Abstract
At 2 and 4 weeks following treatment with phenobarbital (PB), the classical nongenotoxic rodent liver carcinogen, we elucidated unique gene expression changes (both induction and repression) in liver tumor-susceptible B6C3F1 mice, as compared with the relatively resistant C57BL/6. Based on their cancer-related roles, we believe that altered expression of at least some of these genes might underlie PB-induced liver tumorigenesis. Putative constitutive active/androstane (CAR) response elements (CAREs), a subset of PB response elements, were present within multiple genes whose expression was uniquely altered in the B6C3F1 mice, suggesting a role for CAR in their regulation. Additionally, three DNA methyltransferase genes (Dnmt1, Dnmt3a, and Dnmt3b) were repressed uniquely in the tumor-prone B6C3F1 mice, and all possess putative CAREs, providing a potential direct link between PB and expression of key genes that regulate DNA methylation status. Previously, we demonstrated that PB-elicited unique regions of altered methylation (RAMs) in B6C3F1 mice, as compared with the relatively resistant C57BL/6, at 2 and 4 weeks, and annotation of the regions harboring these changes revealed 51 genes. This is extended by the current study, which employed RNA isolated from the same liver tissue used in the earlier investigations. Genes elucidated from both the methylation and expression analyses are involved in identical processes/pathways (e.g., cell cycle, apoptosis, angiogenesis, epithelial-mesenchymal cell transition, invasion/metastasis, and mitogen-activated protein kinase, transforming growth factor-beta, and Wnt signaling). Therefore, these changes might represent very early events that directly contribute to PB-induced tumorigenesis. It is instructive to consider the possibility that, in a hypothesis-driven fashion, these genes are initial candidates that could be utilized to develop a biomarker “fingerprint” of early exposure to PB and PB-like compounds.
Keywords: B6C3F1, CAR, gene expression, mouse liver, phenobarbital, tumors
The accumulation of key heritable modifications, including epigenetic alterations, for example, changes in DNA methylation (Esteller, 2007) which can play a variety of roles in carcinogenesis (Counts and Goodman, 1995) and be involved in the initiation as well as promotion stages (Goodman and Watson, 2002), as well as mutations (Loeb and Harris, 2008), drives the progressive clonal expansions of cells during tumorigenesis (Feinberg et al., 2006; Hanahan and Weinberg, 2000). Although genotoxic carcinogens can interact directly with DNA to form promutagenic adducts, nongenotoxic compounds cause tumorigenesis by other mechanisms, including inflammation, immunosuppression, receptor activation, and epigenetic changes (Luch, 2005). Phenobarbital (PB) is the classical nongenotoxic rodent liver carcinogen (Whysner et al., 1996), and upon treatment with a promoting dose in drinking water (0.05% [wt/wt]) for 12 months, 100% of susceptible C3H/He and B6C3F1 (C3H/He × C57BL/6) mice develop liver tumors, whereas the relatively resistant C57BL/6 mice develop none after 18 months (Becker, 1982).
The mechanism(s) of action of PB remain to be completely elucidated; however, some details have emerged. Connexin32, the major hepatic gap junction-forming protein, is essential for PB-induced liver tumorigenesis (Moennikes et al., 2000), and moreover, PB inhibits gap junction intercellular communication in hepatocytes from sensitive B6C3F1 mice but not the relatively resistant C57BL/6 (Warner et al., 2003). Furthermore, multiple events appear to be linked to PB-associated mouse liver tumorigenesis: mutations in β-catenin (Aydinlik et al., 2001), Ha-ras hypomethylation (Vorce and Goodman, 1991), increased Ha-ras expression (Counts et al., 1997), Ki-ras hypomethylation (Vorce and Goodman, 1991), hypomethylation and increased expression of raf (Ray et al., 1994), and amplification of c-myc (Vorce and Goodman, 1991).
Changes in DNA methylation have been implicated in the ability of PB to promote tumorigenesis. PB treatment resulted in higher levels of regional hypermethylation in two tumor-prone groups (C3H/He >> B6C3F1) versus the relatively resistant C57BL/6 (Watson and Goodman, 2002), as well as a greater decrease in global methylation levels in B6C3F1 versus C57BL/6 mice (Counts et al., 1996). Furthermore, PB-elicited unique regions of altered DNA methylation (RAMs), representing both increases and decreases in methylation levels, were discerned in B6C3F1 versus C57BL/6 mice at 2 and 4 weeks (Bachman et al., 2006). Cloning and annotation of these RAMs resulted in the identification of 51 genes, many of which participate in cancer-related processes, whose methylation statuses were altered uniquely in the susceptible mice (Phillips and Goodman, 2008). These data support the notion that the mechanism of action of PB involves alteration of DNA methylation in key genes which can facilitate tumor formation.
The constitutive active/androstane receptor (CAR) mediates the actions of PB, including induction of xenobiotic-metabolizing enzymes and hepatomegaly (Wei et al., 2000). Microarray analysis of CAR wild-type (WT) and knockout (KO) mice revealed CAR-dependent regulation of approximately half of the 138 gene expression changes (out of 8736 total genes/expressed sequence tags [ESTs]) that PB elicited (Ueda et al., 2002). Therefore, not all of the effects of PB are CAR dependent. Chronic PB treatment culminates in liver tumor development (Huang et al., 2005) and importantly, CAR is necessary for tumor promotion by PB in diethylnitrosamine (DEN)-initiated C3H/He mice (Yamamoto et al., 2004). Phillips et al. (2007) discerned PB-induced unique DNA methylation changes in liver tumor-susceptible CAR WT (both precancerous and liver tumor tissue), as compared with PB-treated resistant CAR KO mice. Annotation of these unique RAMs uncovered multiple genes that conceivably function in cancer-related processes, raising the possibility that at least some of the CAR-mediated alterations in DNA methylation contribute to PB-induced hepatocarcinogenesis (Phillips and Goodman, in press).
We hypothesized that at least some of the changes in hepatic gene expression that occur uniquely in liver tumor-susceptible B6C3F1 mice versus the relatively resistant C57BL/6, following both 2 and 4 weeks of PB treatment, contribute directly to liver tumorigenesis. In the current study, both microarray and quantitative real-time PCR (qRT-PCR) analyses were utilized to evaluate the aforementioned unique expression changes, including those of a subset of genes identified from unique PB-induced RAMs in the susceptible B6C3F1 mice (Phillips and Goodman, 2008), plus three DNA methyltransferase (Dnmt) genes, at very early time points (i.e., 2 and 4 weeks). Additionally, CAR response elements (CAREs), which are a subset of PB response elements, were located within multiple genes that exhibited unique B6C3F1 expression and/or methylation changes in response to PB. Furthermore, we ascertained signaling pathways and processes that the distinctively affected genes might influence. Overall, these results provide support for the notion that at least some of the genes whose expression was uniquely altered in susceptible B6C3F1 mice, as compared with the relatively resistant C57BL/6, might be mechanistically crucial for tumor development, thus enhancing our understanding of how PB promotes hepatocarcinogenesis.
MATERIALS AND METHODS
Animals, treatments and tissue samples.
As described previously, animals were treated, liver tissue was harvested, and RNA was isolated (Bachman et al., 2006). Briefly, B6C3F1 (C57BL/6 × C3H/He) and C57BL/6 mice (ages 29–32 days) were administered PB in drinking water at a concentration of 0.05% (wt/wt), a dose which will cause liver tumors in 100% of the tumor-prone B6C3F1 mice within 12 months, whereas no liver tumors are observed in the relatively resistant C57BL/6 mice after 18 months of treatment (Becker, 1982), for 2 or 4 weeks. Liver tissue was harvested from a total of eight groups (n = 6 per group): B6C3F1, 2 and 4 weeks, control and PB treated, and C57BL/6, 2 and 4 weeks, control and PB treated. RNA was isolated from samples of the same livers employed in our earlier study aimed at identifying PB-induced RAMs (Bachman et al., 2006), and stored at −80°C until its use in the current study.
RNA preparation.
Upon removal from −80°C, RNA samples were purified with the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Samples were eluted in diethyl pyrocarbonate (DEPC)–treated water, and RNA integrity was evaluated on the Agilent 2100 Bioanalyzer using 5 ng sample aliquots and the RNA 6000 Pico Lab-on-a-Chip (Agilent Technologies, Santa Clara, CA). The presence of two distinct peaks, representing 18S and 28S rRNA levels, was indicative of high quality samples. The purity (A260/A280 ratios) and concentrations of the RNA samples were determined via the NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Microarray analysis.
All procedures were performed according to standard protocols found within the Affymetrix Genechip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA).
RNA labeling and fragmentation.
The One-Cycle Target Labeling and Control Reagent kit (Affymetrix) was utilized for first- and second- strand cDNA synthesis plus double-stranded cDNA sample cleanup, and synthesis plus cleanup of biotin-labeled cRNA, of 48 samples (n = 6, for eight groups). To start, 1 μg of total RNA was used for the generation of double-stranded cDNA, which was then used as a template for the synthesis of biotinylated cRNA. The size distribution and yield of the labeled cRNA products were evaluated on the Agilent 2100 Bioanalyzer using 5 ng sample aliquots and the RNA 6000 Pico Lab-on-a-Chip (Agilent Technologies, Santa Clara, CA). Subsequently, 15 μg of labeled cRNA was fragmented to a range of 35–200 bp in a 40 μl of volume reaction (40mM Tris-acetate at pH 8.1, 100mM potassium acetate, and 30mM magnesium acetate) at 94°C for 35 min. The size distribution of the fragmented cRNA was assessed on the Agilent 2100 Bioanalyzer using 5 ng sample aliquots and the RNA 6000 Pico Lab-on-a-Chip (Agilent Technologies, Santa Clara, CA).
Hybridization, washing, staining, and scanning.
Fifteen micrograms of fragmented cRNA was hybridized to a GeneChip Mouse Genome 430 2.0 Array (Affymetrix), containing more than 45,000 probe sets representing over 34,000 genes. The instrumentation utilized for the washing and scanning of the chips is operated by the GeneChip Operating Software (GCOS, Affymetrix), version 3.1. After hybridization cocktails were removed, arrays were washed and stained on an Affymetrix Fluidics 450 station, and subsequently scanned using the Affymetrix GeneChip Scanner 3000 7G, in order to detect hybridization signals. From the resulting image files (DAT file), GCOS computes cell intensity data (CEL file), which is further analyzed to determine differential gene expression patterns.
Data analysis.
Data from Affymetrix GeneChip CEL files were normalized using GC Robust Multi-array Average (Wu et al., 2004). Boxplots and principal components analysis (PCA) were employed to assess data quality. Linear models were built and contrast estimates were performed, comparing treatment to time-matched control samples, using Limma. An empirical Bayes method was used to calculate false discovery rate controlled p values on the contrast estimates from Limma. A CDF file established by Dai et al. (2005), containing more accurate gene/transcript definitions (as compared with those from Affymetrix) based on up-to-date Entrez Gene information, was utilized (Mouse430, version 9.0, from (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download_v9.asp). Based upon these new probe set definitions, data was generated for 16,475 genes (as compared with more than 45,000 features representing over 34,000 genes, as indicated by Affymetrix). All statistical analyses were performed in R (v2.6.1) using Bioconductor (2.1).
Identification of uniquely active genes in liver tumor-prone B6C3F1 mice, as compared with the relatively resistant C57BL/6.
Differentially expressed, that is, active, genes in PB-treated v. control groups were identified based on meeting two criteria: (1) statistical significance, p < 0.01, and (2) a > twofold change in expression (up- or downregulated). Uniquely active genes in the susceptible B6C3F1 groups, as compared with the C57BL/6, were then discerned. At 2 weeks, and separately, at 4 weeks, active genes in the B6C3F1 and C57BL/6 groups were compared with one another. Common genes observed in both the B6C3F1 and C57BL/6 groups, at 2 or 4 weeks, which exhibited expression changes in the same direction (i.e., induced or repressed in both) were not given further consideration. Thus, uniquely active genes in the B6C3F1 mice, as compared with C57BL/6, at 2 or 4 weeks, included (1) common active genes whose expression changes were opposite (e.g., induction in the B6C3F1 and repression in the C57BL/6), and (2) genes which were active only in the B6C3F1. In an analogous manner, uniquely active genes in the C57BL/6 mice, as compared with the B6C3F1, at 2 and 4 weeks, were elucidated.
Uniquely active genes in the B6C3F1 mice at 2 and 4 weeks were compared, in order to ascertain genes that were active only at 2 weeks, only at 4 weeks, and those which “carried forward,” i.e., expression changed in the same direction at both 2 and 4 weeks.
qRT-PCR analysis.
Expression levels of three groups of genes were evaluated by qRT-PCR: (1) a subset of genes which were uniquely active, based on microarray analysis, in the susceptible B6C3F1 mice at 2 (all 18 genes listed in Supplementary Table S4) and/or 4 (all 11 genes listed in Supplementary Table S5) weeks of PB treatment; 2) a subset of genes identified from unique PB-induced RAMs in B6C3F1 mice at 2 (two genes listed in Supplementary Table S9) and/or 4 (17 genes listed in Supplementary Table S9) weeks, as discerned by Phillips and Goodman (2008); and (3) three Dnmt genes (Dnmt1, Dnmt3a, Dnmt3b) at 2 and 4 weeks. The genes in groups 1 and 3, above, were chosen for analysis based on their interesting, potentially cancer-related, documented functions, and the possibility that they affect pertinent signaling pathways (e.g., the three genes from Group 3 are involved in maintaining DNA methylation patterns). The genes in Group 2 also can influence key pathways, and many were selected because they exhibited unique PB-induced RAMs in both the liver tumor-susceptible B6C3F1 and CAR WT mice (Phillips and Goodman, in press).
Reverse transcription of RNA.
RNA, from the same samples used for microarray analysis, was treated with amplification grade DNAse I enzyme (Invitrogen, Carlsbad, CA) to eliminate contaminating genomic DNA. Each reaction, containing 1 μg of RNA, 10× DNase I Reaction Buffer, 2 U DNase I, and DEPC-treated water (Ambion, Inc., Austin, TX) up to 10 μl, was incubated for 15 min at room temperature. DNAse I was inactivated by adding 1 μl of 25mM EDTA solution and subsequently heating the reactions for 10 min at 65°C. Reverse transcription reactions containing DNAse I–treated RNA were prepared with reagents from the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Each reaction contained 1 μg RNA, 10X reverse transcription buffer, 25× dNTP mix (100mM), 10× random primers, 50 U MultiScribe reverse transcriptase, and RNAse-free water, up to 20 μl. Reactions were incubated at 25°C for 10 min, 37°C for 120 min, and 85°C for 5 s. All samples were stored at 4°C until needed.
Primers for qRT-PCR.
Primers were designed using the web-based Primer3 program, v. 0.4.0 (http://frodo.wi.mit.edu/primer3/input.htm) and synthesized by the Macromolecular Structure Facility at Michigan State University. The sizes of the amplicons ranged from 100–140 bp and the primers were 20mers; all other parameters (e.g., melting temperature) remained on the default settings. The majority of primers were designed such that the amplicon spanned an intron-exon junction near the 3′ end of the gene of interest. This was confirmed using GenBank sequence information, and also, in most instances, the UCSC In-Silico PCR web-based tool (July 2007 build, http://genome.ucsc.edu/cgi-bin/hgPcr?command = start). This was done so as to preclude the possibility that the expression data could be attributed to contamination with genomic DNA. Furthermore, we are attempting to not simply confirm the microarray data but are extending this to evaluate proper splicing, that is, this represents a stringent attempt to evaluate changes in expression of functional mRNAs. Names and symbols, accession numbers, and forward and reverse primer sequences of genes chosen for qRT-PCR analysis, plus amplicon sizes, are listed in Supplementary Tables S1A (selected genes that were uniquely active in B6C3F1 mice, based on microarray analysis) and S1B (selected genes identified from unique RAMs, plus three Dnmt's).
qRT-PCR assays.
Reactions were prepared with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), according to the manufacturer's protocol, with each reaction containing 1 μl of cDNA from the aforementioned reverse transcription reaction (with the exception of 18S reactions, which contained 1 μl of 1:100 diluted cDNA), 1× Power SYBR Green PCR Master Mix, 0.3μM of both the forward and reverse primers (with the exception of Dnmt3b, 0.1μM), and DEPC-treated water (Ambion, Inc., Austin, TX) up to 50 μl. PCR amplification of duplicate reactions was conducted in MicroAmp 96-well Optical Plates (Applied Biosystems, Foster City, CA) using the Applied Biosystems 7500 Real-Time PCR System, with the following thermal cycling conditions: 50°C for 2 min, 95°C for 10 min., and 40 cycles of 95°C for 15 s plus 60°C for 1 min. A dissociation protocol was run for each primer pair to ensure that a single product formed, and agarose gel electrophoresis of amplified products was performed to verify amplicon size.
Before samples were analyzed, standard curves for each gene were generated from purified amplicons. Standard curve samples (spanning 102 to 108 copies) for a particular gene were included on the appropriate sample plate so that the absolute quantitation method, which compares the threshold cycle of an unknown sample against a standard curve with known copy numbers, could be used to determine mRNA expression levels. The copy number of the gene of interest for each sample was standardized to that of the 18S rRNA gene to control for differences in RNA quantity, quality, and reverse transcription efficiency. Finally, fold changes in the treatment groups (vs. their time-matched controls) were calculated. Statistical outliers were excluded from the final calculations following their identification by the Grubbs’ test (p < 0.05, http://www.graphpad.com/quickcalcs/Grubbs1.cfm). Significance was determined by a Student's t-test (p < 0.05).
Changes in gene expression in the B6C3F1 versus C57BL/6.
Unique: Based on qRT-PCR analysis, where there is a statistical difference (Student's t-test, p < 0.05) or the change is in an opposite direction.
Confirmed: Microarray data which are confirmed based on qRT-PCR, where the change is unique.
Apparently Confirmed: Circumstances, based on qRT-PCR analysis, where data from at least three of the six samples fell outside the 95% confidence interval (CI) of the control group data were considered an “indication of a change” and thus, apparently confirming the microarray data.
Functional analysis of uniquely active B6C3F1 genes at 2 and 4 weeks of PB treatment.
The Pathway Studio 5.0 (Ariadne Genomics, Rockville, MD) informatics program was utilized to elucidate functions of uniquely active genes. Additionally, the program discerned connections between multiple genes, and linked them to key cellular processes and pathways. The Database for Annotation, Visualization and Integrated Discovery (DAVID), version 6 (Dennis et al., 2003; http://david.abcc.ncifcrf.gov/) was utilized to overlay uniquely active genes on pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Identification of CAREs.
CAREs are a subset of PB-responsive elements (PBREMs) because (1) CAR-independent effects of PB have been observed (Ueda et al., 2002), and thus it is presumed that other classes of cis-elements modulate those effects, and (2) there might exist elements which are necessary, but not sufficient, for transcriptional regulation by CAR. Seven bona fide CAREs were identified from the literature (Chen et al., 2003; Gerbal-Chaloin et al., 2002; Goodwin et al., 2002; Honkakoski et al., 1998; Sueyoshi et al., 1999; Sugatani et al., 2001) and used to construct a position weight matrix. The criteria for bona fide CAREs are that the elements are functional in a reporter gene assay and have been shown to bind to CAR by either gel shift or chromatin immunoprecipitation assays. Additionally, there had to be a match between the sequences of the elements reported in the literature and the most recent build, that is, compilation of genome sequence and annotation data at a particular point in time, of the human, mouse, or rat genome. The matrix similarity score threshold used was 0.80, that is, the identified CAREs were at least 80% similar to the consensus cis element.
The search for putative CAREs was performed on genes from the 3 groups described above, under quantitative real-time PCR (qRT-PCR) Analysis. Potential/possible regulatory regions (−10,000 bp relative to the transcription start site through the 5′-untranslated region), for the genes selected for analysis, were obtained from the UCSC genome database (http://genome.ucsc.edu). These regulatory regions were analyzed using previously described methods (Sun et al., 2004).
RESULTS
The complete microarray data set is provided as Supplementary Table S2. PCA revealed distinct differences in gene expression patterns based on treatment and mouse strain (Supplementary Fig. S1). One animal (from the B6C3F1, 4-week PB group) was omitted from subsequent data analysis after it was deemed to be an outlier by PCA (data point not shown). Additionally, animals within a particular group (e.g., C57BL/6, 4-week PB treated) typically exhibited similar expression patterns (Supplementary Fig. S1). The microarrays (with the exception of the 1 outlier) showed consistent distributions, based on box plots, which is an indication that the experiments are reproducible, while some outlying data points for each individual mouse were observed (Supplementary Fig. S2). The aforementioned outlier (one B6C3F1, 4-week PB-treated animal) is depicted in Figure S2 but was not utilized for data analysis.
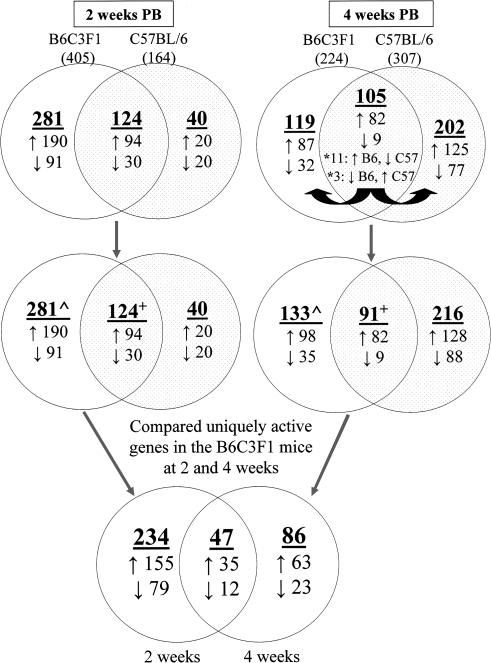
Active genes in the four PB-treated groups, as compared with their time-matched controls, were identified (Fig. 1). Lists of the active genes represented in the Venn diagrams (Fig. 1) can be found in Supplementary Tables S3A–I.
FIG. 1.
Identification of uniquely active genes in tumor-susceptible B6C3F1 mice, as compared with the relatively resistant C57BL/6, at 2 or 4 weeks of phenobarbital (PB) treatment. Microarray analysis was utilized to ascertain genes whose hepatic expression was altered (↑, induced or ↓, repressed) in four groups of PB-treated mice, as compared with their time-matched controls. Genes were considered to be differentially expressed (i.e., active) based on meeting two criteria: (1) statistical significance, p < 0.01, and (2) a > 2-fold change in expression (up- or downregulated). Active genes in the B6C3F1 and C57BL/6 groups were compared, at both 2 and 4 weeks. Genes that exhibited opposite changes (e.g., induced in the B6C3F1 and repressed in the C57BL/6) at a particular time point, depicted by asterisks (*), were considered to be uniquely active in each of the groups, and thus, segregated. Uniquely active genes in the B6C3F1 at 2 and 4 weeks (∧) were discerned by excluding those genes at each time point which overlapped in the B6C3F1 and C57BL/6 mice (overlapping portions of Venn diagrams), and whose changes occurred in the same direction (i.e., induced or repressed in both groups, +). Of the 367 total uniquely active genes elicited by PB in the susceptible B6C3F1 mice, as compared with the relatively resistant C57BL/6, 234 were observed only at 2 weeks, 86 were observed only at 4 weeks, and 47 were observed at both time points of treatment.
At 2 weeks, PB altered the expression of 405 and 164 genes in the B6C3F1 and C57BL/6 mice, respectively (Fig. 1). Comparison of these revealed 124 overlapping active genes and for all of them, the changes occurred in the same direction (i.e., 94 were induced and 30 were repressed) in both groups. These genes were not considered further. Therefore, 281 genes (190 induced and 91 repressed) were uniquely active in the B6C3F1 mice, as compared with the C57BL/6, at 2 weeks of PB treatment. Conversely, 40 genes (20 induced and 20 repressed) were uniquely active in the C57BL/6 mice, as compared with the B6C3F1.
At 4 weeks, PB altered the expression of 224 and 307 genes in the B6C3F1 and C57BL/6 mice, respectively (Fig. 1). Comparison of these revealed 105 overlapping active genes, of which 91 exhibited fold changes in the same direction (i.e., 82 induced and 9 repressed) in both groups, and thus were not considered further. Additionally, 14 active genes exhibited opposite changes: 11 were induced in the B6C3F1 and repressed in the C57BL/6, whereas three were repressed in the B6C3F1 and induced in the C57BL/6. Due to their opposing expression changes, these 14 genes were considered to be uniquely active in the B6C3F1, as well as in the C57BL/6. In total, 133 genes (98 induced and 35 repressed) were uniquely active in B6C3F1 mice, as compared with the C57BL/6, at 4 weeks of PB treatment. In contrast, 216 genes (128 induced and 88 repressed), including the 14 that exhibited opposite changes, were uniquely active in the C57BL/6 mice, as compared with the B6C3F1.
The validity of the microarray data is supported by the observations that, as expected, induction of classical PB-induced genes (e.g., Cyp2b9, Cyp2b10, Cyp2b13, Gsta2, Gstt3) in both the susceptible B6C3F1 and relatively resistant C57BL/6 groups at 2 and 4 weeks was observed. These genes are included among the 124 common active 2-week genes and 91 active common 4-week genes (Fig. 1; Supplementary Tables S3B and S3E, respectively).
Next, uniquely active genes in the susceptible B6C3F1 mice at 2 (281 genes) and 4 (133 genes) weeks were compared (Fig. 1; Supplementary Tables S3G–I): 47 were altered in the same direction (35 induced and 12 repressed) at both time points, 234 (155 induced and 79 repressed) were observed only at 2 weeks, and 86 (63 induced and 23 repressed) were observed only at 4 weeks. In total, PB treatment resulted in 367 total uniquely active genes in B6C3F1 mice at 2 and/or 4 weeks. Twenty-eight of these were selected, as described in the Methods, for confirmation of their expression statuses by qRT-PCR analysis. Of the 17 genes chosen that were uniquely active only at 2 weeks, expression changes of 11 were confirmed (Supplementary Table S4). The expression change of the single uniquely active carry forward gene (i.e., its expression was induced at both 2 and 4 weeks) that was tested, Slco1a4, was confirmed at 2, but not 4, weeks (Supplementary Tables S4 and S5, respectively). Finally, of the 10 genes chosen that were uniquely active only at 4 weeks, expression changes of 8 were confirmed (Supplementary Table S5).
Many of the 367 total uniquely active genes in B6C3F1 mice at 2 and/or 4 weeks were found to be involved in multiple key signaling pathways (Figs. 2–5; Supplementary Tables S6-S7), and emphasis will be placed on those whose expression changes were confirmed by qRT-PCR, plus those genes that were not selected for confirmation.
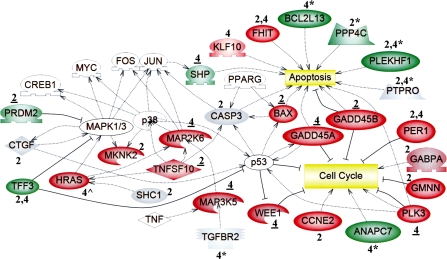
FIG. 2.
The expression and/or methylation statuses of genes involved in the cell cycle, apoptosis, and MAPK signaling were altered by PB at 2 and 4 weeks. Microarray analysis was used to identify uniquely active genes in livers of tumor-susceptible B6C3F1 mice at 2 and/or 4 weeks, as described in the Methods. Functional annotation of 367 uniquely active genes (Fig. 1; Supplementary Tables S3G–I) was performed. Expression is denoted as upregulated (red) or downregulated (green). Regarding qRT-PCR analysis, a change was confirmed if the time point is underlined (data in Supplementary Tables S4–S5); gray indicates either “not confirmed” (genes identified by microarray analysis) or “no expression change” (genes identified from regions of altered DNA methylation, RAMs). Genes noted as red or green that lack both underlining and symbols were not selected for confirmation. Genes identified from unique PB-induced RAMs in the B6C3F1 mice at 2 and/or 4 weeks (Phillips and Goodman, 2008) are denoted by asterisks (*), and their expression was evaluated at the time point directly preceding the asterisk (Supplementary Table S9). Ha-ras (∧) was hypomethylated and upregulated uniquely in B6C3F1 mice at 4 weeks (Bachman et al., 2006). White symbols represent “bridging” genes that were not altered in response to PB. Positive ( ) or negative (
) or negative ( ) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins (
) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins ( ), ligands (
), ligands ( ), kinases (
), kinases ( ), transcription factors (
), transcription factors ( ), phosphatases (
), phosphatases ( ), and membrane receptors (
), and membrane receptors ( ).
).
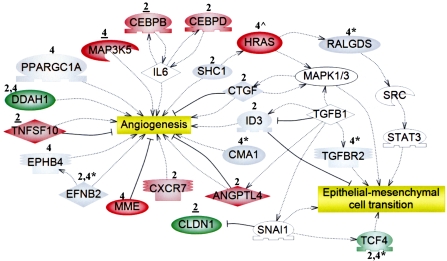
FIG. 3.
The expression and/or methylation statuses of genes involved in angiogenesis and the EMT were altered by PB at 2 and 4 weeks. Microarray analysis was used to identify uniquely active genes in livers of tumor-susceptible B6C3F1 mice at 2 and/or 4 weeks, as described in the “Methods.” Functional annotation of 367 uniquely active genes (Fig. 1; Supplementary Tables S3G–I) was performed. Expression is denoted as upregulated (red) or downregulated (green). Regarding qRT-PCR analysis, a change was confirmed if the time point is underlined (data in Supplementary Tables S4–S5); gray indicates either “not confirmed” (genes identified by microarray analysis) or “no expression change” (genes identified from regions of altered DNA methylation, RAMs). Genes noted as red or green that lack both underlining and symbols were not selected for confirmation. Genes identified from unique PB-induced RAMs in the B6C3F1 mice at 2 and/or 4 weeks (Phillips and Goodman, 2008) are denoted by asterisks (*), and their expression was evaluated at the time point directly preceding the asterisk (Supplementary Table S9). Ha-ras (∧) was hypomethylated and upregulated uniquely in B6C3F1 mice at 4 weeks (Bachman et al., 2006). White symbols represent “bridging” genes that were not altered in response to PB. Positive ( ) or negative (
) or negative ( ) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins (
) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins ( ), ligands (
), ligands ( ), kinases (
), kinases ( ), transcription factors (
), transcription factors ( ), and membrane receptors (
), and membrane receptors ( ).
).
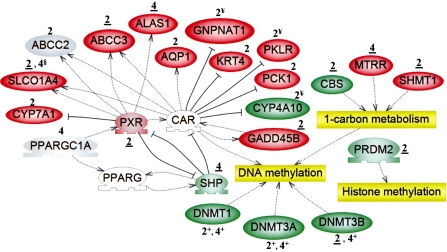
FIG. 4.
The expression of genes involved in epigenetic-related processes, and those that are indicative of nuclear receptor activation, was altered by PB at 2 and 4 weeks. Microarray analysis was used to identify uniquely active genes in livers of tumor-susceptible B6C3F1 mice, as described in the “Methods.” Functional annotation of 367 uniquely active genes (Fig. 1; Supplementary Tables S3G–I) was performed. Regarding qRT-PCR analysis, a change was confirmed if the time point is underlined (data in Supplementary Tables S4–S5); gray indicates “not confirmed.” Expression of Slco1a4 was confirmed at 2, but not 4 (§) weeks. Genes noted as red or green that lack both underlining and particular symbols (+ and §) were not selected for confirmation. A plus sign (+) denotes a gene whose expression was evaluated only by qRT-PCR. White symbols represent “bridging” genes that were not altered in response to PB. Positive ( ) or negative (
) or negative ( ) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins (
) regulation is illustrated, and the shapes of the entities represent the specific class of molecules to which the gene belongs: extracellular proteins ( ) and transcription factors (
) and transcription factors ( ). According to Ueda et al. (2002), two genes (Krt4 and Pck1) exhibited CAR-dependent downregulation in response to PB (while we detected upregulation), and three genes (¥) were under CAR-dependent “blocking,” that is, they were induced or repressed only in CAR KO mice (versus the wild-type).
). According to Ueda et al. (2002), two genes (Krt4 and Pck1) exhibited CAR-dependent downregulation in response to PB (while we detected upregulation), and three genes (¥) were under CAR-dependent “blocking,” that is, they were induced or repressed only in CAR KO mice (versus the wild-type).
FIG. 5.
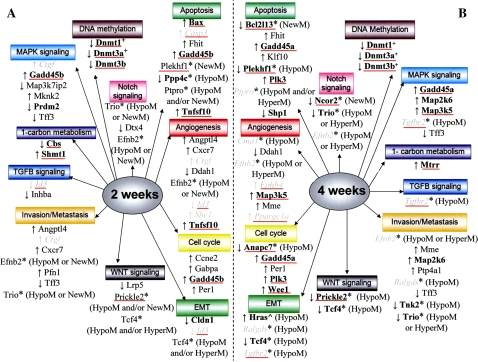
Summary of pathways and processes involving genes that exhibited unique PB-elicited expression and/or methylation changes at 2 and/or 4 weeks. PB induced unique hepatic regions of altered DNA methylation (RAMs), including hypomethylations (HypoM), hypermethylations (HyperM), and new methylations (NewM) in tumor-susceptible B6C3F1 mice at 2 (in A) and/or 4 (in B) weeks (Bachman et al., 2006). Genes, identified from unique RAMs (Phillips and Goodman, 2008a), are depicted by an asterisk (*). Regarding analysis of their expression in B6C3F1 mice by qRT-PCR (Supplementary Table S9): genes that exhibited an expression change (induction, ↑ or repression, ↓), are bolded, those that did not are in italics, and those that were not evaluated are in normal font. Bachman et al. (2006) demonstrated unique hypomethylation and upregulation of Ha-ras (∧) in the B6C3F1 mice at 4 weeks. As described in the “Methods,” microarray analysis was employed to identify uniquely active genes in the B6C3F1 mice at 2 and/or 4 weeks. Expression of uniquely active genes was confirmed (bolded), or evaluated and not confirmed (italics) by qRT-PCR (Supplementary Tables S4–S5). Uniquely active genes that were not selected for confirmation are listed in normal font. A plus sign (+) denotes a Dnmt gene whose unique expression change was revealed only by qRT-PCR. A red line underneath a gene indicates that at least one putative CARE was found within 10 kb upstream of the transcriptional start site.
Genes which can affect mitogen-activated protein kinase (MAPK) signaling were induced (Map3k5, Map2k6, Mknk2, and Tnfsf10) or repressed (Prdm2 and Tff3) by PB at 2 or 4 weeks (Fig. 2). MAPK pathways that involve p38 and Mapk1/3 can eventually result in the activation of key transcription factors, including Myc, Jun, and Fos (Fig. 2).
PB altered the expression of genes that can influence cell cycle progression (Ccne2, Gabpa, Gadd45a, Gadd45b, Gmnn, Per1, Plk3, and Wee1) at 2 and/or 4 weeks (Fig. 2). For Gadd45b, the B6C3F1 PB-treated animals demonstrated a statistically significant increase in expression over their controls at 2 weeks, as determined by qRT-PCR (Supplementary Table S4). The level of expression in the B6C3F1 appears to be higher than that seen in the C57BL/6; however, this does not reach statistical significance due to a particularly low level of expression in one of the B6C3F1 mice (Supplementary Table S4). A complete list of the genes that were uniquely active in the B6C3F1 mice, which have discernable connections to growth/survival, at 2 and/or 4 weeks are listed in Supplementary Tables S6 and S7, respectively. The induction of genes that can affect apoptosis (Bax, Fhit, Gadd45a, Gadd45b, Klf10, Shp, Tnfsf10) was observed at 2 and/or 4 weeks (Fig. 2). Additional genes that are positively or negatively associated with decreased growth/survival (i.e., apoptosis and cell death), and which were altered at 2 or 4 weeks, are listed in Supplementary Tables S6 and S7, respectively.
Figure 3 depicts a subset of the uniquely active genes at 2 and/or 4 weeks which are linked to angiogenesis (Angptl4, Cxcr7, Ddah1, Map3k5, Mme, Tnfsf10) and epithelial-mesenchymal cell transition (EMT) (Cldn1). Complete lists of uniquely active 2- (Table S6) and 4- (Table S7) week genes, including carry forward genes, which are positively or negatively associated with angiogenesis, invasion, and metastasis are included in Supplementary Information.
The pregnane X nuclear receptor (PXR), a xenobiotic sensor, was induced at 2 weeks (Fig. 4). At 2 or 4 weeks, PB altered the expression of several genes which are documented downstream targets of CAR and/or PXR (e.g., Alas1, Abcc3, Gadd45b, Slco1a4: 2 weeks). Additionally, uniquely active kinase, phosphatase, phosphatase activator (regulatory subunit), and phosphatase regulatory (inhibitor) genes are presented in Supplementary Table S8.
The expression of genes involved in epigenetic processes (DNA methylation: Dnmt3b, one-carbon metabolism: Cbs, Mtrr, Shmt1, and histone methylation: Prdm2) was affected by PB at 2 or 4 weeks (Fig. 4). Additionally, unique repression of three Dnmt genes (Dnmt1, Dnmt3a, Dnmt3b) was demonstrated in the B6C3F1 groups at both 2 and 4 weeks, and these changes were strikingly different from what was observed in the C57BL/6 at the corresponding time points (Table 1). At 2 weeks, expression of Dnmt3b decreased to a statistically greater extent (Student's t-test, p < 0.05) in the B6C3F1 mice, as compared with the C57BL/6, and thus, the change is considered to be unique in the B6C3F1.
TABLE 1.
qRT-PCR Analysis of Expression of Three Dnmt Genes
| Gene | Time pointa | B6C3F1 | C57BL/6 | ||
| Fold changeb | No. of mice that exhibited a change outside 95% CI of control groupc | Fold changeb | No. of mice that exhibited a change outside 95% CI of control groupc | ||
| Dnmt1: DNA methyltransferase 1 | 2 weeks | ↓ −1.47d | — | n.c. | ↑ 3 |
| 4 weeks | ↓ −1.49d | — | n.c. | — | |
| Dnmt3a: DNA methyltransferase 3a | 2 weeks | ↓ −1.60d | — | ↑ 1.33 | — |
| 4 weeks | ↓ −1.81d | — | n.c. | ↓ 3 | |
| Dnmt3b: DNA methyltransferase 3b | 2 weeks | ↓ −6.60d,e,f | — | ↓ −4.08e | — |
| 4 weeks | ↓ −2.00d | — | ↑ 1.47 | — | |
PB was administered to B6C3F1 and C57BL/6 mice for 2 or 4 weeks.
A fold change of gene expression (↑, upregulation or ↓, downregulation) is listed if it was statistically significant (Student's t-test, p < 0.05) in a treatment group v. its time-matched control. If it was not significant, no change (n.c.) is listed.
If a fold change was not statistically significant, a gene was considered to show an “indication of a change” if data from at least three of the six samples in the PB-treated group fell outside (above, ↑ or below, ↓) the 95% confidence interval (CI) of the control group data. If there was no indication of a change, or if a fold change was statistically significant, these data are not shown (—).
The expression change was considered to be unique in the B6C3F1 mice, as compared with the C57BL/6.
At 2 weeks, expression of Dnmt3b decreased to a statistically greater extent (Student's t-test, p < 0.05) in the B6C3F1, as compared with the C57BL/6, and thus, the change was considered to be unique in the B6C3F1.
At 2 weeks, Dnmt3b was uniquely active, based on microarray analysis, in the B6C3F1 mice.
In addition to assessing the effects of PB on global gene expression, we were interested in evaluating the expression of genes that were previously shown to exhibit unique RAMs in DNA isolated from the same livers of these tumor-susceptible B6C3F1 mice, as compared with their relatively resistant C57BL/6 counterparts (Phillips and Goodman, 2008). The expression of a subset of these (19 total: 2 at 2 weeks and 17 at 4 weeks) was initially evaluated in the B6C3F1 mice (Supplementary Table S9). If the methylation and expression statuses appeared to be inversely correlated (as described in Supplementary Table S9), expression in the time-matched C57BL/6 group was evaluated. At 4 weeks, three genes (Bcat2, Bcl2l13, Ube2d1) exhibited unique decreases in expression and unique increases in methylation in the B6C3F1 mice, as compared with the C57BL/6. Two genes exhibited decreases in expression that were not statistically different (Student's t-test, p < 0.05) in the B6C3F1 and C57BL/6 mice at 2 (Srms) or 4 (Trio) weeks, and their methylation statuses were ambiguous (either increased and/or decreased). At 4 weeks, Ncor2 exhibited increased methylation levels and decreased expression in the B6C3F1 mice, and the extent of the fold change was statistically less (Student's t-test, p < 0.05) than that observed in the C57BL/6. The expression of 13 genes was only evaluated in the B6C3F1 mice: seven were downregulated and hypomethylated, whereas expression of the other six did not change.
Genes identified from unique RAMs (Phillips and Goodman, 2008) were added to Figures 2–5 in order to depict complementary data from the gene expression and DNA methylation analyses. Also included in Figures 2 and 3 is Ha-ras, which we previously demonstrated to be hypomethylated and induced uniquely in B6C3F1 mice, as compared with the C57BL/6, at 4 weeks of PB treatment (Bachman et al., 2006).
Thirty genes of interest (18 uniquely active genes based on microarray analysis, three Dnmt's, and nine genes identified from unique RAMs) harbored at least one putative CARE within 10-kb upstream of their transcriptional start sites (Table 2; base sequences of the putative CAREs are listed in Supplementary Table S10). All 30 genes were subjected to qRT-PCR analysis (Table 1, Supplementary Tables S4, S5, and S9). Of the 18 which were identified as uniquely active based on microarray analysis, the expression statuses of 13 were confirmed by qRT-PCR (Supplementary Tables S4 and S5).
TABLE 2.
| Genec | Time point (weeks) in B6C3F1 | Microarray analysisd | qRT-PCR analysis | Methylation changee | No. of putative CAREs | |
| Exhibited expression changes based on microarray analysisf | Abcc3 | 2 | ↑ | Cg | 5 | |
| Bax | 2 | ↑ | A.C.h | 6 | ||
| Casp3 | 2 | ↑ | n.c.i | 2 | ||
| Cbs | 2 | ↓ | Cg | 3 | ||
| Cebpb | 2 | ↑ | Cg | 2 | ||
| Gadd45b | 2 | ↑ | A.C.h | 4 | ||
| Id3 | 2 | ↓ | n.c.i | 1 | ||
| Lcn2 | 2 | ↑ | n.c.i | 1 | ||
| Prdm2 | 2 | ↓ | A.C.h | 1 | ||
| Shmt1 | 2 | ↑ | A.C.h | 2 | ||
| Tnfsf10 | 2 | ↑ | Cg | 1 | ||
| Ephb4 | 4 | ↑ | n.c.i | 5 | ||
| Gadd45a | 4 | ↑ | Cg | 2 | ||
| Map3k5 | 4 | ↑ | A.C.h | 4 | ||
| Mtrr | 4 | ↑ | Cg | 1 | ||
| Plk3 | 4 | ↑ | A.C.h | 1 | ||
| Ppargc1a | 4 | ↑ | n.c.i | 1 | ||
| Wee1 | 4 | ↑ | Cg | 5 | ||
| DNA methyltransferasesj | Dnmt1 | 2 | N.D.k | ↓l | 3 | |
| 4 | N.D.k | ↓l | ||||
| Dnmt3a | 2 | N.D.k | ↓l | 2 | ||
| 4 | N.D.k | ↓l | ||||
| Dnmt3b | 2 | ↓ | Cg | 1 | ||
| 4 | N.D.k | ↓l | ||||
| Exhibited unique regions of altered DNA methylation (RAMs)m | Anapc7 | 4 | N.D.k | ↓l | HypoM | 2 |
| Bcat2 | 4 | N.D.k | ↓l | NewM | 2 | |
| Bcl2l13 | 4 | N.D.k | ↓l | NewM | 2 | |
| Ddx54 | 2 | N.D.k | n.a.n | HypoM | 1 | |
| 4 | N.D.k | ↓l | HypoM | |||
| Ncor2 | 4 | N.D.k | ↓l | NewM | 2 | |
| Plekhf1 | 2 | N.D.k | n.a.n | NewM | 3 | |
| 4 | N.D.k | ↓l | HypoM | |||
| Prickle2 | 2 | N.D.k | n.a.n | HypoM and/or NewM∧ | 1 | |
| 4 | N.D.k | n.s.o | HypoM | |||
| Srms | 2 | N.D.k | ↓l | HypoM or HyperM, and/or NewM∧ | 4 | |
| Tgfbr2 | 4 | N.D.k | n.s.o | HypoM | 1 | |
Base sequences of putative CAREs, located 10 kb upstream of the transcriptional start site, are listed in Supplementary Table S10.
We used the same position weight matrix and 80% stringency cut-off (as described in “Methods”) on 25,000 randomly constructed sequences of 10 kb in length and found that, on average, there were only 0.561 CAREs found per sequence (across all 25,000 sequences). The probability of a gene having more than 1 CARE is 10.8%, whereas the probability of having more than 2, 3, or 4 CAREs is 2.2, 0.38, and 0.044%, respectively. The probability of not having any CAREs is 57.4%. We view the informatic approach for identification of CAREs as hypothesis generating.
Genes that exhibited unique PB-elicited changes in expression (Table 1, Supplementary Tables S3G–I and S4–S5) and/or methylation (Phillips and Goodman, 2008a) in B6C3F1 mice at 2 and/or 4 weeks of PB treatment.
Expression changes (↑, upregulation and ↓, downregulation) of uniquely active genes that were identified based on microarray analysis (as described in the Methods). For genes and time points listed in bolded font, the expression change was confirmed, or appeared to be confirmed, by qRT-PCR analysis. The expression of genes listed in normal black font was evaluated but was not confirmed.
PB-induced decreases in methylation (Hypomethylation, HypoM) and increases in methylation (Hypermethylation, HyperM and new methylations, NewM), as determined by Phillips and Goodman (2008). For certain genes (∧), the methylation statuses were ambiguous (i.e., not definitively determined).
Detailed confirmatory qRT-PCR data are found in Supplementary Tables S4 (2 weeks) and S5 (4 weeks).
The expression change, based on microarray analysis, was confirmed (C), as described in the “Methods.”
The expression change, based on microarray analysis, was apparently confirmed (A.C.), as described in the “Methods.”
The expression change, based on microarray analysis, was not confirmed (n.c.).
Detailed qRT-PCR data are found in Table 1.
N.D. (not detected) indicates that the gene was not active.
Direction (↑, upregulation and ↓, downregulation) of the statistically significant (Student's t-test, p < 0.05) fold change.
Detailed qRT-PCR data are found in Supplementary Table S9.
Expression was not evaluated.
Fold change was not significant, n.s. (Student's t-test, p < 0.05).
DISCUSSION
Unique hepatic gene expression changes in tumor-prone B6C3F1, as compared with relatively resistant C57BL/6 mice, were discerned at very early time points following PB treatment. We hypothesize that at least some of these contribute directly to the development of liver tumors which are destined to become evident at later time points, for example, 12 months (Becker, 1982). Strikingly, as early as 2 and 4 weeks, the expression of genes that regulate key cancer-related processes is altered uniquely in tumor-susceptible mice, as compared with their relatively resistant counterparts (Figs. 2–5). These data both complement and extend our previous research, which identified multiple genes that exhibited unique methylation changes in hepatic DNA from the same B6C3F1 mice at 2 and 4 weeks of PB treatment (Phillips and Goodman, 2008). Importantly, genes elucidated from both the methylation and expression analyses participate in several of the same overarching pathways and signaling processes (e.g., cell cycle, apoptosis, angiogenesis, EMT, invasion/metastasis, and MAPK, Tgf-β, and Wnt signaling) (Fig. 5). Additionally, several pathways are represented by genes that exhibited changes only in methylation (Notch signaling) or expression (DNA methylation, 1-carbon metabolism) (Fig. 5). Overall, these data illuminate very early events (alterations in DNA methylation and gene expression) which might facilitate selective clonal expansion, and ultimately, tumor formation, in response to PB.
A subset of the uniquely active B6C3F1 genes at 2 and/or 4 weeks are involved in growth (e.g., cell cycle), survival, and apoptosis (Figs. 2 and 5; Supplementary Tables S6 and S7). Multiple genes identified from unique B6C3F1 RAMs (Phillips and Goodman, 2008) participate in these same processes (Fig. 5), and several of them (Anapc7, Bcl2l13, Ppp4c, Plekhf1) exhibited an expression change (Supplementary Table S9). These observations are consistent with the known ability of PB to enhance hepatocyte proliferation between 1 and 4 weeks of treatment (Counts et al., 1996; Kolaja et al., 1996a) and decrease apoptosis within focal lesions that become evident after prolonged treatment (i.e., 30–60 days) (Kolaja et al., 1996b). In this context, it is important to note that PB treatment for 1–2 weeks also increases hepatocyte proliferation in the resistant C57BL/6 strain (Counts et al., 1996). Chronic PB administration inhibits normal hepatocyte proliferation, whereas a subpopulation of cells escapes the mitoinhibitory effects and may serve as the progenitors for tumor formation (reviewed in Counts et al., 1996). A novel aspect of this research is the identification of several growth regulatory genes which are uniquely active in the tumor-susceptible B6C3F1 mice.
Importantly, PB-elicited unique changes in methylation (Phillips and Goodman, 2008) and expression of angiogenesis-associated genes at 2 and/or 4 weeks in the susceptible B6C3F1 mice (Figs. 3 and 5; Supplementary Tables S6 and S7). Angiogenesis is required for both normal development and tumor growth and metastasis (Folkman, 2002; Papetti and Herman, 2002). It is remarkable to see altered expression of genes involved in this process so soon after commencing PB treatment. Furthermore, genes involved in invasion and metastasis, plus EMT, a developmental program which appears to facilitate the two former processes (Thiery, 2002; Yang et al., 2006), were uniquely active in the susceptible B6C3F1 mice at 2 and/or 4 weeks of PB treatment (Figs. 3 and 5; Supplementary Tables S6 and S7). Genes in which unique PB-induced B6C3F1 RAMs occurred (Phillips and Goodman, 2008) are also involved in these processes (Fig. 5), and notably, PB altered the expression of Tcf4 in the B6C3F1 mice (Fig. 3). Although invasion and metastasis are typically regarded as late events in tumorigenesis, untransformed cells can travel through the bloodstream to a secondary site, where additional molecular alterations leading to malignant growth may occur (Podsypanina et al., 2008). The current report is the first to indicate that PB can start to facilitate EMT, invasion and metastasis at a very early stage of tumor development.
The fundamental genes involved in human and mouse tumorigenesis are likely identical, however, mice are more susceptible than humans (Rangarajan and Weinberg, 2003). The regulation of these genes, of which epigenetic control (e.g., altered DNA methylation) is a component, might be a key species-specific difference that contributes to their disparate sensitivities. For instance, methylation patterns are less stable in rodent, as compared with human, cells (reviewed in Goodman and Watson, 2002). In the B6C3F1 mice, PB uniquely altered the expression of genes which influence methylation status. Changes in genes that function in 1-carbon metabolism (Cbs, Mtrr, and Shmt1; Fig. 4), which generates the Dnmt methyl donor S-adenosylmethionine, occurred. Remarkably, the expression of 3 Dnmt genes (1, 3a, and 3b) was uniquely repressed in the B6C3F1 mice at both 2 and 4 weeks (Table 1), thus linking PB to early expression changes of key genes involved in regulating DNA methylation patterns. Interestingly, all six of these genes harbor CAREs, indicating that CAR might modulate their transcription.
Additional genes whose expression and/or methylation statuses were altered uniquely at these extremely early time points in the susceptible B6C3F1 mice can participate in multiple signaling pathways of development and differentiation that have previously been linked to tumorigenesis: transforming growth factor-beta (Tgf-β), Wnt, and Notch (Fig. 5). Tgf-β signaling functions in a tumor suppressive manner in normal cells and in an oncogenic fashion during later stages of tumorigenesis (Massagué, 2008). Aberrant Wnt signaling is known for its prominent role in colorectal cancer (Klaus and Birchmeier, 2008), as epigenetic silencing of Wnt antagonists (e.g., secreted frizzled-related proteins) and activating mutations in β-catenin occur (Baylin and Ohm, 2006). β-Catenin mutations are also prevalent in DEN-initiated, PB-promoted mouse liver tumors (Schwarz et al., 2003). Finally, abnormal Notch signaling can contribute to a variety of human malignancies in an oncogenic or tumor suppressive manner (Bolós et al., 2007).
We are interested in developing a pattern of gene expression and DNA methylation that could be used as an early biomarker of PB treatment. The current study takes us a step in that direction by focusing on a subset of the uniquely active B6C3F1 genes at 2 and/or 4 weeks of PB treatment. Candidate genes include those with documented roles in cancer-related processes (Ccne2, Cxcr7, Gadd45a, Gadd45b, Inhba, Lrp5, Mknk2, Wee1). Other potential candidates include those genes, possessing tumorigenesis-related functions, that exhibited unique RAMs solely in the B6C3F1 group (Cma1, Ralgds, Tgfbr2; from Phillips and Goodman, 2008), and common RAMs with the same methylation status in both liver tumor-prone B6C3F1 and CAR WT mice (Bcat2, Ddx54, Srms, Wscd1, Zscan22; from Phillips and Goodman, in press).
Efforts have been made to identify expression signatures of nongenotoxic rodent hepatocarcinogenesis. Iida et al. (2005) evaluated hepatic gene expression in B6C3F1 mice treated for 2 weeks with oxazepam. Two of the six genes which were upregulated, excluding those involved in drug metabolism, (Gadd45b and Maff), and one of the three which were downregulated (Tsc-22) matched our results. Fielden et al. (2007) reported a 37 gene signature based on expression data from rats (which are not as responsive to PB-induced tumorigenesis as the susceptible strains/stocks of mice) treated for 5 days with one of a variety of nongenotoxic compounds. Only one of these (Usp2) was active in our study. However, the direction of change was opposite and, thus, there were no real matches. Our approach for elucidating PB-induced gene expression changes that might be useful biomarker candidates is more appropriate than what is in the literature currently because we are “subtracting out” those alterations that occurred in the relatively resistant C57BL/6 mice, and thus, are focusing upon changes in the susceptible B6C3F1 that might truly be related directly to tumorigenesis.
The RNA utilized for gene expression analysis was isolated from whole liver. Thus, the expression patterns are presumably derived from multiple cell types (e.g., hepatocytes, sinusoidal epithelial cells, Kupffer cells), which could partially explain changes that do not appear to obviously correlate with overall enhancement or inhibition of a process (e.g., genes that positively regulate angiogenesis are both induced and repressed, Fig. 3 and Supplementary Tables S6 and S7). This approach is beneficial for ascertaining early changes that accumulate in multiple cells, including potential tumor progenitors, such as stem cells (Reya et al., 2001), or mature cells (e.g., hepatocytes) that have de-differentiated. Indeed, because hepatocytes can proliferate (Counts et al., 1996; Kolaja et al., 1996a; Michalopoulos, 2007) and can be reprogrammed to a stem-like phenotype (Aoi et al., 2008), it is possible that adult liver cells can revert into a de-differentiated state and serve as the tumor progenitor. Importantly, this notion is not incompatible with the aforementioned stem cell theory.
Kinase, phosphatase, phosphatase activator (regulatory subunit), and phosphatase regulatory (inhibitor) genes, which were uniquely active in the tumor-susceptible BbC3F1 mice at 2 and 4 weeks, are listed in Supplementary Table S8. In this context, it is instructive to note that phosphorylation and dephosphorylation events (Kawamoto et al., 1999; Sueyoshi et al., 2008; Yamamoto et al., 2003) are necessary for CAR activation, and we speculate that PB might affect expression levels of genes involved. Therefore, candidate kinases and phosphatases which were uniquely active in the tumor-susceptible B6C3F1 mice at 2 and 4 weeks are listed in Supplementary Table S8. Induction of known CAR target genes was observed (Fig. 4). Furthermore, in a hypothesis generating fashion, putative CAREs were discerned within a subset of uniquely active genes in the B6C3F1 mice, plus multiple genes that harbored unique PB-elicited B6C3F1 RAMs (Supplementary Table S10). These data indicate that CAR might regulate gene transcription at 2 and 4 weeks.
Expression analysis of a subset of genes that exhibited unique RAMs in the B6C3F1 mice (Supplementary Table S9) revealed several (Bcat2, Bcl2l13, Ube2d1) whose levels were unique as compared with the C57BL/6 group, suggesting that they might be important for tumor formation, particularly the apoptosis-inducer Bcl2l13. For others (Srms and Trio), the expression patterns were not statistically different in the B6C3F1 and C57BL/6 mice, and therefore, in the case of Trio, which has been linked to cancer, the expression change might be necessary, but not sufficient, for tumor formation. For numerous genes, decreased methylation was associated with decreased expression. This resembles the situation for the imprinted insulin-like growth factor receptor type-2 (Igf2r) gene, in which hypomethylation of an intronic CpG island is associated with paternal silencing, whereas methylation appears to facilitate maternal expression (Wutz et al., 1997).
There often is not an association between expression and the genomic location of the DNA methylation change (Supplementary Table S9). The approach taken to discern unique B6C3F1 RAMs is unbiased, in the sense that regions to be assayed are not pre-determined (Bachman et al., 2006). Thus, methylation changes were ascertained in various genomic locations (not only promoters) that could potentially influence expression. It is possible that numerous regulatory elements, including regions in which RAMs occurred, function in concert to control expression. Additionally, multiple methylation changes may be required to affect expression of a gene and not all might have accumulated within 4 weeks. In these instances, an obvious link between expression status and the location of a RAM might not be apparent.
In general, there was good agreement between the microarray and qRT-PCR data (Supplementary Tables S4 and S5). However, discrepancies between these two techniques are often encountered (Etienne et al., 2004; Morey et al., 2006), and there were a few instances when the expression statuses of genes, identified as uniquely active in the B6C3F1 mice by microarray analysis, were not confirmed by qRT-PCR (Supplementary Tables S4 and S5). Nonetheless, the overall validity of the microarray data is supported by the observed induction of typical PB-responsive genes in both the B6C3F1 and C57BL/6 groups at 2 and 4 weeks (Supplementary Tables S3B and S3E, respectively). Furthermore, our data reinforce published results that demonstrate PB-elicited induction of Gadd45β (Peffer et al., 2007), Aqp1 and Alas1 (Ueda et al., 2002), and Abcc3, Dio1, Gna14, Lcn2, Por, and Thrsp, plus repression of Arntl (Stahl et al., 2005). All of these were altered in the same directions in the B6C3F1 at 2 or 4 weeks (Supplementary Tables S3G and S3I). Contrastingly, different expression patterns of several uniquely active B6C3F1 genes (Cyp4a10, Gnpnat1, Krt4, Pck1, and Pklr), as compared with Ueda et al. (2002), were reported (Fig. 4). Reasons for these inconsistencies might be differences in our microarray platforms, the number of genes/ESTs on the array plus more recent and accurate annotation information, mouse strain/stock, and/or the method/duration of PB administration.
In summary, this extensive analysis of PB-elicited expression at very early time points has revealed numerous uniquely active genes in tumor-susceptible B6C3F1, as compared with relatively resistant C57BL/6 mice, many of which are involved in key cancer-related processes (Figs. 2–5; Supplementary Tables S6 and S7). Significantly, genes which demonstrated unique RAMs in the B6C3F1 mice (Phillips and Goodman, 2008) also participate in several of these pathways (Figs. 2–5), and for a subset, their expression levels in the B6C3F1 groups were affected by PB treatment. The unique repression of three major Dnmt genes in the B6C3F1 mice at both 2 and 4 weeks provides a potential direct link between PB and the expression of key genes that regulate DNA methylation patterns. Furthermore, we located putative CAREs within numerous genes that were altered by PB, including all three Dnmt's, suggesting that these might be regulated by CAR. We propose that at least some of the genes that exhibited unique changes in expression and/or DNA methylation represent initial key, heritable changes that confer proliferative advantages to cells, resulting in the progressive accumulation of modifications and the perturbation of signaling pathways that facilitate PB-induced tumorigenesis. It is instructive to reflect on the possibility that, in a hypothesis-driven fashion, these genes represent appropriate candidates that could be utilized to develop a biomarker “fingerprint” of early exposure to PB and PB-like compounds.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health/National Institute of Environmental Health Sciences training (grant no. T32-ES-7255) predoctoral fellowship support to J.M.P.; and R.J. Reynolds Tobacco Company research support (unrestricted gift).
Supplementary Material
Acknowledgments
We thank the Michigan State University Research Technology and Support Facility for technical assistance with the microarray analyses.
References
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Aydinlik H, Nguyen TD, Moennikes O, Buchmann A, Schwarz M. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of β-catenin-mutated mouse liver tumors. Oncogene. 2001;22:7812–7816. doi: 10.1038/sj.onc.1204982. [DOI] [PubMed] [Google Scholar]
- Bachman AN, Phillips JM, Goodman JI. Phenobarbital induces progressive patterns of GC-rich and gene-specific altered DNA methylation in the liver of tumor-prone B6C3F1 mice. Toxicol. Sci. 2006;91:393–405. doi: 10.1093/toxsci/kfj155. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Becker FF. Morphological classification of mouse liver tumors based on biological characteristics. Cancer Res. 1982;42:3918–3923. [PubMed] [Google Scholar]
- Bolós V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr. Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol. Pharmacol. 2003;64:316–324. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- Counts JL, Goodman JI. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Counts JL, McClain RM, Goodman JI. Comparison of effect of tumor promoter treatments on DNA methylation status and gene expression in B6C3F1 and C57BL/6 mouse liver and in B6C3F1 mouse liver tumors. Mol. Carcinog. 1997;18:97–106. doi: 10.1002/(sici)1098-2744(199702)18:2<97::aid-mc5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Counts JL, Sarmiento JI, Harbison ML, Downing JC, McClain RM, Goodman JI. Cell proliferation and global methylation status changes in mouse liver after phenobarbital and/or choline-devoid, methionine-deficient diet administration. Carcinogenesis. 1996;17:1251–1257. doi: 10.1093/carcin/17.6.1251. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Etienne W, Meyer MH, Peppers J, Meyer RA., Jr Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques. 2004;36:618–620. doi: 10.2144/04364ST02. 622, 624–626. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol. Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J. Biol. Chem. 2002;277:209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- Goodman JI, Watson RE. Altered DNA methylation: A secondary mechanism involved in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2002;42:501–525. doi: 10.1146/annurev.pharmtox.42.092001.141143. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, D'Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol. Pharmacol. 2002;62:359–65. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg GF. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Iida M, Anna CH, Holliday WM, Collins JB, Cunningham ML, Sills RC, Devereux TR. Unique patterns of gene expression changes in liver after treatment of mice for 2 weeks with different known carcinogens and non-carcinogens. Carcinogenesis. 2005;26:689–99. doi: 10.1093/carcin/bgi005. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Kolaja KL, Stevenson DE, Johnson JT, Walborg EF, Jr, Klaunig JE. Subchronic effects of dieldrin and phenobarbital on hepatic DNA synthesis in mice and rats. Fundam. Appl. Toxicol. 1996a;29:219–228. doi: 10.1006/faat.1996.0025. [DOI] [PubMed] [Google Scholar]
- Kolaja KL, Stevenson DE, Walborg EF, Jr, Klaunig JE. Dose dependence of phenobarbital promotion of preneoplastic hepatic lesions in F344 rats and B6C3F1 mice: Effects on DNA synthesis and apoptosis. Carcinogenesis. 1996b;17:947–954. doi: 10.1093/carcin/17.5.947. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Harris CC. Advances in chemical carcinogenesis: A historical review and prospective. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luch A. Nature and nurture—Lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration. J. Cell. Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moennikes O, Buchmann A, Romualdi A, Ott T, Werringloer J, Willecke K, Schwarz M. Lack of phenobarbital-mediated promotion of hepatocarcinogenesis in connexin32-null mice. Cancer Res. 2000;15:5087–5091. [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papetti M, Herman IM. Mechanisms of normal and tumor-derived angiogenesis. Am. J. Physiol. Cell. Physiol. 2002;282:C947–970. doi: 10.1152/ajpcell.00389.2001. [DOI] [PubMed] [Google Scholar]
- Peffer RC, Moggs JG, Pastoor T, Currie RA, Wright J, Milburn G, Waechter F, Rusyn I. Mouse liver effects of cyproconazole, a triazole fungicide: role of the constitutive androstane receptor. Toxicol. Sci. 2007;99:315–325. doi: 10.1093/toxsci/kfm154. [DOI] [PubMed] [Google Scholar]
- Phillips JM, Goodman JI. Identification of genes that may play critical roles in phenobarbital (PB)-induced liver tumorigenesis due to altered DNA methylation. Toxicol. Sci. 2008;104:86–99. doi: 10.1093/toxsci/kfn063. [DOI] [PubMed] [Google Scholar]
- Phillips JM, Goodman JI. Multiple genes exhibit phenobarbital (PB)-induced constitutive active/androstane receptor (CAR)-mediated DNA methylation changes during liver tumorigenesis and in liver tumors. Toxicol. Sci. doi: 10.1093/toxsci/kfp031. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Yamamoto Y, Negishi M, Maronpot RR, Goodman JI. Orphan nuclear receptor constitutive active/androstane receptor-mediated alterations in DNA methylation during phenobarbital promotion of liver tumorigenesis. Toxicol. Sci. 2007;96:72–82. doi: 10.1093/toxsci/kfl188. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;26:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: Modeling human cancer in mice. Nat. Rev. Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- Ray JS, Harbison ML, McClain RM, Goodman JI. Alterations in the methylation status and expression of the raf oncogene in phenobarbital-induced and spontaneous B6C3F1 mouse liver tumors. Mol. Carcinog. 1994;9:155–166. doi: 10.1002/mc.2940090307. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Schwarz M, Wanke I, Wulbrand U, Moennikes O, Buchmann A. Role of connexin32 and β-catenin in tumor promotion in mouse liver. Toxicol. Pathol. 2003;31:99–102. doi: 10.1080/01926230390173932. [DOI] [PubMed] [Google Scholar]
- Stahl S, Ittrich C, Marx-Stoelting P, Köhle C, Ott T, Buchmann A, Schwarz M. Effect of the tumor promoter phenobarbital on the pattern of global gene expression in liver of connexin32-wild-type and connexin32-deficient mice. Int. J. Cancer. 2005;115:861–869. doi: 10.1002/ijc.20815. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Moore R, Sugatani J, Matsumura Y, Negishi M. PPP1R16A, the membrane subunit of protein phosphatase 1β, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol. Pharmacol. 2008;73:1113–1121. doi: 10.1124/mol.107.042960. [DOI] [PubMed] [Google Scholar]
- Sugatani J, Kojima H, Ueda A, Kakizaki S, Yoshinari K, Gong QH, Owens IS, Negishi M, Sueyoshi T. The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology. 2001;33:1232–1238. doi: 10.1053/jhep.2001.24172. [DOI] [PubMed] [Google Scholar]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Vorce RL, Goodman JI. Hypomethylation of ras oncogenes in chemically induced and spontaneous B6C3F1 mouse liver tumors. Toxicol. Environ. Health. 1991;34:367–384. doi: 10.1080/15287399109531574. [DOI] [PubMed] [Google Scholar]
- Warner KA, Fernstrom MJ, Ruch RJ. Inhibition of mouse hepatocyte gap junctional intercellular communication by phenobarbital correlates with strain-specific hepatocarcinogenesis. Toxicol. Sci. 2003;71:190–197. doi: 10.1093/toxsci/71.2.190. [DOI] [PubMed] [Google Scholar]
- Watson RE, Goodman JI. Effects of phenobarbital on DNA methylation in GC-rich regions of hepatic DNA from mice that exhibit different levels of susceptibility to liver tumorigenesis. Toxicol. Sci. 2002;68:51–58. doi: 10.1093/toxsci/68.1.51. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Whysner J, Ross PM, Williams GM. Phenobarbital mechanistic data and risk assessment: Enzyme induction, enhanced cell proliferation, and tumor promotion. Pharmacol. Ther. 1996;71:53–91. doi: 10.1016/0163-7258(96)00067-8. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kawamoto T, Negishi M. The role of the nuclear receptor CAR as a coordinate regulator of hepatic gene expression in defense against chemical toxicity. Arch. Biochem. Biophys. 2003;409:207–211. doi: 10.1016/s0003-9861(02)00456-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Weinberg RA. Exploring a new twist on tumor metastasis. Cancer Res. 2006;66:4549–4552. doi: 10.1158/0008-5472.CAN-05-3850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.