Abstract
Ultrafast transient absorption spectroscopy of wild-type bacteriorhodopsin (WT bR) and 2 tryptophan mutants (W86F and W182F) is performed with visible light excitation (pump) and UV probe. The aim is to investigate the photoinduced change in the charge distribution with 50-fs time resolution by probing the effects on the tryptophan absorption bands. A systematic, quantitative comparison of the transient absorption of the 3 samples is carried out. The main result is the absence in the W86F mutant of a transient induced absorption band observed at ≈300–310 nm in WT bR and W182F. A simple model describing the dipolar interaction of the retinal moiety with the 2 tryptophan residues of interest allows us to reproduce the dominant features of the transient signals observed in the 3 samples at ultrashort pump-probe delays. In particular, we show that Trp86 undergoes a significant Stark shift induced by the transient retinal dipole moment. The corresponding transient signal can be isolated by direct subtraction of experimental data obtained for WT bR and W86F. It shows an instantaneous rise, followed by a decay over ≈500 fs corresponding to the isomerization time. Interestingly, it does not decay back to zero, thus revealing a change in the local electrostatic environment that remains long after isomerization, in the K intermediate state of the protein cycle. The comparison of WT bR and W86F also leads to a revised interpretation of the overall transient UV absorption of bR.
Keywords: retinal proteins, dipolar interactions, tryptophan, ultraviolet probe
The membrane protein bacteriorhodopsin (bR) is a proton pump, the biological activity of which is triggered by the absorption of light by the protonated retinal cofactor (1, 2). On light excitation, the retinal moiety undergoes an isomerization from 13-cis to all-trans that proceeds with very high speed (≈0.5 ps) and large quantum efficiency (≈0.64). In the excited Franck–Condon state, before isomerization, a large photoinduced charge translocation along retinal increases its dipole moment by as much as 10–30 D (3, 4). Remarkably, the photoisomerization of the same chromophore in any environment other than that of the natural protein scaffold is no longer specific and it is less efficient and slower (5), indicating the enzymatic role of the protein environment that enhances the speed and selectivity of the retinal isomerization. Site-directed mutagenesis has shown that charged amino acid residues closely interacting with the charged retinal moiety (Arg82, Asp85, Asp212) largely influence the isomerization speed (6, 7). Theoretical investigations of a retinal-like model compound (8) rationalize these findings in modeling how a chloride counterion affects the energetics and photoreactivity. In addition, within the protein, the large photoinduced charge translocation along retinal is expected to induce a strong, ultrafast dielectric response of the full protein environment (9, 10), which could also participate in the enzymatic function by driving specifically the isomerization path to all-trans retinal. Besides, the exact nature of the driving force for the biological activity of the protein has long been (11) and still is a subject of intensive investigations. Hence, protein conformational changes similar to those observed in functional wild-type (WT) bR have been observed in artificial proteins with nonisomerizing retinal chromophores (12), suggesting that the initial charge translocation alone would activate the modified proteins. In the initial steps of the photocycle, the protein has to convert and store light energy, so as to drive the primary proton transfer from the protonated retinal to the Asp85 counter ion, and successive proton transfer reactions via small conformational rearrangements of the protein. Several origins have been proposed and debated for the dominating driving force leading to the primary and subsequent proton transfers, among which are (i) energy storage in the strained geometry of the isomerized chromophore (13), and (ii) electrostatic energy storage (14), either mainly due to the modification by the isomerization of the electrostatic interactions between the protein and the donor/acceptor pair (15), or to a light-induced charge separation between primary proton donor and acceptor (16).
To investigate the light-induced electrostatic and/or dielectric modifications along or in the vicinity of retinal, we performed ultrafast absorption spectroscopy on the nearby tryptophan residues. Wild-type bR contains 8 tryptophan residues; two of them (Trp86 and Trp182) are in close interaction with the retinal moiety, as shown in Fig. 1. Spectroscopic properties of tryptophans are very sensitive to the local electrostatic environment (19), because their dipole moments in the ground and lowest-lying excited states are different (20). Our previous experiments of ultrafast transient UV spectroscopy on VIS excitation of bR have revealed new ultrafast dynamics (21, 22) in the 260- to 310-nm range. An exciton-coupling model involving the interacting dipole moments of retinal, Trp86 and Trp182 was introduced to rationalize the data (21). The present study complements our previous work (21, 22) by (i) investigating the important W86F mutant; (ii) repeating the measurements on the WT form and the W86F and W182F mutants in such a way (see Materials and Methods) as to ensure a quantitative comparison between their transients; and (iii) extending the range of probe wavelengths beyond 310 nm. The 2 mutants W86F and W182F are obtained by inserting a spectroscopically silent phenylalanine residue in place of Trp86 or Trp182, respectively. Based on the knowledge of the protein structure, the exciton-coupling model gives a simple physical picture that reproduces the dominant features of the spectra observed immediately after pump excitation in all 3 samples. In particular, it is shown that the spectral contribution of Trp86 can be isolated by direct subtraction of the pump-probe signals of WT bR and W86F mutant obtained under appropriate conditions. The resulting transient signal originates from a Stark shift of the Trp86 absorption band induced by a change in the retinal dipole moment and/or in the protein dielectric and electrostatic environment. Trp86 thus appears as an ultrafast voltmeter probing the local charge reorganization during the isomerization process. Interestingly, the transient Stark shift rises instantaneously (as compared with our experimental finite time resolution) and does not vanish but remains long after the retinal isomerization, indicating a long-lived modification of the local electrostatic (dielectric) environment.
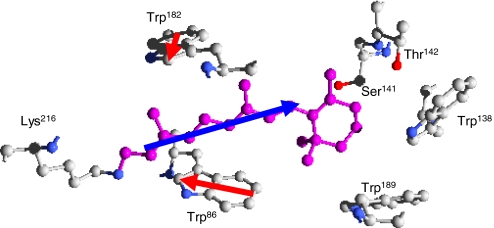
Fig. 1.
Schematic view [obtained with Deepview (17)] of retinal (in purple), together with 6 surrounding amino acids, as they appear in bR, according to the 1C3W X-ray diffraction structure (18). Besides 4 tryptophan residues, 2 polar residues (Ser141 and Thr142) reside near the chromophore, with their hydroxyl group (in red) pointing toward the β-ionone ring of retinal. In this work we investigate the interactions of the retinal dipole moment (blue arrow) with Trp86 and Trp182 dipoles (the red arrows represent the difference between La and ground state dipole moments of Trp182 and Trp86).
Experimental Results.
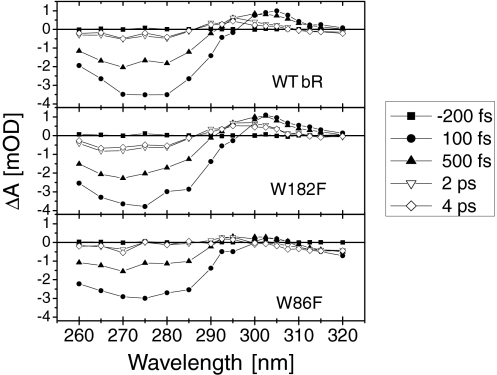
For the WT and the 2 mutant protein samples, Fig. 2 displays transient absorption spectra reconstructed from single wavelength measurements (see Materials and Methods). All 3 samples show a dominant negative signal (bleaching) between 260 nm and 290 nm, with a somewhat weaker amplitude in W86F compared with the other 2 samples. Compared with our previous data (21, 22) the occurrence of a similar bleach signal in the 3 samples now clarifies that it is dominated by a retinal response, rather than a tryptophan response. More differences between the 3 samples are observed at ≈300–310 nm where WT bR and W182F show a positive signal (induced absorption), but not W86F. For W86F an additional weak negative signal is found at wavelengths larger than 310 nm. The induced absorption slightly shifts to the blue at later times (>1 ps) in WT bR and W182F.
Fig. 2.
UV (260–320 nm) absorption spectra measured at 5 selected delay times after light excitation at 550 nm. The spectra for WT bR (Top), and for the W182F (Middle) and W86F (Bottom) mutants are reconstructed from a set of 20 pump-probe experiments with single-wavelength probe. At each wavelength, the transient absorptions of the 3 samples are acquired sequentially.
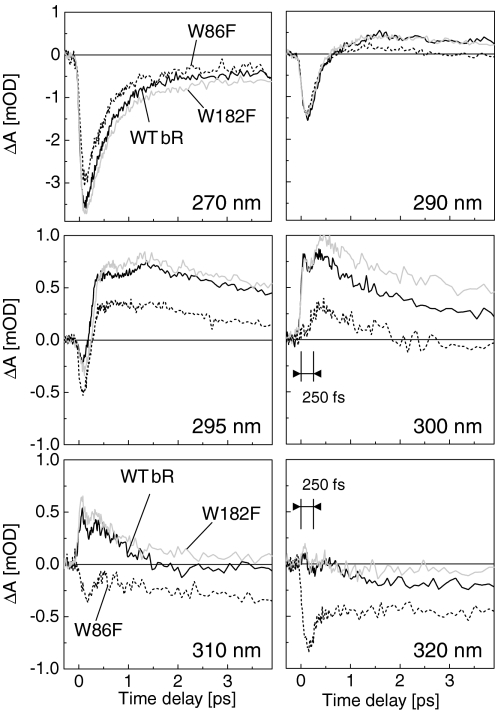
Transient absorption signals of the 3 samples are displayed as a function of time for 6 selected wavelengths between 270 and 320 nm in Fig. 3. At wavelengths shorter than 285 nm (see transients at 270 nm), the mutants show a time behavior very similar to that of WT bR. In that range of wavelength, the dynamics is nearly wavelength-independent [supporting information (SI) Fig. S1], with an instantaneous rise (i.e., much shorter than the experimental time resolution) followed by a decay identical to that previously measured (22). At 290 nm and longer wavelengths, whereas W182F signal remains similar to that of WT bR, the transient signal of the W86F mutant shows new dynamics with systematically less absorption, consistent with the above-mentioned absence of the absorption feature ≈300–310 nm. More interestingly, at 300 nm, the onset of W86F signal shows a 250-fs delay, and returns to below zero after 2 ps. At longer wavelengths (see traces at 310 nm) unlike the other 2 samples, the W86F signal is negative throughout the time range, with an amplitude increasing toward longer wavelengths. At 320 nm the transient signal of W86F displays a pronounced bleaching, whereas the other 2 samples display a signal close to the noise level at all delays. Notice that all transients display a remarkable nonexponential behavior, which itself is the result of the sum of nonexponential population dynamics, as pointed out in ref. 22.
Fig. 3.
Transient absorption signals of WT bR (solid black line), W182F (solid gray line), and W86F (dashed black line) at 6 selected wavelengths from 260 to 320 nm. Note that the vertical scale is different for the first 2 graphs: they correspond to shorter wavelengths at which the induced absorption signal is larger.
To interpret the differences between the transient absorptions observed in the 3 samples, we now discuss the results of the model introduced in ref. 21 to describe the coupling between the 3 chromophores retinal, Trp86, and Trp182, via dipole–dipole interaction.
Modeling Dipole–Dipole Interactions.
The model is based on the knowledge of the state and transition dipole moments of the 3 chromophores (retinal, Trp86, and Trp182), and of the protein structure, i.e., relative positions and orientations of the chromophores. Each of the chromophores is described as a 3-level entity having 3 state dipoles and 2 transition dipoles (0 → 1 and 0 → N for retinal; 0 → La and 0 → Lb for tryptophans). The 0 → N transition of retinal is nearly resonant with the tryptophan transitions (23, 24). A detailed description of the modeling is found in Materials and Methods. The results are as follows. We first compute the interaction of the retinal dipole with the dipoles (state and transition dipoles) of each of the 2 tryptophan residues. We find that, compared with Trp86, the interaction energies of retinal with Trp182 are 3 to 4 times weaker. By modifying appropriately the relative positions and orientations of the tryptophan residues, we also use the same model to compute the interactions of the retinal with the other 6 tryptophans of WT bR. The interaction energies of retinal with Trp138 and Trp189 appear to be 1 order of magnitude weaker than with Trp86, and at least 2 orders of magnitude weaker with the other 4 tryptophans. This justifies a posteriori why we had been focusing only on the influence of Trp86 and Trp182 previously (21) and in the present case. Also, it has been suggested that Trp138 and Trp189 participate in the enzymatic function of the protein in enhancing the light-induced dipole in bR (25). Because we show here that they have relatively weak dipole–dipole interactions with retinal, we rather speculate that other amino acids, namely the polar Ser141 and Thr142 residues (with their hydroxyl group lying, respectively, as close as 3.2 Å and 3.5 Å from the nearest carbon atom of retinal; see Fig. 1), might be responsible for the protein effect around the β-ionone ring of retinal.
Second, coming back to the interaction of retinal with Trp86 and Trp182, we may distinguish 2 types of dipolar interactions. Interactions between state dipoles of 2 different chromophores yield a shift of the energy levels and absorption bands (Stark shifts), but no quantum state mixing (diagonal matrix elements of the dipole–dipole interaction operator). Quantum state mixing, redistribution of oscillator strength and resonance splitting occur when transition dipoles are involved in excitonic couplings (off-diagonal matrix elements). Because of the relative orientation of the 3 chromophores (see Fig. 1), the La transition of Trp86 undergoes a Stark shift that is ≈7 times larger than in the case of Trp182. Typically, assuming that the change of retinal dipole moment on light excitation is Δμ = 15 D and that the dipole–dipole interactions are scaled by a relative dielectric constant εr = 2 (see Materials and Methods), we find that the Stark shift of the La transition of Trp86 is as large as ≈1,400 cm−1. In contrast, even with a 0 → N retinal transition dipole moment of 3 D (which corresponds to an oscillator strength as large as ∼20% of that of the 0 → 1 retinal transition), the excitonic couplings, i.e., off-diagonal matrix elements between quasi-resonant states of retinal and tryptophans, are no more than 70 cm−1 and 200 cm−1 for the La transitions of Trp182 and Trp86, respectively. Also, compared with La, the Lb state and transition dipoles are weaker. Hereafter, although the Lb state and transition dipole moments of both Trp182 and Trp86 are included in the model, we will focus the discussion on the influence of the La state and transition dipoles only.
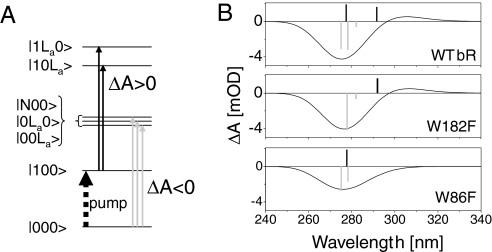
Finally, by diagonalizing the Hamiltonian of the 3 interacting chromophores, we can compute a stick spectrum from the new transition dipoles of the interacting system. Assuming a 3,500 cm−1 broadening for each of these transitions, we model the transient absorption spectrum of the protein just after excitation to the S1 state as in ref. 21. The results for WT bR and the 2 mutants are displayed in Fig. 4. Immediately after light excitation at 550 nm, the transient signal observed between 260 nm and 320 nm is the sum of 2 contributions, as illustrated by Fig. 4A. The first one is the bleaching of the transitions from the ground state to the excitonic manifold of retinal and the 2 tryptophans (ΔA < 0, light-gray arrows). The second contribution is the induced absorption of both tryptophans whereas retinal is excited in S1 (ΔA > 0, black arrows). Because the difference between the dipole moments of the La and ground states of Trp182 is nearly orthogonal to the retinal backbone, the Trp182 La transition is hardly shifted by the retinal dipole moment, so that its induced absorption partially compensates the bleach signal. Mutating Trp182 both reduces the bleach signal of the excitonic manifold, which now misses the Trp182 oscillator strength, and removes its induced absorption. The net effect on the transient signal is almost vanishing. In the case of Trp86, however, its La transition is significantly shifted on retinal excitation, because retinal and Trp86 state dipoles are in attractive interaction. Therefore, the induced absorption and the bleach signal overlap only partially, leading to a positive contribution rising at ≈310 nm. In addition, removing Trp86 again reduces the bleach signal and cancels the induced absorption, but the net effect no longer vanishes because both contributions occur at slightly different wavelengths.
Fig. 4.
Modeling the dipole–dipole interactions between retinal, trp86, and Trp182 in bR. (A) Simplified sketch of the electronic states involved in the model (Lb states of tryptophans are omitted for clarity). Because of the dipolar interactions, each quantum state is a mixture of product states of retinal, Trp86 and Trp182 states (in that order). The transient signals observed immediately after retinal excitation are the superposition of 2 contributions: (i) the bleaching (ΔA < 0) of the transitions from ground state to the excitonic manifold, and (ii) the induced absorption (ΔA > 0) of the tryptophans in interaction with the retinal excited in S1. (B) Modeling of the transient absorption signals (solid lines) of WT bR, W182F, and W86F immediately after excitation of retinal into S1 (see text). The bars are the stick spectra obtained from the model (Lb sticks not shown). Negative bars (gray) reproduce the bleach of the excitonic manifold (3 states in the case of WT bR only). Positive bars (black) are due to the induced absorption of both tryptophans, with the red-most shifted one corresponding to Trp86.
Altogether, the Trp182 mutation hardly affects the modeled spectrum, whereas Trp86 mutation reduces the negative contribution at ≈275 nm and suppresses the positive signal at ≈310 nm. These results are in very good agreement with the experimental observations and rely only on the dipole–dipole interaction and on the protein structure.
Discussion
Experimental results show that mutating Trp86 removes the induced absorption (IA) observed at ≈300–310 nm at short time delays and at slightly bluer wavelengths at later times (see Fig. 2). First, this is not due to a red shift of the bleaching signal that would then overlap and hide this IA, because such a shift would already appear in the static UV absorption band (≈280 nm) of the mutant, and is not observed (see Materials and Methods and Fig. 6). Therefore, 2 more reasons can be invoked: either this IA is a pure Trp86 absorption band that simply disappears when Trp86 is removed, or it is the signature of an excitonic sublevel of the interacting complex of Trp86 and retinal produced via a possible near-resonant 1 → N′ transition of retinal (N′ > N), which was not accounted for in our model. In the latter case, the excitonic coupling must be less than the 200 cm−1 estimated above for the coupling of Trp86 to the 0 → N retinal transition, because the oscillator strength of this hypothetic 1 → N′ transition is certainly not larger than that of the 0 → N transition. Hence, because no induced absorption remains at ≈300–310 nm under Trp86 mutation, we conclude that retinal has no such 1 → N′ transition with significant oscillator strength. Therefore, the Stark-shifted La transition observed here is very likely a signature of pure Trp86, which can be isolated by simple subtraction between the transient signals of W86F and WT bR.
Fig. 6.
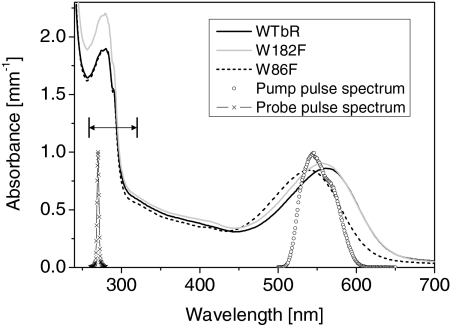
Typical absorption spectra of the protein solutions as used in the experiment, together with the spectrum of the pump and probe pulses. The double arrow shows the range of probe wavelengths scanned (260–320 nm). The sample concentrations and the spectral overlap of the laser pulse and the protein absorbances are carefully controlled so that the excitation probability of all samples is identical to within 10%. Note that the blue shift of the mutants absorption bands indicates that the dipolar interaction with both tryptophans stabilizes more the excited state than the ground state of the retinal cofactor.
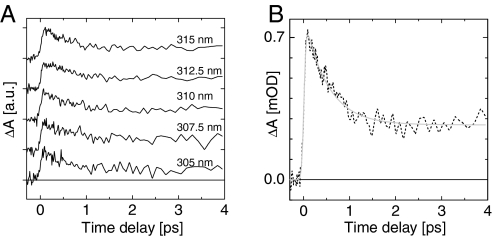
Fig. 5A displays the difference between the WT bR and Trp86 transient signals at 5 selected wavelengths between 305 and 315 nm. Although both samples show very different transient behaviors (see Fig. 3), the differential signals are remarkably identical in that range of probe wavelengths. The average over the 305- to 315-nm range of the differential signals is displayed in Fig. 5B. According to the conclusions of the above model, this signal is the transient signature of the La transition of Trp86, Stark-shifted by the light-induced change of the retinal dipole moment. It shows a sudden rise representing the change of the retinal dipole moment, and it decays on the timescale of the isomerization. Also, the blue shift of the induced absorption initially observed at 300–305 nm in WT bR and W182F is attributed to a reduction of the Stark shift while isomerization proceeds.
Fig. 5.
Transient contribution of Trp86, isolated by subtracting the transient signals of WT bR and W86F. (A) Results obtained from single-wavelength measurements at 305, 307.5, 310, 312.5, and 315 nm. Each trace is shifted vertically for clarity. (B) Average of the 5 traces displayed in A, and its fit to the sum of an exponential decay (with time constant τ = 500 ± 50 fs) and a step-like function, convoluted with a Gaussian function modeling the instrument response function (IRF).
The average trace presented in Fig. 5B is fitted by the sum of a monoexponential decay and a step-like function, convoluted with a Gaussian function modeling the instrument response function (IRF). The resulting decay time is500 ± 50 fs corresponding to the well-known retinal isomerization time, and the plateau is seen to remain constant up to at least 20 ps, which is our largest observation time window (Fig. S2). Interestingly, the fit also yields a FWHM of 95 fs for the IRF, which is independently measured to be at most 90–100 fs (see Materials and Methods). This means that the signal rises in a time significantly shorter than the IRF. Retinal and retinal proteins show a large dipole moment increase already in the Franck–Condon region, which has been measured by Stark spectroscopy (26, 27) and must induce an instantaneous rise of the signal discussed here. In addition to that, a twist-induced charge translocation (28) is predicted to increase dramatically the retinal dipole moment on a sub-20-fs timescale when the molecule reaches the conical intersection (29). This 100-fs delayed event should induce further rise of the signal, but no such contribution is observed here. Note, that events much faster than the time resolution contribute to the signal with an amplitude significantly reduced because of the convolution by the IRF (possibly reduced down to the noise level in the present case).
The amplitudes associated with the fast decay and slow components of the Trp86 transient signal are 66% and 34%, respectively. However, only two-thirds of the light-excited molecules continue to the J and K intermediate states of the protein cycle, after evolving through the conical intersection (30). The remaining third recover their initial state and their associated Trp86 signal should return to zero. Hence, in the Trp86 transient signal effectively produced by the isomerizing molecules, the amplitudes of the fast and slow components are closer to 50% and 50%. This means that in the J (vibrationally unrelaxed) and K (relaxed) states where the isomer form of retinal is in its electronic ground state, Trp86 still undergoes a significant Stark shift. With the present X-ray diffraction resolution, the only measurable structural changes are the “bicycle pedal” movement of retinal into a distorted conformation of the isomerized chromophore and a modification of the near hydrogen-bonding network (13). Hence, the retinal backbone and dipole moments hardly change their orientations, but the dielectric near environment (that is εr in our model) is altered. Given that the Stark shift of the Trp86 absorption band is proportional to Δμ/εr and that the ground state dipole moment of the all-trans retinal photoproduct is very similar to that of the 13-cis reactant (27), there is a strong indication that the remnant Stark shift of Trp86 in the J and K states is a signature of the dielectric response of the near environment of the residue, implied by a modification of the hydrogen-bonding network, a strained retinal or possibly multiple smaller-scale electron density reorganizations in the protein scaffold. In line with that, transient infrared spectroscopy of bR indicates that also the protein binding pocket might show structural dynamics already on the subpicosecond timescale (31), as is observed in the similar case of green-absorbing proteorhodopsin (32). Besides the dielectric environment, the electric field itself produced by the protein at the location of the tryptophan residue may be modified. Finally the long-lived Stark shift observed here may be an indication that a significant amount of energy is stored in the K state as electrostatic energy (15), to be released on longer timescales via structural rearrangements leading to subsequent proton transfer processes.
Let us now come back to the transient signals of the Trp86-free W86F mutant. Based on the above assignment and isolation of the pure Trp86 transient response we conclude that, in the range 300–320 nm, the signal of W86F (see Fig. 3) is dominated by the transient signature of the retinal chromophore. In particular, a delayed induced absorption (IA) is observed at 300 nm. In WT bR and W182F this IA onset arrives ≈250 fs after the instantaneous rise of the Trp86 signal, whereas it arrives with the same delay on top of a close-to-zero background in the W86F signal. At 320 nm, the retinal bleach seen in the W86F transient suddenly reduces, also in a nonexponential way, after ≈250–300 fs. Delayed IA onset and nonexponential signals were already observed in our previous work (22), and we confirm that they indicate an impulsive (coherent) formation of the J state of bR, when excited with short pulse duration (20 fs here), in line with the coherent torsional dynamics observed in the visible light (33, 34). However, the reason why the same reaction mechanism (here, isomerization) can give rise to both exponential (Trp86 transient Stark shift) and nonexponential (retinal transient signature) signals is not clear, and warrants further investigation.
Conclusion
The ultrafast response of WT bR and its W86F and W182F mutants is recorded in the UV on excitation of retinal with visible light. By direct subtraction of the transient signal in the WT and the mutant proteins, we isolate the contribution of the Trp86 residue. Our analysis is strongly supported by a simple, robust theoretical description capturing the dominant features revealed by the experiments, and based on the knowledge of the structure of the protein and on the dipole–dipole interaction. We show that Trp86 (but not Trp182) undergoes a significant (Stark) red shift of its La absorption band in response to the light-induced charge translocation along retinal backbone. Hence, Trp86 operates as a local ultrafast voltmeter. As a consequence we find that (i) the Stark shift of the Trp86 absorption band and therefore the underlying retinal charge translocation occur on a sub-50-fs time constant, correcting previous analysis, and confirming the timescale deduced from optical rectification experiments (4). (ii) On a 500-fs timescale, i.e., the timescale of the isomerization process, the Stark shift reaches a long-lived plateau, indicating a different electrostatic environment compared with that of ground state bR. This points to a long-lived change in the electrostatic environment of Trp86, which lends credence to an “electrostatic conflict” (15) arising already in the early J intermediate of the photocycle of the protein because of the isomerization. (iii) With this assignment of the transient Trp86 contribution, we conclude that the instantaneous negative signal in the range 260–290 nm is dominated by bleaching of retinal, although it is a complex superposition of bleaching and induced absorption of the excitonic manifold of the 3 chromophores.
To the best of our knowledge, tryptophans have been used in studies of photoinduced energy transfer (35) and electron transfer (36) in proteins, processes that are not or only indirectly related to their function, and as a probe of the solvation dynamics of the interfacial water layer on proteins (37). Here, we demonstrate their use as a reporter of the functional electric field changes within the protein in the course of its biological activity. Thus, the generality of such studies should be emphasized, because similar experiments may be applied to all proteins undergoing significant function-related changes in electrostatic and dielectric properties.
Materials and Methods
Experimental Work.
Mutants bR, W86F, and W182F were constructed by using the Kunkel method (38) and expressed homologously as described (39). Isolation of bR and its mutants from cells was carried out according to the standard procedure (40). After lysis of the cells in the presence of DNaseI (Sigma) the resultant membrane fragments were washed with water and purified by centrifugation on a sucrose gradient (25–45% wt/wt). Membrane fractions of buoyant density corresponding to that of wild-type bR (1.18 g/mL) were collected throughout, washed repeatedly with water, and then resuspended in the appropriate buffer for spectroscopic measurements.
Transient absorption spectroscopy is performed based on an amplified Ti:Sa laser system (1-kHz repetition rate, 1 mJ per pulse) that is used to operate 2 identical noncollinear optical parametric amplifiers (22), both delivering 20-fs, 50-nm-broad (FWHM) pulses. One produces a pump beam centered at 550 nm, the other is frequency doubled, yielding 3- to 4-nm-broad probe pulses, at a central wavelength adjustable from 260 nm to 320 nm. The pump intensity is adjusted within the linear regime of excitation probability. The buffered (pH = 7), sonicated suspensions of purple membrane patches containing natural or mutant proteins are circulated in a 100-μm-thick quartz cuvette. With such a thin film of liquid solution, the group velocity mismatch between pump and probe wavelengths does not affect significantly the time resolution of the setup. The latter is determined at each probe wavelength by measuring the rise time of the long-lived transient absorption of a sulforhodamine dye circulated in the same cuvette. Assuming the dye signal should be an instantaneous step function, convoluted by a Gaussian curve modeling the IRF, we find for the IRF an upper boundary value of 90–100 fs FWHM, with hardly any dependence on the probe wavelength.
Fig. 6 displays the stationary absorption spectra of the 3 samples as prepared for the measurements, together with the pump pulse spectrum. The maximum of the S0 → S1 absorption band lies at 565 nm, 556 nm, and 540 nm for WT bR, W182F, and W86F, respectively. The overlap of the pump spectrum with the normalized spectra of the wild-type protein and the mutants is identical to within ±4% for all samples. In addition the concentration of each sample is adjusted to within <5% so that altogether, the excitation probability is identical in the 3 samples within <10% uncertainty. We observed no difference in the transient signals by shifting the pump pulse spectrum by ±15 nm with respect to the maximum S0 → S1 absorption band. Approximately 20 different probe wavelengths are selected successively. At each fixed probe wavelength, the transient absorption signals are spectrally integrated (photodiode) as a function of pump-probe delay for wild-type bR, W86F, and W182F successively in 3 experimental runs in a row. Then, the first sample is again measured to check the stability of the experimental conditions over the full run. When necessary, the full run is repeated until satisfactory reproducibility is achieved. As compared with our previous results with WT bR and W182F (21), acquiring the data in such a controlled way now allows us to compare reliably and quantitatively the relative amplitudes of the transient signals in the 3 samples.
Modeling Dipole–Dipole Interactions.
The model (which is the same as used in ref. 21) is built on the same theoretical grounds as the one used successfully to model excitonic couplings in photosynthetic reaction centers (41). Tryptophan residues are described by a ground state and 2 almost degenerate excited electronic states, namely Lb and La. In the case of retinal in addition to the ground and first excited electronic states, we introduce a higher-lying excited electronic state (denoted state N) that is nearly resonant with the tryptophan La and Lb states (23). The dipole moments of the 3 chromophores interact through the dipole–dipole interaction operator. Within this model, the Hamiltonian is a 27 × 27 matrix, which we compute from the knowledge of the individual properties of the chromophores, and from their relative orientation inside the protein (see supporting online material of ref. 21 for a detailed discussion). In a variant of this model, we place a point charge on the nitrogen atom linking the retinal to the lysine residue Lys216 to account for the positive charge carried by the protonated retinal moiety. Simultaneously we place a negative charge distributed over the 2 nearest oxygens of Asp-85 and Asp-212, usually described as the corresponding counter ions (6, 42). Both models (with or without point charges) give qualitatively and almost quantitatively the same picture, and what we discuss in this work applies to both. By diagonalizing the Hamiltonian of the model system, we get a basis set of eigenstates for the interacting system in which new transition and state dipole moments are computed.
The protein structure and the dipole moments of isolated tryptophans are relatively well known (18, 20, 21). In the case of retinal, all of the dipole moments are aligned along the retinal backbone defined here as the line from C13 to C7. Only 2 critical parameters are adjusted in the model, to reproduce the energy shifts of the S0 → S1 absorption band observed in the 2 mutants by removing the one or the other tryptophan. The first parameter is the location of the origin of the retinal dipole moments, which is a critical constraint to reproduce the ratio between the energy shifts of both mutants. The second parameter is the retinal dipole moment change normalized by the relative dielectric constant: Δμ/εr. We compute the model by using either the 1C3W (18) or the 1AP9 (43) structures. In both cases, the optimal origin for the retinal dipole moments is slightly different but equally good results are obtained in reproducing the mutant spectral shifts assuming Δμ/εr ≃ 6 D. After diagonalization, these parameters reproduce a dipole moment change of retinal in the range 10–30 D for 1.5 < εr < 4. Here, we note, that εr is presumably very model-dependent (44), and that in our simplified description it accounts a priori for an average shielding effect of the electrostatic interactions due to the global dielectric protein environment and to the hydrogen bond network, polar amino acid residues, water molecules, and any other specific polarizable residue in the near environment of retinal. In addition, we emphasize that, compared with the previous discussion of the model (21), forcing it to reproduce the spectral shifts of the S0 → S1 transition of both mutants is an additional major constraint that permits a more reliable definition of the critical parameters. As a matter of fact, the predicted increase of signal in W182F compared with WT bR is not confirmed here, after using additional W86F data to better constrain the model.
Supplementary Material
Acknowledgments.
J.L. thanks J. Helbing and M. Olivucci for fruitful discussions. This work was supported by the DYNA program on “Ultrafast structural dynamics in Physics, Chemistry, Biology and Material Science” of the European Science Foundation (J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812877106/DCSupplemental.
References
- 1.Haupts U, Tittor J, Oesterhelt D. Closing in on bacteriorhodopsin: Progress in understanding the molecule. Annu Rev Biophys Biomol Struct. 1999;28:367–399. doi: 10.1146/annurev.biophys.28.1.367. [DOI] [PubMed] [Google Scholar]
- 2.Mathies RA, Lin SW, Ames JB, Pollard WT. From femtoseconds to biology: Mechanism of bacteriorhodopsin's light-driven proton pump. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- 3.Birge RR, Zhang C-F. Two-photon double resonance spectroscopy of bacteriorhodopsin. assignment of the electronic and dipolar properties of the low-lying 1Ag*−-like and 1Bu*+-like π,π* states. J Chem Phys. 1990;92:7178–7195. [Google Scholar]
- 4.Colonna A, Groma GI, Martin J-L, Joffre M, Vos MH. Quantification of sudden light-induced polarization in bacteriorhodopsin by optical rectification. J Phys Chem B. 2007;111:2707–2710. doi: 10.1021/jp0673462. [DOI] [PubMed] [Google Scholar]
- 5.Zgrablic G, Voïtchovsky K, Kindermann M, Haacke S, Chergui M. Ultrafast excited state dynamics of the protonated schiff base of all-trans retinal in solvents. Biophys J. 2005;88:2779–2788. doi: 10.1529/biophysj.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song L, El-Sayed MA, Lanyi JK. Protein catalysis of the retinal subpicosecond photoisomerization in the primary process of bacteriorhodopsin photosynthesis. Science. 1993;261:891–894. doi: 10.1126/science.261.5123.891. [DOI] [PubMed] [Google Scholar]
- 7.Heyne K, Herbst J, Dominguez-Herradon B, Alexiev U, Diller R. Reaction control in bacteriorhodopsin: Impact of Arg82 and Asp85 on the fast retinal isomerization, studied in the second site revertant Arg82Ala/Gly231Cys and various purple and blue forms of bacteriorhodopsin. J Phys Chem B. 2000;104:6053–6058. [Google Scholar]
- 8.Cembran A, Bernardi F, Olivucci M, Garavelli M. Excited-state singlet manifold and oscillatory features of a nonatetraeniminium retinal chromophore model. J Am Chem Soc. 2003;125:12509–12519. doi: 10.1021/ja030215j. [DOI] [PubMed] [Google Scholar]
- 9.Xu D, Martin C, Schulten K. Molecular dynamics study of early picosecond events in the bacteriorhodopsin photocycle: Dielectric response, vibrational cooling and the j, k intermediates. Biophys J. 1996;70:453–460. doi: 10.1016/S0006-3495(96)79588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennis JTM, et al. Ultrafast protein dynamics of bacteriorhodopsin probed by photon echo and transient absorption spectroscopy. J Phys Chem B. 2002;106:6067–6080. [Google Scholar]
- 11.Lewis A. The molecular mechanism of excitation in visual transduction and bacteriorhodopsin. Proc Natl Acad Sci USA. 1978;75:549–553. doi: 10.1073/pnas.75.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rousso I, et al. Microsecond atomic force sensing of protein conformational dynamics: Implications for the primary light-induced events in bacteriorhodopsin. Proc Natl Acad Sci USA. 1997;94:7937–7941. doi: 10.1073/pnas.94.15.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schobert B, Cupp-Vickery J, Hornak V, Smith SO, Lanyi JK. Crystallographic structure of the K intermediate of bacteriorhodopsin: Conservation of free energy after photoisomerization of the retinal. J Mol Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 14.Edman K, et al. High-resolution x-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature. 1999;401:822–826. doi: 10.1038/44623. [DOI] [PubMed] [Google Scholar]
- 15.Bondar A-N, Fischer S, Smith JC, Elstner M, Suhai S. Key role of electrostatic interactions in bacteriorhodopsin proton transfer. J Am Chem Soc. 2004;126:14668–14677. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]
- 16.Braun-Sand S, Sharma PK, Chu ZT, Pisliakov AV, Warshel A. The energetics of the primary proton transfer in bacteriorhodopsin revisited: It is a sequential light-induced charge separation after all. Biochim Biophys Acta. 2008;1777:441–452. doi: 10.1016/j.bbabio.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guex N, Peitsch MC. SWISS-MODEL and the swiss-pdbviewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 18.Luecke H, Schobert B, Richter H-Th, Cartailler J-Ph, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 19.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callis PR. 1La and 1Lb transitions of tryptophan: Applications of theory and experimental observations to fluorescence of proteins. Methods Enzymol. 1997;228:113–150. doi: 10.1016/s0076-6879(97)78009-1. [DOI] [PubMed] [Google Scholar]
- 21.Schenkl S, van Mourik F, van der Zwan G, Haacke S, Chergui M. Probing the ultrafast charge translocation of photoexcited retinal in bacteriorhodopsin. Science. 2005;309:917–920. doi: 10.1126/science.1111482. [DOI] [PubMed] [Google Scholar]
- 22.Schenkl S, et al. Insights into excited-state and isomerization dynamics of bacteriorhodopsin from ultrafast transient uv absorption. Proc Natl Acad Sci USA. 2006;103:4101–4106. doi: 10.1073/pnas.0506303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becher B, Tokunaga F, Ebrey TG. Ultraviolet and visible absorption spectra of the purple membrane protein and the photocycle intermediates. Biochemistry. 1978;17:2293–2300. doi: 10.1021/bi00605a006. [DOI] [PubMed] [Google Scholar]
- 24.Polland HJ, Franz MA, Zinth W, Kaiser W, Oesterhelt D. Energy transfer from retinal to amino acids - a time-resolved study of the ultraviolet emission of bacteriorhodopsin. Biochim Biophys Acta. 1986;851:407–415. [Google Scholar]
- 25.Aharoni A, Khatchatouriants A, Manevitch A, Lewis A, Sheves M. Protein-β-ionone ring interactions enhance the light-induced dipole of the chromophore in bacteriorhodopsin. J Phys Chem B. 2003;107:6221–6225. [Google Scholar]
- 26.Mathies R, Stryer L. Retinal has a highly dipolar vertically excited singlet state: Implications for vision. Proc Natl Acad Sci USA. 1976;73:2169–2173. doi: 10.1073/pnas.73.7.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locknar SA, Peteanu LA. Investigation of the relationship between dipolar properties and cis-trans configuration in retinal polyenes: A comparative study using stark spectroscopy and semiempirical calculations. J Phys Chem B. 1998;102:4240–4246. [Google Scholar]
- 28.González-Luque R, et al. Computational evidence in favor of a two-state, two-mode model of the retinal chromophore photoisomerization. Proc Natl Acad Sci USA. 2000;97:9379–9384. doi: 10.1073/pnas.97.17.9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frutos LM, Andruniów T, Santoro F, Ferré N, Olivucci M. Tracking the excited-state time evolution of the visual pigment with multiconfigurational quantum chemistry. Proc Natl Acad Sci USA. 2007;104:7764–7769. doi: 10.1073/pnas.0701732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tittor J, Oesterhelt D. The quantum yield of bacteriorhodopsin. FEBS Lett. 1990;263:269–273. [Google Scholar]
- 31.Herbst J, Heyne K, Diller R. Femtosecond infrared spectroscopy of bacteriorhodopsin chromophore isomerization. Science. 2002;297:822–825. doi: 10.1126/science.1072144. [DOI] [PubMed] [Google Scholar]
- 32.Amsden JJ, et al. Subpicosecond protein backbone changes detected during the green-absorbing proteorhodopsin primary photoreaction. J Phys Chem B. 2007;111:11824–11831. doi: 10.1021/jp073490r. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T, Saito T, Ohtani H. Real-time spectroscopy of transition states in bacteriorhodopsin during retinal isomerization. Nature. 2001;414:531–534. doi: 10.1038/35107042. [DOI] [PubMed] [Google Scholar]
- 34.Hou B, Friedman N, Ottolenghi M, Sheves M, Ruhman S. Comparing photoinduced vibrational coherences in bacteriorhodopsin and in native and locked retinal protonated schiff bases. Chem Phys Lett. 2003;381:549–555. [Google Scholar]
- 35.Hochstrassser RM, Negus DK. Picosecond fluorescence decay of tryptophans in myoglobin. Proc Natl Acad Sci USA. 1984;81:4399–4403. doi: 10.1073/pnas.81.14.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu W, et al. Ultrafast quenching of tryptophan fluorescence in proteins: Interresidue and intrahelical electron transfer. Chem Phys. 2008;350:154–164. [Google Scholar]
- 37.Pal SK, Peon J, Zewail AH. Biological water at the protein surface: Dynamical solvation probed directly with femtosecond resolution. Proc Natl Acad Sci USA. 2002;99:1763–1768. doi: 10.1073/pnas.042697899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunkel T, Roberts J, Zakour R. Rapid and sufficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–405. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 39.Pfeiffer M, Rink T, Gerwert K, Oesterhelt D, Steinhoff H-J. Site-directed spin labeling reveals the orientation of the amino acid side chains in the E-F loop of bacteriorhodopsin. J Mol Biol. 1999;287:163–171. doi: 10.1006/jmbi.1998.2593. [DOI] [PubMed] [Google Scholar]
- 40.Oesterhelt D, Stoeckenius W. Isolation of the cell membrane of halobacterium halobium and its fraction into red and purple membrane. Methods Enzymol. 1974;31:667–686. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 41.Koolhaas MHC, van der Zwan G, van Mourik F, van Grondelle R. Spectroscopy and structure of bacteriochlorophyll dimers. 1. Structural consequences of nonconservative circular dichroism spectra. Biophys J. 1997;72:1828–1841. doi: 10.1016/S0006-3495(97)78829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tachikawa H, Kawabata H. Effects of the residues on the excitation energies of protonated schiff base of retinal (PSBR) in br: A TD-DFT study. J Photochem Photobiol B. 2005;79:191–195. doi: 10.1016/j.jphotobiol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 44.Schutz C, Warshel A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.