Abstract
A protein interaction network describes a set of physical associations that can occur between proteins. However, within any particular cell or tissue only a subset of proteins is expressed and so only a subset of interactions can occur. Integrating interaction and expression data, we analyze here this interplay between protein expression and physical interactions in humans. Proteins only expressed in restricted cell types, like recently evolved proteins, make few physical interactions. Most tissue-specific proteins do, however, bind to universally expressed proteins, and so can function by recruiting or modifying core cellular processes. Conversely, most ‘housekeeping' proteins that are expressed in all cells also make highly tissue-specific protein interactions. These results suggest a model for the evolution of tissue-specific biology, and show that most, and possibly all, ‘housekeeping' proteins actually have important tissue-specific molecular interactions.
Keywords: human, protein interaction networks, tissue-specific evolution
Introduction
Nearly all processes in biology are dependent on the precise physical interactions among many individual proteins. These range from the maintenance of cellular architecture and the propagation of the genetic material, to the ability of cells to process and respond to environmental information. Defining a near-complete map of the physical interactions that can occur between human proteins—the human protein ‘interactome'—is an important ambition of current research. Similar to the sequence of the human genome, the human interactome serves as a resource for researchers and can be used to understand how proteins are organized to perform functions within a cell (Bork et al, 2004; Cusick et al, 2005).
Protein interactome mapping projects were pioneered in model organisms (Uetz et al, 2000; Walhout et al, 2000; Ito et al, 2001; Ho et al, 2002; Li et al, 2004; Gavin et al, 2006; Krogan et al, 2006), with initial efforts in humans focused on particular pathways or genomic regions (Bouwmeester et al, 2004; Lehner and Sanderson, 2004; Lehner et al, 2004; Jeronimo et al, 2007). More recently, the cloning of large sets of human open reading frames and improvements in interaction assays have allowed these efforts to be expanded by an order of magnitude to the scale of the human proteome (Rual et al, 2005; Stelzl et al, 2005; Ewing et al, 2007). These data, combined with extensive efforts to collate known interactions from the scientific literature (Bader et al, 2001; Xenarios et al, 2002; Pagel et al, 2005; Persico et al, 2005; Stark et al, 2006; Kerrien et al, 2007; Vastrik et al, 2007; Ruepp et al, 2008), mean that there is now a reasonably extensive resource of known human protein interactions (Hart et al, 2006).
A global interactome network provides an overview of all of the physical interactions that can occur between human proteins. However, very little is known about when and where each of these interactions can occur. Within any particular cell or tissue of the human body not all protein interactions can occur. Most simply, if two genes are not expressed in a cell, then an interaction between their protein products cannot occur.
In unicellular organisms, one approach that has been used to investigate the dynamics of interaction networks between cellular states has been to integrate interactome data with expression data. This approach has been used to identify co-regulated interaction modules (Ihmels et al, 2002; Komurov and White, 2007) or to investigate the relationships between interaction network topology and gene co-expression (Han et al, 2004). Additional studies have used gene expression (Luscombe et al, 2004; de Lichtenberg et al, 2005) or functional information (Rachlin et al, 2006) to investigate the cellular conditions (or ‘context') under which interactions can occur, and to distinguish between condition-dependent and condition-independent interactions.
In the present study, we apply a similar approach to the human protein interaction network, using global gene expression data to identify the human cells and tissues in which each interaction can or cannot occur. By performing this analysis, we are able to investigate the relationship between the tissue specificity of a protein and its number of interaction partners. Moreover, and strikingly, we find extensive communication between universally expressed proteins and those with tissue-specific expression. Even the most tissue-specific proteins normally interact directly with components of the core cellular machinery. Conversely, nearly all universally expressed ‘housekeeping' proteins have protein interactions that can only occur in a restricted subset of cells. Our results suggest a model for the evolution of tissue-specific functions through the modification and re-use of core cellular processes, and that most ‘housekeeping' proteins should probably be considered as important for tissue-specific processes.
Results
Construction of a global human protein interaction network
To construct a global human physical protein interaction network, we integrated data from 21 different sources to define a network of 80 922 physical interactions that can occur between 10 229 human proteins. We only included interactions supported by at least one piece of direct experimental evidence demonstrating physical association between two human proteins (see Materials and methods; Supplementary Table 1). Moreover, to account for differences in interaction assay reliability, throughout this work, we also consider a high-confidence subset of this global network that consists of interactions reported in at least two independent primary research publications. There are a total of 13 102 of these multiple publication-supported interactions that connect 4750 human proteins.
Determining the tissue specificity of human protein interactions
We then used gene expression data (Su et al, 2004) to determine the cells and tissues of the human body in which each of these interactions can occur (Figure 1A). If two genes are co-expressed in a cell, then under some condition their products can physically interact in that cell. However, if two proteins are not expressed in a tissue, then the interaction cannot occur in this tissue. The complete set of interactions, their supporting evidence, and the cells and tissues in which each interaction can occur are provided as Supplementary Table 1 as a resource for researchers interested in the biology of any particular human cell or tissue.
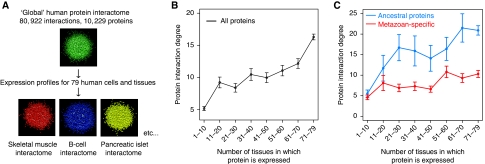
Figure 1.
Tissue-specific and recently evolved proteins make few protein interactions. (A) Integrating protein interaction and expression data to construct ‘local' interactomes for human cells and tissues. (B, C) The relationship between protein interaction degree and protein expression breadth (the number of tissues in which a protein is expressed) for the complete human protein interaction network (B), and (C) for ancestral (pre-metazoan) proteins (blue) and for metazoan-specific proteins (red). P<10e−15 in all cases, Kolmogorov–Smirnov test. Bars indicate one standard error. Interaction degree is the maximum number of co-expressed interaction partners. The same analysis is performed for the multiple-support network and for a network without protein complex-derived interactions in Supplementary Figure 1.
Tissue specific and recently evolved proteins make few protein interactions
We first examined the relationship between the tissue specificity of a protein and the number of interactions that it makes (a protein's interaction degree). We find that more tissue-specific proteins make fewer interactions than widely expressed proteins (Figure 1B, Spearman's rho=0.19, P<2.2e−16). This is true both for the complete and for the multiple-support interaction dataset (Supplementary Figure 1A), and when excluding all protein complexes (Supplementary Figure 1B). It has been shown earlier that tissue-specific proteins are more likely to be recent evolutionary innovations than universally expressed proteins (Lehner and Fraser, 2004b). We find that more-recently evolved proteins have fewer interactions than ancient proteins, but that the relationship between tissue specificity and interaction degree is seen for both sets of proteins (Figure 1C). That is, the older a protein is, and the more tissues in which it is expressed, the more protein interactions it is likely to have.
The most tissue-specific proteins normally interact with core cellular components
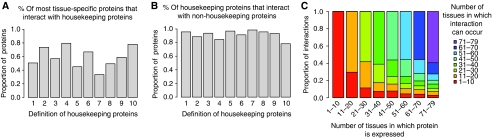
We next analyzed the extent to which tissue-specific proteins interact with the most widely expressed proteins. We find that even when only considering the most tissue-restricted proteins (proteins expressed in ⩽10/79 tissues), most of them are known to interact directly with universally expressed human proteins (Figure 2A). The same result is seen when only considering high-confidence human protein interactions (Supplementary Figure 2A), and when using diverse definitions of universally expressed proteins (Figure 2A). Thus, most tissue-specific proteins can function by directly contacting components of the core cellular machinery.
Figure 2.
Most tissue-specific proteins interact with core cellular components, and most housekeeping proteins have tissue-specific physical interactions. (A) The proportion of the most tissue-specific proteins (proteins expressed in only 1–10/79 tissues) that interact with universally expressed housekeeping proteins. (B) The percentage of housekeeping proteins that interact with non-housekeeping proteins. These data are for the complete network. The same analysis is shown for the high-confidence multiple-support network in Supplementary Figure 2. Housekeeping proteins are defined by 10 criteria: (1) this study 79/79 tissues, (2) this study 71–79 tissues, (3) this study 79/79 tissues with reduced expression stringency, (4) this study 71–79 tissues with reduced stringency, (5) this study 79/79 tissues with increased stringency, (6) this study 71–79 tissues with increased stringency, (7) Zhu et al microarray data 18/18 tissues, (8) Zhu et al microarray data 16–18 tissues, (9) Zhu et al EST data 18/18 tissues, (10) Zhu et al EST data 16–18 tissues (Zhu et al, 2008). (C) Many proteins make interactions that can only occur in a subset of the tissues in which they are expressed. The number of tissues in which the interactions of a protein can occur is compared with the number of tissues in which a protein is expressed for proteins falling into each of the eight bins of tissue specificity. Data are shown for the complete network. Data for the filtered multiple-support network and reduced and increased stringency expression thresholds are shown in Supplementary Figure 3.
Most universally expressed proteins have tissue-specific protein interactions
Constitutively expressed proteins are often considered as important for ‘housekeeping' biological processes that are required in all cells. However, nearly all of the most widely expressed proteins have interactions with other proteins that are not themselves universally expressed (Figure 2B). That is, most universally expressed proteins have physical interactions that can only occur in a restricted subset of cells and tissues. The same result is seen when using the complete interaction dataset, when only considering high-confidence interactions described in multiple independent publications (Supplementary Figure 2B), or when using diverse definitions of universally expressed proteins (Figure 2B). Thus most, and possibly all, universally expressed proteins have tissue-specific molecular interactions.
Proteins that themselves have restricted expression patterns also have many interactions that can only occur in a subset of the tissues in which they are expressed (Figure 2C). That is, as a consequence of interactions between more and less widely expressed proteins, human protein interactions are often more tissue specific than proteins (P<10−16).
Extensive re-use of housekeeping proteins for tissue-specific biological processes
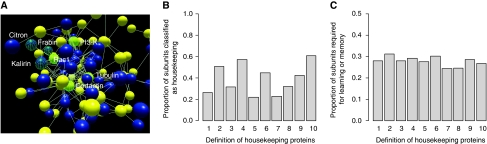
To further illustrate how housekeeping proteins are widely re-used for tissue-specific biological processes, we considered neuronal protein complexes that function in synaptic transmission, learning, and memory. The subunits of these complexes have been identified by extensive proteomic approaches, and the importance of individual subunits for learning and memory have been validated by genetic studies in mice and by clinical studies in humans (Pocklington et al, 2006). We estimate that ∼20–60% of the subunits of these neuronal-specific complexes are actually universally expressed housekeeping proteins (Figure 3A and B). Moreover, in ∼30% of cases, these housekeeping subunits have genetically verified roles in learning and memory (Figure 3C). Thus, universally expressed proteins, through their tissue-specific interactions, can be re-used and essential for highly tissue-specific biological processes.
Figure 3.
The re-use of housekeeping proteins for tissue-specific functions. Here we use the example of neurotransmitter receptor protein complexes identified by affinity purification followed by mass spectrometry (Pocklington et al, 2006). (A) A section of the binary protein interaction network of neurotransmitter receptor complexes, with subunits marked as universally expressed (housekeeping) proteins (yellow) and non-housekeeping (blue). The housekeeping and non-housekeeping interaction partners of the housekeeping protein Rac1 are highlighted and labeled as examples. (B) The percentage of subunits of neurotransmitter receptor protein complexes considered as universally expressed housekeeping proteins is shown for 10 different criteria of housekeeping proteins, as described in Figure 2. Criteria10 is used in panel A. (C) The proportion of these housekeeping subunits that have been experimentally verified as essential for learning and memory in mouse models or that are implicated in psychiatric disease in humans is shown for the same 10 criteria of housekeeping proteins. Protein complex subunits, binary protein–protein interactions, and genetic data are all from Pocklington et al (2006). The network in (A) was visualized using Biolayout Express (3D) (Freeman et al, 2007).
Discussion
The evolution of tissue-specific biological processes
Taken together, our findings suggest the following model for the evolution of tissue-specific functions. Many (but not all) tissue-specific proteins are recent evolutionary innovations (Lehner et al, 2004). In general, these tissue-specific proteins initially make few interactions, and these interactions are frequently with much more widely expressed and ‘housekeeping' components of the cell. Thus, many tissue-specific proteins probably function by directly recruiting or modifying the activities of core cellular components.
There are, however, exceptions to this trend, with some tissue-specific proteins acting as ‘local' hubs in the interaction network of a particular tissue (our unpublished observation).
Frequent re-use of housekeeping proteins for tissue-specific biology
Universally expressed ‘housekeeping' proteins tend to make many interactions. Many of these interactions (∼50–60%, Supplementary Figure 3) are with other housekeeping proteins. However, the majority of universally expressed proteins also make interactions that can only occur in a subset of the tissues in which they are expressed. Therefore, there appears to be very frequent, and possibly universal, re-use of ‘housekeeping' proteins to perform tissue-specific biological processes. That is, most housekeeping proteins can be considered to be important for different (or at least modified) biological processes in different tissues.
In summary, our results suggest that it might be better to consider the biology of any particular tissue in the terms of the particular interactions that can occur in that tissue, rather than simply in the terms of the unique proteins that are expressed there.
The importance of interaction network dynamics
In unicellular yeast, broadly expressed proteins can have precisely temporally regulated activities because of their interactions with proteins with restricted expression profiles (de Lichtenberg et al, 2005). We show here that a similar process may be widely used in multicellular organisms to restrict and modify the activities of a protein to a subset of the tissues in which it is expressed.
Together with earlier analyses in yeast (Han et al, 2004; Luscombe et al, 2004; de Lichtenberg et al, 2005), this work highlights the importance of considering global interaction networks as having dynamic, not static, structures, and topologies. Additional work analyzing how the networks of molecular interactions change between cell types, states, and conditions should prove a fruitful approach for understanding living systems.
Materials and methods
Protein interaction data
We compiled human protein interactions from a total of 21 different databases, as listed in Table I. We required that each interaction be supported by at least one piece of direct experimental evidence demonstrating physical association between two human proteins, and removed all interactions that did not meet these criteria. All interactions were mapped to common Ensembl gene identifiers. The complete network (‘CRG-all'), consists of 80 922 interactions between 10 229 human proteins (approximately half the human proteome) and is available as Supplementary Table S1.
Table 1.
Human protein interaction datasets used to construct or support the integrated human interaction network
| Dataseta | Description | Reference |
|---|---|---|
| BIND | Literature curation | Bader et al (2001) |
| BIOGRID | Literature curation | Stark et al (2006) |
| Bioverse | Data integration | McDermott et al (2005) |
| CCSB-HI1 | Yeast two-hybrid | Rual et al (2005) |
| Co-citation | Text-mining | Ramani et al (2005) |
| Co-expression conservation | Conserved co-expression relationships | Ramani et al (2008) |
| CORUM | Literature curation | Ruepp et al (2008) |
| DIP | Literature curation | Xenarios et al (2002) |
| HPRD | Literature curation | Peri et al (2003) |
| Intact | Literature curation | Kerrien et al (2007) |
| IntNetDB | Data integration | Xia et al (2006) |
| MDC | Yeast two-hybrid | Stelzl et al (2005) |
| MINT | Literature curation, orthology | Persico et al (2005) |
| MIPS | Literature curation | Pagel et al (2005) |
| OPHID | Orthology | Brown and Jurisica (2005) |
| Ottowa | Affinity purification- mass spectrometry | Ewing et al (2007) |
| PC/Ataxia | Yeast two-hybrid | Lim et al (2006) |
| Reactome | Literature curation | Vastrik et al (2007) |
| Sanger | Orthology | Lehner and Fraser (2004a) |
| Transcription complexes | Affinity purification- mass spectrometry | Jeronimo et al (2007) |
| Unilever | Data integration | http://www.cytoscape.org/cgi-bin/moin.cgi/Data_Sets |
aConserved co-expression, co-citation, or evolutionary conservation data are only used in the final network as additional supporting evidence. All interactions must have at least one piece of experimental binding evidence to be included in the final dataset, or physical binding evidence from at least two publications to be included in the multiple-support network.
Filtered interaction dataset
In total, 13 102 of the interactions in our network between 4750 proteins are supported by experimental evidence of physical binding reported in at least two different primary research publications. Given the multiple lines of evidence supporting these interactions, we use this subset of interactions (‘CRG-filtered') as high-confidence interactions to confirm that our conclusions are not affected by interaction data quality or sampling (see Supplementary Figures).
Expression data
To identify which protein interactions can occur in a particular cell or tissue type, we used global gene expression data. Although interactions can be regulated by localization, phosphorylation, etc, we aim to distinguish the proteins that can interact under some condition in a tissue from those that cannot, and mRNA expression is a reasonable indicator of this potential. We used expression data from the GNF Atlas project that measured expression across 79 different human cell or tissue types (Su et al, 2004). The MAS5 normalized expression levels were averaged between experimental replicas, and in cases where more than one probe set was present for a gene, the more sensitive probe set was used. In this dataset, a gene is considered as present in a tissue, if its normalized expression level is >200 (Su et al, 2002). However, our conclusions remain the same when this stringency is increased or decreased (see Supplementary information). At this threshold, >98% of the interaction partners in our global network for which expression information is available are co-expressed in least one human tissue.
Housekeeping proteins
We identified universally expressed housekeeping proteins using a total of 10 different criteria. First, we used the GNF Atlas data, and considered housekeeping proteins as those with an expression level above 200 in all 79 tissues, or in more than 70/79 tissues (i.e. allowing for some false-negatives). Second, we used the same two tissue criteria, but increased (250) or decreased (150) the stringency at which a gene is considered expressed. Third, we used four additional sets defined in an earlier publication—genes identified as expressed in 18/18 or at least 16/18 tissues using microarray data, and genes with the same tissue criteria but defined using expressed sequence tag (EST) data (Zhu et al, 2008).
Neurotransmitter receptor complexes
Components of N-methyl-D-aspartate receptor and metabotropic receptor complexes were identified by extensive proteomic studies as described (Pocklington et al, 2006). We used the 215 subunits of these complexes that could be mapped to human Ensembl gene identifiers, of which 77 have demonstrated roles in learning and memory through genetic studies in mice or are implicated in psychiatric disorders in humans (Pocklington et al, 2006). We used the sets of housekeeping proteins described above to identify how many of these subunits represent universally expressed proteins.
Protein evolution
Proteins were classified as metazoan specific or pre-metazoan using the analysis of Freilich et al (2005).
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Material
Supplementary Figures 1 - 3
Acknowledgments
This work was funded by a European Research Council (ERC) Starting Researcher Grant, the Ministry of Science and Innovation (MICINN-Plan Nacional), the CRG-EMBL Systems Biology Program, ICREA and a Leonardo da Vinci fellowship to AB. We thank three anonymous referees for helpful suggestions.
References
- Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW (2001) BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res 29: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Jensen LJ, von Mering C, Ramani AK, Lee I, Marcotte EM (2004) Protein interaction networks from yeast to human. Curr Opin Struct Biol 14: 292–299 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Brown KR, Jurisica I (2005) Online predicted human interaction database. Bioinformatics 21: 2076–2082 [DOI] [PubMed] [Google Scholar]
- Cusick ME, Klitgord N, Vidal M, Hill DE (2005) Interactome: gateway into systems biology. Hum Mol Genet 14 (Spec No. 2): R171–R181 [DOI] [PubMed] [Google Scholar]
- de Lichtenberg U, Jensen LJ, Brunak S, Bork P (2005) Dynamic complex formation during the yeast cell cycle. Science 307: 724–727 [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y et al. (2007) Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol Syst Biol 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman TC, Goldovsky L, Brosch M, van Dongen S, Maziere P, Grocock RJ, Freilich S, Thornton J, Enright AJ (2007) Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol 3: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Massingham T, Bhattacharyya S, Ponsting H, Lyons PA, Freeman TC, Thornton JM (2005) Relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biol 6: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M (2004) Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Hart GT, Ramani AK, Marcotte EM (2006) How complete are current yeast and human protein-interaction networks? Genome Biol 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B et al. (2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komurov K, White M (2007) Revealing static and dynamic modular architecture of the eukaryotic protein interaction network. Mol Syst Biol 3: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Lehner B, Fraser AG (2004a) A first-draft human protein-interaction map. Genome Biol 5: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Fraser AG (2004b) Protein domains enriched in mammalian tissue-specific or widely expressed genes. Trends Genet 20: 468–472 [DOI] [PubMed] [Google Scholar]
- Lehner B, Sanderson CM (2004) A protein interaction framework for human mRNA degradation. Genome Res 14: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM (2004) Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83: 153–167 [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY (2006) A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125: 801–814 [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431: 308–312 [DOI] [PubMed] [Google Scholar]
- McDermott J, Guerquin M, Frazier Z, Chang AN, Samudrala R (2005) BIOVERSE: enhancements to the framework for structural, functional and contextual modeling of proteins and proteomes. Nucleic Acids Res 33: W324–W325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stumpflen V, Mewes HW, Ruepp A, Frishman D (2005) The MIPS mammalian protein–protein interaction database. Bioinformatics 21: 832–834 [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC et al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13: 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico M, Ceol A, Gavrila C, Hoffmann R, Florio A, Cesareni G (2005) HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics 6(Suppl 4): S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocklington AJ, Cumiskey M, Armstrong JD, Grant SG (2006) The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behaviour. Mol Syst Biol 2: 2006.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachlin J, Cohen DD, Cantor C, Kasif S (2006) Biological context networks: a mosaic view of the interactome. Mol Syst Biol 2: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM (2005) Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol 6: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Li Z, Hart GT, Carlson MW, Boutz DR, Marcotte EM (2008) A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol Syst Biol 4: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, Mewes HW (2008) CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res 36: D646–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B et al. (2005) A human protein–protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB (2002) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 99: 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L (2007) Reactome: a knowledge base of biologic pathways and processes. Genome Biol 8: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122 [DOI] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30: 303–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Dong D, Han JD (2006) IntNetDB v1.0: an integrated protein–protein interaction network database generated by a probabilistic model. BMC Bioinformatics 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, He F, Song S, Wang J, Yu J (2008) How many human genes can be defined as housekeeping with current expression data? BMC Genomics 9: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 - 3