Abstract
The sequencing of individual DNA strands with nanopores is under investigation as a rapid, low-cost platform in which bases are identified in order as the DNA strand is transported through a pore under an electrical potential. Although the preparation of solid-state nanopores is improving, biological nanopores, such as α-hemolysin (αHL), are advantageous because they can be precisely manipulated by genetic modification. Here, we show that the transmembrane β-barrel of an engineered αHL pore contains 3 recognition sites that can be used to identify all 4 DNA bases in an immobilized single-stranded DNA molecule, whether they are located in an otherwise homopolymeric DNA strand or in a heteropolymeric strand. The additional steps required to enable nanopore DNA sequencing are outlined.
Keywords: α-hemolysin, DNA sequencing, genomics, protein engineering, protein pore
A price of $1,000 for a human genome sequence would be a critical development in medicine (http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04-003.html). At that price, many people could afford to have their genomes sequenced and personalized medicine would become a reality (1, 2). However, despite impressive recent price reductions, today's sequencing technologies require the use of costly enzymes, fluorescently labeled molecules, state-of-the-art imaging equipment, and high-capacity data storage devices (3). Therefore, most probably, the cost of DNA sequencing can only be driven down further by using single-molecule technologies, all of which would avoid expensive, error-prone enzymatic DNA amplification techniques (4, 5). One such approach is single-molecule sequencing-by-synthesis, which was initiated in academic laboratories (6, 7) and is now under commercial development (8, 9). However, single-molecule sequencing-by-synthesis still requires expensive imaging and data-handling equipment.
By contrast, single-molecule nanopore sequencing would be “reagent-free” and employ a cheap electrical readout (10). In the most commonly investigated manifestation of nanopore sequencing, single-stranded DNA (ssDNA) is driven electrophoretically through a biological or solid-state pore of a few nanometers diameter (11). The goal is to identify the DNA bases in sequence as they pass a recognition site within the pore by recording the extent to which each base modulates the ionic current driven through the pore by the same applied potential that moves the DNA strand. The nanopore approach requires no DNA amplification and no enzymes or expensive nucleotide analogs. It is amenable to implementation in a highly parallel format and should be capable of long reads and the direct determination of modified bases, notably 5-methylcytosine (12). A 10,000-protein nanopore device could determine a human genome sequence with high coverage in <1 day (4, 10).
Several advances suggest that nanopore sequencing with protein pores is feasible. Notably, Ghadiri and coworkers showed that when ssDNA is immobilized within the α-hemolysin (αHL) protein pore, it is possible to distinguish between 2 bases when one or the other is located at a specific position within the pore (13). Further, with a view toward exonuclease sequencing, in which bases are sequentially cleaved from a DNA strand, all 4 DNA bases can be identified as deoxyribonucleoside 5′-monophosphates by using an engineered αHL pore equipped with a cyclodextrin molecular adapter (12, 14). Wang and colleagues first showed that the direction in which DNA enters the αHL pore (5′ or 3′ threading) affects the extent of current block (15), an observation supported by the data of Mathé et al. (16), and recent studies have shown that the rate of capture of DNA by protein pores is enhanced when the interior surfaces bear a net positive charge (17, 18). Under the high applied potentials required for threading, freely moving DNA is translocated through the wild-type (WT) αHL pore too quickly for bases to be identified, unless the bases are modified with bulky groups (19). In a step toward the management of this problem, the Ghadiri group have shown that ssDNA can be ratcheted one base at a time through the αHL pore by the action of a DNA polymerase (20). In the present work, we return to the problem of base identification and show that all 4 DNA bases can be distinguished in both homopolymeric and heteropolymeric immobilized DNA strands.
Results and Discussion
Translocating homopolymer sequences can be distinguished by protein nanopores (10, 11, 21–24). The transition between 2 homopolymer sequences within a translocating single RNA strand can also be observed (21, 25). Individual base pairs at the end of an immobilized DNA strand can also be identified within a nanopore (26, 27), but it is not clear how this might be adapted for sequencing. Recently, individual modified nucleotide bases have been observed “on the fly” (19), but these structures were very bulky. When DNA is immobilized within the αHL pore, by using a 5′- or 3′-terminal hairpin or biotin–streptavidin complex, better resolution of homopolymer sequences can be achieved because of the prolonged observation time (13, 28–31) and in the present work we have extended the biotin–streptavidin approach.
Improved Discrimination of Oligonucleotides with a Mutant αHL Pore.
ssDNA oligonucleotides with biotin tags at the 3′ terminus were allowed to form complexes with streptavidin [supporting information (SI) Fig. S1]. In this state, the DNAs were captured and immobilized by αHL pores in an applied potential, but they were not translocated into the trans compartment (Fig. 1A) (30–32). The immobilized DNA molecules caused a sequence-dependent decrease in the current flow through the pore (Fig. 1B), and here we quote the residual current (IRES) as a percentage of the open pore current (IO). We examined the WT αHL pore and the pore formed by E111N/K147N. The latter forms stable pores despite the removal of the electrostatic interactions between Glu-111 and Lys-147 residues at the central constriction (33, 34). We hoped that the increased space at the constriction would cause more current to flow in the presence of DNA and hence produce a greater dispersion of IRES values. At +160 mV in 1 M KCl, 25 mM Tris·HCl, pH 8.0, containing 0.1 mM EDTA (the conditions for all of the experiments reported in this article), WT αHL pores have a mean open-pore current level (IOWT) of 171 ± 7 pA (n = 20), whereas pores formed from E111N/K147N gave IOE111N/K147 = 167 ± 7 pA (n = 20). Poly(dA)60 oligonucleotides blocked WT pores to a lesser extent (IRESpoly(dA) = 20.0 ± 1.3%) than poly(dC)60 (IRESpoly(dC) = 19.4 ± 1.4%) (Fig. 1B). The residual current difference between the poly(dA) and the poly(dC) oligonucleotide blockades (ΔIRES = IRESpoly(dA) − IRESpoly(dC)) was +0.6 ± 0.1%. It should be noted that the ΔIRES values showed little experimental variation, whereas the absolute current values showed variation that exceeded ΔIRES (Table S1). In practice, the small ΔIRES values were readily determined from event histograms (Fig. 1B). Although the IO levels of WT and E111N/K147N pores are similar, IRES values, as we had hoped, were higher when oligonucleotides were immobilized within the E111N/K147N pores (Fig. 1C): IRESpoly(dA) = 33.9 ± 0.7% and IRESpoly(dC) = 36.6 ± 0.6%. Remarkably, besides an increase in the residual current, there is also a change in the sign of ΔIRES, with poly(dA) blockades giving a lower IRES than poly(dC) oligonucleotide blockades in the E111N/K147N pores: ΔIRES = −2.7 ± 0.4% (Fig. 1C).
Fig. 1.
Discrimination of immobilized DNA homopolymers by αHL pores. (A) Schematic representation of a homopolymeric DNA oligonucleotide (blue circles, only the first 25 nucleotides of the 60-nucleotide-long sequence are shown) immobilized inside an αHL pore (gray, cross-section) through the use of a biotin (yellow)–streptavidin (red) linkage. The αHL pore can be divided into 2 halves, each ≈5 nm in length; an upper vestibule located between the cis entrance and the central constriction, and a 14-stranded, transmembrane, antiparallel β-barrel, located between the central constriction and trans exit. The central constriction of 1.4 nm diameter is formed by the Glu-111, Lys-147 (shaded green), and Met-113 side chains contributed by all 7 subunits. (B and C Left) Current levels for the WT and E111N/K147N pores when blocked with immobilized poly(dC) and poly(dA) oligonucleotides. (B and C Right) Typical event histograms displaying the residual current levels, caused by poly(dC) and poly(dA) oligonucleotide blockages, for the WT and E111N/K147N pores. The mean residual current levels for each oligonucleotide were determined by performing Gaussian fits to the data.
Nucleic acid homopolymers have been distinguished with the WT αHL pore by several groups on the basis of differences in IRES (summarized in Table S2). Meller et al. (22) found that poly(dA) and poly(dC) were difficult to distinguish during translocation through the pore, in part, because of the broad distributions of IRES values. By contrast, when ssDNA was immobilized in the pore with a 3′ hairpin (5′ threading), Ashkenasy et al. (13) found a ΔIRESpoly(dA)−poly(dC) value of −10.5%. The value for 3′ threading was similar. Interestingly, Purnell et al. (31), using biotin–streptavidin immobilization, found that ΔIRES depends on whether the 5′ or 3′ end of the DNA enters the pore first (5′ entry, ΔIRESpoly(dA)−poly(dC) = +1.2%; 3′ entry, ΔIRESpoly(dA)−poly(dC) = −2.9%). Our results (5′ entry: ΔIRESpoly(dA)−poly(dC) = +0.6%) are in approximate agreement with the latter work. We note that that ΔIRES is voltage-dependent (Fig. S2), and that Purnell et al. (31) worked at a lower applied potential. It is worth noting that IRES is greater when the DNAs are attached to streptavidin (Table S2). Perhaps, DNA is more stretched in the electric field within the pore when it is anchored on the cis side. If this is so, it would be preferable to sequence DNA under similar conditions. This would be the case, for example, when DNA is ratcheted through the pore by an enzyme (20, 35).
Interestingly, the open-pore currents carried by the WT pore and E111N/K147N are similar at +160 mV (Fig. S3), but the residual currents in the presence of ssDNA are almost twice as high in the mutant pore (e.g., Fig. 1 B and C), which may be the basis of why E111N/K147N gives better discrimination between poly(dA) and poly(dC). We suggest that the ring of charged lysine and glutamic acid side chains in the constriction (residues 147 and 111, Fig. 1A), which are replaced with asparagines in the mutant, might have one or more effects, including a coulombic block to ion transport, or a steric block based either simply on the bulk of the large amino acid side chains, which might “grip” the translocating DNA, or a collapse of the barrel around the DNA. In any case, the current, which is carried largely by hydrated K+ ions while the negatively charged DNA strand is in the pore (30, 36), is reduced in the WT pore and so is base discrimination in terms of differences in absolute current or percentages of the open pore current (ΔIRES). The actual current levels that are observed cannot be readily rationalized, especially when it is noted that poly(dA) gives the higher residual current in the WT pore and poly(dC) in the E111N/K147N pore. A simplistic conclusion is that the central constriction (comprising residues Lys-147, Glu-111, and Met-113 in the WT) forms a recognition site. This is interesting because Ashkenasy et al. (13) concluded that recognition occurs at the trans exit. In the latter case, the ssDNA was immobilized by 5′- or 3′-terminal hairpins, which probably enter the pore and perturb recognition that occurs at the constriction. Together, the results imply that more than one recognition element might be present in the β-barrel of the αHL pore. Further experimentation, as described below, supports this view.
Defining Recognition Elements Within the αHL Pore.
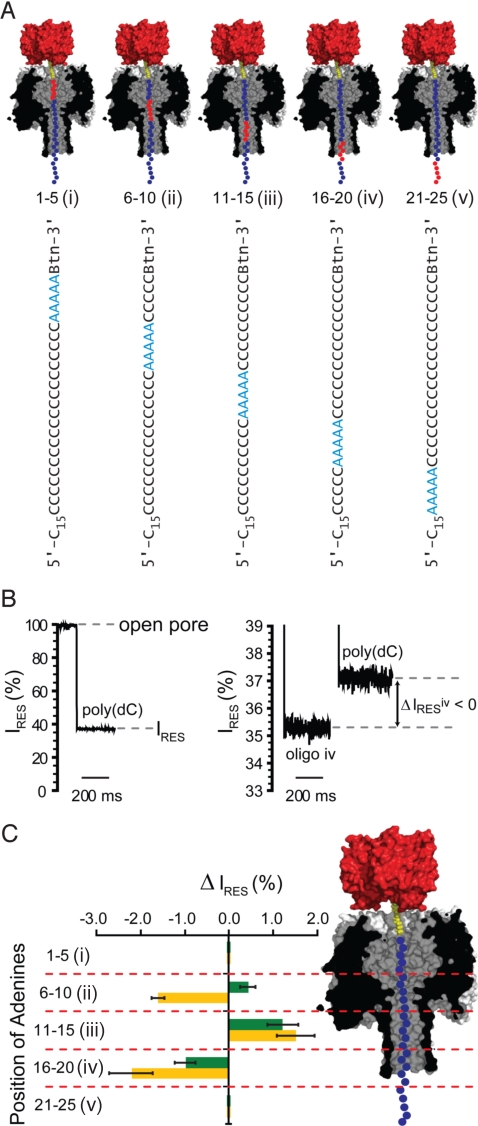
We attempted to better define the regions of the αHL pore that interact with DNA in a base-specific manner (recognition elements) by probing the length of the pore with a set of 5 oligonucleotides, each of which contained a stretch of 5 consecutive adenine nucleotides (A5 oligonucleotides) in an otherwise poly(dC) sequence (Fig. 2A, the locations of the A5 sequences in the figure are justified below). A similar approach for the discovery of base recognition sites was established by Ghadiri and colleagues (13). We determined ΔIRES with respect to a reference poly(dC) oligonucleotide for each of the A5 oligonucleotides (Fig. 2A i–v) for both the WT and E111N/K147N pores (Fig. 2 B and C, Table S3). Our data suggest that when the A5 sequence is closest to the streptavidin anchor (positions 1–5 from the 3′ end), the bases are not recognized by the αHL pore, i.e., ΔIRESA5oligo−poly(dC) = 0, for both WT αHL and E111N/K147N, and the A5 sequence likely lies within the vestibule. However, when the A5 sequence was in positions 6–10, 11–15, and 16–20, the bases were recognized in both pores (Fig. 2C). Importantly, when the A5 sequence was in positions 6–10, the WT and the E111N/K147N pores recognized the DNA in a different way, i.e., for WT αHL, ΔIRESA5oligo−poly(dC) was positive (+0.4 ± 0.2%) and for E111N/K147N, ΔIRESA5oligo−poly(dC) was negative (−1.6 ± 0.1%), suggesting that in this case the A5 sequence lies at the constriction where the mutations are located. Finally, when the A5 sequence was in positions 21–25, no discrimination was seen suggesting that this sequence protrudes through the trans entrance of the pore. Therefore, the sequence bounded by positions 6 and 20 from the 3′ end of the DNA is likely to lie within the narrow confines of the β-barrel, where recognition should be at its strongest. Ashkenasy et al. (13) performed a similar experiment and found that stretches of adenine nucleotides were recognized near the trans entrance, but as noted above they used DNA immobilized with hairpins.
Fig. 2.
Probing DNA recognition by the αHL pore with A5 oligonucleotides. (A) The 5 oligonucleotides (i–v) containing 5 consecutive adenine nucleotides (A5, red circles) at different positions (numbered from the 3′-biotin tag) in an otherwise poly(dC) strand (cytidine nucleotides are shown as blue circles). Only the first 25 of the 40-nucleotide-long sequences are shown. (B Left) The stepwise reduction from the open current value (pore not blocked with DNA) to a residual current (IRES) level of ≈37% when the E111N/K147N pore becomes blocked with a poly(dC) oligonucleotide. (B Right) The IRES levels when a pore is blocked with oligonucleotides of different sequence (oligo iv and poly(dC) are shown). (C) Residual current difference (ΔIRES) between the blockade by oligonucleotides i–v (A) and poly(dC)40 for WT (green bars) and E111N/K147N (orange bars) αHL pores (ΔIRES = IRESi–v − IRESpoly(dC)). The probable location of the adenine (A5) stretch of each oligonucleotide when immobilized with an αHL pore is indicated (Right).
The ssDNA in the pore is elongated compared with its conformation in solution. First, the applied potential produces a force on the DNA, which can be estimated to be ≈8 pN, by the following argument. Let there be ≈30 nt in the entire lumen of the pore (approximately the same as there would be for a strand in a double helix 10 nm in length) and therefore ≈15 nt in the transmembrane β-barrel. The experimentally determined effective charge on each base is ≈0.1e (37, 38). This low value is consistent with the theory of Zhang and Shklovskii (36). Therefore, the overall charge is ≈2.4 × 10−19 C. The field is 0.16 V over the 5 nm of the barrel or 3.2 × 107 Vm−1. Therefore, the force (F = QE) is ≈8 pN. Under this force, ssDNA has a similar extension to the B form of dsDNA (39, 40), so there would indeed be ≈30 nt in the full length of the pore and ≈15 nt in the β-barrel. Second, the effects of enforced confinement would serve to elongate the DNA still further (41–44). Taking into account how streptavidin might dock on the cis surface of the αHL pore, the location of the biotin binding site within streptavidin and the length of the linker between the DNA and the biotinyl group (Fig. S1), the 3′ end of the DNA would be within the lumen and ≈15 Å from the cis entrance (Fig. 1). Therefore, it is reasonable that the DNA strand is located with residues 6 to 20 within the β-barrel (Fig. 2).
Discrimination of Single Adenine Nucleotides.
The results of the A5 scan show that the αHL pore can recognize bases in ssDNA and contains at least 3 recognition sites within the β-barrel. Of course, to be of use in sequencing intact ssDNA strands, the αHL pore must be able to detect single nucleotides. Therefore, we further defined the recognition sites by moving a single A base through a poly(dC) background and comparing the residual current with that of poly(dC) itself. A set of 14 poly(dC) oligonucleotides was made, each containing a single adenine (A1) nucleotide (13). The A1 substitutions were in positions 7–20 relative to the 3′-biotin tag (Fig. 3). ΔIRES [with respect to poly(dC)] was plotted against the position of the adenine nucleotide for both the WT and E111N/K147N pores (Fig. 3, Table S4). Both pores were able to discriminate single adenine nucleotides at multiple positions within the oligonucleotide chain. Remarkably, the pattern of ΔIRES values for the A1 oligonucleotides mirrored the pattern seen with the A5 oligonucleotides (Figs. 2 and 3). Further, the data suggest that there are indeed 3 recognition sites within the barrel, which have been designated R1, R2, and R3 (Fig. 3). These experiments further demonstrate that a single base (A vs. C) can be recognized in an otherwise identical strand at all 3 sites. By contrast, in the hairpin-anchor experiments of Ashkenasy, recognition was confined to the trans entrance (13). When the WT and E111N/K147N pores are compared, the A1 scans appear to be ≈1 nt out of phase, suggesting that the extent of elongation of the ssDNA may differ slightly in the 2 pores.
Fig. 3.
Discrimination of a single adenine nucleotide by αHL. The graph (Middle) indicates the differences in residual current (ΔIRES values) between blockades caused by a poly(dC) oligonucleotide containing a single adenine nucleotide (the sequence of each oligonucleotide is shown to the left) and blockades caused by poly(dC)40 for WT (green) and E111N/K147N (orange) αHL pores (see also Table S4). R1, R2 and R3 represent the 3 proposed recognition sites in the αHL nanopore. Their probable locations are indicated on the cross-section of the β barrel domain of the αHL pore (Right).
Probing the 3 Recognition Sites of αHL for 4-Base Discrimination.
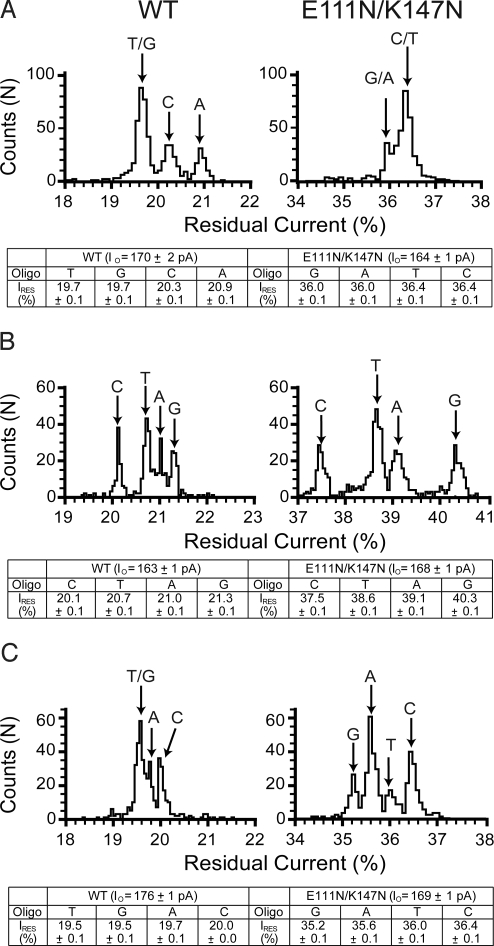
In addition to the detection of individual bases, to sequence ssDNA, αHL pores must also be able to distinguish between G, A, T, and C within a DNA chain. To examine this possibility, the WT and E111N/K147N pores were probed with 3 sets of 4 oligonucleotides. Each oligonucleotide was a homopolymer [poly(dC)], except at a specific position, where it was substituted with either G, A, T, and C [the latter oligonucleotide being poly(dC) itself]. Each of the 3 sets had substitutions at a different position in the sequence, which were designed to probe the R1, R2, and R3 recognition sites.
The first set of oligonucleotides had the G, A, T, or C substitution at position 9 (from the 3′ end) and was designed to probe R1 (Fig. 4A). Although there is some discrimination between the 4 oligonucleotides in this set, neither the WT nor the E111N/K147N pore is able to distinguish all 4 bases. The second set had the G, A, T, or C substitution at position 14 and was designed to probe R2 (Fig. 4B). In this case, both the WT and E111N/K147N pores clearly separated C, T, A, and G, in order of increasing IRES. The span between C and G is far greater for the E111N/K147N pores (ΔIRES = 2.8%) than it is for WT pores (ΔIRES = 1.2%). The final set had the G, A, T, or C substitution at position 18 to probe R3 (Fig. 4C). In this case, only the E111N/K147N pores are able distinguish the 4 bases, but in the reverse order, namely, G, A, T, and C, and the spread of IRES values is not as large as seen with the set substituted at position 14.
Fig. 4.
Recognition of all 4 DNA bases sites by the WT and E111N/K147N αHL pores. Histograms of the residual current levels for WT (Left) and E111N/K147N (Right) pores are shown. Three sets of 4 poly(dC) oligonucleotides were used, with each set containing either a single G, A, T, or C nucleotide at a specific position. All experiments were conducted at least 3 times, and the results displayed in the figure are from a typical experiment. (A) The WT and E111N/K147N pores were interrogated with 5′-CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCNCCCCCCCCBtn-3′ where N represents either G, A, T, or C. Gaussian fits were performed for each peak, and the mean value of the residual current for each oligonucleotide (and the standard deviation) is displayed in the table below the histograms. (B) WT and E111N/K147N pores were interrogated with 4 oligonucleotides with the sequence 5′-CCCCCCCCCCCCCCCCCCCCCCCCCCNCCCCCCCCCCCCCBtn-3′. (C) WT and E111N/K147N pores were interrogated with 4 oligonucleotides with the sequence 5′-CCCCCCCCCCCCCCCCCCCCCCNCCCCCCCCCCCCCCCCCBtn-3′.
For exonuclease sequencing, in which bases are sequentially cleaved from a DNA strand, all 4 DNA bases can be identified as deoxyribonucleoside 5′-monophosphates by using an engineered αHL pore (12, 14). In this case there are no interfering neighboring bases during detection. By contrast, the ability to sequence ssDNA would require the recognition of individual nucleotides in a heteropolymeric background; therefore, we tested this possibility. We were uncertain of the outcome because homopolymeric nucleic acids have been reported to form secondary structures including extended helices (45, 46). Therefore, it was possible that the pronounced differences in residual current that we had observed were the result of disruptions in the DNA structure that caused changes in the conformation of the DNA within the nanopore, which in turn affected current flow.
Single-Nucleotide Discrimination Within a Heteropolymeric Sequence.
To examine discrimination within a heteropolymer, the most promising site, namely R2 in E111N/K147N, was tested. All 4 bases at position 14 in a heteropolymer were recognized with the same order of residual current (C, T, A, and G) as seen in the homopolymeric background (Fig. 5). The immediate context of the identified bases (N) was 5′ CTGNACA 3′, by comparison with 5′ CCCNCCC 3′ in the homopolymer. The span between C and G in the residual current histogram (ΔIRES = 2.9%) was similar to that seen in the homopolymeric background (ΔIRES = 2.8%), although the spacing between the 4 peaks differed in detail (Fig. 5). The sequence we chose does not contain secondary structure such as hairpins, as predicted by the mfold algorithm (47), and is unlikely to form π-stacked helices (45).
Fig. 5.
Probing the E111N/K147N αHL pore for single-nucleotide discrimination in a heteropolymeric oligonucleotide. Histogram (Top) of residual current levels for E111N/K147N pores interrogated with 4 heteropolymeric DNA strands (Middle) that differ at only 1 position (blue). Gaussian fits were performed for each peak, and the mean value of the residual current for each oligonucleotide (and the standard deviation) is displayed (Bottom).
Base Recognition and DNA Sequencing.
Although the ability to identify bases demonstrated here is an advance over previous work and provides a level of understanding that will be useful for further progress, it is reasonable to ask if we can now see whether the approach will lead to DNA sequencing. Obviously, at least 4 pieces of information (i.e., 4 current levels, or the equivalent as outlined below) are required to identify each position in a DNA sequence. A single recognition site in which the neighboring bases do not affect identification, i.e., a “sharp” site, would fulfill this requirement. However, the data we have obtained (Figs. 3, 4B, and 5) suggest that even R2 in E111N/K147N, the most promising site examined in this work, might be too “blunt” to distinguish all 4 bases in a diversity of contexts. For example, the currents in the A1 scan at positions 14 and 15 are similar (Fig. 3), suggesting that it would be difficult to use R2 to distinguish between the sequences CA and AC. If just one recognition site were to be used, it might be possible to sharpen R2 by additional mutagenesis (with both natural and unnatural amino acids) and by site-directed chemical modification, and further blunt the other sites (48). Certainly, the exquisite ability of (context independent) stochastic sensing to distinguish between molecules (14, 49) shows what can be done with nanopores under the most favorable conditions. In the case of solid-state nanopores, it has been suggested that an active property of translocating bases (such as the ability to carry tunneling currents) might be used for sequencing (10, 50–52), but no practical demonstration of such a technique has been made.
Another obvious requirement of a strand sequencing system is an ability to “count” bases. This is most apparent when a homopolymer sequence is considered. Without a modulation of the current between bases, the residual current would remain constant and the length of the homopolymer stretch would be unknown. The time taken to obtain sequence would in itself be an unreliable clock for sequencing because translocation is a stochastic process and the transit times for each base are distributed rather than of a fixed value.
It might be advantageous to use 2 recognition sites within a single pore. Then, even if they are blunt, sufficient information might be gathered to obtain DNA sequence information. For example, let us say that a site B produces a large dispersion of IRES values for G, A, T, and C, whereas a lagging site A, which is say 5 bases away, produces a more modest dispersion that results in the separation of each current level produced by site B into 4 additional levels, so that there are 16 current levels in all. During translocation, a base at site A, will soon move to site B and be read gain. With the information acquired, the sequence would be over-determined. However, the recognition of 16 current levels pushes the limits of the electrical recording technique and the bluntness of the recognition sites would degrade the information content. Nevertheless, sufficient information might remain to determine the DNA sequence. Further, it might be possible to train the system with known sequences, which would enhance its recognition capability. A means of resequencing the same ssDNA would also be advantageous (9), because the stochastic nature of the process dictates that the signal will differ in detail in each run, most conspicuously in the time domain. Of course, the ability to obtain a maximum of useful information will depend on finding a practicable way to slow the ssDNA down to such an extent that filtering of the signal can be used effectively to reduce noise levels (53).
Methods
Full details of experimental procedures can be found in the SI Text.
Protein Preparation.
αHL protein was produced by expression in an Escherichia coli in vitro transcription and translation (IVTT) system and assembled into heptamers on rabbit red blood cell membranes.
Planar Bilayer Recordings.
Lipid bilayers were formed from 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids). Both compartments of the recording chamber contained 0.5 mL of 1 M KCl, 25 mM Tris·HCl, pH 8.0, with 0.1 mM EDTA. Planar bilayer current recordings were performed with a patch clamp amplifier (Axopatch 200B, Axon Instruments) with the cis compartment connected to ground. The αHL pores and the DNA (Table S5) were added to the cis compartment. ssDNA molecules, with a biotinyl group covalently attached to the 3′ end through a linker, were obtained from Sigma-Aldrich (Fig. S1). Solutions of the biotinylated ssDNAs, at 100 μM in 10 mM Tris·HCl, pH 8.0, 0.1 mM EDTA, were mixed with equal volumes of 25 μM streptavidin (SA) (Sigma-Aldrich) in the same buffer. Each oligonucleotide (preincubated with streptavidin for at least 5 min) was added to the cis compartment to a final concentration of 200 nM. Initially, +160 mV was applied to the trans side for 1,800 ms to drive the negatively charged, biotinylated DNA into the pore. The capture of a ssDNA strand by an αHL pore is observed as a stepwise decrease in the open pore current level (IO) to a lower, but stable, current level (IB). A voltage of −140 mV was then applied for 100 ms to eject the immobilized DNA from the pore. The applied potential was then stepped to 0 mV for 100 ms. This 2-s sequence was repeated for at least 100 cycles for each ssDNA species added. The amplified signal (arising from the ionic current passing through the pore) was low-pass filtered at 1 kHz and sampled at 5 kHz with a computer equipped with a Digidata 1440A digitizer (Molecular Devices).
Data Analysis.
Data were analyzed and prepared for presentation with pClamp software (version 10.1, Molecular Devices). Single-channel searches were performed to obtain the average current level for each ssDNA blockade event (IB). The mean IB value for each oligonucleotide was determined by performing a Gaussian fit to a histogram of the IB values. The current blockade for each oligonucleotide was also expressed as the residual current (IRES), wherein the average current level for a DNA blockade (IB) is expressed as a percentage of the open pore current (IO): IRES = (IB/IO) × 100. When comparing several oligonucleotide species, a single-oligonucleotide species was first added to the cis chamber and the current trace required for the determination of IB was recorded. Subsequently, a second (and if required, a third and a fourth) oligonucleotide was added and additional currents recorded. For example, the data in Figs. 4 and 5 come from 4 oligonucleotide species, with sequences that differ by a single nucleotide. When such experiments were repeated, the oligonucleotides were added to the chamber in a different order.
Supplementary Material
Acknowledgments.
This work was supported by the National Institutes of Health, the Medical Research Council, and the European Commission's Seventh Framework Program (FP7) REvolutionary Approaches and Devices for Nucleic Acid Analysis Consortium. D.S. was supported by a BBSRC Doctoral Training Grant. H.B. received a Royal Society Wolfson Research Merit Award.
Footnotes
Conflict of interest statement: Hagan Bayley is the Founder, a Director, and a shareholder of Oxford Nanopore Technologies, a company engaged in the development of nanopore sequencing technology. This article was not supported by Oxford Nanopore Technologies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901054106/DCSupplemental.
References
- 1.Schloss JA. How to get genomes at one ten-thousandth the cost. Nat Biotechnol. 2008;26:1113–1115. doi: 10.1038/nbt1008-1113. [DOI] [PubMed] [Google Scholar]
- 2.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 4.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–637. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 5.Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26:602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 7.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 9.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 10.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009 doi: 10.1038/nnano.2009.12. in press. [DOI] [PubMed] [Google Scholar]
- 13.Ashkenasy N, Sánchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward nanopore DNA sequencing. Angew Chem Int Ed Engl. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Dunning JE, Huang APH, Nyamwanda JA, Branton D. DNA heterogeneity and phosphorylation unveiled by single-molecule electrophoresis. Proc Natl Acad Sci USA. 2004;101:13472–13477. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathé J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maglia G, Rincon Restrepo M, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci USA. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci USA. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell N, Howorka S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew Chem Int Ed Engl. 2008;47:5565–5568. doi: 10.1002/anie.200800183. [DOI] [PubMed] [Google Scholar]
- 20.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid and polyuridylic acid as homopolymers or as segments within single rna molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 24.Butler TZ, Gundlach JH, Troll M. Ionic current blockades from DNA and RNA molecules in the alpha-hemolysin nanopore. Biophys J. 2007;93:3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winters-Hilt S, et al. Highly accurate classification of Watson-Crick basepairs on termini of single DNA molecules. Biophys J. 2003;84:967–976. doi: 10.1016/S0006-3495(03)74913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vercoutere WA, et al. Discrimination among individual Watson-Crick bases pairs at the termini of single DNA hairpin molecules. Nucleic Acids Res. 2003;31:1311–1318. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. Phys Rev Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 29.Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Single molecule DNA rotaxanes of a transmembrane pore protein. Angew Chem Int Ed Engl. 2004;43:3063–3067. doi: 10.1002/anie.200453907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 32.Kasianowicz JJ, Henrickson SE, Weetall HH, Robertson B. Simultaneous multianalyte detection with a nanometer-scale pore. Anal Chem. 2001;73:2268–2272. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 33.Gu L-Q, Cheley S, Bayley H. Prolonged residence time of a noncovalent molecular adapter, β-cyclodextrin, within the lumen of mutant α-hemolysin pores. J Gen Physiol. 2001;118:481–494. doi: 10.1085/jgp.118.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu L-Q, et al. Reversal of charge selectivity in transmembrane protein pores by using non-covalent molecular adapters. Proc Natl Acad Sci USA. 2000;97:3959–3964. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Shklovskii BI. Effective charge and free energy of DNA inside an ion channel. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75 doi: 10.1103/PhysRevE.75.021906. 021906. [DOI] [PubMed] [Google Scholar]
- 37.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in nanopores. Phys Rev Lett. 2003;90:238101–238104. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 38.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr Opin Struct Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 40.Smith SB, Cui Y, Bustamante C. Overstretching b-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Craighead HG. Separation of long DNA molecules in a microfabricated entropic trap array. Science. 2000;288:1026–1029. doi: 10.1126/science.288.5468.1026. [DOI] [PubMed] [Google Scholar]
- 42.Han J, Craighead HG. Characterization and optimization of an entropic trap for DNA separation. Anal Chem. 2002;74:394–401. doi: 10.1021/ac0107002. [DOI] [PubMed] [Google Scholar]
- 43.Wong CT, Muthukumar M. Polymer translocation through a cylindrical channel. J Chem Phys. 2008;128:154903. doi: 10.1063/1.2897932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douville N, Huh D, Takayama S. DNA linearization through confinement in nanofluidic channels. Anal Bioanal Chem. 2008;391:2395–2409. doi: 10.1007/s00216-008-1995-y. [DOI] [PubMed] [Google Scholar]
- 45.Buhot A, Halperin A. Effects of stacking on the configurations and elasticity of single-stranded nucleic acids. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70 doi: 10.1103/PhysRevE.70.020902. 020902. [DOI] [PubMed] [Google Scholar]
- 46.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayley H, Jayasinghe L. Functional engineered channels and pores. Mol Membr Biol. 2004;21:209–220. doi: 10.1080/09687680410001716853. [DOI] [PubMed] [Google Scholar]
- 49.Kang XF, Cheley S, Guan X, Bayley H. Stochastic detection of enantiomers. J Am Chem Soc. 2006;128:10684–10685. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 50.Zwolak M, Di Ventra M. Physical approaches to DNA sequencing and detection. Rev Mod Phys. 2008;80:141–165. [Google Scholar]
- 51.Ohshiro T, Umezawa Y. Complementary base-pair-facilitated electron tunneling for electrically pinpointing complementary nucleobases. Proc Natl Acad Sci USA. 2006;103:10–14. doi: 10.1073/pnas.0506130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J, Lin L, Zhang P, Lindsay S. Identification of DNA basepairing via tunnel-current decay. Nano Lett. 2007;7:3854–3858. doi: 10.1021/nl0726205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapovalov G, Lester HA. Gating transitions in bacterial ion channels measured at 3 microseconds resolution. J Gen Physiol. 2004;124:151–161. doi: 10.1085/jgp.200409087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.