Abstract
A sensitive assay using biotinylated ubiquitin revealed extensive ubiquitination of the large subunit of RNA polymerase II during incubations of transcription reactions in vitro. Phosphorylation of the repetitive carboxyl-terminal domain of the large subunit was a signal for ubiquitination. Specific inhibitors of cyclin-dependent kinase (cdk)-type kinases suppress the ubiquitination reaction. These kinases are components of transcription factors and have been shown to phosphorylate the carboxyl-terminal domain. In both regulation of transcription and DNA repair, phosphorylation of the repetitive carboxyl-terminal domain by kinases might signal degradation of the polymerase.
Keywords: cisplatin, transcription
Ubiquitination of cellular proteins is a covalent modification that has many important physiological functions (1, 2). For example, many ubiquitinated proteins are targeted for degradation by the proteasome, enabling cells to control the level of regulatory proteins. Some ubiquitinated proteins, however, are not targeted for degradation, and in these cases, the modifications may have other functions (3, 4). Proteins become ubiquitinated by the sequential action of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3). Ubiquitination is ATP-dependent because the ubiquitin polypeptide is activated by an ATP-dependent transfer reaction to form a thiol ester on E1.
The large subunit of RNA polymerase II (Pol II LS) becomes ubiquitinated within minutes after exposure of cultured human cells to either UV radiation or the DNA-damaging agent cisplatin (5, 6). These two agents cause DNA lesions that are subject to repair by the nucleotide excision repair system. Several types of lesions induced by either UV radiation or cisplatin arrest elongating RNA polymerase II (Pol II) by creating a physical barrier, and the paused polymerase may signal preferential repair of the transcribed strand. Cells from patients with Cockayne syndrome are deficient in transcription-coupled repair as well as in UV-induced ubiquitination of Pol II LS (see ref. 5). This syndrome has been divided into two complementation groups, CS-A and CS-B. Both types of patients are deficient in UV-induced ubiquitination of Pol II LS. The CS-B protein has been implicated in transcription elongation (7, 8), and there are tentative linkages between DNA repair and the ubiquitin/proteasome pathway (9). There are probably several signals for ubiquitination of Pol II LS. For example, α-amanitin, a specific inhibitor of Pol II, stimulates degradation of Pol II LS in murine fibroblasts (10), probably through the ubiquitin/proteasome system. Although enzymes that catalyze ubiquitination of Pol II LS have not yet been well characterized, Rsp5, an essential hect (homologous to E6-AP carboxyl terminus) protein of the E3 family in Saccharomyces cerevisiae, was shown to bind and signal ubiquitination of both yeast and human Pol II LS through recognition of their carboxyl-terminal domains (CTDs) (11).
The Pol II LS contains a repetitive CTD consisting of tandem repeats of the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser (12). The CTD plays an essential role in mRNA synthesis, with evidence indicating potential roles in initiation, promoter clearance, elongation, and pre-mRNA processing (13–17). SDS/polyacrylamide gel electrophoresis separates Pol II LS into two species: Pol IIA, which has a hypophosphorylated CTD, and Pol IIO, which has a hyperphosphorylated CTD. Three distinct cyclin-dependent kinases (cdks) that can phosphorylate the CTD are associated with Pol II complexes. The mammalian kinase cdk7 and its yeast homolog kin28 are subunits of the general transcription factor TFIIH, which is thought to phosphorylate Pol II subsequent to formation of the preinitiation complex on promoter DNA (18). The yeast kinase Srb10, which is a homolog of cdk8 in mammals, is part of the Pol II holoenzyme and capable of phosphorylating the CTD prior to formation of the initiation complex (19). Finally, the recently discovered transcription factor P-TEFb contains cdk9, which also can phosphorylate CTD (20). The activity of this kinase is required to generate a polymerase that can efficiently elongate (21) and is important for activation of transcription by the HIV protein Tat (22). Some drug inhibitors are relatively specific for certain kinases (23). cdk9 is more sensitive to the drug 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) than is cdk7, whereas H7 and H8 inhibit both kinases to the same extent. Both types of drugs inhibit transcription in vivo and in vitro.
To address how ubiquitination of Pol II LS is regulated, we developed a sensitive system to detect the ubiquitination in vitro. In this system biotinylated ubiquitin was used and ubiquitinated proteins were isolated by chromatography on immobilized avidin. We conclude that phosphorylation of the CTD of Pol II LS can be a signal for ubiquitination of the large subunit.
MATERIALS AND METHODS
Chemical Reagents.
α-Amanitin, 1-(5-isoquinolinylsulfonyl)-2-methylpiperazine (H7), N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide (H8), DRB, and lactacystin were purchased from Calbiochem.
Biotinylation of Ubiquitin.
Two milligrams (2 μmol of NH2 groups) of bovine erythrocyte ubiquitin (Sigma) were dissolved in 1 ml of PBS (pH 7.4) and biotinylated by incubating with 75 μg (0.1 μmol) of EZ-Link Sulfo-NHS-LC-LC-biotin (Pierce) for 30 min at room temperature. Biotinylated ubiquitin was equilibrated with PBS by using HiTrap Desalting (Amersham Pharmacia Biotech), divided into aliquots, and stored at −80°C.
In Vitro Ubiquitination.
Reaction mixtures (25 μl) containing 10 mM Hepes–KOH (pH 7.9), 10% (vol/vol) glycerol, 60 mM KCl, 7 mM MgCl2, 7 mM DTT, 0.1 mM EDTA, 1 mM ATP, 70 μg of HeLa nuclear extract, and 1 μg of biotinylated ubiquitin were incubated for 30 min at 30°C. Ubiquitination reactions were also prepared by using transcription reaction conditions where indicated [ref. 24; 300 ng of poly(I)⋅poly(C), 50 ng of poly(dG)⋅poly(dC), 10 mM creatine phosphate, 1 μl of C/G/UTP mixture (0.125 mM CTP, 5 mM GTP, and 5 mM UTP), and 2 μg of template DNA (pHIV+TAR-G400)].
Ubiquitinated proteins were isolated as follows. The reaction mixtures were diluted with 200 μl of Nonidet P-40 lysis buffer (150 mM NaCl/1% Nonidet P-40/50 mM Tris⋅HCl, pH 7.4) and mixed with 10 μl of avidin-bead (Promega) for 1 hr at 4°C. Bead-bound proteins were washed with Nonidet P-40 lysis buffer and incubated further with 100 μl of Nonidet P-40 lysis buffer containing 1 M NaCl. After incubation for 30 min at 4°C, bead-bound proteins were washed twice with Nonidet P-40 lysis buffer containing 1 M NaCl and once with Nonidet P-40 lysis buffer. Twenty microliters of SDS-sample buffer was added directly to the avidin-bead, and 10-μl aliquots of the bound proteins were resolved by SDS/polyacrylamide gel electrophoresis on 10% or 4–20% gradient polyacrylamide gels and transferred to Immobilon-P membranes (Millipore).
Total ubiquitinated proteins were detected by using HRP-streptavidin-biotin complex (ABC-HRP; Pierce; HRP is horseradish peroxidase) and ECL reagent (Amersham Pharmacia Biotech). Immunoblotting was performed essentially as described by Harlow and Lane (25). The Pol II LS was detected with the N20 antibody, a rabbit polyclonal IgG (Santa Cruz Biotechnology). Phosphorylated CTD was detected with the H14 antibody, a mouse monoclonal IgM that recognizes phosphoserine at position 5 in CTD heptapeptide repeats (26). Ubiquitinated proteins were detected with the 1B3 antibody, a mouse monoclonal IgG (MBL International, Watertown, MA).
Glutathione S-Transferase (GST)-CTD.
The yeast GST-CTD fusion protein (6×His-GST-12CA5-CTD fusion) was expressed in Escherichia coli and purified as described (27). Six hundred nanograms of GST-CTD was added to the ubiquitination reactions as described above.
Assays for kinase activity were performed as follows: GST-CTD was incubated for 30 min at 30°C in the ubiquitination reaction in the presence of 10 μCi of [γ-32P]ATP (1 μCi = 37 kBq). After incubation, GST-CTD was immunoprecipitated with the anti-hemagglutinin (HA) epitope antibody 12CA5 essentially as described by Harlow and Lane (25) and analyzed by SDS/4–20% polyacrylamide gel electrophoresis followed by Western blotting. Phosphorylated GST-CTD was detected by using the H14 antibody or analyzed with the Storm system and imagequant software (Molecular Dynamics).
RESULTS
Pol II LS Is Ubiquitinated in Vitro.
A very sensitive method for detection of ubiquitination of proteins in vitro by using biotinylated ubiquitin and avidin-affinity purification was developed. Proteins associate with the avidin-coated beads dependent upon ubiquitination and can be identified in the eluted fraction by Western blotting with specific antibodies.
Upon incubation with ATP, a large number of proteins in a HeLa nuclear extract were reactive with antibodies specific for ubiquitin (α-Ub 1B3; Fig. 1A, lane 2). When biotinylated ubiquitin (bio-Ub) was added to the reaction, a similarly large number of proteins was ubiquitinated in an ATP-dependent fashion as detected by developing a Western blot with HRP-streptavidin-biotin complex (ABC-HRP; Fig. 1A, lane 4). These ubiquitinated proteins were almost completely bound to the avidin-bead (Fig. 1A, lane 6).
Figure 1.
(A) In vitro ubiquitination of HeLa nuclear extract proteins. HeLa nuclear extract was incubated with 1 mM ATP and 1 μg of biotinylated ubiquitin (bio-Ub) as indicated. Total proteins (lanes 1–4) or avidin-bound proteins (lanes 5 and 6) were analyzed by SDS/4–20% polyacrylamide gel electrophoresis followed by Western blotting. Ubiquitinated proteins were probed either with anti-ubiquitin monoclonal antibody 1B3 (α-Ub 1B3; lanes 1 and 2) or HRP-streptavidin-biotin-complex (ABC-HRP; lanes 3–6). (B) In vitro ubiquitination of Pol II LS. HeLa nuclear extract was incubated with 1 mM ATP and 1 μg of bio-Ub as indicated. Flow-through (lanes 1–4) or avidin-bound proteins (lanes 5–8) were analyzed as described for A. Pol II LS was probed with N20, an anti-Pol II LS antibody (α-Pol II LS N20). Ubiquitinated Pol II LS (Ub-Pol II) was detected in the avidin-bound fraction as slow migrating material (lane 6).
Hypophosphorylated and hyperphosphorylated Pol II LS (designated as Pol IIA and Pol IIO, respectively) can be separated by SDS/polyacrylamide gel electrophoresis (28). Antibodies to Pol II LS, N20, recognize both Pol IIA and Pol IIO. Both types of subunits were present in nuclear extracts after incubation with ATP (Fig. 1B, lanes 2 and 4) as contrasted with the situation where only Pol IIA was observed when the reaction was incubated in the absence of ATP (Fig. 1B, lanes 1 and 3). After incubation with both ATP and bio-Ub, the avidin-bound fraction contained a broad band of N20-reactive material migrating slower than Pol IIO (Fig. 1B, lane 6). In view of the controls, this band represented ubiquitinated forms of Pol II LS. Comparison of lanes 2 and 4 of Fig. 1B suggested that a majority of the Pol IIO generated during incubation of the extract was ubiquitinated.
Transcription-Related Ubiquitination of Pol II LS.
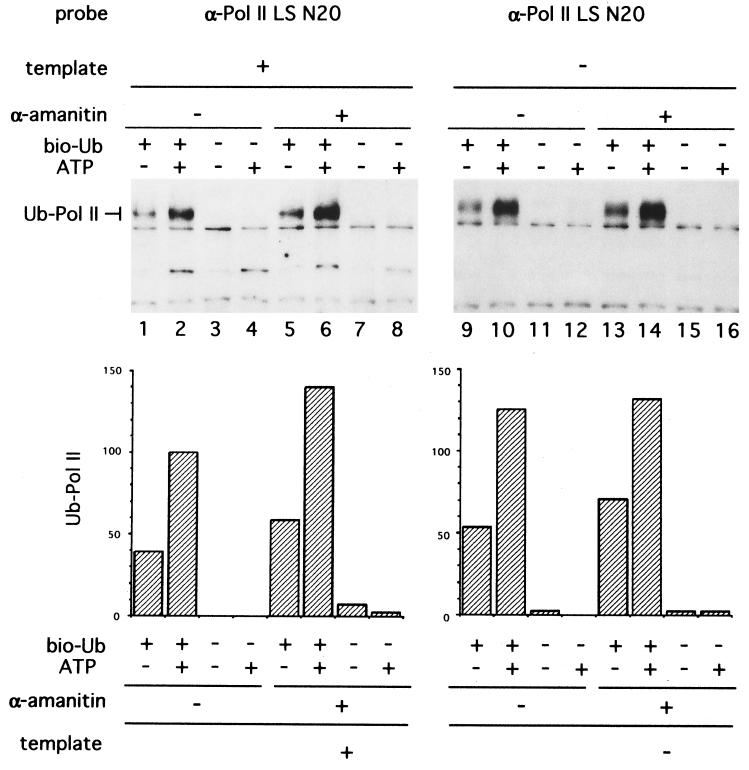
The effect of addition of template DNA on ubiquitination of Pol II was examined. A plasmid containing the HIV promoter was added to reaction mixtures under conditions permitting transcription. The presence of this template reduced the levels of ubiquitination of Pol II LS about 30% as compared with control reactions, respectively (Fig. 2, lanes 2 and 10). A previous study showed that treatment of mouse NIH 3T3 cells with α-amanitin, a Pol II-specific inhibitor, stimulated Pol II LS degradation in vivo (10). It remained unclear whether this degradation was mediated by ubiquitin- and proteasome-dependent mechanisms. We therefore determined the effect of α-amanitin on ubiquitination of the Pol II LS in vitro. Ubiquitination of Pol II LS was stimulated about 1.4-fold by 5 μM α-amanitin in the presence of template DNA (Fig. 2, compare lanes 2 and 6). In contrast, α-amanitin did not stimulate ubiquitination of Pol II LS in the absence of template DNA (Fig. 2, lanes 10 and 14). Addition of 5 μM α-amanitin inhibited transcription of the template completely (data not shown). The degree of template-dependent suppression of ubiquitination is probably limited by the inefficient utilization of polymerases for transcription in these reactions. Typically, only a fraction of the total Pol II engages in transcription and thus could respond to the presence of template DNA. In spite of these limitations, the results suggest that the majority of the ubiquitination of Pol II in these reactions is not dependent upon engagement of the polymerase in transcription.
Figure 2.
Ubiquitination of Pol II LS is reduced by template DNA. (Upper) Ubiquitination reactions were done under in vitro transcription conditions and analyzed by SDS/10% polyacrylamide gel electrophoresis followed by Western blotting. XbaI-cut (2 μg) pHIV+TAR-G400 (24) was added as a template DNA. Reaction mixtures were preincubated with 5 μM α-amanitin for 3 min at 30°C followed by incubation with template and 1 mM ATP as indicated. Pol II LS was probed with anti-Pol II LS antibody N20 (α-Pol II LS N20). (Lower) Ubiquitinated Pol II LS was quantified and normalized to lane 2 as 100.
Pol II LS Ubiquitination Is Correlated to CTD-Phosphorylation.
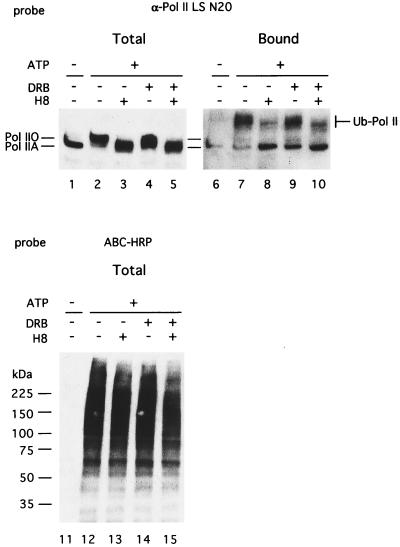
Several kinase inhibitors have been used to investigate regulation of Pol II function through CTD phosphorylation (22, 29, 30). These inhibitors are thought to suppress phosphorylation of the CTD of Pol II and thus affect transcription. Because the degree of ubiquitination of Pol IIO was apparently greater than that of Pol IIA (Fig. 1B), it is possible that phosphorylation of the CTD was a signal for ubiquitination. Hence the effects of kinase inhibitors on Pol II ubiquitination were studied (Fig. 3). H8 at 500 μM, which inhibits both cdk7 and cdk9 activities, inhibited Pol II LS hyperphosphorylation (Fig. 3, lanes 3 and 5) as well as its ubiquitination (Fig. 3, lanes 8 and 10). On the other hand, DRB at 5 μM, which inhibits cdk9 activity in vitro (50% inhibition at 0.65 μM) and Tat-dependent transcription activity in vitro (50% inhibition at 1 μM; ref. 22), only partially inhibited CTD hyperphosphorylation (Fig. 3, lane 4) and did not affect Pol II ubiquitination (Fig. 3, lane 9). These inhibitors did not significantly affect total protein ubiquitination (Fig. 3, lanes 13–15), indicating specificity for the kinase inhibitors.
Figure 3.
Suppression of hyperphosphorylation and ubiquitination of Pol II LS by the kinase inhibitor H8. Ubiquitination reactions were done under conditions similar to those for in vitro transcription, and ubiquitinated proteins were analyzed as described for Fig. 1. Fifty micromolar DRB or 500 μM H8 was added in the reactions where indicated. Total proteins (2.5 μl, lanes 1–5 and 11–15) or avidin-bound proteins (12.5 μl equivalent, lanes 6–10) were probed with anti-Pol II LS antibodies N20 (lanes 1–10) or streptavidin-biotin-HRP-complex (lanes 11–15).
These observations are consistent with the proposal that a hyperphosphorylated CTD could be a signal for Pol II LS ubiquitination. Alternatively, a Pol II LS-specific ubiquitination enzyme(s) might be inhibited directly or indirectly by the H8 drug. Experiments in which reactions were incubated with ATP before addition of H8 and bio-Ub clearly indicated that H8 did not inhibit the ubiquitin transfer reactions directly (data not shown). These results indicated a role for phosphorylation of CTD as a signal for ubiquitination.
CTD-Dependent Ubiquitination Leads to Proteasome-Dependent Protein Degradation.
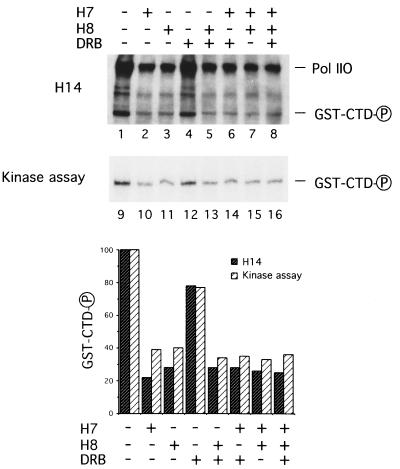
To further address whether CTD phosphorylation is adequate for Pol II LS ubiquitination, we examined the ubiquitination of a fusion protein synthesized by joining sequences encoding GST to those encoding CTD, GST-CTD (27). This chimeric protein was also tagged with a short amino acid sequence, HA, to permit immunoprecipitation with the monoclonal anti-HA antibody 12CA5. Upon incubation of GST-CTD with HeLa nuclear extract and ATP, phosphorylated Pol II LS and GST-CTD were detected by developing a Western blot with the monoclonal antibody H14 (26), which recognizes phosphoserine in position 5 of the CTD heptapeptide repeats (Fig. 4 Upper, lanes 1–8). Radiolabeled GST-CTD was detected (Fig. 4 Upper, lanes 9–16) after incubation of the reactions with [γ-32P]ATP and immunoprecipitation with 12CA5. Phosphorylation of GST-CTD was strongly inhibited by 500 μM H7 and 500 μM H8, but not by 50 μM DRB. Since the extents of reduction of both the phosphoserine epitope and the radiolabeling of GST-CTD were similar (Fig. 4 Lower), phosphorylation of GST-CTD is probably restricted to the CTD. Consistent with this conclusion, incubation of GST alone in the reaction did not result in its phosphorylation (data not shown).
Figure 4.
Suppression of hyperphosphorylation of Pol II LS and GST-CTD by the kinase inhibitors H7 and H8. (Upper) Ubiquitination reactions were done in the presence of 10 μCi of [γ-32P]ATP and 600 ng of GST-CTD. Either 500 μM H7, 500 μM H8, or 50 μM DRB was added in the reactions where indicated. Total proteins (2.5 μl, lanes 1–8) or anti-HA (12CA5)-bound proteins (12.5 μl equivalent, lanes 9–16) were resolved on an SDS/4–20% polyacrylamide gel followed by Western blotting. Phosphorylated CTD was detected by monoclonal antibody H14 (lanes 1–8), and radiolabeled GST-CTD was analyzed as described in the text (Kinase assay, lanes 9–16). (Lower) Phosphorylated GST-CTD, detected by using either the H14 monoclonal antibody or the kinase assay, was quantified and normalized to lane 1 or 9 as 100.
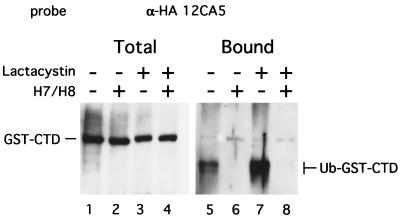
The critical question was whether the GST-CTD protein was ubiquitinated when the reaction was incubated with ATP. Reaction mixtures containing GST-CTD were incubated with bio-Ub and ATP and analyzed by affinity chromatography (Fig. 5). GST-CTD was ubiquitinated only in the absence of the kinase inhibitors (Fig. 5, lanes 5 and 7). In contrast to the situation of ubiquitination of Pol II LS, the ubiquitinated GST-CTD material migrated faster than the input substrate, suggesting that it may be partially degraded. This signal is dependent on the CTD, since substitution of GST alone in place of GST-CTD did not generate a signal in the avidin-bound fraction (data not shown). To determine whether degradation of GST-CTD was mediated by a proteasome-dependent pathway, lactacystin was added as a proteasome inhibitor. Lactacystin is considered specific for protease activities associated with the proteasome but does not inhibit the ubiquitination of proteins (31). The fast-migrating form of ubiquitinated GST-CTD was enhanced 2.5-fold by 20 μM lactacystin (Fig. 5, compare lanes 5 and 7), whereas the levels of unubiquitinated GST-CTDs were the same regardless of the presence of either the kinase inhibitor or lactacystin (Fig. 5, lanes 1–4). These results strongly indicate that the specific phosphorylation of CTD confers a susceptibility to ubiquitination.
Figure 5.
Ubiquitination of GST-CTD is inhibited by kinase inhibitors H7 and H8, and Ub-GST-CTD is stabilized by proteasome inhibitor lactacystin. Ubiquitination reactions were done in the presence of 600 ng of GST-CTD. Total proteins (2.5 μl, lanes 1–4) or avidin-bound proteins (12.5 μl equivalent, lanes 5–8) were resolved on SDS/4–20% polyacrylamide gel electrophoresis followed by Western blotting. GST-CTD was detected with anti-HA (12CA5) antibody. Either 500 μM H7/500 μM H8 or 20 μM lactacystin was added in the reactions where indicated.
DISCUSSION
The large subunit of RNA polymerase II is readily ubiquitinated during incubation of transcription reactions. The signal for this ubiquitination is almost certainly hyperphosphorylation of the CTD. First, attachment of CTD sequences to another protein conferred sensitivity to ubiquitination. Second, addition of inhibitors of cdk-type kinases, which phosphorylate the CTD, dramatically suppressed ubiquitination of the Pol II LS while not blocking modification of other cellular proteins. Similarly, ubiquitination of the chimeric protein formed by fusion with the CTD was also suppressed by the same kinase inhibitors. Not all inhibitors of kinases that phosphorylate the CTD were active in suppressing ubiquitination, suggesting that a subgroup of these kinases generate such signals.
When transcription reactions were incubated with ATP, a significant fraction of Pol II LS was hyperphosphorylated, converting it to the Pol IIO form. In the presence of bio-Ub, a majority of the Pol IIO was modified by ubiquitination. A similar degree of ubiquitination of Pol LS probably occurs in most transcription reactions, since the endogenous level of ubiquitin is significant and this might partially explain changes in the activities of such reactions when preincubated with ATP (32). It is unlikely that the extent of ubiquitination observed in vitro is typical of processes in vivo, as the large subunit of Pol II is very stable under normal conditions. The high levels of ubiquitination observed in vitro probably reflect a heterogeneous set of Pol II-containing complexes generated by the extraction process and the mixing of components from cells at different parts of the cycle—for example, components from mitotic cells with those of G1 cells.
A signal for ubiquitination of the large subunit of Pol II is phosphorylation of the CTD, probably by a specific subgroup of kinases. The drugs H7 and H8, which inhibit cdc2, cdk7, cdk8, and cdk9 kinases, inhibited the ubiquitination of Pol IIO, whereas the drug DRB, which preferentially inhibits cdk9 or P-TEFb, did not inhibit this ubiquitination (23). The specific kinases responsible for phosphorylation of the CTD during reactions in vitro is unclear. The cdc2 kinase will phosphorylate the CTD (33) and could be contributed to the extracts by mitotic cells. Both the cdk7 kinase of TFIIH (18) and the cdk8 kinase of the holoenzyme will also phosphorylate CTD in transcription reactions (19). Phosphorylation by cdk8 (Srb10) occurs before formation of the preinitiation complex and suppresses transcription for a specific subset of genes. It is possible that cdk8 phosphorylation of CTD could stimulate ubiquitination of Pol II. The cdk7 kinase of TFIIH is thought to phosphorylate the CTD after formation of the initiation complex permitting elongation (see, for example, ref. 34). It is possible that cdk7 might also phosphorylate the CTD in its role in transcription-coupled nucleotide excision repair (35). This phosphorylation event could be a signal for ubiquitination and degradation of the polymerase. The cdk9 kinase of the transcription factor P-TEFb primarily phosphorylates the CTD of Pol II after formation of a preinitiation complex (21). This kinase may not be a major contributor to signals for ubiquitination of Pol II LS, as it is sensitive to the drug DRB and probably phosphorylates the CTD only in initiation complexes.
Exposure of cells to either of the DNA-damaging agents UV radiation or cisplatin induces rapid ubiquitination of Pol II LS (6). These DNA lesions block elongation by Pol II and are subject to preferential repair of the transcribed strand of DNA by the nucleotide excision repair system. This system depends on the action of components of the TFIIH transcription factor as well as the Cockayne syndrome A and B proteins, CS-A and CS-B, respectively (5). The CS-B protein enhances elongation, suggesting that it can directly interact with Pol II (7), and the CS-B complex also binds elongating Pol II in vitro (36, 37). Fibroblasts from Cockayne syndrome patients, either CS-A or CS-B, are deficient in ubiquitination of Pol II LS after treatment with either UV radiation or cisplatin, suggesting that this modification of the polymerase might be central to the repair process (5). Since the TFIIH factor is required for transcription-coupled repair and its cdk7 kinase phosphorylates the CTD, it is likely that one of its roles in DNA repair is to phosphorylate the CTD, signaling its ubiquitination and degradation.
Ubiquitination of a protein is commonly mediated by an E3-type factor that binds to both the target protein and the ubiquitinated intermediate E2 factor (1). The nature of the E3 factor recognizing the phosphorylated CTD is unknown. In yeast, an essential hect E3 protein, Rsp5, will bind the CTD and mediate ubiquitination of Pol II LS in vitro. Furthermore, a decrease in the intracellular levels of Rsp5 in vivo results in a 5-fold increase in the levels of Pol II LS in vivo (11). A human homolog of Rsp5 is probably not important for the ubiquitination of Pol II LS in the transcription reactions analyzed here, since the yeast Rsp5 binds to both phosphorylated and nonphosphorylated forms of this protein. There are probably several mechanisms directing ubiquitination of Pol II LS in vivo, one of which depends on phosphorylation of the CTD. The availability of a sensitive assay and the efficiency of ubiquitination of Pol II LS in vitro indicate that it should be possible to biochemically define components mediating this important process.
Acknowledgments
We thank J. B. Kim for stimulating discussions. We also thank B. J. Blencowe for H14 antibody, J. B. Kim for GST-CTD, and M. Siafaca for her help in preparation of the manuscript. We especially thank D. Tantin for help in editing the manuscript. This work was supported by U.S. Public Health Service Grants RO1-AI32486 and PO1-CA42063 from the National Institutes of Health, by Cooperative Agreement CDR-8803014 from the National Science Foundation to P.A.S., and, partially, by National Cancer Institute Cancer Center Support (Core) Grant P30-CA14051.
ABBREVIATIONS
- Pol II
RNA polymerase II
- Pol II LS
large subunit of Pol II
- CS-A and CS-B
Cockayne syndrome complementation groups A and B
- CTD
carboxyl-terminal domain
- cdk
cyclin-dependent kinase
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- H7
1-(5-isoquinolinylsulfonyl)-2-methylpiperazine
- H8
N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide
- HRP
horseradish peroxidase
- GST
glutathione S-transferase
- bio-Ub
biotinylated ubiquitin
References
- 1.Ciechanover A, Schwartz A L. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 3.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 4.Saitoh H, Pu R T, Dasso M. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 5.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg F C, Warren S L. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 7.Selby C P, Sancar A. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tantin D. J Biol Chem. 1998;273:27794–27799. doi: 10.1074/jbc.273.43.27794. [DOI] [PubMed] [Google Scholar]
- 9.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Nature (London) 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen V T, Giannoni F, Dubois M-F, Seo S-J, Vigneron M, Kédinger C, Bensaude O. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huibregtse J M, Yang J C, Beaudenon S L. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young R A. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 13.Dahmus M E. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 14.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 15.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmetz E J. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 17.Hirose Y, Manley J L. Nature (London) 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 18.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Nature (London) 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 19.Hengartner C J, Myer V E, Liao S-M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 20.Peng J, Zhu Y, Milton J T, Price D H. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, Sharp P A. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 26.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 27.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 28.Dahmus M E. Methods Enzymol. 1996;273:185–193. doi: 10.1016/s0076-6879(96)73019-7. [DOI] [PubMed] [Google Scholar]
- 29.Marciniak R A, Sharp P A. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yankulov K, Yamashita K, Roy R, Egly J-M, Bentley D L. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 31.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 32.Marciniak R A, Calnan B J, Frankel A D, Sharp P A. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Corden J L. J Biol Chem. 1991;266:2290–2296. [PubMed] [Google Scholar]
- 34.Kumar K P, Akoulitchev S, Reinberg D. Proc Natl Acad Sci USA. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svejstrup J Q, Vicki P, Egly J M. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 36.Tantin D, Kansal A, Carey M. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You Z, Feaver W J, Friedberg E C. Mol Cell Biol. 1998;18:2668–2676. doi: 10.1128/mcb.18.5.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]