ND-ERS) generates a mischarged Glu-tRNAGln (E-tRNAQ) (Lapointe et al. 1986), then, the glutamate-charged tRNAQ is transamidated into glutamine by a tRNA-dependent amidotransferase (AdT). The AdT enzymes catalyzing this process differ between bacteria and archaea. Bacteria solely use a heterotrimeric enzyme called the GatCAB AdT (Curnow et al. 1997; Becker and Kern 1998), while archaea utilize an archaeal-specific heterodimeric GatDE AdT (Tumbula et al. 2000).
The way Q-tRNAQ is generated in organelles is far from uniform among eukaryotes and is unknown for many organisms. The first report on organellar Q-tRNAQ synthesis came from experiments with barley chloroplast extracts, which unambiguously showed that Q-tRNAQ synthesis proceeds by transamidation of a mischarged glutamyl-tRNAGln (E-tRNAQ) (Schön et al. 1988). In addition, this work suggested that plant and mouse mitochondria use the same transamidation pathway to form mitochondrial glutaminyl-tRNAGln (Q-mtRNAQ), based on the result that QRS activity could not be detected in any of the mitochondrial extracts that were assayed. The general idea that organellar and especially mitochondrial Q-tRNAQ synthesis uniformly proceeds via the transamidation pathway was later reinforced by accumulating sequence data coming from whole-genome sequencing projects. All eukaryotic genomes sequenced so far are deprived of the gene encoding a mitochondrial QRS (mQRS). With the exception of a few protozoans, they concomitantly all display a gene for a GatB ortholog, named PET112, which is always predicted to encode a mitochondrial protein. Since the presence of a gatB gene in a given prokaryotic genome invariably signifies the presence of an AdT in the corresponding organism, the presence of mitochondrial GatB homologs (Pet112p) in eukaryotes should predict the existence of a mitochondrial AdT. Recent studies with Arabidopsis thaliana validated this assumption by showing that mitochondrial and chloroplastic Q-tRNAQ are actually made by a unique dual-targeted GatCAB AdT (Pujol et al. 2008) that transamidates both mitochondrial and chloroplastic E-tRNAQ formed by a unique mischarging ERS also addressed to both cellular compartments. This paradigm is, however, contradicted by two studies reporting the existence of a mQRS in trypanosomatidae (Nabholz et al. 1997; Rinehart et al. 2004). Likewise, a recent study reported that in the yeast Saccharomyces cerevisiae, the cQRS and two nuclear-encoded tRNAsQ are imported in the mitochondria and used to generate Q-tRNAQ that participates to mitochondrial protein synthesis (Rinehart et al. 2005). This result is somewhat surprising since it contradicts several earlier studies suggesting that yeast mitochondrial Q-tRNAQ was not generated by direct charging of mitochondrial tRNAQ by a mQRS, but rather by a mitochondrial transamidation pathway. For example, it had been shown that the yeast gatB ortholog, PET112 (Mulero et al. 1994), is essential for respiratory metabolism and that the respiratory deficiency induced by its alteration can be rescued by expression of a mitochondrially targeted version of the Bacillus subtilis GatB subunit (Kim et al. 1997). This suggests that a mitochondrial AdT could well exist and be responsible for Q-tRNAQ formation in yeast mitochondria. However, Rinehart et al. (2005) demonstrated that the yeast mitochondrial glutamyl-tRNA synthetase (mERS) is incapable of charging with glutamate any of the tRNAQ species present in yeast mitochondria. This constitutes a strong argument against the existence of a mitochondrial transamidation pathway in yeast. Indeed, if there is no ND-ERS in mitochondria, then the mischarged E-tRNAQ, which is the obligatory substrate intermediate for the AdT, cannot be generated. Consequently, with no means of supplying the AdT with its substrate, the existence of a mitochondrial tRNA-dependent transamidation pathway makes no sense. Yet, the S. cerevisiae genome encodes a mitochondrial GatB ortholog. The logical deduction one would make is that the yeast mitochondrial GatB protein, Pet112p, has deviated from its canonical function in tRNA-dependent transamidation and exhibits an essential alternative mitochondrial function. We therefore designed a series of experiments to identify the functional role of this protein.
Using a systems-based approach and despite expectations to the contrary, we characterized a novel type of trimeric AdT in yeast mitochondria. Biochemical, genetic, and subcellular localization experiments unambiguously prove that this AdT is mitochondrial and is responsible for Q-mtRNAQ formation. We confirm that yeast mERS is indeed a discriminating ERS and cannot produce E-mtRNAQ. However, to our surprise, we found that the cytosolic ERS (cERS) can synthesize the mischarged E-tRNAQ substrate for the AdT. In agreement with this result, we found that a fraction of cERS localizes to mitochondria and is in fact the missing nondiscriminating ERS. By trying to understand how the cERS can be dual localized, we found that the major portion of cERS is sequestered in the cytoplasm by binding to Arc1p, a protein that serves as a cytoplasmic anchoring platform for cERS but also for cytosolic methionyl-tRNA synthetase (cMRS). Finally, we also found that the level of Arc1p expression is decreased when the cells are switched from fermentation to respiratory metabolism, which results in increased cERS import to allow higher levels of Q-mtRNAQ formation for elevated mitochondrial protein synthesis. We propose that Arc1p constitutes a relay in the molecular pathway that allows yeast cells to switch from fermentation to respiration. This constitutes a novel mechanism of regulation of dual localization of a protein that is used and essential in both the cytoplasm and mitochondria.
Results
The yeast mitochondrial GatB protein (Pet112p) participates in the assembly of an AdT-like trimeric particle
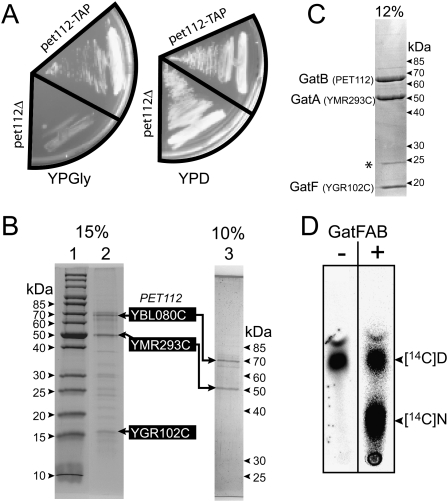
To identify Pet112p interactants, we engineered a yeast strain in which a tandem affinity purification (TAP) tag coding sequence was chromosomally fused, in-frame, to the 3′-end of PET112 (Puig et al. 2001). Figure 1A shows that fusion of this tag does not interfere with the mitochondrial function of the recombinant protein, since unlike the pet112 deletion strain (pet112Δ) (Supplemental Table 1), the PET112-TAP cells still grow on a respiratory medium. The Pet112-TAP fusion protein and associated components were recovered from cell extracts by TAP. Analysis of the final pull-down fraction by SDS-PAGE showed several proteins associated with the Pet112 bait (Fig. 1B). Mass spectrometry analysis of the band recovered from the gel identified seven Pet112-interacting proteins, among which two were annotated as mitochondrial and were present in stoichiometric amounts when compared with the Pet112-TAP fusion protein. These two interactants are Ymr293c and Ygr102c. Ymr293c is annotated as being an amidase that displays an amidohydrolase signature motif and is homologous to bacterial GatA proteins (Saccharomyces Genome Database). In addition, YMR293C has recently been renamed HER2 (Hmg2p ER remodeling) because it has been reported to be involved in proliferation or remodeling of the endoplasmic reticulum that is caused by overexpression of Hmg2p (Federovitch et al. 2008). Ygr102c is a small protein of unknown function that has been detected in highly purified mitochondria in high-throughput studies (Reinders et al. 2006). When we crossed the available functional and structural data, we realized that we purified a mitochondrial heterotrimer composed of a GatB ortholog (Pet112), a GatA homolog (Ymr293c), and a small protein like GatC (Ygr102c), matching almost perfectly with the bacterial GatCAB AdT. To confirm the existence of this trimeric particle, we performed TAP on extracts of a strain expressing C-terminally TAP-tagged Ymr293c. Analysis of the purified fraction shows that Pet112p and Ygr102c coelute with the Ymr293c fusion protein (Supplemental Fig. 1). The three genes encoding Pet112p, Ymr293c, and Ygr102c were then synthetically reconstructed with codon optimization to allow efficient expression in Escherichia coli. For coexpression purposes, the three genes were subcloned into pET-20b in an operonal arrangement, and to facilitate identification and purification of the putative AdT, sequences encoding the V5 viral epitope and a 6xHis tag were fused to the 3′-end of PET112. E. coli strain BL21 was transformed with the overexpressing plasmid, and operon expression was induced. Figure 1C shows that Ymr293c and Ygr102c coelute with Pet112p from an immobilized Co2+ resin, showing that the three proteins can assemble into a trimer in the heterologous E. coli expression system. Having purified a particle that structurally resembles an AdT, we wanted to check whether this putative AdT can indeed catalyze tRNA-dependent transamidation. Since no mischarging ERS could be identified in yeast, and having shown that the plant mitochondrial GatCAB AdT (Pujol et al. 2008), like all GatCAB AdTs studied so far, can transamidate both E-tRNAQ and aspartyl-tRNAAsn (D-tRNAN), we attempted transamidation of a heterologous bacterial D-tRNAN with the purified putative yeast AdT. Figure 1D shows that the yeast GatCAB-like AdT very efficiently transamidates Thermus thermophilus D-tRNAN. This amidation is strictly tRNA-dependent since pure yeast AdT does not convert free [14C]Asp into [14C]Asn (data not shown). While Pet112p and Ymr293c are highly similar to the bacterial GatB and GatA subunits, respectively, and can therefore be considered as real orthologs of GatB and GatA, Ygr102c has no similarity to any bacterial GatC subunit; we therefore named this subunit GatF. These lines of experiments show that using a systems-based approach we isolated a GatFAB trimeric AdT able to catalyze tRNA-dependent amidation of a mischarged D-tRNAN.
Figure 1.
Identification of a GatCAB-like AdT in yeast. (A) Growth phenotypes of the pet112Δ and pet112-TAP strains obtained in fermentation (YPD) and respiratory (YPGly) conditions after 48 h of incubation at 30°C. Origin and engineering of the strains are described in the Materials and Methods and Supplemental Table 1. (B, lanes 2,3) G250 Colloidal Blue-stained SDS-PAGE separation of the Pet112-TAP interactants purified by TAP. (Lane 1) Molecular weight standards. The three putative mitochondrial proteins, identified by mass spectrometry, are highlighted. (C) Coomassie Blue-stained SDS-PAGE of the overexpressed S. cerevisiae GatFAB AdT purified on an immobilized Co2+ column. The band labeled with an asterisk (*) corresponds to contaminating E. coli prolyl-isomerase (25 kDa; Mukherjee et al. 2003). (D) Autoradiogram of the TLC plate analyzing the conversion of 10 μM tRNAAsn-bound [14C]aspartate ([14C]D) into [14C]asparagine ([14C]N) catalyzed by 0.1 μM pure S. cerevisiae GatFAB AdT in the presence (+) or absence (−) of glutamine as an amide group donor. Aspartylation and transamidation reactions of the Thermus thermophilus tRNAAsn were performed as described previously (Bailly et al. 2007).
Existence of a yeast mitochondrial tRNA-dependent transamidation pathway that generates glutaminyl-mtRNAGln
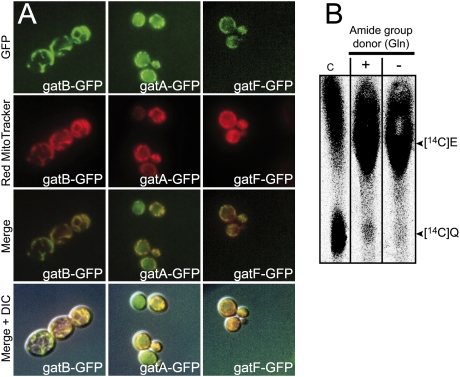
Although genetic experiments showed that Pet112p is a mitochondrial protein, evidence for a mitochondrial localization of Ymr293c and Ygr102c comes from whole high-throughput localization studies (http://www.yeastgenome.org; Huh et al. 2003). Moreover, no mitochondrial targeting sequence can be detected in Ymr293c using standard bioinformatics tools (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html). We therefore examined the intracellular localization of the three subunits of our putative trimeric AdT by expressing in yeast versions of Pet112p, Ymr293c, and Ygr102c fused to GFP at their C termini. Adding the GFP tag does not interfere with the mitochondrial function of the recombinant proteins, since the yeast strains expressing the Gat-GFP fusion proteins instead of the normal Gat subunits do not display any growth defect when plated on respiratory medium (Supplemental Fig. 2). Figure 2A shows that all three proteins are localized within the mitochondria as demonstrated by the colocalization (merge) of GFP fluorescence with that of the red-MitoTracker. The observed mitochondrial GFP fluorescence is solely due to the fact that GFP is fused to mitochondrial-targeted proteins, since GFP alone has been reported to be exclusively cytosolic (Bordonné 2000; Navarro et al. 2004). Having confirmed by direct visualization that the three subunits localize to mitochondria, we checked whether mitochondrial extracts can catalyze transamidation of mitochondrial E-tRNAQ into Q-tRNAQ. Total mitochondrial tRNA and total mitochondrial protein extract were assayed for the capacity of the protein extract to catalyze mischarging of mtRNAQ with [14C]glutamate ([14C]E) and subsequent amidation of the [14C]E into [14C]glutamine ([14C]Q). Figure 2B shows that the mitochondrial extract is able to generate a mischarged mitochondrial [14C]E-tRNAQ species and to convert the [14C]E into [14C]Q only in the presence of an amide group donor. The above results demonstrate that there exists a yeast mitochondrial tRNA-dependent transamidation pathway responsible for the formation of Q-mtRNAQ. This suggests that the mitochondrial GatFAB AdT we identified may well catalyze transamidation of E-tRNAQ. However, while the transamidation that we observed using mitochondrial extracts undoubtedly shows that E-tRNAQ can be formed in yeast mitochondria, the nondiscriminating glutamyl-tRNA synthetase (ND-ERS) that generates this intermediate inside mitochondria remained to be identified.
Figure 2.
Mitochondrial localization and activity of the S. cerevisiae GatFAB AdT. (A) Visualization of the mitochondrial localization of the C-terminally GFP-tagged GatA, GatB, and GatF subunits by fluorescence microscopy. Yeast gatA-GFP, gatB-GFP, and gatF-GFP strains were logarithmically grown on synthetic medium as described in the Materials and Methods. Living cells were stained with Red MitoTracker and examined with a phase interference microscope as described in the Materials and Methods. GFP fluorescence can be overlapped with the Red MitoTracker staining of mitochondria. Note that a fraction of Ymr293c localizes also in the cytosol in agreement with a recent report on a possible additional function of this protein in this compartment (Federovitch et al. 2008). (B) Autoradiogram of the TLC plate showing the conversion of yeast mitochondrial tRNA-bound [14C]E into [14C]Q catalyzed by a yeast mitochondrial extract. Coupled aminoacylation and transamidation of 16 μg of unfractionated mitochondrial tRNA, using 10 μg of a tRNA-free mitochondrial protein extract, was performed in 50 μL of transamidation buffer supplemented with 20 μM of [14C]E and with (+) or without (−) 1 mM of glutamine (amide group donor). After 40 min of incubation, samples were treated as described in the Materials and Methods.
The cytosolic glutamyl-tRNA synthetase is nondiscriminating and charges the mitochondria-encoded tRNAGln with glutamate
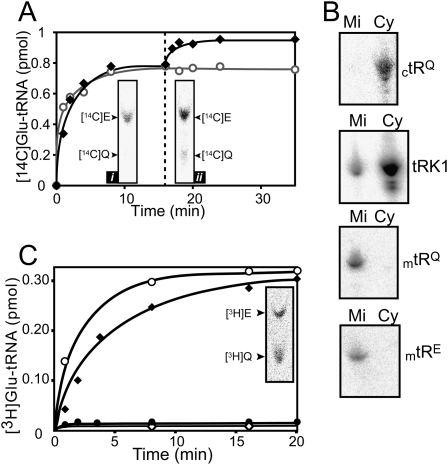
It has been established that the S. cerevisiae genome encodes only two distinct ERSs, one being mitochondrial and the other cytosolic (Tzagoloff and Shtanko 1995; Gagnon et al. 1996). Furthermore, it has been shown that the mERS is of a discriminating nature and cannot generate the E-mtRNAQ obligatory substrate of the AdT (Rinehart et al. 2005). Our data fully support this result since the [14C]E-tRNA formed by charging of total mitochondrial tRNA with purified yeast mERS and [14C]E does not yield any tRNA-dependent synthesis of [14C]Q when incubated with pure yeast GatFAB AdT (Fig. 3A, inset i). Moreover, mtRNAE is perfectly charged by mERS, while mtRNAQ is not (Fig. 3C). We then examined the possibility that the cytosolic glutamyl-tRNA synthetase (cERS) can carry out mischarging of mtRNAQ, despite the fact that this ERS has so far never been reported to be localized in mitochondria and does not display any identifiable mitochondrial targeting sequence (MTS). If cERS is capable of mischarging mtRNAQ, then when the aminoacylation plateau of total mitochondrial tRNA with mERS is reached, addition of cERS should promote an increase in the aminoacylation plateau level because a mtRNA species different from mtRNAE would additionally be aminoacylated. Figure 3A shows that upon addition of cERS there is an increase in the amount of mtRNA that can be glutamylated. This increase is cERS-dependent since addition of the same amount of mERS resulted in an unchanged plateau level. The fact that the additional glutamylated mtRNA formed by cERS can be transamidated by purified yeast GatFAB AdT confirms that cERS mischarges mtRNAQ (Fig. 3A, inset ii). However, it has been reported that three tRNAQ species can be found in yeast mitochondria: one mitochondrial-encoded species and two nuclear-encoded species (ctRNAsQ) that are mainly cytosolic but that are also imported to some extent into these organelles (Rinehart et al. 2005). To identify the tRNAQ species charged by cERS and subsequently transamidated by the AdT, we probed, by Northern-blotting, our total mitochondrial tRNA preparation for the presence of the three tRNAQ species using a probe specific for mtRNAQ and one that hybridizes to both ctRNAsQ that are believed to be imported from the cytoplasm. Figure 3B shows that the total mitochondrial tRNA contains both mtRNAQ and mtRNAE and that our probes are highly specific since they do not cross-hybridize to any cytosolic tRNA species. While we could, as expected, detect the presence of both ctRNAsQ species in the total cytosolic tRNA sample, we could not detect any trace of these two tRNAs in our mitochondrial tRNA sample (Fig. 3B). We confirmed that absence of ctRNAsQ in our mitochondrial tRNA sample was not biased by the extraction procedure, since it contained tRK1 (Fig. 3B), a yeast cytosolic tRNALys that has been shown to be imported at a level of 5% in yeast mitochondria (Martin et al. 1979; Entelis et al. 1998). Figure 3B shows that the amount of tRK1 present in our mitochondrial tRNA corresponds to the expected quantity.
Figure 3.
Identification of the missing nondiscriminating ERS and of the AdT tRNAQ substrate. (A) Aminoacylation curves of unfractionated mtRNA with mERS and subsequent addition of mERS (-○-) or cERS (-◆-) when the aminoacylation plateau with mERS was reached (16 min, dashed line). Autoradiogram of the TLC plate analyzing transamidation by purified yeast GatFAB AdT of [14C]E-mtRNA (40 μL) generated at plateau level by mERS (inset i) or generated at plateau level by combined charging with mERS and cERS (inset ii). (B) Identification by Northern blotting of the tRNAQ and tRNAE species present in our unfractionated mtRNA preparations. 15 μg of unfractionated mtRNA (mi) was hybridized with probes specific for mitochondrial-encoded tRNAQ (mtRQ), tRNAE (mtRE), and nuclear-encoded tRNAQ (ctRQ) and tRNAK (tRK1) shown previously to be imported into mitochondria. As a control, 15 μg of unfractionated ctRNA (cy) was probed with the same probes. (C) Charging curves of purified mtRNAQ and mtRNAE by cERS (respectively, -◆- and -●-) and mERS (respectively, -◇- and -○-). (Inset) Transamidation of the pure mischarged [14C]E-mtRNAQ by pure yeast GatFAB AdT.
These data show that cERS has the potential to be the missing nondiscriminating ERS used for the mitochondrial transamidation pathway, since it is capable of charging the mtRNAQ (Fig. 3C). In addition, the inset in Figure 3C shows that the E-mtRNAQ generated by cERS can subsequently be transamidated by the recombinant yeast GatFAB. Notably, while cERS is able to charge ctRNAE, it is unable to charge the mtRNAE (Fig. 3C).
A fraction of the cytoplasmic glutamyl-tRNA synthetase is imported into mitochondria
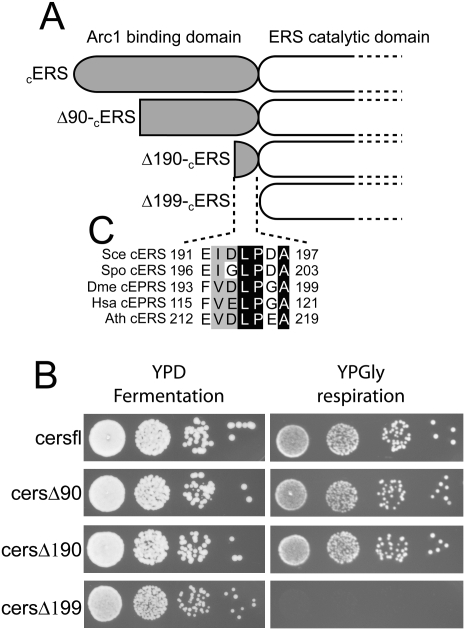
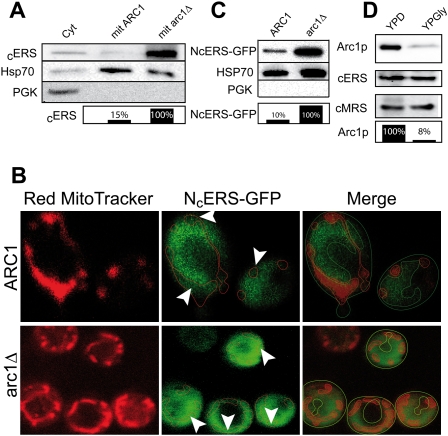
While the yeast cERS possesses all the biochemical features of the missing ND-mERS that catalyzes the first step of the transamidation pathway, it was not known whether this enzyme can be imported into mitochondria. When compared with prokaryotic ERSs, the yeast cERS possesses 200 additional residues appended to the N terminus. While this additional domain does not display any recognizable MTS, we hypothesized that a nonconventional signaling sequence might be embedded in this part of the protein. We therefore constructed a series of cERS N-terminal deletion mutants with the expectation of cleaving off the signaling sequence without impairing the cytoplasmic catalytic activity of this enzyme (Fig. 4A). Such mutant cERSs should still be able to promote growth on fermentable medium (glucose) but not on respiratory medium (glycerol). We used a yeast shuffling strain in which a plasmid-borne wild-type cERS gene complements the chromosomal deletion of this gene. This strain was transformed with the recombinant plasmids expressing our three deletion mutants and subsequently plated on 5-FOA to select for loss of the plasmid encoding wild-type cERS. Figure 4B shows that growth on glucose and glycerol media of the strains expressing either ΔN90-cERS (cersΔ90) or ΔN190-cERS (cersΔ190) is comparable with that of the wild-type strain (cersfl). However, growth of the cersΔ199 strain that expresses the ΔN199-cERS mutant is severely impaired in respiratory conditions but is almost unchanged on fermentable medium when compared with the wild-type strain (Fig. 4B). The petite phenotype observed for the cersΔ199 mutant strain demonstrates that cERS has an essential mitochondrial function that is lost upon removal of its N-terminal extension. Since the catalytic activity of the mutant ΔN199-cERS seems unaltered in the cytoplasm, the deletion very likely hindered the import of the mutant cERS into mitochondria. To confirm that the cERS is imported into mitochondria, we probed protein extracts from purified mitochondria for the presence of cERS using antibodies directed against yeast cERS. Figure 5A shows that cERS is present in mitochondria of a wild-type yeast strain. When compared with the amount found in a cytosolic fraction from the same strain, it appears that a significant amount of cERS is imported into the mitochondria. Our mitochondrial extracts were free of any cytosolic contaminants as shown by the absence of a cytosolic marker (PGK) in the mitochondrial proteins. To further confirm that the MTS of cERS is embedded within its first 199 residues, we subcloned into pRS315 the part of the cERS gene encoding the first 199 amino acids in-frame and upstream of the gene encoding GFP (NcERS-GFP). The chimeric gene was cloned under the control of its own promoter in order to not bias the amount of NcERS-GFP that would be targeted to the mitochondria by overexpression of the fusion protein. The transformed yeast cells were grown in respiratory conditions, and the subcellular localization of the NcERS-GFP was analyzed by confocal laser scanning microscopy. Figure 5B (top panels) shows that NcERS-GFP, as expected, is abundantly localized in the cytoplasm. However, a visible amount of NcERS-GFP is undoubtedly localized in mitochondria, as shown by overlapping GFP and red MitoTracker signals (Fig. 5B, white arrows). Since we used a focal plane as thin as 900 nm, the portion of the GFP fluorescence that colocalizes with the red MitoTracker emission cannot be due to interfering cytosolic GFP fluorescence that would overlap mitochondria. This demonstrates that the MTS of cERS is indeed localized within the 199 N-terminal residues. Taking into account that the ΔN190-cERS variant did not induce respiratory deficiency whereas the ΔN199-cERS variant did, the MTS is very likely localized between residues 191 and 199.
Figure 4.
A noncanonical MTS located after the first 190 residues of CERS is responsible for its mitochondrial import. (A) Schematic representation of the different cERS variants. (B) Growth phenotypes on fermentative (YPD) or respiratory (YPGly) media of haploid S. cerevisiae cersfl, cersΔ90, cersΔ190, and cersΔ199 strains (see Supplemental Table 1). (C) Multiple alignment of S. cerevisiae cERS with other eukaryotic cERSs and glutamyl-prolyl-tRNA synthetases (EPRS). (Sce) S. cerevisiae, (Spo) Schizosaccharomyces pombe, (Dme) Drosophila melanogaster, (Ath) A. thaliana, (Hsa) Homo sapiens. The region analyzed corresponds to the residues 190–199 of the S. cerevisiae cERS. Black shading denotes 100% amino acid identity, while gray signifies at least 80% homology (amino acid conservation in at least four out of the five proteins).
Figure 5.
cERS is maintained in the cytosol by binding to Arc1p, a cytosolic retention platform, whose expression is down-regulated in response to a switch from fermentation to respiration. (A) Immunodetection, by Western blot, of cERS in purified cytosolic (cyt) and mitochondrial (mit) protein extracts of ARC1 (wild type, RS354) or arc1Δ strains. Amounts of total protein and cross-contamination of each mitochondrial extract were controlled using antibodies against Hsp70 (Hsp70) as a mitochondrial marker and against phosphoglycerokinase (PGK) as a cytosolic marker. (B) Analysis of the subcellular localization of NcERS-GFP in ARC1 (wild type, RS354) or arc1Δ yeast strains transformed with pRS315/NtercERS-GFP. Living cells were stained with Red MitoTracker and examined with a confocal microscope as described in the Materials and Methods. Cellular areas where fluorescence of NcERS-GFP overlaps with the Red MitoTracker staining of mitochondria are indicated by white arrowheads. (C) Western blot quantification of import of NcERS-GFP in mitochondria of ARC1 (wild type, RS354) or arc1Δ strains. Amounts of total protein and level of cytosolic contamination of each mitochondrial extract were controlled using anti-Hsp70 (Hsp70) and anti-PGK (PGK) antibodies, respectively. (D) Western blot quantification of the variation in expression, by wild-type yeast strain (ARC1), of Arc1p, cERS, and cMRS during fermentation (YPD) or respiration (YPGly). Histograms representing quantifications of the relative amounts of cERS (A), NcERS-GFP (C), and Arc1p (D) present in extracts were performed with ImageJ software.
These lines of experimental evidence demonstrate that cERS, in addition to being nondiscriminating, is also mitochondrial. Therefore, upon import into the organelle, cERS is able to synthesize the mischarged E-mtRNAQ that will subsequently be transamidated by the mitochondrial GatFAB AdT, generating the Q-mtRNAQ necessary for mitochondrial protein synthesis.
Arc1p acts as a cytosolic anchoring platform for regulated retention of cERS in the cytoplasm
An important issue raised by our findings is the understanding of the mechanism that regulates the dual localization of cERS, since its activity is essential for both cytosolic and mitochondrial protein synthesis. It is already known from previous studies that both yeast cERS and cytosolic methionyl-tRNA synthetase (cMRS) are associated with Arc1p, a cofactor of nuclear export (Simos et al. 1996). Interaction of cMRS and cERS with Arc1p, mediated by their N termini (Deinert et al. 2001), increases their tRNA aminoacylation efficiencies and prevents nuclear translocation of the two aaRS (Galani et al. 2001). While nuclear localization of cMRS has been confidently shown, the nuclear localization of yeast cERS is somewhat less clear (Golinelli-Cohen and Mirande 2007). Since Arc1p entraps yeast cERS in the cytosol, we surmised that this aaRS is preferentially a mitochondrial protein, and that Arc1p captures and holds in the cytoplasm the portion of cERS required for cytosolic translation. To verify this hypothesis, we checked by Western blotting whether removal of Arc1p directly impacts the fraction of cERS addressed to the mitochondrion. Figure 5A shows that, indeed, in an Arc1p deletion strain (arc1Δ), the pool of cERS imported into mitochondria is drastically increased when compared with the amount found in mitochondria purified from the wild-type strain (ARC1). The absence of PGK and presence of HspP70 in comparable amounts in both the arc1Δ and wild-type (ARC1) mitochondrial extracts shows that there is no contamination by a cytosolic portion of cERS and that comparable amounts of total mitochondrial protein extracts were subjected to PAGE and transferred onto the membrane. A similar result was observed when comparing, by confocal laser scanning microscopy, the amount of the NcERS-GFP fusion protein addressed to mitochondria in wild type (ARC1) and an arc1Δ strain transformed with the plasmid expressing NcERS-GFP. Figure 5B shows that the portion of NcERS-GFP addressed to the organelle, in the absence of Arc1p, is increased in such a manner that the intensity of the mitochondrial fluorescence signal equals that of the cytosolic one. We observed a comparable increased import of NcERS-GFP in the arc1Δ mutant as compared with the wild type by Western blotting using anti-GFP antibodies (Fig. 5C). These experiments confirm that Arc1p retains in the cytoplasm a pool of cERS that otherwise would naturally be imported by mitochondria. Taking into account that the mitochondrial pool of cERS is increased by more than one order of magnitude upon depletion of Arc1p, we conclude that, in an arc1Δ background, cERS is preferentially a mitochondrial protein.
Increased import of cERS into mitochondria is needed only when yeast cells are grown on a respiratory carbon source, suggesting that Arc1p regulates the subcellular sorting of cERS in response to metabolic changes. Figure 5D indeed shows that the amount of Arc1p is drastically decreased when yeast uses respiration as compared with fermentation, while the content of cMRS and cERS does not vary. In addition, in cells grown on glucose, cERS, cMRS, and Arc1p seem to be present in stoichiometric amounts, in agreement with previous studies reporting that Arc1p always copurifies with equivalent amounts of cERS and cMRS (Graindorge et al. 2005). During respiration, however, there is an excess of free cERS and cMRS that can be addressed to their target subcellular compartments, the mitochondria and the nucleus, respectively. These results demonstrate that Arc1p is a cytosolic retention platform for cERS and cMRS, whose amounts are regulated in response to the switch from fermentation to respiration to allow relocalization of these cytosolic aaRSs into organelles.
Discussion
The work described herein reopens the debate on how Q-tRNAQ synthesis is achieved in yeast mitochondria and more generally in other eukaryotic species. Recent studies have reported that import of both ctRNAQ and cQRS was needed for mitochondrial Q-tRNAQ synthesis in yeast (Rinehart et al. 2005) and that mammalian mitochondria also import ctRNAQ (Rubio et al. 2008). This led to a change of paradigm concerning organellar Q-tRNAQ synthesis that was, until these reports, assumed to uniformly proceed via tRNA-dependent transamidation (Schön et al. 1988). Our work unambiguously demonstrates that in yeast, mitochondria use the transamidation pathway to generate Q-mtRNAQ (Fig. 6), and that this pathway is essential and sufficient for mitochondrial translation since deletion of any of the AdT subunit genes or suppression of the mitochondrial targeting capacity of the ND-ERS induces a respiratory deficiency. Beyond the discovery of a mitochondrial trimeric AdT in S. cerevisiae, we also found orthologs of the PET112 and YMR293C genes in all fungal genomes that are complete and accessible (http://cbi.labri.fr/Genolevures). However, recognizable orthologs of YGR102C (encoding GatF) could only be found in the Kluyveromyces lactis (gi:50307515) and Candida glabrata (gi:50288545) genomes. The human genome encodes, in addition to orthologs of the gatA (YMR293C) and gatB (PET112) genes, an ortholog of the bacterial gatC gene (gatA: gi:19923522, gatB: gi:4758893, gatC: gi:50978623). Two of these gene products, GatB and GatC, like in yeast, display the sequence of a canonical MTS. We are therefore confident that most, if not all, eukaryotes use the transamidation pathway to generate Q-mtRNAQ. It has been recently confirmed that this is the case in A. thaliana mitochondria (Pujol et al. 2008).
Figure 6.
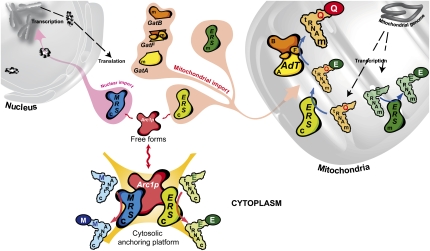
Subcellular localization, ligand binding, and activities of the proteins and RNA involved in S.cerevisiae Q-mtRNAQ formation. In S. cerevisiae, the mitochondrial (m) tRNAQ is glutamylated by a fraction of the cytosolic (c) ERS imported into mitochondria. The mitochondrial E-tRNAQ is then transamidated into Q-tRNAQ by a mitochondrial GatFAB AdT. Formation of the mitochondrial E-tRNAE is mediated solely by the mitochondrial ortholog (mERS) of the cERS. A portion of the cERS is maintained in the cytosol by binding to Arc1p that additionally by capturing the cytosolic MRS hinders its nuclear addressing. Both enzymes are essential to the cytosolic translation since they catalyze formation of the cytosolic E-tRNAE and M-tRNAM, respectively.
In addition, our experiments indicate that the yeast mitochondrial-encoded tRNAQ is the only tRNAQ used by the mitochondrial translation apparatus, since our mitochondrial tRNA preparations were always deprived of any trace of nuclear-encoded tRNAQ. Indeed, in conditions where the cytoplasmic lysine tRNA isoacceptor (tRK1) was well detected in our mitochondrial tRNA preparations, the ctRNAQ were undetectable. It has been proposed that the mitochondrial import of nuclear-encoded ctRNAQCUG is required for the decoding of CAG glutamine codons in mitochondrial translation because the uridine modification, cmnm5s2U, in the wobble position of the anti-codon of mtRNAQ would restrict the reading to only CAA glutamine codons (Rinehart et al. 2005). However, all other reports published so far point out that cmnm5s2U is able to pair with both A and G in the codon's third position (Yokoyama and Nishimura 1995; Umeda et al. 2005; Kurata et al. 2008), implying that mtRNAQ (cmnm5s2U) is able to decode both CAA and CAG glutamine codons. In this respect, we may notice that the yeast mitochondrial-coded mtRNALys, which bears the same cmnm5s2U wobble modification as mtRNAQ, was shown to be fully functional in mitochondrial protein synthesis (i.e., capable of decoding both AAA and AAG lysine codons), at least at normal temperature (28°C) for growth on respiratory media (Kamenski et al. 2007). At higher growth temperatures (37°C), the mitochondrial import of ctRNALysCUU (tRK1) becomes essential for the reading of AAG codons because the cmnm5s2U wobble modification of mtRNALys is impaired at 37°C. However, this defect in cmnm5s2U modification is specific for mtRNALys, since formation of cmnm5s2U in mtRNAQ was not affected in the same conditions (Kamenski et al. 2007). We therefore believe that import of ctRNAQ for CAG codon translation purposes in mitochondria cannot be regarded as an absolute requirement. Furthermore, assuming that the two ctRNAsQ would nevertheless be imported into yeast mitochondria, our data show that these species would be imported at a very low level, at least in the strains and growth conditions used in our work, which would question their involvement in mitochondrial translation.
Along the same lines, the report that cQRS is essential for mitochondrial protein synthesis is inconsistent with data obtained by us and others (Nabholz et al. 1997). If we assume that cQRS is responsible for Q-mtRNAQ synthesis, then blocking the mitochondrial import of cERS or deletion of any of the genes encoding GatFAB should not induce a respiratory-deficient phenotype (this work; Mulero et al. 1994; Kim et al. 1997). These considerations raise two possibilities: Either cQRS is not imported into yeast mitochondria, or the fraction of cQRS that is imported does not participate to Q-mtRNAQ formation. To verify the latter, we engineered a cQRS variant fused to a canonical MTS and tried to complement an AdT-defective strain (Supplemental Table 1; Materials and Methods). Drop tests showed that this cQRS variant is unable to complement the respiratory deficiency of a pet112 mutant, suggesting that even if cQRS is addressed to mitochondria, it is unable to supply the mitochondrial translation apparatus with Q-mtRNAQ (Supplemental Fig. 3).
One of the most intriguing findings of our work is the fact that yeast cERS is preferentially localized in mitochondria in the absence of Arc1p that acts as a cytosolic retention platform (Fig. 6). Dual localization of cERS is not the first example of a dual, cytosolic- and mitochondrial-localized protein in yeast and more generally in eukaryotes (Karniely and Pines 2005). Among proteins that are distributed between different subcellular compartments, four yeast aaRSs have already been shown to be dual-localized (Turner et al. 2000; Tang et al. 2004). However, in all cases studied so far, dual localization of a protein is almost always the result of translation of two different isoforms either from alternative splice variants or because translation begins at alternative start sites. Thus, both isoforms have different sequences, and since only one has a mitochondrial targeting signal, each isoform is restrained to a single cellular compartment. In the case of cERS, both the cytosolic and the mitochondrial fractions have the same exact amino acid sequence. We could find only a few other examples whereby a single translation product is distributed between membrane-separated subcellular compartments. One is a S. cerevisiae adenylate kinase, Aky2, for which a portion of the enzyme is targeted to the intermembrane space of mitochondria while a fraction folds before the MTS accesses the mitochondrial import receptors and is therefore retained in the cytoplasm (Strobel et al. 2002). Another example is S. cerevisiae fumarase, for which all molecules are targeted to mitochondria and processed, but only a subset of the mature molecules is further translocated into the mitochondrial matrix while another fraction goes back to the cytoplasm by a retrograde movement (Stein et al. 1994; Knox et al. 1998). However, the strategy used to control dual, cytosolic and mitochondrial, localization of cERS is unprecedented and distinct from any example reported so far (Karniely and Pines 2005). The mechanism by which the cERS is retained in the cytoplasm is entrapment by binding to a cytosolic anchoring protein. We suspect that retention is very likely not achieved by masking of the cERS MTS. The recent crystal structure of the binary complex made between the N termini of cERS and Arc1p favors our assumption, since it shows that only residues 122–131 and 157–173 of cERS are involved in the interaction with Arc1p (Simader et al. 2006). The entire Arc1p-binding region has been deleted in the ΔN190-cERS that is still imported and functional in mitochondria, as shown by unaffected growth on glycerol plates of the cersΔ190 strain (Fig. 4B). What was also unexpected is that although both subcellular fractions of the cERS have the same sequence, they catalyze two different reactions; in the cytosol, cERS produces E-ctRNAE, while the mitochondrial subset forms E-mtRNAQ. We suspect that cERS has an intrinsic capacity to be nondiscriminating for the tRNA, but its association with Arc1p in the cytosol restricts its specificity to only tRNAE charging, in agreement with the increased affinity of cERS for tRNAE mediated by binding to Arc1p (Graindorge et al. 2005).
This study also shows that the role of Arc1p is not confined to a catalytic enhancer, but that it constitutes a subcellular sorting platform whose expression is regulated in response to which metabolism—fermentation or respiration—is used by yeast. We showed that when yeast uses respiration, there is a decrease in the expression of Arc1p that leads to release of cERS and enhanced import of this aaRS into the mitochondria in order to sustain increased levels of translation in this compartment. However, the molecular signaling pathway that controls the expression of Arc1p in response to the switch from fermentation to respiration has yet to be deciphered. Since Arc1p has also been shown to restrain cMRS from going into the nucleus, our work suggests that under respiratory conditions there should also be an increase of nuclear targeting of cMRS. The role of cMRS inside the nucleus remains yet to be identified. Reports on high-throughput analyses of S. cerevisiae protein–protein interactions show that cMERS interacts with several nuclear proteins involved in gene transcription regulation and initiation (Gavin et al. 2002; Krogan et al. 2006). One of these interactants is Yll022c (HIF1), which is a component of the HAT-B histone acetyltransferase complex (Poveda et al. 2004), and another is Ygl112c (TAF6), a subunit of TFIID and SAGA complexes involved in transcription initiation by RNA polymerase II and in chromatin modification (Grant et al. 1998). One could easily speculate that cMRS could be involved in transcription regulation of certain genes, and since we suspect the nuclear localization of cMRS to be increased when there is a higher need for mitochondrial activity, one would speculate that the genes under the dependence of cMRS could encode mitochondrial proteins.
Beyond subcellular localization of an aaRS, our data suggest that, in eukaryotes, proteins that have been identified so far to be cytosolic with certainty might nevertheless be capable of being addressed to mitochondria and display an essential function therein, even if a functional organellar ortholog of this cytosolic protein has already been identified. One can wonder whether Arc1p could belong to a broader family of proteins that play the role of cytosolic anchors for proteins that so far have not been considered as being able to reach multiple subcellular destinations. In the light of our work, we suggest a re-examination of the pioneering studies on whole-cell protein–protein interaction networks that led to identification of many unsuspected multienzymatic complexes that are still waiting to be attributed a function. Perhaps gain of function is not the driving force that led to some of these protein–protein interactions, but rather dynamic control of subcellular distribution.
To conclude, our work describes three original aspects of organellar molecular biology. We prove that yeast Q-mtRNAQ synthesis is formed by a bacterial-like AdT, displaying besides two canonical GatA and GatB subunits, a new type of fungi-specific subunit (GatF). We describe a new mechanism of dual—cytoplasmic and mitochondrial—localization of a protein whereby a preferentially mitochondrial protein is also maintained in the cytoplasm by binding to a cytosolic anchoring platform. And finally, we show that the dynamic sorting of this protein between the two compartments is mediated through regulation of the expression of the cytosolic anchoring protein upon the switch from fermentation to respiration.
Materials and methods
Strains and oligonucleotides
Yeast strains used and engineered for this study are described in Supplemental Table 1. Oligonucleotides and biotinylated oligonucleotides used as primers for PCR amplifications and as bait for tRNA affinity purification, respectively, are described in Supplemental Table 2.
Engineering of PET112-TAP strain and TAP purification procedure
The pet112-TAP strain was constructed as described previously (Puig et al. 2001), by homologous replacement of the chromosomal PET112 C terminus with the PCR product obtained by using oligonucleotides 1 and 2 and the pBS1479 plasmid for the amplification reaction. Twenty grams of cells of the pet112-TAP strain, obtained after 8 h of culture in YPD at 30°C, were suspended in 20 mL of disruption buffer (Supplemental Material) and cells were disrupted using a cell disruptor working at 37 psi. The clear lysate obtained after 20 min of centrifugation at 12,000g, was subjected to the TAP procedure as described previously (Puig et al. 2001). The gel bands corresponding to each individual protein present in the final eluate were analyzed by mass spectrometry (MALDI-TOF).
Preparation of mitochondria, total mitochondrial tRNA, and mitochondrial protein extracts
Yeast mitochondria were isolated following the procedure described previously (Entelis et al. 2002; Boldogh and Pon 2007). Total mitochondrial tRNA was extracted from freshly purified mitochondria using the procedure described by Martin et al. (1977, 1979). Protein extracts of the mitochondrial matrix were prepared as follows: Fresh mitochondria, suspended in lysis buffer (Supplemental Material), were disrupted by sonication (VibraCell BioblockScientific) and centrifuged 20 min at 12,000g to remove broken mitochondria and 90 min more at 105,000g in order to pellet all cytosolic contaminants adsorbed on tiny membrane fragments. For Western blot purposes, the supernatant was directly transferred at −80°C. For aminoacylation and transamidation purposes, removal of contaminating mitochondrial tRNA from the supernatant was obtained by DEAE-cellulose chromatography (DE52, Whatman) following the manufacturer's instructions. Proteins eluting with 400 mM NaCl were dialyzed against lysis buffer and used directly.
Cloning, overxpression, and purification of the yeast GatFAB AdT, mERS, and cERS
We designed the yeast gatFAB operon as described previously (Bailly et al. 2008) except that the 3′-end of the gatB gene was extended in-frame by the sequence encoding the V5 epitope and a 6xHis tag. The gatFAB operon was synthesized by Genscript with codon optimization, subcloned into pET20b (Novagen) between the NdeI and XhoI restriction sites, and the recombinant plasmid was transformed into E. coli BL21 λDE3 pLysS. Cytoplasmic ERS (GUS1) and mERS (MSE1) genes, deprived of their stop codons, were PCR-amplified from S. cerevisiae genomic DNA using oligonucleotide pairs 3, 4 and 5, 6 as primers, respectively. The PCR products were directly ligated into the T/A expression vector pBAD-TOPO (Invitrogen) that allows expression of the corresponding proteins C-terminally fused to the V5 epitope and to a 6xHis tag and were transformed into the E. coli Rosetta 2 strain. Expression and purification of soluble GatFAB AdT, mERS, and cERS were achieved as described in the Supplemental Material.
Engineering of yeast expression plasmids of cERS and GatB variants
Yeast strains cersfl, cersΔ90, cersΔ190, and cersΔ199 expressing wild-type cERS and the N-terminally truncated variants ΔN90-cERS, ΔN190-cERS, and ΔN199-cERS, respectively, were obtained by PCR amplification using S. cerevisiae genomic DNA as a template and the following primer pairs, respectively: oligonucleotides 7 and 8, oligonucleotides 9 and 8, and oligonucleotides 10 and 8. The PCR products were digested with HindIII and BamHI and ligated into pRS315 digested with the same restriction enzymes. Genes of the truncated variants were cloned into pRS315 under the control of the NOP1 promoter. Transformation of the cersfl shuffle strain with the recombinant plasmids and plasmid shuffling experiments are described in the Supplemental Material. The gene encoding the first 199 amino acids of cERS fused to GFP (NcERS-GFP) was obtained, using S. cerevisiae genomic DNA as a template and oligonucleotide pair 11 and 12, by PCR amplification of a fragment of the cERS gene (GUS1) encompassing the 5′ untranslated region (350 base pairs) and the first 603 nucleotides of the ORF flanked by the HindIII/BamHI restriction sites. The PCR product was ligated into pRS315-GFP. The resulting pRS315/NcERS-GFP plasmid was transformed into yeast BY4741 (ARC1, wild-type) or arc1Δ strains. The yeast strain gatB-GFP, expressing GatB C-terminally fused to GFP (GatB-GFP), was obtained using the same PCR and cloning strategy and oligonucleotide pair 13 and 14.
Fluorescence and microscopy analysis
Yeast strains gatA-GFP and gatF-GFP expressing the GatA and GatF subunits C-terminally fused to GFP, respectively, were grown in synthetic medium SCG until A600nm = 0.3, while the gatB-GFP strain was grown in SGC lacking leucine. A 1-mL aliquot of the cells was stained with 1 μL of MitoTracker (Invitrogen) according to the manufacturer's instructions. After 20 min of incubation, cells were harvested by centrifugation, washed with fresh culture media, mounted on coverslips, and examined with a DIC Zeiss Axiovert 200M phase interference microscope using a 100× oil immersion objective (NO 1.4). Images were captured with a CoolSnap HQ2 camera and processed with Axiovision software. The same procedure was used for the visualization of the yeast arc1Δ and BY4741 (ARC1) strains transformed with pRS315/NtercERS-GFP except that they were grown in synthetic medium SCG lacking leucine and visualized with a Zeiss LSM510 confocal microscope, using a 63× oil immersion objective (NO 1.4). Images were processed with the AIM3.2 software.
Immunodetection analysis
RS453 S100 extracts were used as controls to visualize the proportion of the cytosolic fraction of cERS. For all extracts, protein concentration was determined using Bradford, and 10–15 μg of proteins were separated by SDS-PAGE on a 12% gel prior to electroblotting to Hybond-P membrane (Amersham). Detection was carried out using HRP-conjugated goat anti-rabbit antibodies (Bio-Rad) and ECL reagents (Amersham) according to the manufacturer's instructions. We used anti-cERS primary antibody (1:5000) to quantify the amount of cERS in each extract. To control the relative quantity and purity of our mitochondrial protein preparations, we used as a mitochondrial matrix protein marker HSP70, which was detected using anti-HSP70 (1:1000), and as a cytosolic marker PGK, which was revealed using anti-PGK (1:1000). For the quantification of Arc1p and cMRS, we used anti-Arc1p (1:5000), anti-cMRS (1:2000).
Northern analyses of tRNA
Detection of the mtRNAQ, mtRNAE, tRNAK1, and ctRNAQ was performed following a procedure previously described (Frugier et al. 2005) using 15 μg of unfractionated yeast mtRNA prepared as described above or total yeast ctRNA commercially available (Boehringer). We used 5 pmol of the specific α[32P]-radiolabeled oligonucleotide probes 17, 18, 19, and 20, respectively (see Supplemental Table 2).
tRNA purification
Purification of mtRNAQ and mtRNAE from 1 mg of unfractionated mitochondrial tRNA, prepared as described above, was performed using 5′-biotinylated specific oligonucleotides 15 and 16, respectively (see Supplemental Table 2), immobilized on streptavidine-agarose beads (Novagen) following the procedure described by Rinehart et al. (2005).
Aminoacylation and transamidation assays
The standard aminoacylation mixture (100 μL) containing 100 mM Na-Hepes (pH 7.2), 30 mM KCl, 10 mM ATP, 12 mM MgCl2, 30 μM L-[14C]glutamate ([14C]E) (330 cpm/pmol, Amersham) or 1 μM L-[3H]glutamate ([3H]E) (3000 cpm/pmol, Amersham), 0.1mg/mL BSA, 0.15 μM pure mtRNAQ or pure mtRNAE or 10 μM total mtRNA, and 0.01 μM mERS or cERS was incubated at 30°C. For determination of the aminoacylation plateau, after incubation times ranging from 0 to 20 min, 10-μL aliquots were withdrawn and TCA-precipitated, and the radiolabeled Glu-tRNA formed was determined by scintillation liquid counting. For transamidation purposes, the [14C]E- or -[3H]E-tRNA formed when plateaus were reached in a 40- to 50-μL aminoacylation mixture was extracted using acid-buffered phenol and chloroform; the radiolabeled E-tRNA was precipitated with ethanol in the aqueous layer recovered by centrifugation, sedimented, and redissolved in water. The standard transamidation mixture (50 μL) containing 100 mM Na-Hepes, 30 mM KCl, 12 mM MgCl2, 10 mM ATP, 1 mM L-Gln (except for controls) as amide group donor, 0.03–0.1 μM radiolabeled Glu-tRNA, and 100 nM S. cerevisiae pure GatCAB AdT was incubated for 15 min at 30°C. The [14C]- and [3H]aa-tRNA was ethanol-precipitated after phenol-chloroform extraction, dissolved in 50 μL of water, and deacylated by 30 min of incubation at 80°C in the presence of 25 mM KOH, followed by neutralization with HCl. The hydrolysate was dried in a Speed-Vac, dissolved in 3 μL of water, and fractionated by TLC on cellulose plates (20 × 20 cm, Merck) extended by a 3MM Whatman paper sheet (20 × 5 cm), with a solvent containing 2-propanol/formic acid/water (80/4/20, v/v/v). The [14C]- and [3H]-aa were revealed by scanning the 2-h-exposed image plate with a Fuji Bioimager.
Acknowledgments
We express our gratitude to Drs. I. Tarassov, N. Entelis, S. Friant, and J. Muterer for technical assistance and sharing of materials and equipment, and to Professor C. Stathopoulos and Dr. M. Blaise for critical reading. This work was supported by the University Louis Pasteur (Strasbourg), the Centre National de la Recherche Scientifique (CNRS), and the Association pour la Recherche sur le Cancer (ARC). M.F. was a recipient of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.518109.
Supplemental material is available at http://www.genesdev.org.
References
- Bailly M., Blaise M., Lorber B., Becker H.D., Kern D. The transamidosome: A dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Bailly M., Blaise M., Roy H., Deniziak M., Lorber B., Birck C., Becker H.D., Kern D. tRNA-dependent asparagine formation in prokaryotes: Characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNA(Asn) Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Becker H.D., Kern D. Thermus thermophilus: A link in the evolution of the tRNA-dependent amino acid amidation pathway. Proc. Natl. Acad. Sci. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I.R., Pon L.A. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2007;80:45–64. doi: 10.1016/S0091-679X(06)80002-6. [DOI] [PubMed] [Google Scholar]
- Bordonné R. Functional characterization of nuclear localization signals in yeast Sm proteins. Mol. Cell. Biol. 2000;20:7943–7954. doi: 10.1128/mcb.20.21.7943-7954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnow A.W., Hong K., Yuan R., Kim S., Martins O., Winckler W., Henkin T.M., Söll D. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinert K., Fasiolo F., Hurt E.C., Simos G. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- Entelis N.S., Kieffer S., Kolesnikova O.A., Martin R.P., Tarassov I.A. Structural requirements of tRNALys for its import into yeast mitochondria. Proc. Natl. Acad. Sci. 1998;95:2838–2843. doi: 10.1073/pnas.95.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis N., Kolesnikova O., Kazakova H., Brandina I., Kamenski P., Martin R.P., Tarassov I. Import of nuclear encoded RNAs into yeast and human mitochondria: Experimental approaches and possible biomedical applications. Genet. Eng. 2002;24:191–213. doi: 10.1007/978-1-4615-0721-5_9. [DOI] [PubMed] [Google Scholar]
- Federovitch C.M., Jones Y.Z., Tong A.H., Boone C., Prinz W.A., Hampton R.Y. Genetic and structural analysis of Hmg2p-induced endoplasmic reticulum remodeling in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:4506–4520. doi: 10.1091/mbc.E07-11-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier M., Ryckelynck M., Giegé R. tRNA-balanced expression of a eukaryal aminoacyl-tRNA synthetase by an mRNA-mediated pathway. EMBO Rep. 2005;6:860–865. doi: 10.1038/sj.embor.7400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon Y., Lacoste L., Champagne N., Lapointe J. Widespread use of the glu-tRNAGln transamidation pathway among bacteria. A member of the α purple bacteria lacks glutaminyl-tRNA synthetase. J. Biol. Chem. 1996;271:14856–14863. doi: 10.1074/jbc.271.25.14856. [DOI] [PubMed] [Google Scholar]
- Galani K., Grosshans H., Deinert K., Hurt E.C., Simos G. The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J. 2001;20:6889–6898. doi: 10.1093/emboj/20.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Golinelli-Cohen M.P., Mirande M. Arc1p is required for cytoplasmic confinement of synthetases and tRNA. Mol. Cell. Biochem. 2007;300:47–59. doi: 10.1007/s11010-006-9367-4. [DOI] [PubMed] [Google Scholar]
- Graindorge J.S., Senger B., Tritch D., Simos G., Fasiolo F. Role of Arc1p in the modulation of yeast glutamyl-tRNA synthetase activity. Biochemistry. 2005;44:1344–1352. doi: 10.1021/bi049024z. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz D., Pray-Grant M.G., Steger D.J., Reese J.C., Yates J.R., III, Workman J.L. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ibba M., Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Ibba M., Becker H.D., Stathopoulos C., Tumbula D.L., Söll D. The adaptor hypothesis revisited. Trends Biochem. Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- Kamenski P., Kolesnikova O., Jubenot V., Entelis N., Krasheninnikov I.A., Martin R.P., Tarassov I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol. Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Karniely S., Pines O. Single translation–dual destination: Mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Stange-Thomann N., Martins O., Hong K.W., Söll D., Fox T.D. A nuclear genetic lesion affecting Saccharomyces cerevisiae mitochondrial translation is complemented by a homologous Bacillus gene. J. Bacteriol. 1997;179:5625–5627. doi: 10.1128/jb.179.17.5625-5627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C., Sass E., Neupert W., Pines O. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 1998;273:25587–25593. doi: 10.1074/jbc.273.40.25587. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kurata S., Weixlbaumer A., Ohtsuki T., Shimazaki T., Wada T., Kirino Y., Takai K., Watanabe K., Ramakrishnan V., Suzuki T.2008Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U·G wobble pairing during decoding J. Biol. Chem. 28318801–18811. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Duplain L., Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.P., Schneller J.M., Stahl A.J., Dirheimer G. Study of yeast mitochondrial tRNAs by two-dimensional polyacrylamide gel electrophoresis: Characterization of isoaccepting species and search for imported cytoplasmic tRNAs. Nucleic Acids Res. 1977;4:3497–3510. doi: 10.1093/nar/4.10.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.P., Schneller J.M., Stahl A.J., Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Shukla A., Guptasarma P. Single-step purification of a protein-folding catalyst, the SlyD peptidyl prolyl isomerase (PPI), from cytoplasmic extracts of Escherichia coli. Biotechnol. Appl. Biochem. 2003;37:183–186. doi: 10.1042/ba20020044. [DOI] [PubMed] [Google Scholar]
- Mulero J.J., Rosenthal J.K., Fox T.D. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Curr. Genet. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- Nabholz C.E., Hauser R., Schneider A. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl-tRNA synthetase activities. Proc. Natl. Acad. Sci. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B., Rubino L., Russo M. Expression of the cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 2004;78:4744–4752. doi: 10.1128/JVI.78.9.4744-4752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda A., Pamblanco M., Tafrov S., Tordera V., Sternglanz R., Sendra R. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Pujol C., Bailly M., Kern D., Maréchal-Drouard L., Becker H., Duchêne A.M. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc. Natl. Acad. Sci. 2008;105:6481–6485. doi: 10.1073/pnas.0712299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Zahedi R.P., Pfanner N., Meisinger C., Sickmann A. Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Rinehart J., Horn E.K., Wei D., Söll D., Schneider A. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J. Biol. Chem. 2004;279:1161–1166. doi: 10.1074/jbc.M310100200. [DOI] [PubMed] [Google Scholar]
- Rinehart J., Krett B., Rubio M.A., Alfonzo J.D., Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes & Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio M.A., Rinehart J.J., Krett B., Duvezin-Caubet S., Reichert A.S., Söll D., Alfonzo J.D. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A., Kannangara C.G., Gough S., Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- Sheppard K., Yuan J., Hohn M.J., Jester B., Devine K.M., Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simader H., Hothorn M., Köhler C., Basquin J., Simos G., Suck D. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic Acids Res. 2006;34:3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Segref A., Fasiolo F., Hellmuth K., Shevchenko A., Mann M., Hurt E.C. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Stein I., Peleg Y., Even-Ram S., Pines O. The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:4770–4778. doi: 10.1128/mcb.14.7.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G., Zollner A., Angermayr M., Bandlow W. Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell. 2002;13:1439–1448. doi: 10.1091/mbc.01-08-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H.L., Yeh L.S., Chen N.K., Ripmaster T., Schimmel P., Wang C.C. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 2004;279:49656–49663. doi: 10.1074/jbc.M408081200. [DOI] [PubMed] [Google Scholar]
- Tumbula D.L., Becker H.D., Chang W.-Z., Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- Turner R.J., Lovato M., Schimmel P. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J. Biol. Chem. 2000;275:27681–27688. doi: 10.1074/jbc.M003416200. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Shtanko A. Mitochondrial and cytoplasmic isoleucyl-, glutamyl- and arginyl-tRNA synthetases of yeast are encoded by separate genes. Eur. J. Biochem. 1995;230:582–586. doi: 10.1111/j.1432-1033.1995.tb20599.x. [DOI] [PubMed] [Google Scholar]
- Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Nishimura S. Modified nucleosides and codon recognition. In: RajBhandary U., Söll D., editors. tRNA: Structure, biosynthesis and function. American Society for Microbiology Press; Washington, DC: 1995. pp. 207–233. [Google Scholar]