Abstract
Tuberous sclerosis complex (TSC) is an autosomal-dominant disease that is caused by mutations in either the TSC1 or TSC2 gene. Smooth muscle-like cells (ASMs) were isolated from an angiomyolipoma of a patient with TSC. These cells lacked tuberin, were labeled by both HMB45 and CD44v6 antibodies, and had constitutive S6 phosphorylation. The cells bear a germline TSC2 intron 8-exon 9 junction mutation, but DNA analysis and polymerase chain reaction amplification failed to demonstrate loss of heterozygosity. Testing for an epigenetic alteration, we detected methylation of the TSC2 promoter. Its biological relevance was confirmed by tuberin expression and a reduction in HMB45 labeling and S6 constitutive phosphorylation after exposure to the chromatin-remodeling agents, trichostatin A and 5-azacytidine. These cells were named TSC2−/meth ASMs. Their proliferation required epidermal growth factor in the medium as previously described for TSC2−/− ASMs. Blockade of epidermal growth factor with monoclonal antibodies caused the death of TSC2−/meth ASMs. In addition, rapamycin effectively blocked the proliferation of these cells. Our data show for the first time that methylation of the TSC2 promoter might cause a complete loss of tuberin in TSC2 cells, and that the pathogenesis of angiomyolipomas might also originate from epigenetic defects in smooth muscle cells. Additionally, the effect of chromatin-remodeling agents in these cells suggests a further avenue for the treatment of TSC as well as lymphangioleiomyomatosis.
Tuberous sclerosis complex (TSC) is an autosomal-dominant disease characterized by hamartomas, in a wide array of tissues and organs, such as brain, kidney, skin, heart, and lungs.1 Abdominal angiomyolipomas are often present in TSC patients; they may cause life-threatening hemorrhages and in such conditions their surgical resection is required.2 The tumor suppressor genes, TSC1 and TSC2, are associated with the development of TSC, and mutations in either gene are responsible for familial and sporadic forms of the disease.1 The TSC2 gene is located on chromosome 16p13 whereas TSC1 on chromosome 9q34.3,4 Hamartin, the TSC1 gene product, stabilizes tuberin, the TSC2 gene product, through binding with it, thereby preventing tuberin from ubiquitination and degradation.5 Tuberin acts as a GTPase-activating protein to regulate Rheb function through the conversion of Rheb from the active GTP-bound form to the inactive GDP-bound form.6,7 Active Rheb activates mTOR, and the up-regulation of the TSC/mTOR signaling pathway leads to increased protein synthesis, cell proliferation, and ultimately to tumorigenesis.8
TSC occurs because of a germline mutation in either TSC1 or TSC2. In most hamartomas, TSC follows a second hit inactivating the wild-type allele. The loss of heterozygosity (LOH) in TSC1 or TSC2 has been documented in angiomyolipomas (AMLs), cardiac rabdomiomas, and lymphangioleiomyomatosis (LAM) cells, but it has only rarely been found in cerebral cortical tubers and skin lesions.9,10,11 Therefore, it is not clear whether inactivation of both alleles is the necessary step for hamartoma pathogenesis. Various explanations have been raised to define the inability to find a second somatic event in TSC lesions, and the failure to demonstrate such events has been attributed to either different genetic and epigenetic deficits in TSC genes or cell heterogeneity in TSC hamartomas.12,13 DNA methylation is an epigenetic change that induces chromatin modifications and repression of transcription via a methyl CpG binding protein MeCP2, and recruitment of a Sin3A/HDAC co-repressor complex.14,15 Twenty-four hamartomas from 10 patients were analyzed by Niida and colleagues11 for second-hit mutations by promoter methylation of TSC2, but no evidence of such inactivation of the second allele was detected.
Here we report the isolation and characterization of a homogenous population of smooth muscle-like cells from AML cells (ASM cells) of a TSC2 patient. The ASM cells were mostly positive for HMB4516 and CD44v6,17 that are markers of TSC and LAM cells, and showed a germline TSC2 intron 8-exon 9 junction mutation with no LOH. However, tuberin was undetectable by immunochemistry and Western blotting. We found that these cells were methylated in the TSC2 promoter, and the involvement of methylation in the inhibition of TSC2 gene was confirmed by the cellular expression of tuberin after exposure to the chromatin remodeling agent, trichostatin A. Thus, ASM cells were named TSC2−/meth ASM cells. The proliferative, morphological, and biochemical characteristics of TSC2−/meth ASM cells were very similar to TSC2−/− smooth muscle cells with LOH that we previously isolated from an AML of a female TSC2 patient (TSC2−/− ASM cells).18,19 The growth of TSC2−/meth ASM cells requires the addition of epidermal growth factor (EGF) to the culture medium, whereas the exposure to specific monoclonal antibody raised against EGFR causes the blockade of proliferation and their death. Our data show for the first time that the methylation of the TSC2 promoter might cause loss of tuberin in TSC2 cells, and that such epigenetic alteration of smooth muscle cell function may underlie their abnormal growth and likely lead to AML development.
Materials and Methods
Establishment of the Angiomyolipoma Culture
The renal angiomyolipoma sample was obtained during total nephrectomy from a 36-year-old man with a history of TSC who had given his informed consent according to the Declaration of Helsinki. The cells were obtained as shown in Lesma and colleagues.18 Briefly, the tumor tissue was manually dissociated using collagenase type II (Sigma, St. Louis, MO) by means of repetitive pipetting. The collagenase was neutralized with a serum-containing medium (50:50 mixture of Dulbecco’s Eagle’s medium/Ham F12; Euroclone, Paignton, UK) supplemented with hydrocortisone (2 × 10−7mol/L) (Sigma-Aldrich, St. Louis, MO), EGF (10 ng/ml) (Sigma-Aldrich), sodium selenite (5 × 10−8 mol/L) (Sigma-Aldrich), insulin (25 μg/ml) (Sigma-Aldrich), transferrin (10 μg/ml) (Sigma-Aldrich), ferrous sulfate (1.6 × 10−6 mol/L) (Sigma-Aldrich), and 15% fetal bovine serum (Euroclone) as indicated by Arbiser and colleagues.20 Vascular smooth muscle-like cells (VSMCs) and A549 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum 10%.
Cell Immunofluorescence Microscopy
The cells were cultured on glass slides, permeabilized with Cytoskelfix (Cytoskeleton, Denver, CO) and dried in air. The primary antibody against α-actin (1:100, Sigma-Aldrich), vimentin (1:70; Santa Cruz Biotechnology, Santa Cruz, CA), S100 (1:8000; DAKO, Carpinteria, CA) keratin 8/18 (1:100; New Marker, Fremont, CA), HMB45 (1:100, DAKO), CD44v6 (1:100; Invitrogen, Carlsbad, CA), hamartin (1:100, Santa Cruz Biotechnology), tuberin (C-20) (1:100, Santa Cruz Biotechnology) were applied overnight at 4°C. The samples were incubated for 3 hours at room temperature with fluorescein isothiocyanate-conjugated anti-mouse antibody (Alexa, Eugene, OR) for α-actin, HMB45, and keratin 8/18, with fluorescein isothiocyanate-conjugated anti-goat antibody (Alexa) for vimentin and hamartin, with rhodamine-conjugated anti-rabbit antibody (Alexa) for tuberin.
HMB45 positivity was evaluated by immunofluorescence. Cells were incubated for 72 hours with trichostatin (3.3 mmol/L). HMB45 labeling was defined as strong (+), and negative (−). Quantitation of HMB45 intensity was achieved with a confocal microscopy (TCS-SP2; Leica, Wetzlar, Germany) and analyzed by Leica confocal software using the profile through stack/series methods as follows: 30 equal polygonal areas covering the cells were randomly selected on each image. The fluorescence intensity values of the selected cells were quantified and the mean and the SD of the data were calculated by GraphPad software (La Jolla, CA). The level of statistical significance was determined by Student’s t-test.
Western Blotting
The cells were lysed in lysis buffer (5 mmol/L ethylenediaminetetraacetic acid, 100 mmol/L deoxycholic acid, 3% sodium dodecyl sulfate), boiled, electrophoretically run on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and transferred to nitrocellulose membranes (Amersham, Arlington Heights, IL). After being blocked at room temperature for 3 hours with 5% dry milk (Merck, Darmstadt, Germany), the membranes were incubated overnight at 4°C with antibodies against tuberin, phospho-tuberin (Thr1462) (1:1000; Cell Signaling, Beverly, MA), hamartin (a gift from Dr. Mark Nellist and Dr. Dicky Halley, Erasmus University, Rotterdam, The Netherlands), phospho-Akt (Ser473) (1:1000, Cell Signaling), phospho-S6 ribosomal protein (Ser235/236) (1:1000, Cell Signaling), S6 ribosomal protein (1:1000, Cell Signaling), phospho-extracellular signal-regulated kinase (Erk) (Thr202/Tyr204) (1:1000, Cell Signaling), Erk (1:1000; Cell Signaling), EGF receptor (1:200; Santa Cruz Biotechnology), β-actin (1:10,000, Sigma-Aldrich), p53 (1:100, Santa Cruz Biotechnology), p21 (1:1000, Cell Signaling). The membranes were washed and incubated for 1 hour with anti-rabbit antibody (1:10,000; Chemicon, Temecula, CA). The reaction was revealed using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL). Densitometric analysis was performed by Kodak MJ project program (Eastman-Kodak, Rochester, NY). Data were expressed as optical density.
Mutation Study
The DNAs were extracted from peripheral blood, angiomyolipoma, and isolated cultured cells using the Wizard Genomic DNA purification kit (Promega, Madison, WI). All of the exons and intron-exons junctions of TSC1 and TSC2 from the genomic DNAs were amplified by means of standard polymerase chain reaction (PCR) and described primers.21 The sequencing reactions were performed as previously reported by Lesma and colleagues18 and repeated at least twice.
LOH Analysis
The panel of microsatellite markers near the TSC2 locus on chromosome 16p13.3 consisted of D16S525, D16S3024, D16S3394, D16S291, D16S664, and D16S663. The 5′ sense primers were labeled with 6-FAM fluorescent dyes (M-Medical, Cornaredo, Italy). The primer sequences were obtained from the Genome Database (http://www.ncbi.nlm.nih.gov/). Microsatellite orientation and position were obtained from UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). LOH was analyzed as previously described in Lesma and colleagues.18
Methylation Assays
The methylation status of the TSC2 and ZFX promoter regions was analyzed by PCR using as template the genomic DNA previously digested with the methylation-sensitive enzyme HpaII and with its isoschizomer MspI, insensitive to methylation (New England Biolabs, Beverly, MA). The primers used for amplifications spanned several methylation-sensitive restriction sites (seven for TSC2 gene) in the 5′ of the tested genes. The UCSC Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) and the CpG island searcher (http://cpgislands.usc.edu/) were consulted to identify TSC2 promoter region and CpG island.
The primers used to TSC2 promoter analysis were sense 5′-atgccgctaccggaagtgc-3′ (chr16: 2037,961-2037,979, UCSC Genome Browser) and antisense 5′-tggtctagagagctctctact-3′ (chr16:2038,391-2038,371, UCSC Genome Browser). After denaturation at 94°C for 5 minutes, 35 cycles of PCR were done; each cycle consisted of 45 seconds at 95°C, 45 seconds at 54°C, and 45 seconds at 72°C. ZFX primers and methods are fully described in Gilbert and Sharp.22
To quantify the methylation levels of TSC2 promoter, pyrosequencing technology was used to analyze the same region studied by methylation-sensitive digestion assay. The bisulphite conversion of genomic DNA (1 μg) was obtained using EZ DNA methylation kit (Zymo Research, Orange, CA). After bisulphite treatment, PCR was performed in a final volume of 50 μl with 2.5 U of Promega Go-Taq Hot Start Polymerase (Promega). The primer sequences for modified sequences are: forward primer 5′-ttygttagagggyggtatagaat-3′ (chr16: 2037,899-2037,921, UCSC Genome Browser) and biotinylated reverse primer 5′-acactacraaatccrcctctc-3′ (chr16: 2038,141-2038,121, UCSC Genome Browser). The PCR conditions were: step 1: 95°C, 2 minutes; step 2: 95°C, 30 seconds; step 3: 61.2°C decrease 0.5°C per cycle, 30 seconds; step 4: 72°C, 30 seconds; step 5: repeat steps 2 to 4 14 more times; step 6: 95°C, 30 seconds; step 7: 54°C, 30 seconds; step 8: 72°C, 30 seconds; step 9: repeat steps 6 to 8 31 more times; step 10: 72°C, 5 minutes. Forty μl of PCR product were used for pyrosequencing assay using the sequencing primers 5′-atyggaagtgygggt-3′ (chr16: 2037,969-2037,983, UCSC Genome Browser).
Pyrosequencing reactions were performed in the PSQ HS 96 system (Biotage, Uppsala, Sweden) by using Pyro Gold reagent kits (Biotage). Methylation was quantified using Pyro Q-CpG Software (Biotage), which calculates the ratio of converted Cs (Ts) to unconverted Cs at each CpG and expresses this as a percentage of methylation. To assess the methylation pattern in normal condition we analyzed different normal DNA from peripheral blood lymphocytes.
Evaluation of Cell Growth and Proliferation
The proliferation growth factor dependence of the smooth muscle-like cells and VSMCs were assayed in the presence or absence of EGF (10 ng/ml) or by replacing EGF with insulin-like growth factor 1 (IGF-1) (50 ng/ml), by counting the cells after 4, 7, 17, 21, and 25 days of culture in a Neubauer chamber. Anti-EGFR antibody (clone 225; Calbiochem, Darmstadt, Germany), and IGF-1R (clone αIR3, Calbiochem) were added at a concentration of 5 μg/ml to the complete medium, and cell proliferations were evaluated after 4, 7, 17, 21, and 25 days of culture. Each data point of the proliferation experiments is the mean of four independent experiments.
Cell Treatments
Trichostatin A (Sigma-Aldrich) was incubated for 72 hours at the concentration of 3.3 mmol/L. 5-Azacytidine (Sigma-Aldrich) was incubated for 96 hours at the concentration of 1 or 10 μmol/L. In the Western blotting experiments to study S6 and Erk phosphorylation, cells were incubated with rapamycin (1 ng/ml) or anti-EGFR antibody (5 μg/ml) for 24 or 48 hours. Cells were incubated for 2 hours with IGF-1 (50 ng/ml) with or without LY294002 (20 μmol/L) (Sigma).
The action of rapamycin (Rapamune-Sirulimus; Wyeth Europa, Maidenhead, UK) was evaluated by adding 1 ng/ml and 5 ng/ml to the TSC2−/meth cells at plating time and 3 hours after plating, and measuring cell proliferation after 5, 8, and 10 days or evaluating the apoptotic effect by terminal dUTP nick-end labeling (TUNEL) analysis after a 10-day incubation. For TUNEL analysis VSMCs were incubated with staurosporin (100 mmol/L) for 1 hour 30 minutes.
TUNEL Analysis
Apoptotic cells were examined by TUNEL analysis using an in situ cell death DeadEnd colorimetric TUNEL system (Promega) following the manufacturer’s instruction. Briefly, cells were fixed for 25 minutes in 4% paraformaldehyde, washed twice in PBS for 5 minutes, permeabilized in 0.2% Triton X-100 for 5 minutes, and washed twice in PBS for 5 minutes. Slides were incubated with rTdT reaction mix for 1 hour at 37°C, then endogenous peroxidases were blocked with 0.3% hydrogen peroxide for 5 minutes at room temperature. After incubation with streptavidin horseradish peroxidase, apoptotic cells were localized using 3′,3′-diaminobenzidine tetrahydrochloride. The number of TUNEL-positive cells was counted and the data presented are mean of the percentage of TUNEL-positive cells.
Statistical Analysis
The data are expressed as mean values ± SEM, and were statistically analyzed using Student’s t-test; significance is indicated for P values of *<0.05, **<0.01, and ***<0.001.
Results
Cellular Characterization
From the angiomyolipoma of a male TSC patient, a homogenous cellular population was isolated and cultured in monolayers. The characteristic elongated shape strongly resembled TSC2 smooth muscle (TSC2−/− ASM) cells isolated from an angiomyolipoma of a female TSC2 patient and previously described by our group.18 The majority of these isolated cells were strongly labeled by α-actin antibody, the marker for smooth muscle cells (Figure 1), whereas the lack of detection of vimentin, keratin 8-18, and S100 suggested that fibroblasts, epithelial-like cells, or adipocytes were absent from this cellular population (Figure 1). The angiomyolipoma-derived smooth muscle (ASM) cells were also labeled by the typical markers of TSC and LAM cells, HMB45 antibody,16 and CD44v6 antibody17 (Figure 1).
Figure 1.
Immunocytochemical characterization of TSC angiomyolipoma-derived cells. Characterization was performed using a panel of antibodies related to the AML cellular constituents. TSC cells were positively labeled by α-actin antibody, a marker of smooth muscle cells. Cells were negative for S100, a marker of lipid-containing cells; vimentin, a marker of fibroblasts; and keratin 8-18, a marker for epithelial-like cells. Antibodies to HMB45 and CD44v6, markers of TSC and LAM cells, positively immunolabeled these TSC cells. Scale bars = 50 μm.
Mutation Analysis
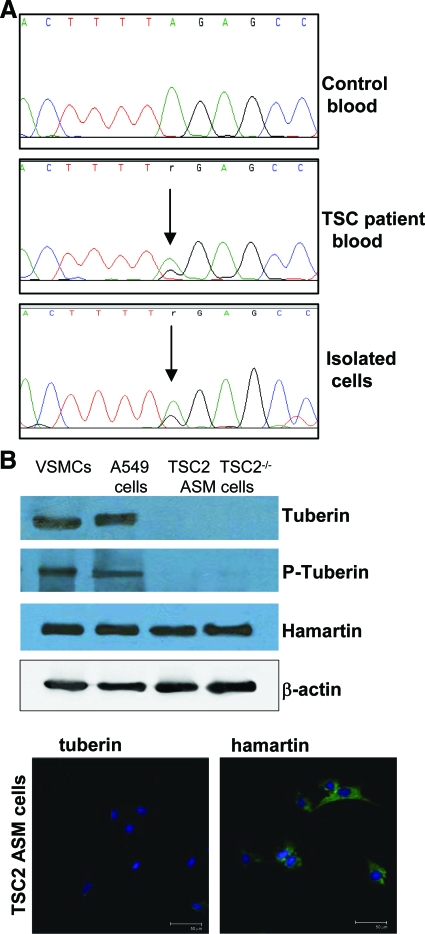
A germline TSC2 intron 8-exon 9 junction heterozygous mutation (c.867-2A>G) was detected by DNA sequencing in patient blood and angiomyolipoma (data not shown) and in angiomyolipoma-derived ASM cells (Figure 2A). The LOH of these ASM cells was tested by means of PCR amplification, using a panel of microsatellite markers near the TSC2 locus on chromosome 16p13.3, but we failed to detect LOH (data not shown). On the other hand, these newly isolated ASM cells lacked tuberin as immunofluorescence and Western blotting failed to reveal any positive reactivity to specific antibodies. This negative result can be compared with what observed in TSC2−/− ASM cells, whereas VSMCs and A549 cells, used as control, expressed tuberin, as expected (Figure 2B). Differently, hamartin was present at comparable levels in all tested cells and β-actin was evaluated as protein control (Figure 2B).
Figure 2.
Genetic and biochemical analysis of TSC angiomyolipoma-derived cells. A: The shown mutation was determined by direct sequence analysis of blood from a control and the TSC patient, and the isolated cells. The mutation site is indicated by a vertical arrow. Cell sequencing revealed the mutation in TSC2 intron 8-exon 9 junction mutation (c.867-2A>G). B: The expression of tuberin and hamartin in the ASM cells was determined by Western blotting and immunofluorescence. Tuberin was undetectable in the purified ASM cells and TSC2−/− ASM cells, whereas it was expressed in VSMCs and A549 cells. Hamartin was expressed in all cell types. The blots are representative of four separate experiments. The immunocytochemical staining is referred only to the purified ASM cells. Scale bars = 50 μm.
Methylation of the TSC2 Promoter Region
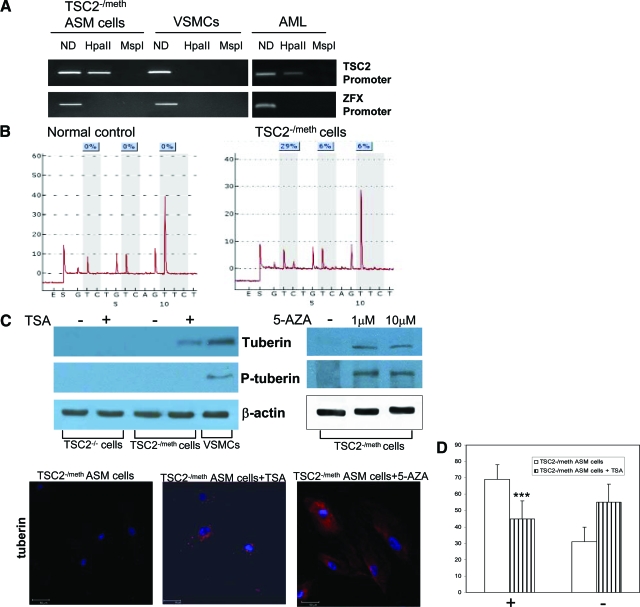
The absence of LOH and the lack of tuberin expression in ASM cells led us into assessing whether the Knudson second hits could be epigenetic. We analyzed the DNA methylation of the CpG island within the TSC2 promoter region, using methylation-sensitive digestion with HpaII restriction enzyme. Methylation (presence of PCR amplification after HpaII digestion) of the promoter in ASM cells was found while normal VSMCs cells were unmethylated (absence of PCR amplification after HpaII digestion) (Figure 3A).
Figure 3.
DNA methylation in TSC2 promoter and effect of chromatin remodeling agents, trichostatin A (TSA) and 5-azacytidine (5-AZA). A: The lack of tuberin in TSC2 ASM cells was evaluated by DNA methylation of the CpG island within the TSC2 and ZFX promoter regions, using methylation-sensitive digestion with HpaII restriction enzyme and its isoschizomer MspI. The determination was performed three times. Methylation occurs at the promoter site in the TSC2 ASM cells (left) and AML tissue (right). B: Pyrograms indicating the percentage of methylation (light blue boxes) of three explicative CpGs within TSC2 promoter region of the normal control (left) and TSC2−/meth ASM cells (right). These CpGs are a part of the methylation sites analyzed by methylation-sensitive digestion assay. C: After incubation with TSA (3.3 μmol/L) for 72 hours (left) or 5-azacytidine (5-AZA) (1 and 10 μmol/L) for 96 hours (right), tuberin and phospho-tuberin (Thr1462) expressions were promoted in TSC2−/meth ASM cells as evaluated by Western blotting. The phosphorylated form was detectable only after 5-AZA incubation. As expected TSA did not have any effect on tuberin expression in TSC2−/− ASM cells. Experiments were done four times. The expression of tuberin induced by treatments with TSA or 5-AZA was also confirmed by immunocytochemical staining. D: Effect of trichostatin A on HMB45 labeling of TSC2−/methASM cells. The positive cells are indicated with +, and the negative with −. The exposure to trichostatin A reduced HMB45 labeling very significantly. Mean values ± SEM. Significant differences (***P < 0.001) versus control were evaluated by Student’s t-test. Scale bars = 50 μm.
To control the enzyme cleavage efficiency we chose two different approaches. In the first case, it was used the digestion with MspI, an isoschizomer of HpaII that, unlike HpaII, can cleave the sequence when the internal C of the restriction site CCGG is methylated. In the second case the cleavage efficiency was tested with the amplification of ZFX promoter, which is always unmethylated. As expected, we found a complete DNA digestion in both controls, and this resulted in no PCR amplification in all samples (Figure 3A). In addition we observed the TSC2 promoter methylation in the original AML tissue, although there was a reduced extent probably because of the heterogeneity of AML (Figure 3A). To confirm the above reported qualitative methylation data we performed the pyrosequencing quantitative assay on the same promoter regions analyzed with methylation-sensitive digestion. This method allows us to reveal quantitatively the methylation of all CpGs position. In normal samples TSC2 resulted demethylated in all CpGs analyzed (Figure 3B). In the TSC2−/meth ASM cells, consistently with our previous results, we found a promoter methylation pattern indicating an epigenetic silencing. The low methylation levels (29%, 6%, 6%) obtained with the pyrosequencing analysis were indicative of methylation of only one allele.
In the isolated ASM cells the epigenetic silencing of TSC2 was confirmed by transcriptional reactivation with consequent tuberin expression after treatment with the histone deacetylase inhibitor trichostatin A for 72 hours or with the DNA methylase inhibitor 5-azacytidine for 96 hours (Figure 3C). Trichostatin A and 5-azacytidine are chromatin-remodeling agents able to reactivate genes epigenetically silenced.23,24,25 After the demonstration of the DNA methylation of the CpG island, the angiomyolipoma-derived ASM cells were named TSC2−/meth ASM cells, and thereafter we shall use this name. After 5-azacytidine exposure we monitored for a week the proliferation of TSC2−/meth cells; the rate of proliferation was comparable with control. With trichostatin A the overproliferation was eliminated (data not shown).
To evaluate the effects of trichostatin A-induced tuberin expression on the phenotype of TSC2−/meth ASM cells, gp100 expression was quantified by exposure to HMB45 antibody, a marker of TSC and LAM cells.16,18 After 72 hours of trichostatin A exposure, the labeling was greatly reduced. HMB45 reactivity was quantified as positive (+) or negative (−). Normally, ∼70% of untreated TSC2−/−ASM cells were strongly labeled by HMB45 (Figure 3C), whereas after incubation with trichostatin A, HMB45-labeled cells were reduced to 45%, and the negative ones increased from 30 to 55% (Figure 3D).
Effect of Trichostatin A on TSC2−/methASM Cell Phenotype
To further investigate the effects of trichostatin A in TSC2−/meth ASM cells, we evaluated S6 and Erk phosphorylation. After a 72-hour incubation with trichostatin A there was a reduction of S6 and Erk phosphorylation in TSC2−/meth ASM cells (Figure 4). This was not observed in TSC2−/− ASM cells (Figure 4). The expression of S6 and Erk were not changed by trichostatin A in both cell lines (Figure 4). Thus the induced expression of tuberin by trichostatin A exposure correlated with down-regulation of the constitutive phosphorylation of S6. As expected, trichostatin A led to a slowing proliferation rate even within these 72 hours (data not shown).
Figure 4.
Effect of trichostatin A (TSA) (3.3 μmol/L) incubation (72 hours) on S6 and Erk phosphorylation in TSC2−/− and TSC2−/meth ASM cells. In TSC2−/meth ASM cells, trichostatin A markedly reduced the constitutive phosphorylation of S6 (Ser235/236) and also Erk (Thr202/Tyr204) phosphorylation. No effect by trichostatin A was observed in TSC2−/− ASM cells. The blots are representative of three independent experiments. These data were confirmed by the densitometric analysis. Mean values ± SEM. Significant differences (***P < 0.001) versus control were evaluated by Student’s t-test.
Role of PI3K Pathway in TSC2−/meth ASM Cells
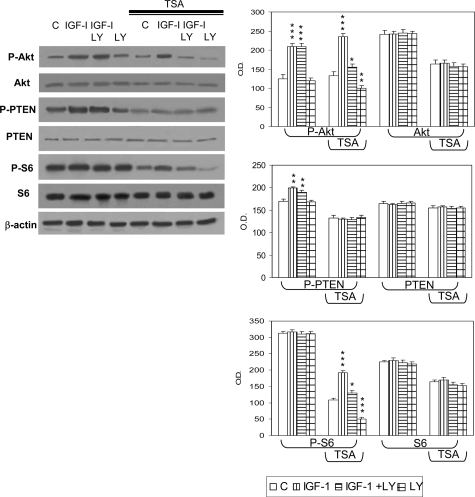
We had previously shown that the specific inhibitor LY294002 was unable to inhibit PI3K in TSC2−/− ASM cells.26 Here we report that a similar phenomenon was also present in TSC2−/meth ASM cells (Figure 5). Addition of IGF-1 increased Akt and PTEN phosphorylation without affecting constitutive S6 phosphorylation and the co-incubation with LY 294002 failed to modify the extent of both basal and IGF-1-mediated phosphorylations (Figure 5). After incubation with trichostatin A for 72 hours, the LY294002 addition was able of reduce the extent of both basal and IGF-1-stimulated Akt phosphorylation in TSC2−/meth ASM cells (Figure 5). In this latter condition, also the IGF-1-promoted increase of S6 phosphorylation was inhibited by LY294002 (Figure 5).
Figure 5.
Effect of trichostatin A (TSA) on the PI3K pathway. TSC2−/meth ASM cells were incubated with or without trichostatin A (3.3 μmol/L) for 72 hours and then with IGF-1 (50 ng/ml) and LY294002 (LY) (20 μmol/L) for 2 hours. IGF-1 exposure of TSC2−/methASM cells induced Akt (Ser473) and PTEN (Ser380/Thr382/Thr383) phosphorylation without affecting S6 (Ser235/236) phosphorylation. The co-incubation with IGF-1 and PI3K inhibitor, LY294002, did not modify the extent of phosphorylation of Akt, PTEN, and S6. The expression of Akt and S6 was unaffected by any treatment. After trichostatin A exposure, LY290042 was capable of antagonizing IGF-1-mediated and basal phosphorylation of both Akt and S6 whereas the phosphorylation of PTEN was unaffected as reported for TSC2−/− ASM cells after retroviral transfection of the TSC2 gene.26 The expression of Akt and S6 proteins was unaltered. The blots are representative of four independent experiments. These data were confirmed by the densitometric analysis (graphs at the right). Mean values ± SEM. Significant differences (*P < 0.05; **P < 0.01; ***P < 0.001) versus control were evaluated by Student’s t-test.
Regulation of Growth of TSC2−/meth ASM Cells
We have detected EGFR by Western blotting analysis of TSC2−/meth ASM cells, and the level of expression is comparable with that of TSC2−/− ASM cells, vascular smooth muscle cells (VSMCs) and A549 cells (Figure 6A, inset). Also, the proliferation of TSC2−/meth ASM cells required EGF addition to the culture medium and the substitution with another growth factor, such as IGF-1, could not substitute for EGF-proliferative action (Figure 6A). Moreover, the exposure to anti-EGFR and anti-IGF-1R antibodies reduced the proliferation and caused the progressive loss of TSC2−/meth ASM cells. The rate of proliferation of the TSC2−/meth ASM cells in presence or absence of EGF and the ability of anti-EGFR or anti-IGF-1R antibodies to kill them are comparable with those observed for TSC2−/− ASM cells (Figure 6B).
Figure 6.
Evaluation of EGF and IGF-1 role in TSC2−/meth ASM proliferation. Cells were counted after 4, 7, 17, 21, and 25 days of culture. A: The growth and proliferation of TSC2−/meth ASM cells required the addition of EGF (10 ng/ml) in the culture medium; IGF-1 (50 ng/ml) failed to promote any increase in cell number. The treatment with either anti-EGFR or anti-IGF-1R antibodies limited the growth rate in the early days of drug exposure and then started to die. Experiments were done four times. Inset: Expression of EGF receptor (EGFR). B: Comparison of the proliferation of the TSC2−/− (full line) and TSC2−/meth (dashed line) ASM cells in presence or absence of EGF, and with anti-EGFR or anti-IGF-1R antibodies. The beginning of the treatment was considered 100% of the percentage of the cells. The growth rate of the two cell lines was similar. Experiments were done three times each. Mean values ± SEM. Significant differences (***P < 0.001) versus control were evaluated by Student’s t-test.
Rapamycin was applied at 1 and 5 ng/ml at time of plating or 3 hours later. Rapamycin significantly reduced the rate of cell proliferation either when applied at plating time or 3 hours after plating causing a significant loss of cells at the concentration of 5 ng/ml (Table 1). The percentage of attached cells was 80 ± 1,6% and 96 ± 0.7% at 3 and 16 hours after plating, respectively. The effect of the delayed addition is different to what was observed in TSC2−/− ASM cells, which were affected by rapamycin only when the drug was applied at plating time.26
Table 1.
Cell Proliferation Rate and Rapamycin Effect
| Days | 5 | 8 | 10 |
|---|---|---|---|
| TSC2−/meth ASM cells | 180% | 230% | 286% |
| TSC2−/meth ASM cells with rapamycin 1 ng/ml at plating time | 143% | 143% | 125% |
| TSC2−/meth ASM cells with rapamycin 1 ng/ml 3 hours after plating | 152% | 160% | 168% |
| TSC2−/meth ASM cells with rapamycin 5 ng/ml at plating time | 89% | 89% | 45% |
| TSC2−/meth ASM cells with rapamycin 5 ng/ml 3 hours after plating | 116% | 125% | 45% |
Percentage increase in cell number. We plated 2.8 × 104 cells and the number of cells was evaluated at the indicated time.
Effect of Anti-EGFR Antibody and Rapamycin on S6 and Erk Phosphorylation in TSC2−/meth ASM Cells
The cellular effects of anti-EGFR antibody on TSC2−/meth ASM cell proliferation were also evaluated biochemically. It is well known that TSC protein abnormalities may lead to dysregulation of mTOR activity with increased p70 S6 kinase activity, and, as expected, TSC2−/meth ASM cells presented constitutive phosphorylation of S6. After 24 or 48 hours of incubation with rapamycin or anti-EGFR antibody the extent of such phosphorylation was reduced without changes in S6 expression (Figure 7). Because the Erk signaling pathway is activated by EGF and is a major contributor to cellular growth, we have studied the phosphorylation of this kinase after exposure to anti-EGFR antibody or rapamycin. The extent of Erk phosphorylation was reduced by anti-EGFR, mainly after a 48-hour incubation while the Erk levels were unchanged (Figure 7). The effect of rapamycin on Erk phosphorylation was, however, rather minor (Figure 7). The rapamycin-mediated reduced ability of TSC2−/meth cells to proliferate was not accompanied by enhancement of apoptosis. Rapamycin was added to the medium at 1 or 5 ng/ml at plating time or 3 hours after plating for 10 days; this did not change the expression of p21 and p53 (Figure 8); moreover, the number of TUNEL-positive cells remained comparable with control untreated cells (Table 2). Differentially, 57.3% of VSMCs, used as control, underwent apoptosis after exposure to 100 mmol/L staurosporin for 1 hour and 30 minutes (Table 2).
Figure 7.
Effect of rapamycin (1 ng/ml) and anti-EGFR antibody (5 μg/ml) on phosphorylation of S6 (Ser235/236) and ERK in TSC2−/meth ASM cells. The blots are representative of three experiments. Incubation with rapamycin inhibited constitutive S6 phosphorylation within 24 hours, whereas inhibition of Erk (Thr202/Tyr204) phosphorylation required 48 hours. The effect of anti-EGFR on Erk phosphorylation was superior to that of rapamycin. These data were confirmed by the densitometric analysis. Mean values ± SEM. Significant differences (***P < 0.001) versus control were evaluated by Student’s t-test.
Figure 8.
Exposure to rapamycin (1 and 5 ng/ml) added at plating time or 3 hours after plating failed to modify the expression of p21 and p53. The levels of β-actin were evaluated as protein control.
Table 2.
Effect of Rapamycin on Apoptosis
| Percent of TUNEL-positive cells | |
|---|---|
| TSC2−/meth ASM cells | 4.5 |
| TSC2−/meth ASM cells + 1 ng/ml rapamycin at plating time | 6.1 |
| TSC2−/meth ASM cells + 5 ng/ml rapamycin at plating time | 6.9 |
| TSC2−/meth ASM cells + 1 ng/ml rapamycin 3 hours after plating | 4.4 |
| TSC2−/meth ASM cells + 5 ng/ml rapamycin 3 hours after plating | 5.2 |
| VSMCs | 5.1 |
| VSMCs + staurosporin | 57.3 |
Percentage of TUNEL-positive cells. Rapamycin was incubated for 10 days at the indicated conditions and TUNEL-positive and -negative cells were counted. VSMCs were incubated with 100 mmol/L staurosporin for 1 hour and 30 minutes.
Discussion
TSC is a tumor suppressor gene disorder associated with benign and malignant tumors, and angiomyolipomas are the most common renal tumors. They are composed of smooth muscle, fat, and vascular cells.27 Tuberin is the TSC2 gene product; it functions as a renal tumor suppressor gene and regulates cell growth and cell cycle progression.28 The lack of tuberin may lead to abnormal cell proliferation.29 LOH in TSC cells occurs in 60% of the angiomyolipomas of women with sporadic LAM.30 We have previously reported the isolation and the characterization of TSC2−/− ASM cells from an angiomyolipoma with a mutation in exon 18 consisting of a stop codon associated to LOH of TSC2 locus.18 The lack of tuberin appears to be strictly related to their EGF requirement for growth.18 In the current study we report that the lack of tuberin in TSC2 cells may also be caused by an epigenetic silencing such as the methylation of the TSC2 gene promoter.
Blood and ASM cells of a TSC2 patient bear the somatic mutation of TSC2 gene in intron8-exon9 junction (867-2AG), on the other hand the newly purified angiomyolipoma cells did not express tuberin and LOH was not detected. This inability to find a deletion of TSC2 locus in the newly isolated angiomyolipoma cells could have been explained by a number of different possibilities such as a second mutations, as shown for the PDK1 and PDK2 genes in autosomal-dominant polycystic kidney disease,31 or the methylations of the wild-type allele frequently observed in human cancers.32,33 In these TSC2 ASM cells we detected a methylation of the CpG island leading to the inhibition of tuberin expression. Thus the methylated purified ASM cells were named TSC2−/meth ASM cells.
DNA methylation at CpG dinucleotides is a common and important epigenetic mechanism of inactivation of tumor suppressor in cancers, and such a genetic modification is performed by DNA methyltransferase.34 Several human genes contain a CpG island within their promoter region, and unmethylated CpG islands are generally protected from methylation in normal cells.35 However, the de novo methylation of CpG islands that overlaps with promoter regions is a common feature in human cancer, and results in the loss of gene function, similar to other mechanisms such as deletion or point mutation.33,36 The methylation of the TSC2 promoter is biologically very relevant in the case of TSC2−/meth ASM cells because the functional effect on tuberin transcription is a complete blockade because of the present germline mutation of TSC2 intron 8-exon 9 junction on the other allele.
In a previous report no evidence of methylation within TSC2 promoter region of the wild-type allele was found in hamartomas obtained from TSC patients.11 Failure to demonstrate promoter methylation may be attributed to heterogeneity of tumor tissues whereas in the case of TSC2−/meth ASM cells we analyzed a pure population. On the other hand, there are several examples in cancer of functional modification of TSC gene by methylation. One is the clinical outcome of patients with breast cancer, that turns unfavorable, where the TSC promoter is methylated.37 Also the down-regulation of TSC gene function in oral squamous cell carcinoma can be ascribed to an epigenetic alteration by methylation of the TSC promoter.38
The role of promoter methylation of the wild-type allele, in the abnormal development of TSC2−/meth cells is confirmed by the normalizing effects of chromatin-remodeling agents, trichostatin A and 5-azacytidine. These agents promoted the expression of tuberin and down-regulated the constitutive phosphorylation of S6 and the labeling by HMB45 antibody. Histone deacetylase (HDAC) inhibitors, such as trichostatin A, are a new class of targeted anticancer agents, which are potent inducers of cellular differentiation, growth arrest, and cell death, and are effective in vitro and in vivo.39 By reversible acetylation, HDAC inhibitors modify the structure and function of histones and proteins in transcription factor complexes, which are involved in the regulation of gene expression. The inhibition of the HDAC could increase the remission rate of many kinds of solid tumors, obviously by cooperation with other anticancer drugs.40 It is thus, conceivable that, in cases such as this, the use of HDAC inhibitors may result in an important additional therapeutic approach to the treatments for TSC now in development.18,41
As described for TSC2−/− ASM cells,18 TSC2−/meth ASM cell growth required the addition of EGF in the growth medium, and the exposure to anti-EGFR antibody caused the blockade of cell proliferation and, then, death. The growth rate of the two types of TSC2 cells is similar, thus the lack of tuberin, resulted from two different genetic lesions, leads to EGF dependence in both cases. The EGFR signaling pathways regulate cell differentiation, proliferation, and migration and, in the case of TSC2 cells, might also be involved in the constitutive S6 phosphorylation. Anti-EGFR antibody has been shown to be efficacious in several types of cancer, such as colorectal and head and neck cancers42 and we suggest that might be also of therapeutic value in TSC.18
Rapamycin, a mTOR inhibitor, has been identified as a potential therapeutic agent for TSC and LAM43,44 and a recently reported clinical trial shows its potential efficacy in promoting the regression of TSC angiomyolipomas.41 The exposure to rapamycin down-regulated the constitutive S6 phosphorylation in TSC2−/meth ASM cells and it was capable of inhibiting cellular proliferation either when added at plating time or in a delayed application. In TSC2−/− ASM cells, rapamycin regulated proliferation only when added at plating time, and this is a clear pharmacological difference with TSC2−/meth ASM cells.18 The inhibitory effects of rapamycin on TSC2−/meth ASM cell proliferation did not require any activation of apoptosis.
In addition to the proliferative effects caused by the lack of tuberin, we observed that LY294002 failed to inhibit IGF-1-mediated Akt phosphorylation in TSC2−/meth ASM cells. When trichostatin A was added to the growth medium, the ability of LY294002 to inhibit IGF-1-mediated and basal Akt phosphorylation was apparent. A similar phenomenon was observed in TSC2−/− ASM cells and the introduction of the TSC2 gene restored the normal pharmacological activity of this drug.26 Thus, in both TSC2 cell types the lack of tuberin affects also the sensitivity to drugs.
In conclusion, this study provides the first evidence that the methylation of the TSC2 promoter as a second hit may cause the blockade of tuberin expression and full deployment of TSC2 cellular phenotype. The demethylation ability of an HDAC inhibitor, trichostatin A, and a DNA methylase inhibitor, 5-azacytidine, allowed the expression of tuberin and reverted the biochemical, pharmacological, and phenotypical characteristics. The growth of these cells requires EGF in the medium, and the blockade of EGFR leads to their death. These methylated cells, differently from TSC2−/−ASM cells18 are very sensitive to the cytostatic action of rapamycin.
Footnotes
Address reprint requests to Elena Lesma, Laboratory of Pharmacology, Dept. of Medicine, Surgery, and Dentistry, Via A. di Rudinì 8, 20142 Milano, Italy. E-mail: elena.lesma@unimi.it.
Supported by the Ministero Superiore della Sanita (IRCCS grant of rare diseases), the Italian Tuberous Sclerosis Association, and the Italian Lymphangioleiomyomatosis Association.
References
- Young J, Povey S. The genetic basis of tuberous sclerosis. Mol Med Today. 1998;4:313–319. doi: 10.1016/s1357-4310(98)01245-3. [DOI] [PubMed] [Google Scholar]
- Neumann HP, Schwarzkopf G, Henske EP. Renal angiomyolipomas, cysts, and cancer in tuberous sclerosis complex. Semin Pediatr Neurol. 1998;5:269–275. doi: 10.1016/s1071-9091(98)80005-3. [DOI] [PubMed] [Google Scholar]
- European Chromosome 16 Tuberous Sclerosis Consortium Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Jansen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JB, Ward S, Green AJ, Yates JR, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- Benvenuto G, Li S, Brown SJ, Braverman R, Vass WC, Cheadle JP, Halley DJ, Sampson JR, Wienecke R, DeClue JE. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Chan JA, Zhang H, Roberts PS, Jozwiak S, Wieslawa G, Lewin-Kowalik J, Kotulska K, Kwiatkowski DJ. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol. 2004;63:1236–1242. doi: 10.1093/jnen/63.12.1236. [DOI] [PubMed] [Google Scholar]
- Henske EP, Scheithauer BW, Short MP, Wollmann R, Nahmias J, Hornigold N, van Slegtenhorst M, Welsh CT, Kwiatkowski DJ. Allelic loss is frequent in tuberous sclerosis kidney lesions but rare in brain lesions. Am J Hum Genet. 1996;59:400–406. [PMC free article] [PubMed] [Google Scholar]
- Niida Y, Stemer-Rachamimow AO, Logrip M, Tapon D, Perez R, Kwiatkowski DJ, Sims K, MacCollin M, Louis DN, Ramesh V. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am J Hum Genet. 2001;69:493–503. doi: 10.1086/321972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KS, Hebert AA, Roach ES, Northrup H. Complete inactivation of the TSC2 gene leads to formation of hamartomas. Am J Hum Genet. 1999;65:1790–1795. doi: 10.1086/302648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle J, Reeve M, Sampson J, Kwiatkowski D. Molecular genetic advances in tuberous sclerosis complex. Hum Genet. 2000;107:97–104. doi: 10.1007/s004390000348. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hashemite N, Walker V, Kwiatkowski DJ. Estrogen enhances whereas tamoxifen retards development of Tsc mouse liver hemangioma: a tumor related to renal angiomyolipoma and pulmonary lymphangioleiomyomatosis. Cancer Res. 2005;65:2474–2481. doi: 10.1158/0008-5472.CAN-04-3840. [DOI] [PubMed] [Google Scholar]
- Pacheco-Rodriguez G, Steagall WK, Crooks DM, Stevens LA, Hashimoto H, Li S, Wang JA, Darling TN, Moss J. TSC2 loss in lymphangioleiomyomatosis cells correlated with expression of CD44v6, a molecular determinant of metastasis. Cancer Res. 2007;67:10573–10581. doi: 10.1158/0008-5472.CAN-07-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesma E, Grande V, Lesma E, Carelli S, Brancaccio D, Canevini MP, Alfano RM, Coggi G, Di Giulio AM, Gorio A. Isolation and growth of smooth muscle-like cells derived from tuberous sclerosis complex-2 human renal angiomyolipoma: EGF is the required growth factor. Am J Pathol. 2005;167:1093–1103. doi: 10.1016/S0002-9440(10)61198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli S, Lesma E, Paratore S, Grande V, Zadra G, Di Giulio AM, Gorio A. Survivin expression in tuberous sclerosis complex cells. Mol Med. 2007;13:166–177. doi: 10.2119/2006-00091.Carelli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbiser JL, Yeung R, Weiss SH, Arbiser ZK, Amin MB, Cohen C, Frank D, Mahajan S, Herron GS, Yang J, Onda H, Zhang HB, Bai X, Uhlmann E, Loehr A, Northrup H, Au P, Davis I, Fisher DE, Gutmann DH. The generation and characterization of a cell line derived from a sporadic renal angiomyolipoma. Am J Pathol. 2001;159:483–491. doi: 10.1016/S0002-9440(10)61720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Sampson JR, Hoogendoorn B, Cohen D, Cheadle JP. Application and evaluation of denaturing HPLC for molecular genetic analysis in tuberous sclerosis. Hum Genet. 2000;106:663–668. doi: 10.1007/s004390000316. [DOI] [PubMed] [Google Scholar]
- Gilbert SL, Sharp PA. Promoter-specific hypoacetylation of X-inactivated genes. Proc Natl Acad Sci USA. 1999;96:13825–13830. doi: 10.1073/pnas.96.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirchia SM, Ferguson AT, Sironi E, Subramanyan S, Orlandi R, Sukumar S, Sacchi N. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor β2 promoter in breast cancer cells. Oncogene. 2000;19:1556–1563. doi: 10.1038/sj.onc.1203456. [DOI] [PubMed] [Google Scholar]
- Sirchia SM, Ren M, Pili R, Sironi E, Somenzi G, Ghidoni R, Toma S, Nicolò G, Sacchi N. Endogenous reactivation of the RAR beta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res. 2002;62:2455–2461. [PubMed] [Google Scholar]
- Sirchia SM, Ramoscelli L, Grati FR, Barbera F, Cordini D, Rossella F, Porta G, Lesma E, Ruggeri A, Radice P, Simoni G, Miozzo M. Loss of the inactive X chromosome and replication of the active X in BRCA1 effective and wild-type breast cancer cells. Cancer Res. 2005;65:2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- Lesma E, Grande V, Ancona S, Carelli S, Di Giulio AM. Anti-EGFR antibody efficiently and specifically inhibits human TSC2-/- smooth muscle cell proliferation. Possible treatment options for TSC and LAM. PLoSONE. 2008;3:e3558. doi: 10.1371/journal.pone.0003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske EP, Wessner LL, Golden J, Scheithauer BW, Vortmeyer AO, Zhuang Z, Klein_Szanto AJ, Kwiatkowski DJ, Yeung RS. Loss of tuberin in both subependymal giant cell astrocytomas and angiomyolipomas supports a two-hit model for the pathogenesis of the tuberous sclerosis tumors. Am J Pathol. 1997;151:1639–1647. [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Hengstschläger M, Rodman DM, Miloloza A, Hengstschlager-Ottnad E, Rosner M, Kubista M. Tuberous sclerosis gene products in proliferation control. Mutat Res. 2001;488:233–239. doi: 10.1016/s1383-5742(01)00058-8. [DOI] [PubMed] [Google Scholar]
- Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci USA. 2000;97:6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koptides M, Mean R, Demetriou K, Pierides A, Deltas CC. Genetic evidence for a trans-heterozygous model for cytogenesis in autosomal dominant polycystic kidney disease. Hum Mol Genet. 2000;9:447–452. doi: 10.1093/hmg/9.3.447. [DOI] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Toyooka KO, Miyajima K, Reddy JL, Toyota M, Sathyanarayana UG, Padar A, Tockman MS, Lam S, Shivapurkar N, Gazdar AF. Epigenetic down-regulation of death-associated protein kinase in lung cancers. Clin Cancer Res. 2003;9:3034–3041. [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanism and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics come to age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Sampson J, Martin TA, Lee-Jones L, Watkins G, Douglas-Jones A, Mokbel K, Mansel RE. Tuberin and hamartin are aberrantly expressed and linked to clinical outcome in human breast cancer: the role of promoter methylation of TSC genes. Eur J Cancer. 2005;41:1628–1636. doi: 10.1016/j.ejca.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Mohiyuddin SMA, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signalling pathway in oral squamous cell carcinoma. BMC Cancer. 2008;8:163–175. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Jiang G. The molecular mechanism of HDAC inhibitors in anticancer effects. Cell Mol Immunol. 2006;3:285–290. [PubMed] [Google Scholar]
- Marks PA, Breslow R. Dimethyl sulfoxide to varinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- Bissler J, McCormack FX, Young LR, Elwing JM, Chuck G. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes EE, Chu E. Anti-EGFR therapies: clinical experience in colorectal, lung, and head and neck cancers. Oncology. 2006;20(5 Suppl 2):15–25. [PubMed] [Google Scholar]
- Lee L, Sudentas P, Donohue B, Asrican K, Worku A, Walker V, Sun Y, Schmidt K, Albert MS, El-Hashemite N, Lader AS, Onda AS, Zhang H, Kwiatkowski DJ, Dabora SL. Efficacy of a rapamycin analog (CCI-779) and IFN-gamma in tuberous sclerosis mouse model. Genes Chromosom Cancer. 2005;42:213–227. doi: 10.1002/gcc.20118. [DOI] [PubMed] [Google Scholar]
- Meikle L, Pollizzi K, Egnor A, Kramvis I, Lane H, Sahin M, Kwiatkowski DJ. Response of a neural model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci. 2008;28:5422–5432. doi: 10.1523/JNEUROSCI.0955-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]