Abstract
Statistical dependencies in the responses of sensory neurons govern both the amount of stimulus information conveyed and the means by which downstream neurons can extract it. Although a variety of measurements indicate the existence of such dependencies1–3, their origin and importance for neural coding are poorly understood. Here we analyse the functional significance of correlated firing in a complete population of macaque parasol retinal ganglion cells using a model of multi-neuron spike responses4,5. The model, with parameters fit directly to physiological data, simultaneously captures both the stimulus dependence and detailed spatio-temporal correlations in population responses, and provides two insights into the structure of the neural code. First, neural encoding at the population level is less noisy than one would expect from the variability of individual neurons: spike times are more precise, and can be predicted more accurately when the spiking of neighbouring neurons is taken into account. Second, correlations provide additional sensory information: optimal, model-based decoding that exploits the response correlation structure extracts 20% more information about the visual scene than decoding under the assumption of independence, and preserves 40% more visual information than optimal linear decoding6. This model-based approach reveals the role of correlated activity in the retinal coding of visual stimuli, and provides a general framework for understanding the importance of correlated activity in populations of neurons.
How does the spiking activity of a neural population represent the sensory environment? The answer depends critically on the structure of neuronal correlations, or the tendency of groups of neurons to fire temporally coordinated spike patterns. The statistics of such patterns have been studied in a variety of brain areas, and their significance in the processing and representation of sensory information has been debated extensively2,3,7–13.
Previous studies have examined visual coding by pairs of neurons11 and the statistics of simultaneous firing patterns in larger neural populations14,15. However, no previous approach has addressed how correlated spiking activity in complete neural populations depends on the pattern of visual stimulation, or has answered the question of how such dependencies affect the encoding of visual stimuli.
Here we introduce a model-based methodology for studying this problem. We describe the encoding of stimuli in the spike trains of a neural population with a generalized linear model (Fig. 1a), a generalization of the well-known linear–nonlinear–Poisson (LNP) cascade model4,5,16,17. In this model, each cell’s input is described by a set of linear filters: a stimulus filter, or spatio-temporal receptive field; a post-spike filter, which captures dependencies on spike-train history (for example, refractoriness, burstiness and adaptation); and a set of coupling filters, which capture dependencies on the recent spiking of other cells. For each neuron, the summed filter responses are exponentiated to obtain an instantaneous spike rate. This is equivalent to exponentiating the filter outputs and then multiplying; the exponentiated post-spike and coupling filters (as plotted in Fig. 1) may therefore be interpreted as spike-induced gain adjustments of the neuron’s firing rate.
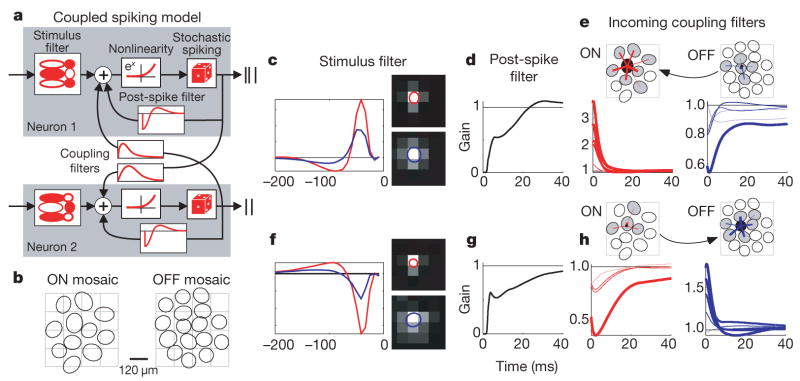
Figure 1. Multi-neuron encoding model and fitted parameters.
a, Model schematic for two coupled neurons: each neuron has a stimulus filter, a post-spike filter and coupling filters that capture dependencies on spiking in other neurons. Summed filter output passes through an exponential nonlinearity to produce the instantaneous spike rate. b, Mosaics of 11 ON and 16 OFF retinal ganglion cell receptive fields, tiling a small region of visual space. Ellipses represent 1 s.d. of a Gaussian fit to each receptive field centre; the square grid indicates stimulus pixels. c–e, Parameters for an example ON cell. c, Temporal and spatial components of centre (red) and surround (blue) filter components, the difference of which is the full stimulus filter. d, Exponentiated post-spike filter, which may be interpreted as multiplying the spike rate after a spike at time zero. It produces a brief refractory period and gradual recovery (with a slight overshoot). e, Connectivity and coupling filters from other cells in the population. The black filled ellipse is this cell’s RF centre, and blue and red lines show connections from neighbouring OFF and ON cells, respectively (line thickness indicates coupling strength). Below, exponentiated coupling filters show the multiplicative effect on this cell’s spike rate after a spike in a neighbouring cell. f–h, Analogous plots for an example OFF cell.
Although this model is strictly phenomenological, its components can be loosely compared to biophysical mechanisms: the stimulus filter approximates the spatio-temporal integration of light in the outer retina and passive dendritic filtering; the post-spike filter mimics voltage-activated currents following a spike; coupling filters resemble synaptic or electrical interactions between cells (and can mimic the effects of shared input noise); and the exponential nonlinearity implements a ‘soft threshold’, converting membrane potential to instantaneous spike probability. Note that the post-spike and coupling filters, which allow stochastic spiking in one cell to affect subsequent population activity, give rise to shared, non-Poisson variability in the model response.
We fit the model to data recorded in vitro from a population of 27 ON and OFF parasol ganglion cells (RGCs) in a small patch of isolated macaque monkey retina, stimulated with 120-Hz spatio-temporal binary white noise. The receptive fields of each of the two cell types formed a complete mosaic covering a small region of visual space (Fig. 1b), indicating that every parasol cell in this region was recorded15,18. Such complete recordings, which have not been achieved elsewhere in the mammalian nervous system, are essential for understanding visual coding in neural populations.
The model contains many parameters that specify the shapes of all filters, but fitting by maximizing likelihood remains highly tractable5. A penalty on coupling filters was used to obtain a minimally sufficient set of coupling filters, which yields an estimate of the network’s functional connectivity19,20.
Figure 1 shows the estimated filters describing input to example ON and OFF cells. The stimulus filters exhibit centre-surround receptive field organization consistent with previous characterizations of parasol cells. Post-spike filters show the time course of recovery from refractoriness after a spike, and coupling filters show the effects of spikes from nearby cells: for the ON cell (top), spikes in neighbouring ON cells elicit a large, transient excitation (increasing the instantaneous spike rate by a factor of three), whereas spikes in nearby OFF cells elicit suppression. These effects are reversed in the OFF cell, which is excited/suppressed by spikes in neighbouring OFF/ON cells. Both populations exhibit approximate nearest-neighbour connectivity, with coupling strength falling as a function of distance between receptive field centres15. We found that fitted stimulus filters have smaller surrounds than the spike-triggered average, indicating that a portion of the classical surround can be explained by interactions between cells21 (see Supplementary Information).
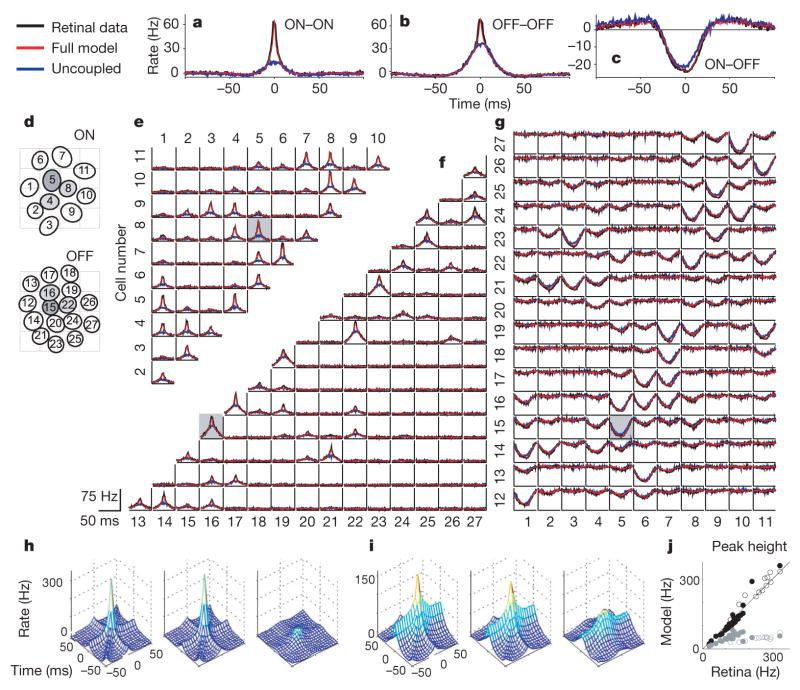
To assess accuracy in capturing the statistical dependencies in population responses, we compared the pairwise cross-correlation function (CCF) of RGCs and simulated model spike trains (Fig. 2). For nearby ON–ON and OFF–OFF pairs, the CCF exhibits a sharp peak at zero, indicating the prevalence of synchronous spikes; however, for ON–OFF pairs, a trough at zero indicates an absence of synchrony. For all 351 possible pairings, the model accurately reproduces the CCF (Fig. 2a–c, e, f).
Figure 2. Analysis of response correlations.
a–c, Example CCFs of retinal responses, and simulated responses of the full and uncoupled models, for two ON cells (a), two OFF cells (b) and an ON–OFF pair (c). The baseline is subtracted so that units are in spikes per s above (or below) the cell’s mean rate. d, Receptive field mosaic overlaid with arbitrary labels. Dark grey indicates cells shown in Fig. 1; light grey indicates cells used for triple correlations (h, i). e, CCFs between all ON pairs, where the i,jth plot shows the CCF between cell i and cell j. The grey box indicates the CCF plotted in a. f, g, CCFs between all OFF–OFF pairs (f) and all ON–OFF pairs (g; abscissa height is 30 Hz). h, Third-order (triplet) CCF between three adjacent ON cells, showing the instantaneous spike rate of cell 5 as a function of the relative spike time in cells 4 and 8 (left, RGCs; middle, full model; right, uncoupled model). i, Analogous triplet CCF for OFF cells 15, 16 and 22. j, Comparison of the triplet CCF peak in RGC and model responses (full model, black; uncoupled, grey) for randomly selected triplets of adjacent ON (open) and OFF (filled) cells.
To examine whether inter-neuronal coupling was necessary to capture the response correlation structure, we re-fitted the model without coupling filters (that is, so that each cell’s response depends only on the stimulus and its own spike-train history). This ‘uncoupled model’ assumes that cells encode the stimulus independently, although correlations may still arise from the overlap of stimulus filters. However, the uncoupled model fails to reproduce the sharp CCF peaks observed in the data. These peaks are also absent from CCFs computed on trial-shuffled data, indicating that fast-timescale correlations are not stimulus-induced and therefore cannot be captured by any independent encoding model.
Higher-order statistical dependencies were considered by inspecting correlations in three-neuron groups: triplet CCFs show the spike rate of one cell as a function of the relative time to spikes in two other cells (Fig. 2e–g)15. For adjacent neurons of the same type, triplet CCFs have substantial peaks at zero (‘triplet synchrony’), which are well matched by the full model.
Although the full and uncoupled models differ substantially in their statistical dependencies, the two models predict average light responses in individual cells with nearly identical accuracy, capturing 80–95% of the variance in the peri-stimulus time histogram (PSTH) in 26 out of 27 cells (Fig. 3a–c). Both models therefore accurately describe average single-cell responses to new stimuli. However, the full model achieves higher accuracy, predicting multi-neuronal spike responses on a single trial (8 ± 3% more bits per spike, Fig. 3d). This discrepancy can be explained by the fact that noise is shared across neurons. Shared variability means that population activity carries information about a single cell’s response (owing to coupling between cells) beyond that provided by the stimulus alone. Individual neurons therefore appear less noisy when conditioned on spiking activity in the rest of the population than they appear in raster plots.
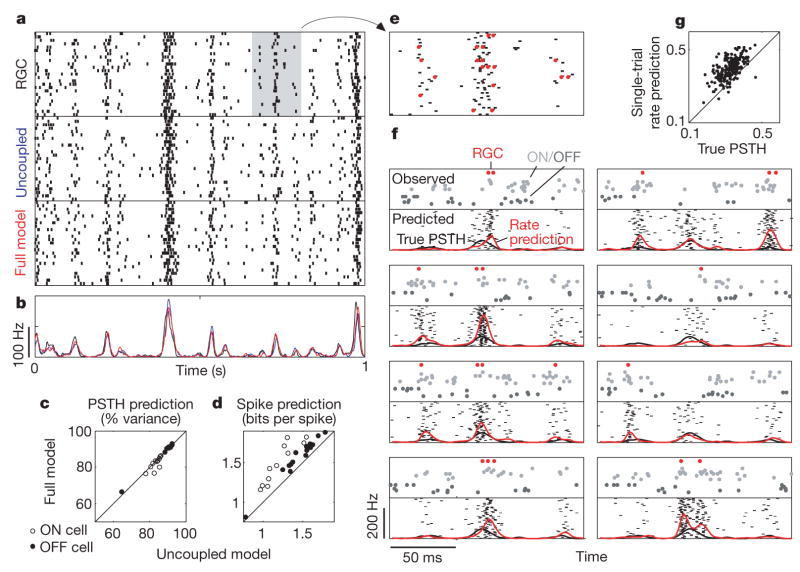
Figure 3. Spike-train prediction comparison.
a, Raster of responses of an ON RGC to 25 repeats of a novel 1-s stimulus (top), and responses of uncoupled (middle) and full (bottom) models to the same stimulus. b, PSTH of the RGC (black), uncoupled (blue) and coupled (red) model; both models account for ~84% of the variance of the true PSTH. c, PSTH prediction by full and uncoupled models, showing that coupling confers no advantage in predicting average responses. d, Log-likelihood of novel RGC spike responses under full and uncoupled models; the full model provides 8% more information about novel spike trains. e, Magnified 150-ms portion of RGC raster and PSTH (grey box in a). Red dots highlight RGC spike times on selected individual trials, which are replotted in f. f, Single-trial spike-train prediction using the coupled model. The top half of each plot shows the population activity on a single trial: true spike times of the cell (red dots), coupled ON cells (light grey dots) and coupled OFF cells (dark grey dots; each line in the raster shows the spike times of a different cell). The bottom half of each plot shows a raster of 50 predicted responses of the cell in question, using both the stimulus and coupled responses (shown above) to predict spike trains. The red trace shows the single-trial rate prediction (population-conditioned PSTH), compared with true PSTH of the cell (black trace, identical in all plots). g, Correlation coefficient of true spike trains with the PSTH (ordinate) and with population-conditioned predictions (abscissa); the full model predicts single-trial responses with higher accuracy than the true PSTH.
We measured the effect of correlations on single-trial, single-cell spike-train prediction by using the model to draw samples of a single cell’s response given both the stimulus and the spiking activity in the rest of the population on a single trial (Fig. 3e, f). Averaging the resulting raster plot gives a prediction of the cell’s single-trial spike rate, or ‘population-conditioned’ PSTH for a single trial. We compared these predictions with the cell’s true spike times (binned at 2 ms) across all trials and found that on nearly every trial, the model-based prediction is more highly correlated with the observed spikes than the neuron’s full PSTH (Fig. 3g). Note that the full PSTH achieves the highest correlation possible for any trial-independent prediction. Thus, by exploiting the correlation structure, the coupled model predicts single-neuron spike times more accurately than any independent encoding model.
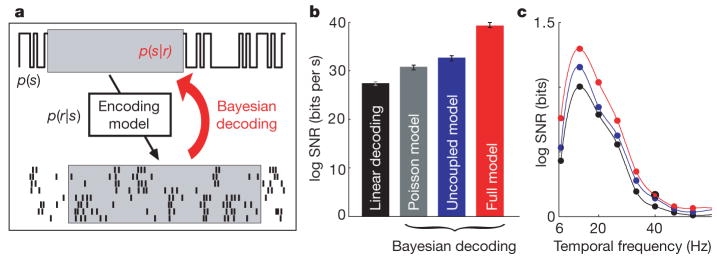
Although the full model accurately captures dependencies in the activity of RGCs, it is not obvious a priori whether these dependencies affect the amount of sensory information conveyed by RGC responses. In principle, the correlation structure could be necessary to predict the responses, but not to extract the stimulus information that the responses carry13. To examine this issue directly, we used the full and uncoupled models to perform Bayesian decoding of the population response (Fig. 4a), which optimally reconstructs stimuli given an accurate description of the encoding process. For comparison, we also performed Bayesian decoding under a Poisson (that is, LNP) model and optimal linear decoding6.
Figure 4. Decoding performance comparison.
a, Shown is a Bayesian decoding schematic: to estimate an unknown stimulus segment from a set of observed spike times (highlighted in boxes), the stimulus prior distribution p(s) is multiplied by the model-defined likelihood p(r|s) to obtain the posterior p(s|r). The posterior mean is the Bayes’ least-squares stimulus estimate. b, Log of the SNR for linear decoding, as well as for Bayesian decoding under the Poisson, uncoupled and full models6. The full model preserves 20% more information than the uncoupled model, which indicates that there is additional sensory information available from the population response when correlations are taken into account. Error bars show 95% confidence intervals based on 2,000 bootstrap resamplings of 3,000 decoded stimulus segments. c, Log SNR decomposed as a function of temporal frequency for various decoding methods (Poisson omitted for clarity).
Each decoding method was used to estimate short (150-ms) segments of the stimulus given all relevant spike times from the full population (Fig. 4b). Bayesian decoding under the coupled model recovers 20% more information than Bayesian decoding under the uncoupled model, indicating that knowledge of the correlation structure is critical for extracting all sensory information contained in the population response. This improvement was invariant to enhancements of the model’s stimulus filters and nonlinearities (see Supplementary Information), indicating that the difference in performance arises specifically from the coupled model’s ability to incorporate the correlation structure. Our results also show that spike history is relevant for decoding (a Poisson model preserves 6% less information than the uncoupled model22) and that restricting to a linear decoder further reduces the information that can be recovered from RGC responses.
Decoding analysis can also be used to examine the coding fidelity of specific stimulus features. As a simple illustration, we examined the temporal frequency spectrum of reconstructed stimuli and found that the response correlation structure is most important for decoding those stimulus frequencies (6–20 Hz) that are encoded with highest fidelity (Fig. 4c).
These results demonstrate that the responses of a population of retinal ganglion cells are well described by a generalized linear model, and that correlations in the response can be exploited to recover 20% more visual information than if responses are regarded as independent given the stimulus. In contrast, previous studies have reported this information gain to be less than 10% for pairs of neurons9,12. However, pairwise analyses provide little evidence about the importance of correlations across an entire population. Second-order correlations between pairs of neurons could give rise to either much larger (scaling with the number of neurons n) or much smaller (falling as 1/n) gains for a full population (see Supplementary Information). To compare more directly with previous findings, we performed Bayesian decoding using isolated pairs of neurons from the same population; we found a ≤10% gain in sensory information when correlations were included (see Supplementary Information). This is consistent with previous findings, and shows that the information gain for a complete population is larger than that observed for pairs. We also compared the model to a pairwise maximum-entropy model, which has recently been shown to capture the instantaneous spiking statistics of groups of retinal ganglion cells14,15. The coupled model exhibits similar accuracy in capturing these statistics, but has the advantage that it accounts for the temporal correlation structure and stimulus dependence of responses, which are essential for assessing the effect of correlations on sensory coding.
Although it provides an accurate functional description of correlated spike responses, the generalized linear model does not reveal the biophysical mechanisms underlying the statistical dependencies between neurons: coupling does not necessarily imply anatomical connections between cells, but could (for example) reflect dependencies due to shared input noise1. The model also lacks several mechanisms known to exist in retinal ganglion cells (for example, contrast gain-control23), which may be required for characterizing responses to a wider variety of stimuli. One additional caveat is that Bayesian decoding provides a tool for measuring the sensory information available in the population response, but it does not reveal whether the brain makes use of this information. Physiological interpretations of the model and mechanisms for neural read-out of sensory information in higher brain areas are thus important directions for future research.
Nevertheless, the generalized linear model offers a concise, computationally tractable description of the population encoding process, and provides the first generative description of the space–time dependencies in stimulus-induced population activity. It allows us to quantify the relative contributions of stimulus, spike history and network interactions to the encoding and decoding of visual stimuli, and clarifies the relationship between single-cell and population variability. More generally, the model can be used to assess which features of the visual environment are encoded with highest and lowest fidelity, and to determine how the structure of the neural code constrains perceptual capabilities. We expect this framework to extend to other brain areas, and to have an important role in revealing the information processing capabilities of spiking neural populations4,19,24,25.
METHODS SUMMARY
Data
Multi-electrode extracellular recordings were obtained in vitro from a segment of isolated, peripheral macaque monkey (Macaca mulatta) retina, and analysis was restricted to two cell types (ON and OFF parasol)15,26,27. A standard spike-sorting procedure, followed by a specialized statistical method for detecting simultaneous spikes, was used to sort spikes (see ref. 28). The retina was stimulated with a photopic, achromatic, optically reduced spatio-temporal binary white noise stimulus refreshing at 120 Hz, with a root-mean-square contrast of 96%.
Fitting
Model parameters were fitted to 7 min of spike responses to a non-repeating stimulus. Each cell’s parameters consisted of a stimulus filter (parametrized as a rank-2 matrix), a spike-history filter, a set of incoming coupling filters and a constant. Temporal filters were represented in a basis of cosine ‘bumps’22. Parameters for the uncoupled and Poisson (LNP) models were fitted independently. Parameters were fitted by penalized maximum likelihood4,5, with an L1 penalty on the vector length of coupling filters to eliminate unnecessary connections.
Encoding
Spike prediction was cross-validated using the log-likelihood of 5 min of novel spiking data (scaled to units of bits per s). Repeat rasters were obtained using 200 presentations of a novel 10-s stimulus. Population-conditional rasters were obtained from the coupled model by sampling the model-defined probability distribution over the neuron’s response given the stimulus and surrounding-population activity on a single trial29.
Decoding
Population responses were decoded using the Bayes’ least-squares estimator (posterior mean) to reconstruct 18-sample single-pixel stimulus segments (cross-validation data). Linear decoding was performed using the optimal linear estimator6. Decoding performance was quantified using the log signal-to-noise ratio (SNR) of each technique, which gives an estimate of mutual information. Breakdown by temporal frequency was obtained by computing the Fourier power spectra of the stimuli and residuals and then computing log SNR.
Supplementary Material
Acknowledgments
We thank M. Bethge, C. Brody, D. Butts, P. Latham, M. Lengyel, S. Nirenberg and R. Sussman for comments and discussions; G. Field, M. Greschner, J. Gauthier and C. Hulse for experimental assistance; M. I. Grivich, D. Petrusca, W. Dabrowski, A. Grillo, P. Grybos, P. Hottowy and S. Kachiguine for technical development; H. Fox, M. Taffe, E. Callaway and K. Osborn for providing access to retinas; and S. Barry for machining. Funding was provided a Royal Society USA/Canada Research Fellowship (J.W.P.); NSF IGERT DGE-03345 (J.S.); NEI grant EY018003 (E.J.C., L.P. and E.P.S.); Gatsby Foundation Pilot Grant (L.P.); Burroughs Wellcome Fund Career Award at the Scientific Interface (A.S.); US National Science Foundation grant PHY-0417175 (A.M.L.); McKnight Foundation (A.M.L. and E.J.C.); and HHMI (J.W.P., L.P. and E.P.S.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Mastronarde DN. Correlated firing of retinal ganglion cells. Trends Neurosci. 1989;12:75–80. doi: 10.1016/0166-2236(89)90140-9. [DOI] [PubMed] [Google Scholar]
- 2.Meister M, Lagnado L, Baylor DA. Concerted signaling by retinal ganglion cells. Science. 1995;270:1207–1210. doi: 10.1126/science.270.5239.1207. [DOI] [PubMed] [Google Scholar]
- 3.Shadlen M, Newsome W. The variable discharge of cortical neurons: implications for connectivity, computation, and information coding. J Neurosci. 1998;18:3870–3896. doi: 10.1523/JNEUROSCI.18-10-03870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. J Neurophysiol. 2004;93:1074–1089. doi: 10.1152/jn.00697.2004. [DOI] [PubMed] [Google Scholar]
- 5.Paninski L. Maximum likelihood estimation of cascade point-process neural encoding models. Network Comp Neural Syst. 2004;15:243–262. [PubMed] [Google Scholar]
- 6.Warland D, Reinagel P, Meister M. Decoding visual information from a population of retinal ganglion cells. J Neurophysiol. 1997;78:2336–2350. doi: 10.1152/jn.1997.78.5.2336. [DOI] [PubMed] [Google Scholar]
- 7.Dan Y, Alonso JM, Usrey WM, Reid RC. Coding of visual information by precisely correlated spikes in the lateral geniculate nucleus. Nature Neurosci. 1998;1:501–507. doi: 10.1038/2217. [DOI] [PubMed] [Google Scholar]
- 8.Panzeri S, Golledge H, Zheng F, Tovee MP, Young MJ. Objective assessment of the functional role of spike train correlations using information measures. Vis Cogn. 2001;8:531–547. [Google Scholar]
- 9.Nirenberg S, Carcieri S, Jacobs A, Latham P. Retinal ganglion cells act largely as independent encoders. Nature. 2001;411:698–701. doi: 10.1038/35079612. [DOI] [PubMed] [Google Scholar]
- 10.Schneidman E, Bialek W, Berry MJ. Synergy, redundancy, and independence in population codes. J Neurosci. 2003;21:11539–11553. doi: 10.1523/JNEUROSCI.23-37-11539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nirenberg S, Latham PE. Decoding neuronal spike trains: how important are correlations? Proc Natl Acad Sci USA. 2003;100:7348–7353. doi: 10.1073/pnas.1131895100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Averbeck BB, Lee D. Coding and transmission of information by neural ensembles. Trends Neurosci. 2004;27:225–230. doi: 10.1016/j.tins.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Latham P, Nirenberg S. Synergy, redundancy, and independence in population codes, revisited. J Neurosci. 2005;25:5195–5206. doi: 10.1523/JNEUROSCI.5319-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneidman E, Berry M, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440:1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shlens J, et al. The structure of multi-neuron firing patterns in primate retina. J Neurosci. 2006;26:8254–8266. doi: 10.1523/JNEUROSCI.1282-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plesser H, Gerstner W. Noise in integrate-and-fire neurons: From stochastic input to escape rates. Neural Comput. 2000;12:367–384. doi: 10.1162/089976600300015835. [DOI] [PubMed] [Google Scholar]
- 17.Simoncelli EP, Paninski L, Pillow J, Schwartz O. In: The Cognitive Neurosciences. 3. Gazzaniga M, editor. MIT; 2004. pp. 327–338. [Google Scholar]
- 18.Frechette ES, et al. Fidelity of the ensemble code for visual motion in primate retina. J Neurophysiol. 2005;94:119–135. doi: 10.1152/jn.01175.2004. [DOI] [PubMed] [Google Scholar]
- 19.Okatan M, Wilson M, Brown E. Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput. 2005;17:1927–1961. doi: 10.1162/0899766054322973. [DOI] [PubMed] [Google Scholar]
- 20.Rigat F, de Gunst M, van Pelt J. Bayesian modelling and analysis of spatio-temporal neuronal networks. Bayes Anal. 2006;1:733–764. [Google Scholar]
- 21.DeVries SH. Correlated firing in rabbit retinal ganglion cells. J Neurophysiol. 1999;81:908–920. doi: 10.1152/jn.1999.81.2.908. [DOI] [PubMed] [Google Scholar]
- 22.Pillow JW, Paninski L, Uzzell VJ, Simoncelli EP, Chichilnisky EJ. Prediction and decoding of retinal ganglion cell responses with a probabilistic spiking model. J Neurosci. 2005;25:11003–11013. doi: 10.1523/JNEUROSCI.3305-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapley RM, Victor JD. The effect of contrast on the transfer properties of cat retinal ganglion cells. J Physiol. 1978;285:275–298. doi: 10.1113/jphysiol.1978.sp012571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris K, Csicsvari J, Hirase H, Dragoi G, Buzsaki G. Organization of cell assemblies in the hippocampus. Nature. 2003;424:552–556. doi: 10.1038/nature01834. [DOI] [PubMed] [Google Scholar]
- 25.Paninski L, Fellows M, Shoham S, Hatsopoulos N, Donoghue J. Superlinear population encoding of dynamic hand trajectory in primary motor cortex. J Neurosci. 2004;24:8551–8561. doi: 10.1523/JNEUROSCI.0919-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litke AM, et al. What does the eye tell the brain? Development of a system for the large scale recording of retinal output activity. IEEE Trans Nucl Sci. 2004;51:1434–1440. [Google Scholar]
- 27.Watanabe M, Rodieck RW. Parasol and midget ganglion cells of the primate retina. J Comp Neurol. 1989;289:434–454. doi: 10.1002/cne.902890308. [DOI] [PubMed] [Google Scholar]
- 28.Segev R, Goodhouse J, Puchalla J, Berry MJ. Recording spikes from a large fraction of the ganglion cells in a retinal patch. Nature Neurosci. 2004;7:1155–1162. doi: 10.1038/nn1323. [DOI] [PubMed] [Google Scholar]
- 29.Pillow JW, Latham P. In: Advances in Neural Information Processing Systems. Platt JC, Koller D, Singer Y, Roweis S, editors. Vol. 20. MIT; 2008. pp. 1161–1168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.