Abstract
The simple structure of Arabidopsis roots provides an excellent model system to study epidermal cell fate specification. Epidermal cells in contact with 2 underlying cortical cells differentiate into hair cells (H cells; trichoblasts), whereas cells that contact only a single cortical cell differentiate into mature hairless cells (N cells; atrichoblasts). This position-dependent patterning, in combination with the constrained orientation of cell divisions, results in hair and nonhair cell files running longitudinally along the root epidermis. Here, we present strong evidence that steroid hormones called brassinosteroids (BRs) are required to maintain position-dependent fate specification in roots. We show that BRs are required for normal expression levels and patterns of WEREWOLF (WER) and GLABRA2 (GL2), master regulators of epidermal patterning. Loss of BR signaling results in loss of hair cells in H positions, likely as a consequence of reduced expression of CAPRICE (CPC), a direct downstream target of WER. Our observations demonstrate that in addition to their well-known role in cell expansion, BRs play an essential role in directing cell fate.
Keywords: brassinosteroids, cell fate specification, positional cues, root epidermis
Molecular genetic studies in Arabidopsis have defined a number of genes that influence root cell fate patterning (1–3). A transcriptional complex composed of the MYB transcription factor WEREWOLF (WER), a WD-40 repeat protein called TRANSPARENT TESTA GLABRA (TTG), and 2 basic helix–loop–helix transcription factors called GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) promotes hairless cell (N cell) differentiation. This complex induces expression of GL2, encoding a homeodomain transcription factor, and CAPRICE (CPC), encoding a single-repeat MYB protein. CPC moves laterally to the adjacent epidermal cells, where it inhibits WER and GL2 expression, thus promoting hair cell (H cell) differentiation (4). Recent mathematical simulations of root epidermal cell specification lend support to a lateral inhibition mechanism relying on the lateral movement of CPC and GL3 (5). The identification of a receptor-like kinase, SCRAMBLED (SCM), has led to a model where an asymmetrically distributed positional signal activates SCM in epidermal cells located between 2 cortical cells (6). Activated SCM promotes hair cell fate by decreasing the abundance of WER (7). Laterally translocated CPC promotes the preferential accumulation of SCM in the H cell position, thus reinforcing hair cell fate in epidermal cells located between 2 cortical cells (8).
The role of hormones in the regulation of root epidermal cell identity program is not well established. Studies using auxin and ethylene have demonstrated that these 2 hormone pathways control root epidermis development at a relatively late stage, after the completion of the cell differentiation program (9). A recent study has demonstrated that auxin transport through the nonhair cells controls the elongation of root hairs in the hair cell position (10). Global transcriptome analysis of whole seedlings revealed that expression of WER can be induced by treatment with brassinosteroids (BRs) (11), a class of steroid hormones known to promote growth in diverse plant species (12). Although BRs are known to affect root elongation (13), no previous work has connected them to the root hair formation program. This study reveals a role for plant steroid hormones in the regulation of key transcription factors required for acquisition of cell fate during root hair development. Before this study, SCRAMBLED, a leucine-rich repeat receptor-like kinase, was the only known mediator of positional cues in the root epidermis (6). The present study points to a cell type-specific role for BRs in root hair formation and provides evidence for an additional mechanism for interpreting cell position.
Results and Discussion
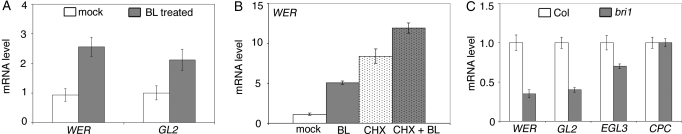
To investigate a potential role for BRs in cell fate specification, we examined the effect of brassinolide (BL), the most biologically active BR, on root tissue. RNA was extracted from roots of seedlings grown in mock or BL treatments. Quantitative RT-PCR experiments demonstrated that WER and GL2 were induced ≈2 and 2.5 times, respectively, in response to 10 nM BL treatment (Fig. 1A), consistent with the results from a previous global transcriptome analysis using whole seedlings (11). WER is likely a BR early-response gene, because the effects of BL could still be detected in the presence of the protein synthesis inhibitor cycloheximide. Although cycloheximide treatment alone led to increased WER expression, an additional effect of BL treatment was clearly observable (Fig. 1B). The effects of reduced BR response were also examined by using the strong BR mutant brassinosteroid insensitive 1 (bri1). BRI1 encodes the BR receptor, a plasma membrane-localized, leucine-rich repeat receptor kinase (14). We found that bri1 plants showed a significant reduction in both WER and GL2 expression (Fig. 1C). Thus, both normal levels and response to BRs are required for the wild-type expression of WER and GL2.
Fig. 1.
BRs regulate expression levels of root epidermal cell markers. (A) Quantitative RT-PCR using wild-type roots treated with 10 nM BL shows increased expression of WER and GL2. (B) Treatment with cycloheximide (CHX) does not inhibit the induction of WER by BL. (C) The bri1 roots show decreased expression of WER, GL2, and CPC. Results are represented as mean ± standard error (n = 3 replicates).
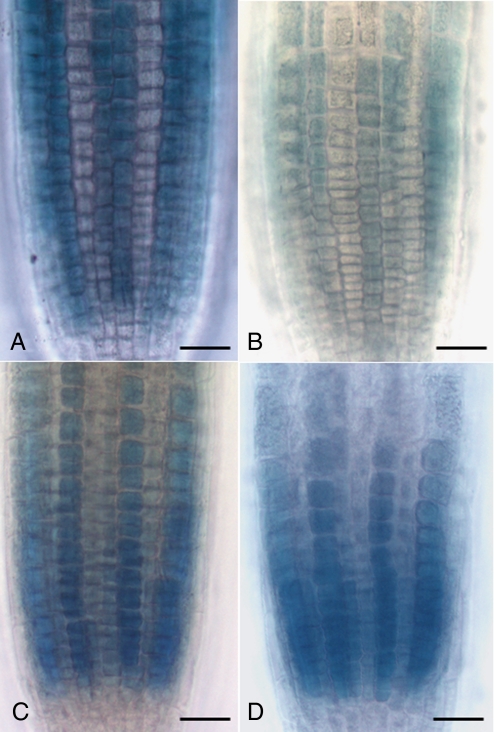
Experiments using the GL2∷GUS reporter, a central tool in studying root epidermal cell fate (6, 9, 15), revealed that BRs are required for the levels of expression and position-dependent patterning of GL2. In contrast to wild-type roots, GL2∷GUS expression in bri1 roots was significantly reduced and was no longer restricted to orderly files (Fig. 2 A and B). This aberrant patterning of GL2∷GUS expression could be more easily observed if staining reactions were left to develop for approximately twice as long as those used for wild-type roots (Fig. 2C). Transverse sections made clear that in many cases, cells in contact with 2 cortical cells aberrantly expressed the GL2∷GUS reporter (Fig. 2H). A loss of GL2∷GUS expression in the nonhair files was never observed in these roots. Similar changes in expression levels and patterns of WER reporters were also seen in bri1 mutants (Fig. S1). When BR levels were reduced by treatment with 100 nM brassinazole (BRZ), a BR biosynthetic inhibitor (16), GL2∷GUS expression resembled what was seen in bri1 mutants (Fig. 2E). In contrast, treatment with BL had no effect on the patterning of GL2 expression (Fig. 2F). Changes in expression levels were not seen consistently in BL-treated roots, likely reflecting an insufficiently large effect to influence the long-lived GUS reporter. In combination, these results suggest that BRs are necessary for proper epidermal patterning but are not sufficient when added exogenously to scramble the SCM-EGL3-CPC-specified program.
Fig. 2.
Disruptions in the BR pathway lead to aberrant levels and patterning of GL2. (A–C) GL2∷GUS expression in roots of wild type (A) and bri1 mutants (B and C). (B) The bri1 roots were stained for 20 min, the same duration as shown for wild type. (C) The bri1 roots stained for 40 min. (D and E) GL2 shows an increased randomness of expression pattern on roots treated with BRZ (E) compared with the control roots (D). (F) BL treatment does not affect the pattern of GL2 expression. (G and H) Cross-section of GL2∷GUS plants in either wild-type (G) or bri1 background (H). (Scale bars: A–F, 50 μm; G and H, 25 μm.)
Although the overall level of WER and GL2 expression was reduced when the BR pathway was compromised, additional pathways must also be promoting N cell fate, because both reporters were clearly detectable in these cells. The effects of BRs were more pronounced in the H cells, where ectopic expression of both WER and GL2 reporters was observed. To assess whether this ectopic expression was sufficient to change the fate of cells, we performed a quantitative analysis of epidermal cell fate relative to cell position. Experiments documenting the number of hair or nonhair cells in the N and H positions of bri1 roots showed that 23% of cells in the H position were nonhair cells (Table 1) compared with 5% in wild-type roots. When wild-type plants were grown on 100 nM BRZ, 18% of cells in the H position were nonhair cells. In contrast, loss of BRs had only a modest effect on the number of hair cells formed in N positions (Table 1), suggesting that BRs are more important for suppressing nonhair fate in H cells than for promoting nonhair fate in N cells. To assess how early BRs may be exerting their effects on cell fate, we compared the number of cells in the N and H positions in wild-type and bri1 root tips. As reported previously (17), we found that in wild-type root tips, there were ≈30% more cells in the H position. In bri1, the number of cells in H and N positions were nearly the same (H/N ratio for wild type, 1.3 ± 0.09; bri1, 1.01 ± 0.02). This indicates that BRs act at an early stage of root development.
Table 1.
Distribution of root hair and nonhair cells in the root epidermis
| H position |
N position |
|||
|---|---|---|---|---|
| Hair, % | Nonhair, % | Hair, % | Nonhair, % | |
| Wild type (Col) | 95 ± 2 | 5 ± 2 | 0 ± 0 | 100 ± 0 |
| bri1 | 77 ± 4 | 23 ± 4 | 4 ± 2 | 96 ± 1 |
| Col and BRZ | 82 ± 3 | 18 ± 3 | 3 ± 2 | 97 ± 2 |
| scm | 76 ± 4 | 24 ± 4 | 41 ± 7 | 59 ± 7 |
| scm bri1 | 57 ± 6 | 43 ± 6 | 9 ± 4 | 91 ± 4 |
Values are expressed as the mean ± standard error of at least 40 roots for each line.
Because epidermal cell fate is correlated with cortical arrangement, we tested the possibility that defective epidermal patterning in the bri1 mutant is due to an abnormality in the root structure. Examination of the cross-sections of the wild-type and bri1 mutant roots revealed no significant difference in the root structure. In wild-type roots, the epidermal cell number was 19.3 ± 1.2, whereas in bri1 roots, it was 19.1 ± 1.0. Both wild-type and bri1 roots displayed 8 cortical cell files and showed no obvious difference in the organization of the cell files.
The root epidermal phenotype observed in bri1 and BRZ-treated plants was reminiscent of cpc mutants, where a large number of cells in the H position adopt the N cell fate (7, 18). Like GL2, CPC expression is WER-dependent (4, 19). Reduced WER expression caused by the loss of the BR pathway could lead to lower expression of CPC, which might allow ectopic expression of WER (and GL2) in H position cells. Consistent with this model, we found a decreased expression of CPC in bri1 roots (Figs. 1B and 3 A and B). To assess whether these changes on gene expression reflected a general reorganization of the root epidermis, we examined the level and pattern of expression of EGL3. EGL3 is a component of the WER transcriptional complex that induces GL2 but is not itself under the transcriptional control of WER (1, 2). Loss of BR signaling showed little to no effect on levels or pattern of expression of EGL3 (Figs. 1B and 3 C and D), strongly suggesting that BRs act specifically through regulation of WER and its downstream targets.
Fig. 3.
BRI1 regulates expression of CPC but not EGL3. (A and B) CPC∷GUS expression is reduced significantly in bri1-116 (B) in comparison with wild type (A). (C and D) The patterning expression of EGL3, a marker of H cell fate, is similar in wild type (C) and in bri1 (D). (Scale bars: 50 μm.)
SCM activity is a major determinant of position-dependent expression of WER and GL2 (6, 8). To determine the relationship between the SCM and BR pathways, scm bri1 double mutants were analyzed (Table 1). In scm bri1 mutants, there was a largely additive effect on conversion of H cells to nonhair cell fate. Approximately 43% of cells in the H position adopted nonhair fate compared with 24% and 23% in the scm and bri1 single-mutant lines, respectively. In the N cell position, scm bri1 double mutants showed a significant reduction in hair cell fate compared with the scm single mutant. This finding is consistent with a model for BR action via modulation of CPC expression. Genetic and molecular studies demonstrate that CPC plays a largely additive role with SCM in repressing WER expression and promoting hair cell fate in H cells (7). The conversion of N cells to hair cell fate observed in scm single mutants is almost completely eliminated in scm cpc double mutants (7), suggesting that ectopic hair cell fate requires CPC activity.
In summary, we have shown that: (i) WER is an early BR response gene, (ii) normal levels and patterning of both WER and its downstream target GL2 are dependent on BR production and response, and (iii) ectopic expression of these master regulators in H cells when BR synthesis or response is compromised is correlated with a change of H cell fate. These results, in combination with previous findings, suggest that 2 leucine-rich receptor-like kinases, SCM and BRI1, convey positional information to root epidermal cells. In such a model, BR-activated BRI1 induces WER expression in cells overlying 1 cortical cell, resulting in the accumulation of CPC. CPC would then translocate to neighboring epidermal cells, where it would inhibit WER and GL2 and promote SCM accumulation. The preferential accumulation of SCM further represses WER activity in these cells, reinforcing the hair cell identity. Earlier studies have indicated that BR pathway components, such as BRI1, BIN2, and BZR1, are localized uniformly to cells within the root epidermis (20–22). Detailed temporal analysis as to the cellular and subcellular localization of these components is required to determine the mechanism of cell type-specific responses.
Recent studies have demonstrated that BRs acting in the epidermis are essential for growth of aerial tissues (23, 24). In this study, we show that BRs are required for normal expression and patterning of master regulators of epidermal cell fate. Unlike any other hormone examined, BRs affect early stages of cell fate by manipulating expression of WER, GL2, and CPC. Collectively, these studies suggest that BRs play an essential role in morphogenesis throughout the plant.
Methods
Plant Materials and Growth Conditions.
Arabidopsis accession Columbia-0 (Col-0) was the wild-type control for all experiments. The mutants, bri1-116 (25) and scm-2 (6), have been described previously. The GL2∷GUS (15), EGL3∷GUS (26), and WER∷GUS (19) lines were obtained from J. Schiefelbein (University of Michigan, Ann Arbor, MI). The CPC∷GUS line was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Seedlings were grown vertically on plates containing 0.5× Linsmaier and Skoog (LS) medium (Caisson Laboratories) with 0.6% agarose (Invitrogen). For hormone assays, seedlings were grown in medium containing BL (Wako Chemicals) or BRZ (a gift from Tadao Asami, Riken, Japan), as described previously (27).
Cycloheximide Assay.
The seedling plates were either flooded with 10 μM cycloheximide (Acros Organics) in 0.5× LS medium. Mock treatment consisted of submersion in 0.5× LS. BL plus cycloheximide treatments were performed by addition of 10 μM cycloheximide and 10 nM BL in 0.5× LS. All treatments were carried out for 2 h, and after treatment, tissues were frozen for quantitative PCR analysis.
Microscopy.
The position of hair and nonhair cells relative to the underlying cortical cells was determined on the roots of 5-day-old seedlings (4). This was done by using an Olympus BX40 microscope with 2 replicates each of ≈20 roots from each genotype and each treatment. For GUS assays, 2 replicates of 25–30 seedlings from mutant lines carrying the reporter construct and the corresponding wild-type lines were examined (28). For plastic sections, roots were embedded in 1.0% agarose, fixed in 4% paraformaldehyde, dehydrated in an ethanol series, embedded in Technovit 7100 (Heraeus Kulzer, EBSciences), and sectioned at 10 μm. The GFP expression in the WER∷GFP plants was examined in seedlings by using a BioRad Radiance 2000 confocal laser scanning microscope (Carl Zeiss MicroImaging) with a 488-nm excitation mirror and 515- to 530-nm emission filter to record images. The images were acquired by using Lasersharp 2000 v.6 (Carl Zeiss MicroImaging). The relative cell division rate in the H and N epidermal cell positions was deduced from the number of cells in the 2 files by using a previously described method (17).
Quantitative RT-PCR.
Total RNA was extracted from roots of 5-day-old seedlings (Spectrum Plant RNA Kit; Sigma), DNase-treated (Ambion Inc.), and reverse-transcribed (iScript cDNA synthesis kit; Bio-Rad). cDNAs were combined with iQSYBR Green Supermix (Bio-Rad) for PCR on a Chromo 4 real-time PCR machine (Bio-Rad). Gene-specific primers were as follows: At1g9840 (5′-ACTCTCTCGGAGTTACAAC-3′; 5′-ATTCAGCTTTGCTGCAGG-3′), At1g11130 (5′-GTCTGAACTGTCATTGGGA-3′; 5′-GTTAGACGATAAGTCCAG-3′), At5G14750 (5′-AGTAAGTAGTAGTGGTGACG- 3′; 5′-TGTCCATCTATAAAGTCCAT-3′), and At1G63650 (5′-CTGAAACCGCCGATAGC-3′; 5′-AACCGTTGAATCCTACTC-3′). At1g13320 was used for normalization (5′-AACGTGGCCAAAATGATGC-3′, 5′-AACCGCTTGGTCGACTATCG-3′).
Supplementary Material
Acknowledgments.
We thank John Schiefelbein and Su Hwan Kwan for helpful discussions, technical advice, and seed stocks. We also thank Keiko Torii and Lynn Pillitteri for sharing their expertise and facilities. We are grateful to Tadao Asami for generously providing brassinazole and Pang Chan for assistance with confocal microscopy. Members of the Nemhauser laboratory provided useful feedback throughout the project. This work was supported by funds from the University of Washington.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811633106/DCSupplemental.
References
- 1.Dolan L. Positional information and mobile transcriptional regulators determine cell pattern in the Arabidopsis root epidermis. J Exp Bot. 2006;57:51–54. doi: 10.1093/jxb/erj037. [DOI] [PubMed] [Google Scholar]
- 2.Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu Rev Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- 3.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 4.Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell. 2002;14:611–618. doi: 10.1105/tpc.010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage NS, et al. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 2008;6:e235. doi: 10.1371/journal.pbio.0060235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 7.Kwak SH, Schiefelbein J. The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol. 2007;302:118–131. doi: 10.1016/j.ydbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Kwak SH, Schiefelbein J. A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr Biol. 2008;18:1949–1954. doi: 10.1016/j.cub.2008.10.064. [DOI] [PubMed] [Google Scholar]
- 9.Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones AR, et al. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol. 2009;11:78–84. doi: 10.1038/ncb1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:e258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mussig C. Brassinosteroid-promoted growth. Plant Biol (Stuttgart) 2005;7:110–117. doi: 10.1055/s-2005-837493. [DOI] [PubMed] [Google Scholar]
- 13.Mussig C, Shin GH, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133:1261–1271. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 15.Masucci JD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 16.Asami T, et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger F, Haseloff J, Schiefelbein J, Dolan L. Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr Biol. 1998;8:421–430. doi: 10.1016/s0960-9822(98)70176-9. [DOI] [PubMed] [Google Scholar]
- 18.Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 19.Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 20.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert S, et al. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci USA. 2008;105:8464–8469. doi: 10.1073/pnas.0711650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savaldi-Goldstein S, Peto C, Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- 24.Reinhardt B, et al. Restoration of DWF4 expression to the leaf margin of a dwf4 mutant is sufficient to restore leaf shape but not size: the role of the margin in leaf development. Plant J. 2007;52:1094–1104. doi: 10.1111/j.1365-313X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- 25.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sessions A, Weigel D, Yanofsky MF. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.