SUMMARY
Prions are proteins that convert between structurally and functionally distinct states, one or more of which is transmissible. In yeast, this ability allows them to act as non-Mendelian elements of phenotypic inheritance. To further our understanding of prion biology, we conducted a bioinformatic proteome-wide survey for prionogenic proteins in S. cerevisiae, followed by experimental investigations of 100 prion candidates. We found an unexpected amino acid bias in aggregation-prone candidates and discovered that 19 of these could also form prions. At least one of these prion proteins, Mot3, produces a bona fide prion in its natural context that increases population-level phenotypic heterogeneity. The self-perpetuating states of these proteins present a vast source of heritable phenotypic variation that increases the adaptability of yeast populations to diverse environments.
Keywords: prion, amyloid, aggregation, non-Mendelian inheritance
INTRODUCTION
The prion hypothesis posits that biological information can be replicated solely through self-propagating conformations of proteins. Though it was initially conceived to explain baffling neurodegenerative diseases in mammals (Griffith, 1967; Prusiner, 1982), it has since grown to encompass a number of non-Mendelian traits in fungi (Ross et al., 2005b; Shorter and Lindquist, 2005; Shkundina and Ter-Avanesyan, 2007). All known prions, except for the initially discovered disease-causing prion PrP, are benign, and in some cases can confer selectable advantages (Saupe et al., 2000; True and Lindquist, 2000; True et al., 2004). The self-templating property of prions makes them both conformationally and epigenetically dominant, and positions prion-forming proteins as metastable cellular switches of protein function.
The realization that protein conformational switches could provide a means for inheritance of phenotypes dates back 15 years (Wickner, 1994), yet only a few proteins with this capacity have been discovered (Shorter and Lindquist, 2005; Du et al., 2008). Most of these have been found in the yeast S. cerevisiae, with the [PSI+] element being the best understood.
[PSI+] is caused by an amyloid-like aggregated state of the translation-termination factor Sup35p. In the prion state, the majority of Sup35p molecules are inactive, resulting in increased levels of nonsense suppression (Liebman and Sherman, 1979; Patino et al., 1996) and programmed frameshifting (Namy et al., 2008). This gives rise to RNA stability changes and functionally altered polypeptides and consequently to phenotypes that can be advantageous under diverse conditions (Eaglestone et al., 1999; True et al., 2004). Remarkably, the ability of Sup35p to switch into a prion conformation, and the regulation of that switch by the protein remodeling factor Hsp104p, have been conserved for over 800 million years of fungal evolution (Chernoff et al., 2000; Zenthon et al., 2006).
Three other amyloid-based prions, formed by the functionally diverse proteins Ure2p, Rnq1p, and Swi1p, have been described in S. cerevisiae. Ure2p regulates nitrogen catabolism; its prion state, [URE3], attenuates this activity resulting in the constitutive utilization of poor nitrogen sources (Aigle and Lacroute, 1975; Wickner, 1994). The Rnq1p protein in its prion state, [RNQ+] (also called [PIN+]), enhances the inducibility of other prions (Derkatch et al., 2000; Bradley et al., 2002). [SWI+], the most recently discovered prion, is caused by an inactive state of the chromatin remodeling factor Swi1p (Du et al., 2008). Intriguingly, [SWI+] represents the first established link between chromatin-based and prion-based epigenetics, although a biological relevance of this connection remains to be elucidated. Indeed, for all of these prion proteins, the putative functionality of their prion forms is highly debated (Nakayashiki et al., 2005).
The conformational duality of amyloid-based prions resides in structurally independent prion-forming domains (PrDs) (Edskes et al., 1999; Li and Lindquist, 2000; Santoso et al., 2000; Sondheimer and Lindquist, 2000). These PrDs are modular and can be transferred to other proteins to create novel prions (Li and Lindquist, 2000). They have a very unusual amino acid composition: enriched for polar residues such as glutamine (Q) and asparagine (N) and depleted of hydrophobic and charged residues. This composition promotes a disordered molten-globule-like conformational ensemble, within which amyloid-nucleating contacts can be made (Serio et al., 2000; Wang et al., 2006; Mukhopadhyay et al., 2007).
The ability of prions to propagate is afforded by the inherent and extremely efficient self-templating capacity of amyloid, a highly ordered β-sheet-rich protein aggregate. The amyloid-like prion state nucleates in cells at a low frequency de novo, but once formed, efficiently propagates this change to soluble conformers (Patino et al., 1996; Glover et al., 1997). Prion domains also form self-propagating amyloid in vitro under physiological conditions (Glover et al., 1997; Taylor et al., 1999), and remarkably, the resulting fibers alone can transform cells to the corresponding prion state (Sparrer et al., 2000; Maddelein et al., 2002; Brachmann et al., 2005; Patel and Liebman, 2007), firmly establishing the protein-only nature of prion inheritance.
However, the relationship between amyloid polymerization and prion propagation is still poorly understood. In fact, most known amyloid-forming proteins are not prions, and even amyloids of prion proteins are not always transmissible (Diaz-Avalos et al., 2005; Salnikova et al., 2005; Baskakov and Breydo, 2007; Sabate et al., 2007). While specific sequence elements ultimately determine the intrinsic amyloidogenic properties of polypeptides (Liu and Lindquist, 1999; Lopez de la Paz and Serrano, 2004; Alexandrov et al., 2008), there are multiple trans-acting factors within the cell, including molecular chaperones, the cytoskeletal machinery, and nucleating factors such as [PIN+], that interact with amyloid prions at every stage of propagation (Chernoff, 2007; Perrett and Jones, 2008). These observations, and our lack of knowledge of the pervasiveness of amyloid-based biological phenomena, create a need to elucidate the amyloid-prion relationship on a comprehensive, genome-wide level.
Such a genome-wide analysis could also reveal new prion-based phenotypes, which would support the idea that prion-mediated phenotypic variation is functionally significant. Prion-like Q/N-rich proteins are abundant in the proteomes of lower eukaryotes, with 100 to 170 such sequences in S. cerevisiae (Michelitsch and Weissman, 2000; Harrison and Gerstein, 2003). However, the experimental tools needed to determine the prion properties of these proteins in a systematic manner have been lacking. Consequently, other than the four aforementioned prions, only one additional yeast protein, New1p, has been shown to harbor a domain capable of forming a prion, albeit in an artificial context (Osherovich and Weissman, 2001).
We bioinformatically scanned the yeast genome for proteins with prion-like character. We then subjected the highest-scoring candidates to genetic, cell biological, and biochemical assays to discern their prion-forming capacity, ultimately determining that at least 24 yeast proteins contain a prion-forming domain. We further evaluated one of these, Mot3p, confirming that it is a bona fide prion with a phenotype that is likely to be advantageous under certain environmental conditions.
RESULTS
A bioinformatics screen reveals multiple prion candidates in yeast
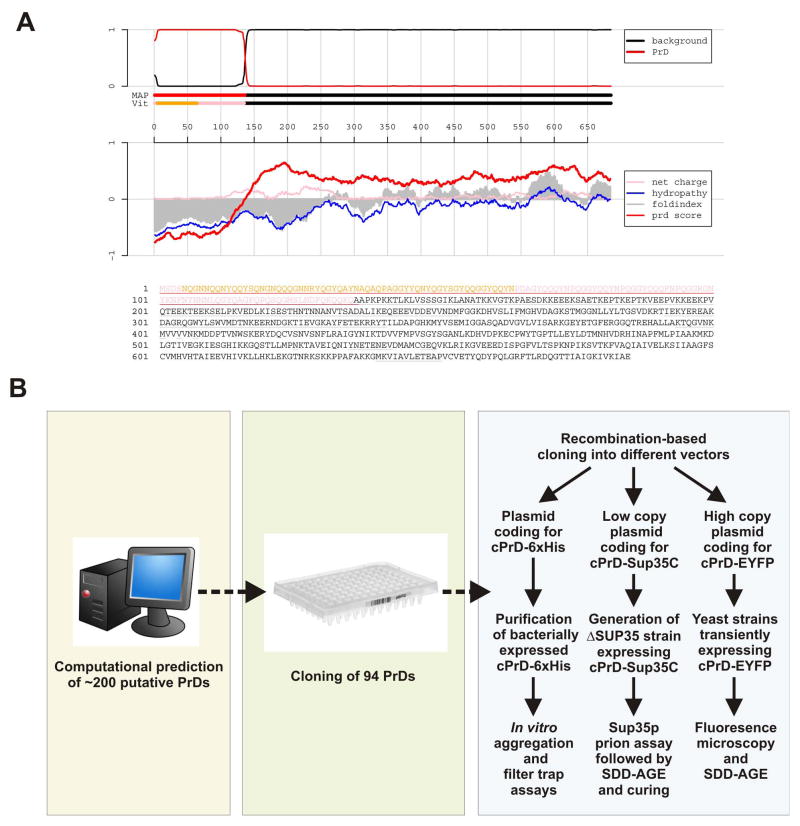
We developed a hidden Markov Model (HMM)-based approach for predicting prions, using the experimentally determined prion domains (PrDs) of Sup35p, Ure2p and Rnq1p, and the prion candidate New1p as positive training examples (at the time, Swi1p had not yet been shown to be a prion). We did not incorporate the other known fungal prion protein, HET-s, nor the mammalian prion protein, PrP, into our model because these proteins have unique sequences that are dissimilar in amino acid composition from the other prion proteins and thus would decrease the predictive power of the model. We acknowledge that our approach is thus necessarily biased towards a particular class of prions, but is nevertheless merited by the large number of Q/N-rich yeast proteins with unknown prion potential. All yeast protein sequences were parsed into prion-like regions and non-prion (background) regions. Proteins with prion-like regions at least 60 amino acids long (denoted “cores”) were considered to be prion candidates, based on the lower size limit of previously characterized yeast prion domains (Masison and Wickner, 1995; King and Diaz-Avalos, 2004). These proteins were then ranked by their core scores. Figure 1A shows an example of the output format of our prediction for the PrD of Sup35p.
Figure 1. Computational prediction and outline of the prion screen.
(A) Output format of the cPrD prediction algorithm for Sup35p. The core region of the cPrD is highlighted in orange and additional predicted regions in pink. The top panel shows the probability of each residue belonging to the HMM state “cPrD” (red) and “background” (black); the tracks “MAP” and “Vit” illustrate the Maximum a Posteriori and the Viterbi parses of the protein into these two states. The lower panel shows sliding averages over a window of width 60 of net charge (pink), hydropathy (blue), and predicted disorder (gray) as in FoldIndex (Prilusky et al., 2005), along with a sliding average based on cPrD amino acid propensities (red).
(B) Overview of the experimental procedures employed to screen for new Q/N-rich prions in yeast. Based on our computational prediction, we generated a cPrD library that was shuttled into a panel of expression vectors for analysis. Experiments were performed with cPrDs expressed in bacteria and yeast and included biochemical assays, cell biological assays and different aggregate visualization techniques.
Our query revealed ~ 200 proteins that have candidate PrDs (cPrDs) in the S. cerevisiae genome (see Table S1 for the complete set of predictions). The list of candidate prions includes Sup35p and Rnq1p as well as the prion candidate New1p in the group of the top 20 candidates. Although the recently discovered Swi1p prion (Du et al., 2008) was not used for training of the algorithm, it ranks at position 21, indicating that our prediction is a valuable tool to uncover new prions. To evaluate these candidates, we tested the amyloid and prion-forming properties of the 100 highest-scoring cPrDs (Figure 1).
Many cPrDs form foci in the cytosol
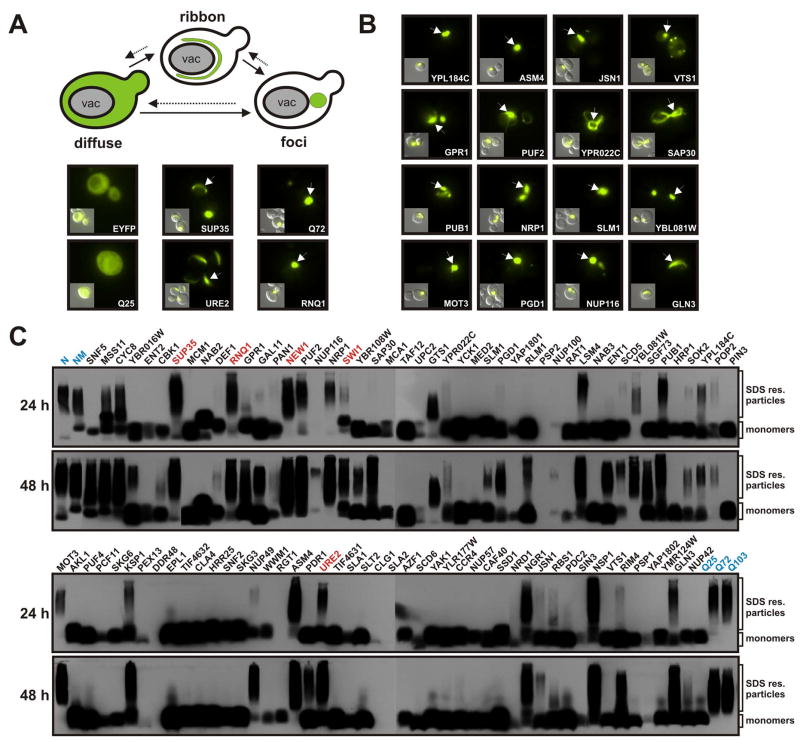
As a first step to experimentally characterize the cPrDs, we investigated their propensity to form foci in living yeast cells by transiently expressing them as chimeras with yellow fluorescent protein (cPrD-EYFP). Several studies have described two morphologically distinct forms of protein aggregation in the yeast cytosol (Derkatch et al., 2001; Ganusova et al., 2006; Taneja et al., 2007). One of these structures has a ring- or ribbon-like appearance and is localized around the vacuole and/or adjacent to the plasma membrane, whereas the other is more punctate and preferentially resides close to the vacuole. We performed pilot experiments with known prions and amyloidogenic proteins to gain a better understanding of the aggregation structures that can be visualized by fluorescence microscopy. We transiently expressed these proteins as EYFP fusions using a galactose-regulatable promoter (Figure 2A). The PrDs of Sup35p and Ure2p proceeded through a characteristic maturation pathway that included an early stage with ribbon-like aggregation patterns and a later stage with puncta (Tyedmers and Lindquist, manuscript in preparation; Derkatch et al., 2001; Ganusova et al., 2006). In contrast, the PrD of Rnq1p and an expanded Q-rich region of the huntingtin protein almost exclusively formed puncta, when expressed from the same promoter.
Figure 2. Prion domains form intracellular aggregates detectable by microscopy and SDD-AGE.
(A) Expression of amyloidogenic proteins in the yeast cytosol leads to the formation of ribbon and dot-like structures. cPrD-EYFP fusion proteins were expressed from a galactose-regulatable plasmid in yeast cells containing the [RNQ+] prion. The yeast cells were subjected to fluorescence microscopy after 24 h of expression. Representative fluorescence microscopy images are shown together with DIC images (insets). Arrows point to aggregates in the yeast cytosol.
(B) A selected set of cPrD candidates forming fluorescent foci after 24 h of expression. Conditions were as described in (A).
(C) Detection of SDS-resistant aggregates by SDD-AGE in cell lysates of yeast strains expressing cPrD-EYFP fusions. Expression of the proteins was induced for 24 h (top gels) or 48 h (bottom gels). Control proteins (highlighted in blue) were the N or NM domains of Sup35p (top left) and the huntingtin protein length variants Q25, Q72 and Q103 (bottom right). Proteins were detected with a GFP-specific antibody. Previously identified prions and the prion candidate New1p are highlighted in red.
We transiently expressed the cPrD-EYFP fusions in yeast cells using a galactose-regulatable plasmid. Despite the intrinsically unfolded nature of the cPrDs, the expression levels of most of the fusions were robust, with a few outliers that expressed at only low levels (Figure S1). A large fraction (69%) of the cPrD-EYFP fusions formed fluorescent foci (Figure 2B and S2). The proteins produced a surprising diversity of patterns with a large variety of ribbon-like structures and punctate foci. Overall, punctate patterns were much more abundant than ribbon structures and ranged from one large focus to multiple small foci distributed all over the cytoplasm. Since foci formation is also a feature of many non-prion proteins, we conducted additional experiments to assess the biochemical properties of the cPrDs.
Multiple cPrDs form highly stable aggregates
Protein aggregates can differ substantially in terms of detergent stability and can adopt either highly ordered (amyloid fibril) or disordered (amorphous) superstructures. To determine whether the cPrDs formed aggregates, and if so, whether they formed prion-like structures, we used semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE). SDD-AGE allows for the resolution of a wide size-range of SDS-resistant (amyloid-like) aggregates, ranging from oligomeric species to polymers assembled from hundreds of individual polypeptides (Bagriantsev et al., 2006; Halfmann and Lindquist, 2008).
Lysates from cells expressing cPrD-EYFP chimeras were analyzed by SDD-AGE after 24 hours of expression (Figure 2C). Remarkably, about one third of the cPrD-EYFP fusions had the SDS-resistance properties of known amyloidogenic proteins (the N and NM domains of Sup35p and the 72Q and 103Q fragments of huntingtin), including all four experimentally verified prions and the prion candidate New1p.
Since amyloid formation is time- and concentration-dependent, we increased the time of transient expression to 48 hours. Despite only modest increases in the cellular levels of the cPrDs (~ 2–3 fold), additional proteins became SDS-resistant, now including almost half the set of investigated cPrDs, with an enrichment for those that scored highly in our algorithm. The ratio of aggregated to soluble protein differed in a time-dependent manner for each candidate and was strongly reproducible. Some cPrD-EYFP fusions were completely aggregated after 24 hours, whereas others initially exhibited a small fraction of aggregated protein that increased after 48 hours of expression (see Table S2 for a classification). In addition, we observed substantial variation in the range of particle sizes, and again these were reproducible for individual cPrDs.
Multiple cPrDs form amyloid in vitro
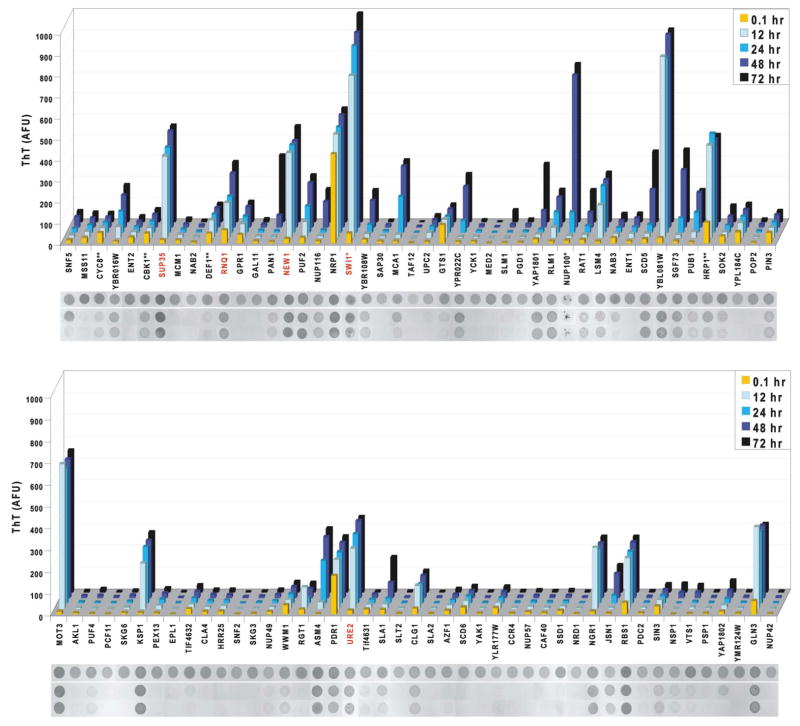
Purified PrDs of known prions form amyloid in vitro. To assess amyloid propensities of our candidate prion domains in the absence of cellular factors, we purified bacterially expressed cPrDs under denaturing conditions, and then analyzed amyloid formation following dilution into a physiological buffer containing thioflavin-T (ThT). ThT is a dye that does not interfere with amyloid assembly, but changes its fluorescence properties upon amyloid binding (LeVine, 1993, 1997).
Overall, the cPrDs displayed tremendous diversity in amyloid propensities (Figure 3). Many, including the four known prions, largely completed amyloid formation within 12 hours. Others did not acquire ThT fluorescence until well over 24 hours after dilution from denaturant (e.g. Pan1p and Yap1801p). Most positive samples initially had very little fluorescence, consistent with nucleated polymerization that is a hallmark of amyloid assembly. Finally, roughly half of the proteins were unable to nucleate within the time-frame examined.
Figure 3. Prion domains have diverse amyloid propensities.
Amyloid formation of cPrD-M-His proteins was followed by ThT fluorescence (arbitrary units) measured at the indicated time points. Shown are means of three replicate assemblies (coefficients of variation were generally < 30; exceptions were Pan1, Nup116, and Yap1802, which each had highly variable lag phases). After the final measurement, reactions were analyzed for detergent-resistant aggregation. Aliquots from one experiment were spotted directly onto nitrocellulose (“total”), or treated with either 0.1% Tween 20 or 2% SDS and filtered through a non-binding membrane. Retained protein was visualized with Ponceau S. *These cPrDs were purified with a polyHis-tag only (no M). **These cPrDs were purified with the M-His tag at their N-terminus. Red labels indicate known prion proteins.
PrDs can follow both amyloid and non-amyloid aggregation pathways (Liu et al., 2002; Vitrenko et al., 2007; Douglas et al., 2008), and some non-amyloid β-sheet structures can alter ThT fluorescence (LeVine, 1993). As an additional characterization, we therefore assayed the SDS-resistance of samples after 72 hours using a filter retardation assay (Scherzinger et al., 1999). In this assay, a non-binding membrane is used to detect protein aggregates, which, due to their size, are specifically retained on the membrane surface. Treating the impeded aggregates with SDS then distinguishes amyloid from non-amyloid, since non-amyloid aggregates become solubilized and flow through.
We found remarkable agreement between the ThT and filter retardation amyloid assays (Figure 3). Acquisition of moderate to strong ThT fluorescence (>100 AFU) coincided with SDS-stable aggregation in every case. Several cPrDs (e.g. those of Snf5p and Nab3p) formed aggregates that were retained by the filter in non-denaturing conditions (0.1% Tween 20) but were eliminated by SDS. Interestingly, this type of aggregate was only observed when ThT fluorescence was absent or greatly delayed, and only with proteins that were more enriched for glutamines than asparagines. In total, the amyloid propensities of isolated cPrDs were in good agreement with the aggregation of these cPrDs in vivo (see Figure S3 and Table S2 for a comparison).
A Sup35p-based prion assay identifies phenotypic switching behavior
To determine whether the cPrDs can confer a heritable switch in the function of the protein to which they are attached, we employed an assay based on the well-characterized prion phenotypes of the translation termination factor Sup35p. The Sup35p protein consists of an N-terminal PrD (N), a highly charged middle domain (M) and C-terminal domain (C), which provides the translation termination function. The PrDs of Sup35p and other prions are modular and can be transferred to non-prion proteins, thereby creating new protein-based elements of inheritance (Li and Lindquist, 2000). This property allowed us to generate cPrD-SUP35C chimeras (under the control of the constitutive ADH1 promoter) that could be tested for their ability to generate [PSI+]-like states, as previously reported for Rnq1p and New1p (Sondheimer and Lindquist, 2000; Osherovich and Weissman, 2001).
Because Sup35p is an essential protein, we first generated a strain in which a deletion of the chromosomal SUP35 was covered by a Sup35p-expressing plasmid (Figure S4). When these cells were transformed with cPrD-SUP35C expression plasmids, a URA3 marker on the covering SUP35 plasmid allowed it to be selected against in 5-FOA-containing medium (plasmid shuffle). The resulting strains contained cPrD-Sup35C fusions as their only source of functional Sup35p. Each was tested for the ability to form a heritable [PSI+] state, using an ADE1 allele with a premature stop codon. Read-through of this allele in [PSI+] cells creates two easily monitored phenotypes, the ability to growth on adenine-deficient medium, and on complete medium, a white colony color, due to the restoration of the adenine biosynthesis pathway, which prevents accumulation of the red byproduct in [psi−] cells.
We obtained 90 viable cPrD-SUP35C strains upon loss of the covering SUP35 plasmid (see Figures S5, S6 and S7 and Supplemental Discussion). Interestingly, several strains spontaneously switched to a white colony color at a high frequency (e.g. New1p, Lsm4p and Nrp1p in Figure S5). Switching rates of prions can be as low as 10−6 to 10−7 (Lund and Cox, 1981; Liu and Lindquist, 1999; Tuite and Cox, 2003). Therefore, in cases where spontaneous switching was not observed, we took advantage of a characteristic of all known prions to induce switching: increased expression of the PrD increases the likelihood of conformational conversion to the prion state. Therefore, we introduced an additional plasmid with a cPrD-EYFP fusion under the control of a galactose-inducible promoter. We identified 22 cPrD-SUP35C strains that showed increased growth on adenine-deficient medium after transient expression of cPrD-EYFP (Figure 4 and S8, see Table S2 for a list of positive cPrDs).
Figure 4. A SUP35-based prion assay is used to detect switching behavior.
(A) A schematic overview of the genetic manipulations deployed to identify cPrDs with prion properties (top) and an example of the used selection procedure (bottom). In the bottom half, the prion state was induced by expressing NM-EYFP for 24 h and the cells were subsequently plated on complete (YPD) and adenine-deficient medium (SD-ade). The same strain grown under non-inducing conditions served as a control. Note that the number of cells plated on SD-ade plates was 200 times the number on YPD plates. Arrows point to colonies that switched to a white colony color.
(B) A selected set of positive candidates identified with the SUP35C-based prion assay. Conditions were as described in (A).
A hallmark of prion proteins is that once the conformational conversion has occurred it is self-sustaining. Thus, transient over-expression of the protein is sufficient to induce a heritable change in phenotype. We tested the ability of the Ade+ colonies to maintain that state on non-selective medium after loss of the overexpression plasmid. Indeed, on complete medium all of the 22 cPrD-SUP35C strains displayed a colony color change from red to white or pink that was maintained over several rounds of re-streaking (Figure S9).
Sequence features that drive prionogenesis – asparagines vs. glutamines
To gain a better understanding of PrD-mediated prion formation, we compared the sequences of the cPrDs that scored positive in each of our assays with those that scored negative (Table S5 and Figure S11). Aggregation prone cPrDs were strongly enriched for asparagines, whereas glutamines, charged residues and prolines were more abundant in non-aggregating cPrDs. This difference was observed in all four assays: foci-formation, SDD-AGE, in vitro amyloid formation, and cPrD-SUP35C switching. The striking difference in the distribution of Qs and Ns was unexpected, as these have largely been considered to be functionally equivalent drivers of prionogenesis (Michelitsch and Weissman, 2000).
Our analysis on these prionogenic sequences also shows that the spacing of “amyloid breaking” prolines and charges (Lopez de la Paz and Serrano, 2004) is an important contribution to prion formation (Figure S11). Other studies have argued that prionogenesis is independent of the polypeptide sequence, provided that amino acid composition is unchanged (Ross et al., 2004; Ross et al., 2005a). Our findings call for a reinterpretation of this conclusion. Together, the composition and sequence biases we have delineated will strongly improve future predictions of amyloid and prion proteins.
Phenotype switches involve an Hsp104p-dependent conformational change
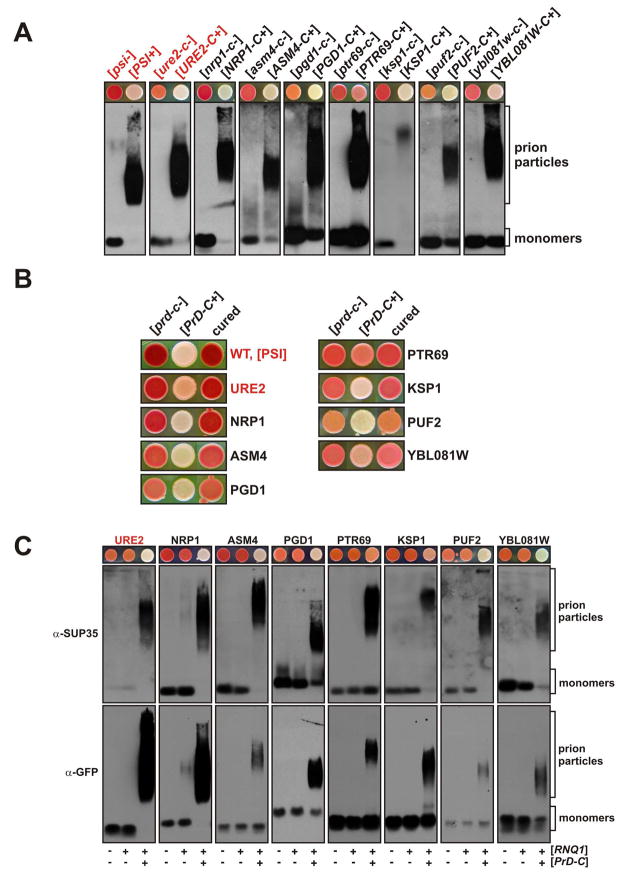
We used SDD-AGE analysis to examine cell lysates of prion-positive and prion-negative strains for changes in the aggregation state of the cPrD-Sup35C fusions (Figure 5A and S10A). In accordance with standard nomenclature, these are hereafter designated [PrD-C+] and [prd-c−], with brackets designating the non-Mendelian character of prion inheritance and capital letters signifying genetic dominance of the trait. All 22 fusions showed a high amount of SDS-resistant aggregation in the [PrD-C+] strains, whereas aggregation was lower or not detectable in the [prd-c−] strains.
Figure 5. The phenotypic switches of cPrD-Sup35C chimeras involve amyloid and are curable.
(A) Cells lysates were prepared from [prd-c−] and [PrD-C+] strains and analyzed by SDD-AGE. The cPrD-Sup35C fusion proteins were detected by using a C domain-specific anti-Sup35p antibody. Corresponding [prd-c−] and [PrD-C+] strains growing on YPD are displayed above the SDD-AGE Western blots.
(B) [PrD-C+] strains were passaged three times on YPD plates containing 5 mM GdnHCl and then spotted onto YPD plates (“cured”). The [prd-c−] and [PrD-C+] strains are shown for comparison.
(C) cPrD-SUP35C strains with a coding sequence for Cerulean integrated at the 3′ end of the corresponding chromosomal gene were subjected to an SDD-AGE analysis in the respective [prd-c−] and [PrD-C+] states. cPrD-Sup35C particles were detected with a C domain-specific anti-Sup35p antibody and the particles containing Cerulean-tagged endogenous protein were detected with an anti-GFP antibody.
All known fungal prions are dependent on chaperones to induce and maintain a prion state. Prions vary in their dependence on different classes of chaperones, but all amyloid-based fungal prions are critically dependent on the protein remodeling factor Hsp104p. Therefore, yeast cells can be cured of prions by genetic manipulations that ablate HSP104 gene function or chemical inhibition of Hsp104p activity (Shkundina and Ter-Avanesyan, 2007). Indeed, passaging of the [PrD-C+] strains on plates containing 5 mM of the Hsp104p inhibitor guanidine hydrochloride (GdnHCl) eliminated the prion in all but one case, New1p (Figure 5B and S10B and Supplemental Discussion).
The prion state of the Rnq1p protein enhances the induction of other yeast prions, most likely by providing an imperfect template on which other aggregation-prone proteins can nucleate (Salnikova et al., 2005). We investigated the role of Rnq1p in prion induction by applying our Sup35p-based system in strains cured of [RNQ+] and in strains carrying a deletion of the gene encoding Rnq1p. In all 10 cases examined (data not shown) we observed a strong reduction in the number of Ade+ colonies, indicating that the de novo formation of [PrD-C+] strains requires the presence of the [RNQ+] prion.
The prion state is transferable to the endogenous protein
To investigate whether the [PrD-C+] states can propagate to the corresponding endogenous protein, we inserted a C-terminal tag at the chromosomal genetic locus. We limited our analysis to the subset of 9 candidates that tolerated C-terminal tagging and whose PrDs were N-terminal or internal. The cell lysates of the resulting [prd-c−] and [PrD-C+] strains were analyzed by SDD-AGE and probed for both the endogenously tagged and chimeric versions of each protein. We detected simultaneous aggregation of cPrD-Sup35C chimeras and the corresponding endogenous proteins in all cases (Figure 5C). The fact that we observed co-aggregation strongly supports the modularity concept of PrDs and the sequence-specific templating of prions. These observations suggest that the endogenous proteins can switch to a prion state, with phenotypic consequences that could impact the survival and adaptation of yeast cells.
[MOT3+], a newly discovered prion conferred by Mot3p
Preliminary studies of the refined list of 29 cPrDs (candidates highlighted in red in Table S2) indicated that several of the full-length candidate prion proteins are capable of self-sustained prion aggregation (data not shown). To rigorously establish that one of these candidates operates as a prion in a physiologically relevant manner, we focused on one, Mot3p, for which robust prion-selective assays could be readily predicted. Mot3p is a globally acting transcription factor that modulates a variety of processes, including mating, carbon metabolism, and stress response (Grishin et al., 1998).
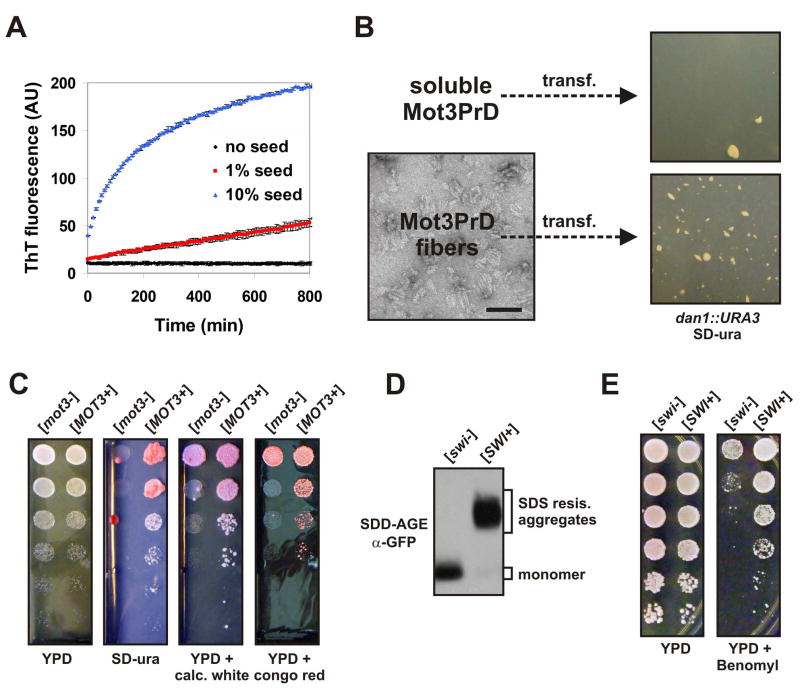
Mot3p tightly represses anaerobic genes, such as DAN1, during aerobic growth. To facilitate analysis of Mot3 transcriptional activity, we created a Mot3p-controlled auxotrophy by replacing the DAN1 ORF with URA3. The resulting strains normally could not grow without supplemental uracil. URA3 expression, and uracil-free growth, should be restored upon reduction of Mot3p activity, as with MOT3 deletion (Figure 6A) or, potentially, sequestration of Mot3p by prion formation. We transiently overexpressed Mot3PrD-EYFP for 24 hours, via a galactose-inducible promoter, and plated the cells onto glucose media lacking uracil. We found that even this transient elevation of intracellular Mot3PrD levels increased the number of Ura+ dan1::URA3 cells by two to three orders of magnitude (Figure 6D). We observed similar results in both S288C and W303 strain backgrounds (data not shown). The phenotype persisted even after the inducing plasmid had been lost (after multiple passages on non-selective media; Figure 6B). As expected for a prion-based phenotype, uracil auxotrophy was dominant in matings to ura− cells (data not shown). We then analyzed diploid ura− and Ura+ dan1::URA3 strains by SDD-AGE, taking advantage of a naturally occurring 6xHis motif in Mot3p that allowed for its immunodetection. The Ura+ state, but not the ura− state, corresponded to the presence of SDS-resistant aggregates of Mot3p (Figure 6C). We hereafter refer to this heritable state of Mot3p as “[MOT3+]”, using standard prion nomenclature, to reflect its causal determinant.
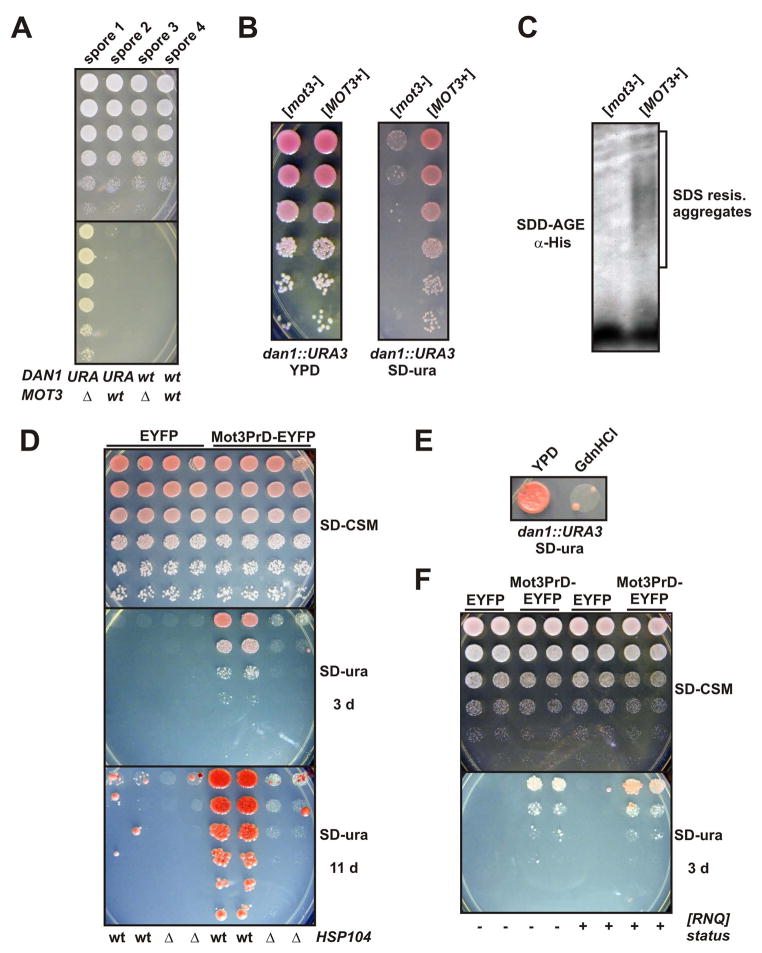
Figure 6. The transcription factor Mot3p is a prion.
(A) A Mot3p-reporter strain is Ura+ when Mot3p is inactive. A dan1::URA3/DAN1 mot3::KanMX4/MOT3 diploid was sporulated and tetrads dissected. Shown are five-fold serial dilutions of four spores from a tetratype tetrad, plated onto SD-CSM and SD-ura. Spore genotypes are as indicated.
(B) 5-fold serial dilutions of [mot3−] and [MOT3+] dan1::URA3 cells were spotted onto YPD and SD-ura.
(C) Lysates of diploid [mot3−] and [MOT3+] cells were investigated by SDD-AGE and Western blotting. Mot3p was detected via its naturally occurring 6xHis motif using an anti-His antibody.
(D) Wildtype HSP104 yeast cells and HSP104-deleted yeast cells, each carrying plasmids for galactose-inducible expression of either Mot3PrD-EYFP or control protein EYFP, were compared for [MOT3+] induction. Two transformants each were grown over night in galactose media, washed once in water, then plated at five fold serial dilutions to SD-CSM or SD-ura plates.
(E) A [MOT3+] isolate was passaged three times on YPD plates or YPD plates containing GdnHCl, and then grown over night in liquid YPD prior to spotting onto SD-ura plates.
(F) A [RNQ+] dan1::URA3 strain was converted to [rnq−] by four passages on GdnHCl-containing plates. The [RNQ+] and [rnq−] strains were transformed with a Mot3PrD-EYFP plasmid and assessed for [MOT3+] induction as in (D).
The strongest evidence that a phenotype is prion-based is the ability to induce it in prion-free cells by transformation with recombinant prion amyloid generated in vitro (Sparrer et al., 2000; Maddelein et al., 2002; Brachmann et al., 2005; Patel and Liebman, 2007). We produced Mot3PrD amyloid fibers from bacterially-expressed recombinant protein in vitro. These fibers strongly stimulated amyloid assembly of soluble Mot3PrD protein in vitro (Figure 7A). In both the W303 and S288C strain backgrounds, transformation with Mot3PrD fibers increased the frequency of Ura+ colonies by ~50 fold relative to control transformations with soluble Mot3PrD (Figure 7B and data not shown). These experiments prove not only that [MOT3+] is a protein-only heritable phenotype, but also that intracellular Mot3p readily engages in self-perpetuating prion aggregation.
Figure 7. Mot3p and Swi1p form amyloid-based prions that can be beneficial.
(A) Mot3PrD amyloid fibers used for fiber transformation experiments were added at 1% or 10% (w/w) to fresh 20 μM Mot3PrD amyloid assembly experiments and monitored for acquisition of ThT fluorescence. Shown are means +/− STDV.
(B) Mot3PrD-M-His protein was polymerized and examined for a fibrillar amyloid morphology by transmission electron microscopy (bar = 100 nm). [mot3−] spheroplasts were transformed with either soluble (freshly diluted) or amyloid Mot3PrD-M-His protein and plated directly onto SD-ura plates containing 1 M sorbitol.
(C) [MOT3+] and [mot3−] isolates were grown over night in YPD, then spotted (serial 5 fold dilutions) to SD-ura, YPD, or YPD containing calcofluor white (50 μg/ml) or congo red (500 μg/ml).
(D) Lysates of diploid [swi−] and [SWI+] cells carrying an integration of the coding sequence for EGFP at one of the two chromosomal loci of the SWI1 gene were investigated by SDD-AGE and Western blotting. Swi1p-EGFP fusion proteins were detected using an anti-GFP antibody.
(E) 5-fold serial dilutions of [swi−] and [SWI+] cells were spotted on YPD and YPD containing benomyl (5 mg/l).
Does this newly identified prion share other characteristics with well-known yeast prions? Well-characterized yeast prions rely, at least partially, on the presence of other prions like [RNQ+] for their efficient appearance. We compared [MOT3+] induction by transient Mot3PrD-EYFP expression in [RNQ+] and GdnHCl-treated, [rnq−] strains. Surprisingly, the appearance of Ura+ colonies was essentially unaffected by the prion status of Rnq1p (Figure 6F). The [RNQ+]-independence of [MOT3+] is unique among characterized yeast prions and suggests that [MOT3+] may appear frequently in diverse yeast strains. The ability of Mot3p to bypass a common regulatory barrier to prion nucleation also indicates that [MOT3+] induction may have elevated sensitivity to environmental stresses.
Amyloid-based yeast prions also rely on Hsp104p, the only known cellular factor capable of shearing amyloid fibers, in order to propagate efficiently. We tested if inactivation of Hsp104p eliminated [MOT3+], using GdnHCl treatment. We found that passaging otherwise stable [MOT3+] strains on medium containing 5 mM GdnHCl restored them to the original [mot3−] state (Figure 6E). We further confirmed that the Hsp104p-dependence of [MOT3+] was not unique to this particular isolate, by assaying the de novo inducibility of [MOT3+] in the absence of HSP104. In these cells, the appearance of Ura+ colonies was severely diminished (Figure 6D). This effect was observed both for over expression-induced and spontaneously-appearing Ura+ colonies (the latter arise among mock-induced cells after extended incubation times). These results indicate not only that [MOT3+], like all other known yeast prions, critically requires Hsp104p-mediated fiber shearing for inheritance, but also that [MOT3+] naturally appears at a detectable frequency.
Mot3p regulates a complex cell-wall remodeling program during adaptation to anaerobiosis, through predominantly gene-repressive activities (Grishin et al., 1998; Abramova et al., 2001; Hongay et al., 2002). We reasoned that a prion state of Mot3p would likely perturb this activity, giving rise to cell wall-related [MOT3+] phenotypes. Accordingly, we treated [mot3−] and [MOT3+] cells with the commonly used cell wall stressors, calcofluor white and congo red. Hypersensitivity to these agents is indicative of cell wall defects (Ram et al., 1994; Mrsa et al., 1999). We found that [MOT3+] isolates were relatively resistant to these agents (Figure 7C), consistent with a modified cell wall resulting from derepression of Mot3p-repressed cell wall proteins.
Do other candidates behave as prions? During the course of our studies, Du and coworkers presented strong genetic evidence for Swi1p as a prion protein (Du et al., 2008). Swi1p was one of eleven Q/N-rich proteins reported to allow for [PSI+]-induction when overexpressed (Derkatch et al., 2001). We used the [PSI+] co-induction methodology (Du et al., 2008) to confirm these findings for Swi1p. Previous studies speculated that the prion-like properties of Swi1p are based on amyloid aggregation, but direct biochemical evidence had been lacking (Derkatch et al., 2001; Du et al., 2008). We tested the physical status of the endogenous full-length Swi1 protein on SDD-AGE gels. It formed SDS-resistant aggregates in [SWI+] cells but not in [swi−] cells (Figure 7D). Finally, we asked if the [SWI+] state might have beneficial consequences under some circumstances by subjecting cells to a condition previously shown to interact synthetically with mutations in SWI1 – exposure to the microtubule-inhibiting fungicide benomyl (Hillenmeyer et al., 2008). Indeed, [SWI+] cells exhibited strong resistance (Figure 7E).
DISCUSSION
We conducted the first comprehensive study of prion-like Q/N-rich domains in yeast, using three different criteria to investigate their ability (1) to form amyloid in vivo under physiological conditions; (2) to self-assemble in vitro in the absence of other factors; and (3) to replicate indefinitely in cells as self-perpetuating epigenetic elements. We identified 24 protein domains that satisfied the stringent third criterion for prion behavior. This group includes the known prions Ure2p, Sup35p, Rnq1p, and Swi1p, the previously identified prion candidate New1p, and a functionally diverse set of 19 new candidates.
All but one prion candidate (New1p, see Supplemental Discussion) were strictly dependent on the protein remodeling factor Hsp104p. Interestingly, most proteins that satisfied the stringent third criterion also passed criteria one and two (Figure S3), underscoring the importance of amyloid’s distinctive self-templating properties for prion phenomena. In addition, the in vitro aggregation results compare extremely well with the aggregation of these cPrDs in vivo, a remarkable finding given the absence in vitro of the prion regulators [RNQ+] and Hsp104p. Q/N-rich sequences, despite having overtly similar amino acid compositions, have biochemical differences that give rise to a range of amyloid propensities. These differences affect amyloidogenesis intrinsically, rather than simply by controlling interactions with intracellular prion-promoting factors. Thus, prion-forming proteins are predisposed to form amyloid even in the absence of factors that govern prionogenesis in vivo. The implication is, then, that factors like [RNQ+] and Hsp104p may have evolved, in part, to regulate the frequency of spontaneous prion appearance and to promote the stable propagation of prions once they appear. The dependence of prions on these factors as well as their interaction with other chaperones and stress proteins links them intricately with cellular stress pathways (Chernoff, 2007; Shorter and Lindquist, 2008; Tuite et al., 2008) and makes them likely to respond to environmental changes that create even minor perturbances in protein homeostasis (Tyedmers et al., 2008).
Interestingly, the number of cPrDs forming fluorescent foci was higher than the number of cPrDs forming SDS-resistant aggregates in vivo. This finding, in combination with our detection of SDS-sensitive aggregates in vitro, indicates that some cPrDs are able to generate non-amyloid aggregates, with a low structural stability that prevents detection by SDD-AGE. A sequence analysis of our candidates suggests one explanation for these different behaviors. Highly amyloidogenic proteins were generally more N-rich, whereas non-amyloidogenic proteins were more Q-rich. A direct analysis of the distinct contributions of each of these residues to prion formation is underway and will be reported elsewhere.
The refined set of candidate yeast prions is strongly enriched for proteins involved in gene expression such as transcription factors (p = 5.3 × 10−5) and RNA-binding proteins (p = 5.1 × 10−4), a finding that supports the idea that PrDs function as epigenetic switches influencing important cellular pathways. The discoveries of [SWI+] (Du et al., 2008) and [MOT3+] lend further support to the idea that prion-based phenomena are biologically significant. The causal agents of these prions are each global transcription factors; consequently their prion states are likely to have far-reaching phenotypic effects. Our in depth analysis of one of these prions, [MOT3+], reveals a very appreciable spontaneous induction frequency (~10−4, Figure 6 and data not shown) that is further dramatically enhanced by overexpression or the introduction of preformed amyloid seeds. We speculate that the prion domain, and expression level, of Mot3p are partial products of selection for prion bistability. The prion properties of these transcription factors may generate an optimized phenotypic heterogeneity that buffers yeast populations against diverse environmental insults.
The role of prions has expanded considerably since their public inception as agents of disease. Prions in yeast and filamentous fungi drive heritable switches that increase phenotypic diversity. Conversely, higher organisms may have harnessed prion-like conformational templating to initiate stable switches involved in, for instance, neuronal synapse activation (Si et al., 2003). The regulation and frequency of prion switching may have been honed by selective pressures unique to each protein. Self-perpetuating prion and prion-like processes that serve an adaptive role may generally undergo increased switching under stressful conditions that perturb protein homeostasis, as has been established for the [PSI+] prion (Tyedmers et al., 2008). We suspect that many of our confirmed prion domains, in their endogenous contexts, will have switching rates that similarly respond to homeostatic cues. The heterogeneity theoretically possible with over twenty different prion switches amplifies the phenotype space of a single proteome, creating an advantageous scenario for any isogenic population under duress. Further, since prions are uniquely self-perpetuating yet metastable, any beneficial phenotype they produce can be easily maintained or lost in subsequent generations depending on selective pressures. This differs fundamentally both from genetic mutations, which are relatively permanent, and from non-heritable phenotypic changes, such as those arising from transcriptional noise.
Self-templating aggregates like yeast prions constitute a paradigm-shifting mechanism for the replication of biological information. Prion-based biological information arises spontaneously yet specifically within select proteins, is metastable, and is intricately linked to stress-response pathways. These features position prions as ideal bet-hedging devices (King and Masel, 2007) capable of responding to environmental stimuli. Our studies provide the tools for future genome-wide investigations of protein-based self-perpetuating changes in diverse organisms. We predict that these studies will profoundly impact our future understanding of phenotypic variation and will help to unravel the complex relationship between genotype and phenotype.
EXPERIMENTAL PROCEDURES
Computational determination of prion candidates
A Hidden Markov Model was used to identify candidate prions, followed by a Viterbi algortithm-based approach to parse proteins into prion and non-prion segments, as detailed in Supplemental Data.
Cloning and vector construction
Recombination-based cloning was used to generate cPrD reporter chimeras for yeast and bacterial expression (Alberti et al., 2007). See Table S3 for primers and the supplement for cloning specifics.
Yeast strains and media
The yeast strains used in this study (Table S4) were derived from YJW509 and YJW584 (Osherovich et al., 2004), and S288C. Standard genetic manipulations and media conditions were used, as detailed in Supplemental Data.
In vivo aggregation assays
cPrDs were fused to Sup35C for prion switching assays (Sondheimer and Lindquist, 2000), and EYFP for fluorescence microscopy. These experiments and SDD-AGE confirmation of SDS-resistant aggregation (Halfmann and Lindquist, 2008) were performed with modifications as detailed in Supplemental Data.
In vitro aggregation assays
cPrDs were expressed in E. coli strain BL21AI as cPrD-Sup35M-His7 fusions, except where noted, and purified under denaturing conditions. Proteins were diluted to 20 μM into physiological buffer containing ThT and agitated continuously, with fluorescence measurements taken at indicated time points. Additional details are found in Supplemental Data.
Fiber transformation
In vitro-generated Mot3PrD-M-His7 aggregates were examined by transmission electron microscopy to confirm a fibrillar amyloid appearance, and transformed into dan1::URA3 reporter strains using methodology as described (Tanaka et al., 2006).
Supplementary Material
Acknowledgments
This work was supported in part by the G. Harold & Leila Y. Mathers Foundation, NIH grant GM025874 and HHMI. SA was supported by a fellowship of the Deutsche Forschungsgemeinschaft (DFG). RH was supported by a fellowship of the NSF. We are grateful to Ndubuisi Azubuine and Tsering Nanchung for constant supply of plates and media. We thank Sherwin Chan and Gerald Fink for contributing the URA3-HIS5 plasmid. We thank Jijun Dong for assisting with TEM, and members of the Lindquist lab for critical reading of the manuscript.
Footnotes
Supplemental Experimental Procedures, Discussion and References, five tables, eleven figures and can be found with this article online.
References
- Abramova N, Sertil O, Mehta S, Lowry CV. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. Journal of bacteriology. 2001;183:2881–2887. doi: 10.1128/JB.183.9.2881-2887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigle M, Lacroute F. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem. 2008;283:15185–15192. doi: 10.1074/jbc.M802071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods in enzymology. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Baskakov IV, Breydo L. Converting the prion protein: what makes the protein infectious. Biochimica et biophysica acta. 2007;1772:692–703. doi: 10.1016/j.bbadis.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. The EMBO journal. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proceedings of the National Academy of Sciences of the United States of America 99 Suppl. 2002;4:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO. Stress and prions: lessons from the yeast model. FEBS letters. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Molecular microbiology. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? The EMBO journal. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DL. Strain-specific morphologies of yeast prion amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. The EMBO journal. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Molecular and cellular biology. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- Grishin AV, Rothenberg M, Downs MA, Blumer KJ. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics. 1998;149:879–892. doi: 10.1093/genetics/149.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science (New York, NY. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay C, Jia N, Bard M, Winston F. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. The EMBO journal. 2002;21:4114–4124. doi: 10.1093/emboj/cdf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- King OD, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theoretical population biology. 2007;72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H., 3rd Stopped-flow kinetics reveal multiple phases of thioflavin T binding to Alzheimer beta (1–40) amyloid fibrils. Archives of biochemistry and biophysics. 1997;342:306–316. doi: 10.1006/abbi.1997.0137. [DOI] [PubMed] [Google Scholar]
- Li L, Lindquist S. Creating a protein-based element of inheritance. Science (New York, NY. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- Liebman SW, Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. Journal of bacteriology. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proceedings of the National Academy of Sciences of the United States of America 99 Suppl. 2002;4:16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de la Paz M, Serrano L. Sequence determinants of amyloid fibril formation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science (New York, NY. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. Journal of bacteriology. 1999;181:3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset JP. Epigenetic control of polyamines by the prion [PSI(+)] Nature cell biology. 2008 doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] Journal of molecular biology. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science (New York, NY. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Perrett S, Jones GW. Insights into the mechanism of prion propagation. Curr Opin Struct Biol. 2008;18:52–59. doi: 10.1016/j.sbi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics (Oxford, England) 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Ram AF, Wolters A, Ten Hoopen R, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast (Chichester, England) 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Molecular and cellular biology. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nature cell biology. 2005b;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- Sabate R, Baxa U, Benkemoun L, Sanchez de Groot N, Coulary-Salin B, Maddelein ML, Malato L, Ventura S, Steven AC, Saupe SJ. Prion and non-prion amyloids of the HET-s prion forming domain. Journal of molecular biology. 2007;370:768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Saupe SJ, Clave C, Begueret J. Vegetative incompatibility in filamentous fungi: Podospora and Neurospora provide some clues. Current opinion in microbiology. 2000;3:608–612. doi: 10.1016/s1369-5274(00)00148-x. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates GP, Lehrach H, Wanker EE. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington’s disease pathology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science (New York, NY. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Shkundina IS, Ter-Avanesyan MD. Prions. Biochemistry. 2007;72:1519–1536. doi: 10.1134/s0006297907130081. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nature reviews. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. The EMBO journal. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Molecular cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sparrer HE, Santoso A, Szoka FC, Jr, Weissman JS. Evidence for the prion hypothesis: induction of the yeast [PSI+] factor by in vitro- converted Sup35 protein. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Molecular cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science (New York, NY. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Tuite M, Stojanovski K, Ness F, Merritt G, Koloteva-Levine N. Cellular factors important for the de novo formation of yeast prions. Biochemical Society transactions. 2008;36:1083–1087. doi: 10.1042/BST0361083. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- Wang X, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science (New York, NY. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Zenthon JF, Ness F, Cox B, Tuite MF. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot Cell. 2006;5:217–225. doi: 10.1128/EC.5.2.217-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.