Abstract
The Rad52 pathway has a central function in the recombinational repair of chromosome breaks and in the recovery from replication stress. Tolerance to replication stress also depends on the Mec1 kinase, which activates the DNA replication checkpoint in an Mrc1-dependent manner in response to fork arrest. Although the Mec1 and Rad52 pathways are initiated by the same single-strand DNA (ssDNA) intermediate, their interplay at stalled forks remains largely unexplored. Here, we show that the replication checkpoint suppresses the formation of Rad52 foci in an Mrc1-dependent manner and prevents homologous recombination (HR) at chromosome breaks induced by the HO endonuclease. This repression operates at least in part by impeding resection of DNA ends, which is essential to generate 3′ ssDNA tails, the primary substrate of HR. Interestingly, we also observed that the Mec1 pathway does not prevent recombination at stalled forks, presumably because they already contain ssDNA. Taken together, these data indicate that the DNA replication checkpoint suppresses genomic instability in S phase by blocking recombination at chromosome breaks and permitting helpful recombination at stalled forks.
Keywords: checkpoints, DNA replication, genomic instability, homologous recombination, S. cerevisiae
Introduction
The genome of eukaryotic cells is particularly vulnerable during the S phase of the cell cycle, when replication forks encounter DNA lesions or natural obstacles such as tightly bound protein complexes and highly expressed genes. Stalled forks are unstable structures that can induce chromosomal rearrangements if they are not properly processed and restarted (Labib and Hodgson, 2007; Tourriere and Pasero, 2007). Arrested forks are detected by a surveillance pathway called the intra-S checkpoint, which coordinate fork repair mechanisms and arrest cell cycle progression (Branzei and Foiani, 2008). In budding yeast, this checkpoint response is initiated with the recruitment of the checkpoint kinase Mec1 onto RPA-coated single-strand DNA (ssDNA) in a Ddc2-dependent manner (Rouse and Jackson, 2002). Phosphorylation of the checkpoint mediator Mrc1 by Mec1 promotes the hyperactivation of the effector kinase Rad53 (Alcasabas et al, 2001; Osborn and Elledge, 2003). Mec1 and Rad53 execute multiple functions, the most important being the maintenance of arrested forks (Lopes et al, 2001; Tercero et al, 2003). To this end, checkpoint kinases regulate the activity and/or the association of several fork components, including RPA (Brush et al, 1996), DNA polymerases (Pellicioli et al, 1999; Cobb et al, 2003), MCMs (Cobb et al, 2005) and Exo1 (Cotta-Ramusino et al, 2005; Segurado and Diffley, 2008). These modifications prevent the collapse of arrested forks and the formation of abnormal replication intermediates, including double-strand breaks (DSBs).
DSBs can be repaired by different pathways, the most important being homologous recombination (HR) and non-homologous end joining (NHEJ). Although NHEJ operates throughout the cell cycle, HR is restricted to S and G2/M phases. Recombination is initiated at DSBs with the nucleolytic degradation of DNA ends to generate 3′-ended ssDNA. This resection process depends on the coordinated action of Mre11, Exo1, Sgs1 and Sae2 (Paques and Haber, 1999; Krogh and Symington, 2004; Llorente and Symington, 2004; Clerici et al, 2005; Mimitou and Symington, 2008; Zhu et al, 2008) and requires high cyclin-dependent kinase (CDK) activity (Aylon et al, 2004; Ira et al, 2004; Huertas et al, 2008; Zierhut and Diffley, 2008). Homology search and DNA-strand exchange is catalysed by the assembly of Rad51–ssDNA nucleoprotein filaments on RPA-coated ssDNA, a process that is mediated by Rad52 and by the Rad55–57 heterodimer. Strand invasion, promoted by the dsDNA translocase Rad54, leads to the synthesis of the DNA sequence disrupted at the DSB (San Filippo et al, 2008).
Recombination-related processes have a central function in the recovery of stalled or collapsed replication forks. Broken forks can be repaired by a single-ended invasion recombination mechanism called break-induced replication (Wang et al, 2004; Llorente et al, 2008). Arrested forks can also be restarted by recombinational repair, a process that has been extensively studied in Escherichia coli (Michel et al, 2007). However, the importance of recombination-mediated fork restart mechanisms remains to be established in eukaryotic cells. Indeed, unlike bacteria, eukaryotic genomes contain a large excess of replication origins, which can be used to compensate for stalled forks (Ge et al, 2007; Ibarra et al, 2008). Moreover, although cells mutated for members of the RAD52 epistasis group are hypersensitive to replication inhibitors (Chang et al, 2002; Lundin et al, 2005), it has been difficult to determine whether this pathway is necessary for the recovery of stalled forks in S phase or for the repair of residual replication-dependent DSBs in G2.
Homologous recombination must be tightly regulated to prevent genomic instability in S phase. Central to this regulation are specialized DNA helicases such as Srs2, Mph1 and Sgs1, which modulate different steps of the HR process (Gangloff et al, 2000; Krejci et al, 2003; Veaute et al, 2003; Branzei and Foiani, 2007; Prakash et al, 2009). Some of these mechanisms depend on the ubiquitinylation and/or sumoylation of PCNA (Pfander et al, 2005; Branzei et al, 2006; Davies et al, 2008). The DNA replication checkpoint has also been implicated in the regulation of HR. However, it is not clear whether the Mec1 pathway has a positive or a negative role in this process. Thus, recent reports indicate that Mec1 promotes HR in budding yeast by phosphorylating recombination proteins such as Rad55 and Slx4 in response to MMS (Bashkirov et al, 2000; Herzberg et al, 2006; Flott et al, 2007). Similarly, ATR and Chk1 activate HR in mammalian cells by phosphorylating Rad51, Nbs1 and FANCD2 (Sorensen et al, 2005; Branzei and Foiani, 2008). In contrast, the checkpoint kinase Cds1 prevents the activity of Mus81 and Rad60 at stalled forks in fission yeast (Boddy et al, 2003; Kai et al, 2005; Raffa et al, 2006). Moreover, live cell imaging studies in budding and fission yeast indicate that Rad52–GFP recombination foci are absent from wild-type cells exposed to hydroxyurea (HU), but accumulate in checkpoint-deficient cells (Lisby et al, 2004; Meister et al, 2005). These data suggest that HR foci are actively suppressed by the replication checkpoint. Alternatively, recombination foci could be absent from wild-type cells because checkpoint-proficient cells do not accumulate recombinogenic substrates in HU.
To help clarify the effect of the replication checkpoint on recombinational repair during S phase, we have monitored how Saccharomyces cerevisiae cells respond to genotoxic agents that generate different types of replication stress. HU is an inhibitor of ribonucleotide reductase that slows down fork progression by reducing dNTP pools (Koc et al, 2004; Alvino et al, 2007). Methyl methanesulphonate (MMS) induces a stable fork arrest by methylating DNA on N7-deoxyguanine and N3-deoxyadenine (Vázquez et al, 2008). Importantly, both HU and MMS elicit a robust replication checkpoint response but do not generate DSBs in checkpoint-proficient cells (Lundin et al, 2005; Tourriere and Pasero, 2007). In contrast, the topoisomerase I inhibitor camptothecin (CPT) induces checkpoint-blind lesions that are not detected by the replication checkpoint but generate DSBs upon passage of the fork (Redon et al, 2003). Finally, the radiomimetic agent Zeocin induces chromosome breaks independently of DNA replication. Using different combinations of these drugs, we show here that both the formation of recombination foci and the recombinational repair of DSBs are actively repressed by drugs that activate the DNA replication checkpoint—such as HU and MMS—but not by CPT. Moreover, we provide direct evidence that recombination occurs during S phase to promote the recovery of stalled forks, regardless of checkpoint activity. Finally, we show that HU and MMS impede the resection of DNA ends in an Mrc1-dependent manner. As ssDNA is already present at stalled forks, we propose that inhibition of resection under replication stress prevents toxic recombination events in S phase without interfering with helpful recombination at stalled replication forks.
Results and discussion
HR foci assemble in cells exposed to Zeocin or CPT, but not to HU or MMS
To monitor the cellular response to different types of genotoxic insults, we first examined the ability of S. cerevisiae cells to assemble Rad52 subnuclear foci at Zeocin-induced DSBs. Haploid cells carrying a GFP-tagged version of Rad52 (Lisby et al, 2001) were arrested in G1 with α-factor and were exposed or not to Zeocin. Cells were then either released into S phase or maintained in G1 for the indicated period of time (Figure 1A). The percentage of cells presenting HR foci was determined microscopically. As reported earlier (Lisby et al, 2001, 2004), we measured a 5- to 10-fold increase of spontaneous Rad52 foci upon entry into S phase (Figure 1B), which form at chromosome breaks occurring during normal DNA replication. After Zeocin treatment, we observed a further five-fold increase of Rad52 foci in S-phase cells (Figure 1B). However, Zeocin did not induce Rad52 foci in G1-arrested cells, which is consistent with the fact that high CDK activity is required for the resection of DNA ends and the formation of RPA-coated ssDNA, the substrate recognized by Rad52 (Lisby et al, 2001; Aylon et al, 2004; Ira et al, 2004).
Figure 1.
Genotoxic agents differentially affect the formation of Rad52 foci in S phase. (A) Overview of the assay. Wild-type cells (PP534) expressing Rad52–GFP were arrested in G1 with α-factor for 120 min. Half of the culture was exposed to the radiomimetic agent Zeocin (ZEO) for 30 min to generate DSBs. Cells treated with or without Zeocin were either maintained in G1 or released into S phase for the indicated time. (B) The percentage of G1- and S-phase cells forming spontaneous and Zeocin-induced Rad52 foci was scored as described in Materials and Methods. Representative images of cells showing Rad52–GFP foci are shown. (C) Percentage of wild-type cells forming Rad52 foci in response to genotoxic drugs. Rad52–GFP cells were arrested in G1 and were released into S phase in the absence of drugs (untreated) or in the presence of 200 mM hydroxyurea (HU), 0.033% methyl methanesulphonate (MMS), 20 μM camptothecin (CPT) or 100 μg/ml Zeocin (ZEO). The fraction of cells showing Rad52–GFP foci was determined for the indicated time after release from α-factor. (D, E) Cells arrested at the G1/S transition are competent to form Rad52 foci. Thermosensitive cdc7-4 cells (PP544) expressing Rad52–GFP were arrested in G1 with α-factor for 120 min and exposed to 100 μg/ml Zeocin for 30 min in the presence of α-factor. Cells were then released into S phase at the permissive (24°C) or restrictive (37°C) temperature for the cdc7-4 mutation and the percentage of cells forming Rad52 foci was scored 90 min after release from G1. Flow cytometry analysis indicates that although cdc7-4 cells are blocked at the G1/S transition (D), they are able to assemble Rad52 foci (E).
Next, we investigated whether genotoxic drugs that affect replication fork progression such as HU, MMS and CPT are also able to induce Rad52 foci in S phase. CPT blocks the Top1–DNA cleavable complex and generates nicks that are converted into DSBs upon passage of the replication fork (Pommier, 2006). As expected, we found that CPT induces the formation of Rad52 foci to a level comparable to Zeocin (Figure 1C). In contrast, the fraction of cells presenting Rad52 foci after HU or MMS exposure was even lower than in untreated cells (Figure 1C). The lack of Rad52 foci could reflect the fact that HU- and MMS-treated cells are arrested too early in S phase to be competent for HR. To address this possibility, we monitored the ability of cells arrested at the CDC7 execution point to assemble HR foci at DSBs induced by Zeocin. The Cdc7 kinase is essential for the activation of replication origins and cdc7-4 mutants accumulate at the G1/S transition at 37°C with an unreplicated genome and elevated CDK activity (Bousset and Diffley, 1998). Interestingly, we found that cdc7-4 cells are equally able to form Rad52 foci when arrested at the G1/S transition or released into S phase in the presence of Zeocin (Figure 1D and E). Taken together, these data indicate that exit from G1, but neither the initiation of DNA replication nor the presence of homologous sequences, is required to form recombination foci at Zeocin-induced DSBs.
HU and MMS prevent the formation of HR foci at DSBs induced by Zeocin
Our data indicate that CPT induces Rad52 foci in S phase, but HU or MMS do not (Figure 1C). This difference could be due to the fact that unlike CPT, HU and MMS do not generate DSBs in checkpoint-proficient cells (Sogo et al, 2002; Lundin et al, 2005). To test this hypothesis, we induced DSBs with Zeocin prior to the addition of HU, MMS or CPT and we monitored cells' ability to assemble Rad52 foci. As shown in Figure 2A, addition of CPT did not prevent HR foci assembly in cells previously exposed to Zeocin. In contrast, formation of Rad52 foci at Zeocin-induced breaks was strongly suppressed by HU or MMS. As HU and MMS, but not CPT, elicit a robust activation of the replication checkpoint (Redon et al, 2003), these data suggest that the DNA replication checkpoint prevents the formation of Rad52 foci at DSBs.
Figure 2.
HU and MMS prevent the assembly of Rad52 foci at DSBs. (A) Wild-type cells (PP534) expressing Rad52–GFP were arrested in G1, exposed to Zeocin as described in Figure 1 and released into S phase for 90 min in fresh medium containing no drug (S), 200 mM HU, 0.033% MMS or 20 μM CPT. The fraction of cells showing Rad52 foci was scored. (B) Wild-type cells expressing Rad52–GFP were arrested in G1 for 120 min and cells were maintained in G1 for another 30 min in the presence or absence of 100 μg/ml Zeocin (grey box). Cells were then released into S phase and HU was added to the culture at the indicated time (white boxes). (C) Percentage of cells presenting Rad52–GFP foci. (D) PFGE analysis of chromosome mobility in Zeocin-treated cells exposed to HU at the indicated time points after release from G1. Averaged values for a representative set of chromosomes are shown. (E) cdc7-4 mutants (PP544) were arrested in G1 for 120 min at 24°C and exposed to 100 μg/ml Zeocin for another 60 min at 24°C. Cells were then released into S phase for 90 min in the presence (ZEO+HU) or the absence (ZEO) of 200 mM hydroxyurea. The percentage of cells presenting Rad52 foci is shown.
Next, we investigated whether HU treatment is sufficient to induce a disassembly of preformed Rad52 foci. G1-arrested cells were exposed to Zeocin and were released into S phase as described above. HU was either added to the culture immediately after release from G1 or was added 50 or 90 min later (Figure 2B). Cells were harvested 130 min after release and the fraction of cells presenting Rad52–GFP foci was determined. As shown above, addition of HU immediately after release from G1 suppressed the formation of HR foci and induced a two-fold reduction of cell viability (Supplementary Figure 1A). However, Rad52 foci were not affected when HU was added 50 min later (Figure 2C). To verify that HU is able to block S-phase progression when added 50 min after release from G1, we monitored the electrophoretic mobility of chromosomes by pulsed-field gel electrophoresis (PFGE) (Figure 2D). The fact that this mobility was still altered reflects the persistence of unreplicated chromosomes. We therefore conclude that replication fork arrest prevents the formation of HR foci but does not affect their maintenance once formed.
HU inhibits DNA replication by depleting dNTP pools. As dNTPs are also required for HR, we checked whether HU suppresses Rad52 foci by blocking fork progression or by altering dNTP pools. To this end, we generated DSBs with Zeocin in cells arrested at the G1/S transition with the cdc7-4 mutation and we measured the capacity of HU treatment to prevent Rad52 foci formation in the absence of DNA replication. Under these conditions, Rad52 foci formed even in the presence of HU, indicating that HU-arrested forks—and not HU itself—inhibit recombination foci (Figure 2E).
HU- and MMS-arrested forks prevent the repair of HO-induced DSBs
As MMS and HU prevent the formation of Rad52 foci, we next investigated whether these drugs also inhibit HR at DSBs. A unique DSB was generated at the MAT locus with the HO endonuclease and the ability of HU- or MMS-treated cells to repair this break was monitored by Southern blot hybridization after StyI digestion (Sugawara and Haber, 2006). Cleavage of MATa with a galactose-inducible HO generates a small HO-cut fragment (C) that is further converted into a larger repair product (R) by gene conversion using the HMLα locus as a template (Figure 3A). In untreated or CPT-treated cells, this gene conversion product was detected 2–3 h after HO induction (Figure 3B and C). In contrast, gene conversion was strongly delayed in HU-exposed cells and was completely suppressed after MMS treatment (Figure 3B and C). This repression was further confirmed by semiquantitative PCR analysis of the gene conversion product (Supplementary Figure 2). These data indicate that MMS and HU prevent both the formation of Rad52 foci and the recombinational repair of chromosome breaks.
Figure 3.
Cells exposed to HU or MMS are unable to repair HO-induced DSBs. (A) Overview of the assay. Early log-phase cultures of wild-type cells (PP723) bearing a GAL-HO construct were exposed to 200 mM HU for 4 h, 0.033% MMS for 2 h or 20 μM CPT for 3 h prior to HO induction (see Supplementary Figure 1B for DNA content profiles). Expression of the HO endonuclease was induced for 60 min with 2% galactose and repressed with 2% glucose. Genomic DNA was extracted at the indicated time points and was digested with StyI (S). The smallest of the two StyI fragments (*) generates fragment C (cut) upon cleavage with HO. Homologous recombination with HMLα generates fragment R (repair). (B) Southern blot analysis of DSB repair in untreated cells (ctrl) or in cells exposed to HU, MMS or CPT. (C) Relative intensity of the repair band in untreated control cells and in cells exposed to HU, MMS and CPT.
HU and MMS impede resection at HO-induced DSBs
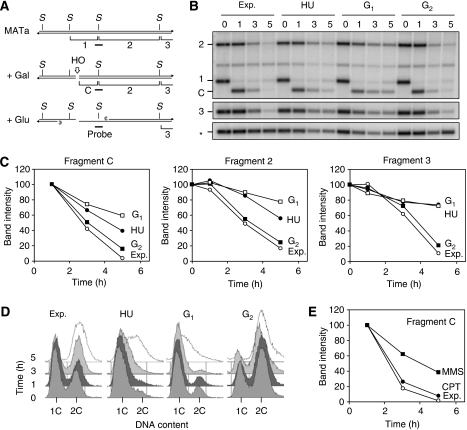
Homologous recombination is initiated with the resection of DNA ends to generate a 3′ ssDNA overhang. To check whether this process is affected by genotoxic drugs, a non-reparable HO cut was induced at the MATa locus of donorless cells (Figure 4A) and the 3′–5′ resection of DSB ends was monitored on Southern blot hybridization (Ira et al, 2004). During the resection process, the intensity of the cut fragment (Figure 4B, fragment C) and of adjacent StyI fragments (Figure 4B, fragments 2 and 3) diminishes as StyI restriction sites are inactivated (Figure 4B; Exp.). To determine whether resection activity varies during the cell cycle and whether it is affected by replication stress, cells were arrested in G1 with α-factor and were released in the presence of nocodazole or HU to arrest cells in G2 or in S phase, respectively (Figure 4D). As shown earlier (Ira et al, 2004), we observed an efficient resection of the HO DSB in exponentially growing and G2-arrested cells, but not in cells blocked in G1 (Figure 4B and C). We also observed a strong inhibition of resection in cells exposed to HU (Figure 4B and C) and to MMS, but not in cells treated with CPT (Figure 4E; Supplementary Figure 3). These data are consistent with the fact that HU and MMS exposure prevent DSB repair in S phase (Figures 2 and 3), presumably by impeding the formation of 3′ssDNA tails.
Figure 4.
HU and MMS impede resection of HO-induced DSB. (A–C) Exponentially growing (Exp.) wild-type cells (PP1123) bearing a GAL-HO construct were arrested in G1 with α-factor, in G2 with 15 μg/ml nocodazole or in S phase with 200 mM HU. HO expression was induced as described above and the resection of HO-induced DSB at the MATa locus was monitored by Southern blot hybridization. The rate of disappearance of the HO-cut band (fragment C) and of adjacent StyI fragments (2 and 3) was expressed for each time point as a percentage of the intensity of the initial band. The intensity of resection bands was normalized to the intensity of the RAD9 gene (*). (D) Flow cytometry analysis of DNA content in exponentially growing cells and in cells arrested in G1, G2 or with HU. (E) Southern blot analysis of the resection rate of HO-induced DSB in the presence of 0.033% MMS or 20 μM CPT.
Mrc1-dependent inhibition of resection in response to replication stress
Activation of Rad53, the effector kinase of the Mec1 pathway, depends on two mediator proteins called Rad9 and Mrc1 (Alcasabas et al, 2001; Gilbert et al, 2001). Although Rad9 functions as a general mediator of the DNA damage response, Mrc1 is active only at stalled replication forks (Tourriere and Pasero, 2007). To determine which branch of the intra-S-phase checkpoint is implicated in the regulation of HR, we next monitored Rad52 foci formation in mrc1Δ and rad9Δ cells. Wild-type and mutant cells were arrested in G1 with α-factor and treated with Zeocin, as described in Figure 2A. Cells were then released into S phase in the presence or absence of HU and their ability to form Rad52 foci was scored. In wild-type cells and in rad9Δ mutants, we observed a sharp induction of Rad52 foci that was largely reduced upon addition of HU (Figure 5A). In contrast, mrc1Δ cells were unable to repress the formation of HR foci in response to HU (Figure 5A) and to MMS (Figure 5B), indicating that the assembly of Rad52 foci is primarily repressed by the Mrc1-dependent branch of the Mec1 pathway.
Figure 5.
Mrc1 suppresses the formation HR foci and ssDNA overhangs in response to replication stress. (A) Wild-type (PP534), mrc1Δ (PP388), rad9Δ (PP1154), mrc1AQ (PP471) and mrc1AQ rad9Δ (PP1064) cells were arrested in G1 and were exposed or not to Zeocin (ZEO) as indicated in Figure 2A. Cells were then released into S phase for 90 min in the presence or absence of 200 mM HU. The percentage of cells containing Rad52–GFP foci is indicated. (B) Percentage of wild-type (PP534) and mrc1Δ (PP388) cells forming Zeocin-induced Rad52 foci in S phase after exposure to HU, MMS or CPT. Cells were arrested in G1, treated with Zeocin and were released into S phase for 90 min in the presence (HU, MMS and CPT) or absence (S) of drugs as described in Figure 1B. (C) Analysis of DSB resection at MATα in untreated mrc1Δ (PP736) cells (Exp.) or in cells exposed to HU, MMS or CPT. Experiment was performed as described in Figure 4. (D, E) Resection rate at the HO-cut band (fragment C) and at the adjacent StyI fragment (fragment 2) in untreated mrc1Δ cells (Exp.) and in cells exposed to drugs (HU, MMS and CPT). The ratio of intensity of fragments in the presence or absence of HU is indicated for wild-type and mrc1Δ cells.
It has recently been reported that Mrc1 has a structural function at replication forks that is distinct from its checkpoint function (Szyjka et al, 2005; Tourriere et al, 2005). To determine which function of Mrc1 is important to regulate HR, we analysed Rad52 foci formation in the separation-of-function allele mrc1AQ, which is only defective for the checkpoint function of Mrc1 (Osborn and Elledge, 2003; Szyjka et al, 2005). Surprisingly, Rad52 foci were still repressed in HU-treated mrc1AQ mutants (Figure 5A), indicating either that Mrc1 prevents HR independently of its checkpoint function or that the replication checkpoint is still partially active in mrc1AQ cells. We favour the second hypothesis as we observed that mrc1AQ cells still repress a significant fraction of their late replication origins, including ARS1212 and ARS1413 (CA, unpublished results). As Rad9 is partially able to compensate for the checkpoint defect of mrc1Δ mutants (Alcasabas et al, 2001), we monitored the assembly of Rad52–GFP foci in mrc1AQ rad9Δ double mutants. We observed a derepression of HR foci formation in HU comparable to mrc1Δ mutants (Figure 5A). We therefore conclude that the checkpoint function of Mrc1 is required to prevent the formation of Rad52 foci in response to replication stress, this function being complemented by Rad9 in mrc1AQ cells.
Next, we checked whether Mrc1 is also implicated in the HU-dependent repression of resection. Wild-type and mrc1Δ cells were exposed to HU and resection was monitored at the MATα locus after induction of HO, as described in Figure 4 (see Supplementary Figure 4A for a map of StyI fragments at the MATα locus). Interestingly, we found that the HU- and MMS-dependent inhibition of DSB resection was strongly reduced in mrc1Δ mutants, to a level comparable to CPT-treated cells (Figure 5C–E). Taken together, these data indicate that the DNA replication checkpoint prevents HR under replication stress by impeding the formation of 3′ ssDNA overhangs at DSBs.
Rad51 is required for the recovery of stalled replication forks
Our observation that HR is suppressed under replication stress is in apparent contradiction to the fact that mutants of the Rad52 pathway are exquisitely sensitive to agents inducing fork arrest (Chang et al, 2002). It has been proposed that HR is repressed at stalled forks in checkpoint-proficient cells but is still required to repair residual DSBs after the completion of S phase. Alternatively, the Rad52 pathway could have a function at stalled forks that is distinct from DSB repair and that is not repressed by the replication checkpoint. The view that recombination is active at stalled forks under replication stress is supported by a recent report showing that replication forks progress at a very slow rate in MMS-treated rad52Δ cells (Vázquez et al, 2008). To test whether recombination is essential to complete S phase after MMS exposure, DNA replication was monitored in rad51Δ cells using a variety of assays, including PFGE, 2D gel electrophoresis and DNA combing. Wild-type and rad51Δ cells were arrested in G1 and released synchronously into S phase in the presence of BrdU and MMS. Cells were then released from MMS, and DNA samples were processed as described earlier (Luke et al, 2006; Zaidi et al, 2008). As shown in Figure 6A, chromosomes do not migrate into the PFGE gel after release from G1 due to the presence of replication intermediates but complete replication in wild-type cells ∼90 min after removal of the drug. In rad51Δ cells, chromosomes failed to reenter the gel even 130 min after release (Figure 6A and B), indicating that Rad51 is essential to complete DNA replication after MMS exposure.
Figure 6.
Rad51 is required for the recovery of MMS-induced replication fork stalling. Wild-type (PP633) and rad51Δ (PP484) cells were arrested in G1 with α-factor and released synchronously into S phase in the presence of 400 μg/ml BrdU and 0.033% MMS for 60 min. Cells were then collected and resuspended in fresh medium containing BrdU for the indicated time after MMS release (t=0). (A) PFGE analysis of chromosome mobility in wild-type and rad51Δ cells after release from MMS arrest. (B) Quantitation of chromosome mobility during release from MMS. (C) DNA fibres from wild-type and rad51Δ cells treated as indicated above were analysed by DNA combing. Representative DNA fibres from MMS-treated cells are shown. Red: DNA; green: BrdU; white: BrdU channel alone; brackets: unreplicated gaps. Scale bar is 50 kb. (D) Quantitation of the percentage of replication and the number of unreplicated gaps per megabyte of genomic DNA in wild-type and rad51Δ cells released from MMS arrest. (E) 2D gel analysis of replication intermediates in wild-type, rad51Δ and sgs1Δ cells exposed for 60 min to 0.1% MMS. 2D gel electrophoresis was performed as described, using a probe against the early origin ARS305 (Tourriere et al, 2005). X, X-spike; B, bubble arc; Y, Y arc. Closed and open arrowheads point to the presence or the absence of X-spikes, respectively.
To confirm that the altered electrophoretic mobility of chromosomes in rad51Δ cells reflects the persistence of stalled forks, we followed the kinetics of fork recovery at the level of individual molecules by DNA combing. DNA fibres were stretched on silanised coverslips, and BrdU-labelled sites were visualized by immunofluorescence (Figure 6C). The percentage of replication was determined for each individual DNA fibre and the number of unreplicated gaps was scored. This analysis confirmed that rad51Δ cells are unable to complete S phase after MMS treatment because of the persistence of unreplicated gaps (Figure 6D). After 130 min, ∼40 gaps per haploid genome were detected in rad51Δ cells, which is sufficient to explain the altered electrophoretic mobility of these chromosomes. Using 2D gel electrophoresis, we confirmed the presence of Rad51-dependent recombination intermediates in MMS-treated cells (Figure 6E). These intermediates generate a so-called ‘X-spike', which accumulate in MMS-treated sgs1Δ cells used here as a positive control (Branzei and Foiani, 2007), but are not detected in rad51Δ cells. The presence of this X-spike correlates also with an increase of unequal sister-chromatid recombination events (Supplementary Figure 4B) in a Rad51-dependent manner (Fasullo et al, 2001; Duro et al, 2008). Taken together, these data indicate that recombination mechanisms operate at stalled forks and are essential to complete S phase after replication stress.
Conclusion
Here, we show that activation of the Mrc1-dependent branch of the intra-S-phase checkpoint prevents HR at DNA DSBs, presumably to suppress HR-mediated genomic instability under replication stress. Yet, we also found that Rad51-dependent recombination events occur at stalled forks and are essential to complete DNA replication after MMS treatment. To reconcile these apparently conflicting observations, we propose that the DNA replication checkpoint differentially regulates recombination at DSBs and at stalled forks. The major difference between these two types of recombination is that DSB repair depends on the formation of 3′ ssDNA overhangs, whereas ssDNA is already present at stalled forks and can be used to initiate recombination with the sister chromatid (Fabre et al, 2002). By impeding resection, the replication checkpoint would therefore function as a switch to prevent potentially dangerous HR at DSBs and to allow helpful SCR at stalled forks (Figure 7). It has recently been reported that SCR depends on specific factors such as Rtt109, Asf1 and Rtt101, which are fully dispensable for HR (Duro et al, 2008). It is therefore possible that SCR and HR take place in different subnuclear compartments and/or operate with different kinetics, which would explain why cells exposed to MMS do not show Rad52 foci. Finally, it is now important to determine how the replication checkpoint prevents the formation of ssDNA at DSBs. Recent evidence indicates that resection of DNA ends is a complex process that depends on the regulation of multiple DNA helicases and exonucleases (Raynard et al, 2008). One of these enzymes, Exo1, is repressed by Rad53 to protect stalled forks and uncapped telomeres (Cotta-Ramusino et al, 2005; Morin et al, 2008; Segurado and Diffley, 2008). Whether Exo1 or other resection factors are also inhibited by Rad53 to suppress HR is an important question that remains to be addressed.
Figure 7.
Differential regulation of homologous recombination at DNA double-strand breaks and stalled forks under replication stress. (A) DSBs are efficiently repaired by homologous recombination (HR) during S phase after the formation of 3′ ssDNA overhangs. (B) In the presence of HU or MMS, stalled replication forks induce the activation of the DNA replication checkpoint in an Mrc1-dependent manner through the recruitment of the Mec1–Ddc2 complex on RPA-coated ssDNA. Hyperphosphorylated Rad53 functions in trans to prevent HR at DSBs, presumably by blocking the formation of ssDNA tails. In contrast, sister-chromatid recombination occurs at stalled forks, which already contain ssDNA (see text for details). SCC, sister-chromatid cohesion.
Materials and methods
Strains and growth conditions
Strains used in this study are listed in Supplementary Table I. Cells were grown at 25°C in synthetic complete medium and were arrested in G1 for 2.5 h with 2 μg/ml α-factor (GenePep, France). Cells were released from G1 by the addition of Pronase and were resuspended in fresh medium containing 200 mM HU (Sigma), 0.033% methyl methanesulphonate (Sigma) or 20 μM CPT (Sigma). DSBs were induced in G1 with the addition of 100 μg/ml Zeocin (InvivoGen). Thermosensitive cdc7-4 cells were arrested at the G1/S transition by shifting the culture to 37°C 1 h before release from α-factor. Cell cycle progression was monitored by flow cytometry (FACScan).
Microscopy
All experiments were performed at 25°C to allow the GFP chromophore to form efficiently. Cells were immediately fixed with 4% paraformaldehyde and immobilized on a glass slide with 1.2% low melting agarose (Lisby et al, 2004). Images were acquired by using a CoolSNAP HQ CCD camera mounted on a Leica DM6000B microscope. For each field of view, eight fluorescent images were acquired at 0.5-μm intervals along the z-axis. Rad52–GFP foci were counted by inspecting all focal planes for at least 300 cells. To stain genomic DNA, 10 μg/ml DAPI was added to the culture 30 min before imaging.
Analysis of HO cleavage, repair and DNA resection
DNA was extracted from cells before and after HO induction and digested with StyI as described (Sugawara and Haber, 2006). The digested DNA was separated on 1% agarose gel, blotted to GeneScreen nylon membranes (NEN) and probed with radiolabelled probe as indicated in Figure 3A. Bands were quantitated using a Typhoon Trio (Amersham GE) and signals were normalized to a StyI fragment containing the RAD9 gene. The same protocol was used for the analysis of ssDNA resulting from strand resection, except that StyI fragments were run on a 1% agarose gel in denaturing conditions (50 mM NaOH and 1 mM EDTA) as described (Sugawara and Haber, 2006).
Dynamic molecular combing
DNA combing was performed as described earlier (Michalet et al, 1997; Versini et al, 2003). BrdU was detected with a rat monoclonal antibody (BU1/75; AbCys; 1/20) and a secondary antibody-coupled Alexa 488 (Molecular Probes; 1/50). DNA molecules were counterstained with a mouse monoclonal antibody against ssDNA (MAB3034; Chemicon; 1/500) and an anti-mouse coupled to Alexa 546 (Molecular Probes; 1/50). Images were recorded on a Leica DM6000B microscope and were processed as described (Pasero et al, 2002). For each experiment, the total length of unreplicated gaps (red-only stretches larger than 5 kb) was measured for >20 Mb of randomly selected DNA fibres and was expressed as an average number of gaps per megabyte of total DNA. The percentage of replication was determined as the ratio of the total length of replicated stretches to the total length of unreplicated DNA gaps.
Unequal sister-chromatid exchange assay
Unequal sister-chromatid exchange was measured as described earlier (Fasullo et al, 2001). Briefly, cells were grown in rich medium with glucose and adenine (YPAD) to 5 × 106 cells per ml and then exposed for 45 min to 0.02% MMS. Here, 2 × 106 cells were plated on solid synthetic medium without histidine (SC-His) agar and 200 cells on YPAD plates. After 3 days at 30°C, colonies were counted. Relative unit represents the number of His+ colony per 200 YPAD colonies. Values represent the average of three independent experiments.
Supplementary Material
Supplementary Figures 1–4
Supplementary Table SI
Supplementary Figure Legends
Acknowledgments
We thank L Crabbé, A Lengronne, H Tourrière and S Schmidt for critical comments on the paper. We are also grateful to G Ira, S Jackson, D Lydall, M Peter, J Rouse and E Schwob for strains. We thank the DNA Combing Facility of Montpellier for silanised coverslips. Work in the PP lab is supported by FRM (Equipe FRM), CNRS (ATIP) and INCa. CA thanks ARC (Association pour la Recherche contre le Cancer) for fellowship.
References
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK (2007) Replication in hydroxyurea: it's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD (2000) DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol 20: 4393–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Shanahan P, McDonald WH, Lopez-Girona A, Noguchi E, Yates IJ, Russell P (2003) Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol Cell Biol 23: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JF (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2007) RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev 21: 3019–3026 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2008) Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9: 297–308 [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M (2006) Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Brush GS, Morrow DM, Hieter P, Kelly TJ (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA 93: 15075–15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW (2002) A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA 99: 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell 29: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J (2008) Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 7: 811–818 [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T (2001) Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics 158: 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J (2007) Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ (2007) Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 8: 129–136 [DOI] [PubMed] [Google Scholar]
- Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, Bashkirova EV, Yates JR III, Heyer W-D (2006) Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol 26: 8396–8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J (2008) Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M, Boddy MN, Russell P, Wang TS (2005) Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev 19: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Wheeler LJ, Mathews CK, Merrill GF (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279: 223–230 [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Labib K, Hodgson B (2007) Replication fork barriers: pausing for a break or stalling for time? EMBO Rep 8: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response; spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M (2006) The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 16: 786–792 [DOI] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res 33: 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A, Vernis L, Poidevin M, Gasser SM, Baldacci G (2005) Temporal separation of replication and recombination requires the intra-S checkpoint. J Cell Biol 168: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R (2007) Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst) 6: 967–980 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D (2008) Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 27: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18: 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Pommier Y (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6: 789–802 [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa GD, Wohlschlegel J, Yates JR III, Boddy MN (2006) SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J Biol Chem 281: 27973–27981 [DOI] [PubMed] [Google Scholar]
- Raynard S, Niu H, Sung P (2008) DNA double-strand break processing: the beginning of the end. Genes Dev 22: 2903–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9: 857–869 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JFX (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T (2005) The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Haber JE (2006) Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol 408: 416–429 [DOI] [PubMed] [Google Scholar]
- Szyjka SJ, Viggiani CJ, Aparicio OM (2005) Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell 19: 691–697 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Vázquez MV, Rojas V, Tercero JA (2008) Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair (Amst) 7: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P (2003) The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J 22: 1939–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ira G, Tercero JA, Holmes AM, Diffley JFX, Haber JE (2004) Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol 24: 6891–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B (2008) Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 9: 1034–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF (2008) Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–4
Supplementary Table SI
Supplementary Figure Legends