Abstract
Herein we demonstrate the use of a novel dimerization-based molecular beacon (MB) probe consisting of two metallo-phthalocyanine (Pc) fluorophores that use near-IR fluorescence, appropriate for highly specific and sensitive in-vivo and/or in-vitro DNA/RNA detection. Pc’s possess a propensity to form non-fluorescent H-dimers that is utilized as the molecular “off” switch in the closed MB conformation. The “on” switch, which is generated when the solution target binds to the loop of the MB forming the open form, also provides two fluorophores for transduction resulting in a doubling of the extinction coefficient and improving the resulting fluorescence yield compared to a classical single-fluorophore/quencher MB system. In addition, the Pc-based MBs possess high thermal, photo and chemical stabilities that are essential for many highly sensitive applications, such as molecular imaging. The dimer-based MBs were obtained using a simple single-step synthesis procedure and demonstrated excellent quenching efficiencies (98%) as well as a high signal-to-background ratio (~60) exceeding the performance characteristics of many conventionally-available MB probes.
Since their 1996 inception by Tyagi and Kramer,1 molecular beacons (MBs) have become powerful tools for a variety of applications in DNA/RNA detection (e.g. real-time PCR, SNP detection, in vivo cellular imaging, cancer imaging etc.). MB-based detection systems are separation-free and highly specific however, their sensitivity (defined as the ratio of the fluorescence in the open versus closed form; signal-to-background, SB) is limited due to incomplete quenching of the closed form while signal in an open form is confined to one fluorophore. Various approaches for SB enhancement have been pursued including introduction of multiple fluorophores and/or quenchers,2 eximer emission,3, 4 FRET,5 conjugated-polymers,6 electrochemiluminescence7 or quantum-dots.8 The quest for more sensitive probes is still necessary along with the need to increase the MB reporting fluorophore’s photostability to prolong observation times in imaging applications and conducting assays in the near-IR, where significant autofluorescence can be avoided.9
Water soluble metallo-phthalocyanines (Pc’s) are near-IR fluorophores that possess high extinction coefficients, favorable quantum yields, narrow absorption/emission envelopes and display extraordinarily high photostabilities.10, 11These qualities make Pc’s particularly attractive for MB applications along with other unique characteristics, such as: (i) Intrinsic propensity to form non-fluorescent H-type dimers that can be utilized as a molecular “off” switch for the closed MB form while two flourophores instead of one would be available for readout in the open form; (ii) diminished Pc dimerization when it is part of a relatively large oligonucleotide construct (e.g. MB hybridized to a complementary target as has been demonstrated by us), but the lack of fluorescence when bound to a relatively short DNA construct.10 These characteristics of Pc dyes could provide dimer-based MB probes (DBMB) with unique properties and high SB ratios.
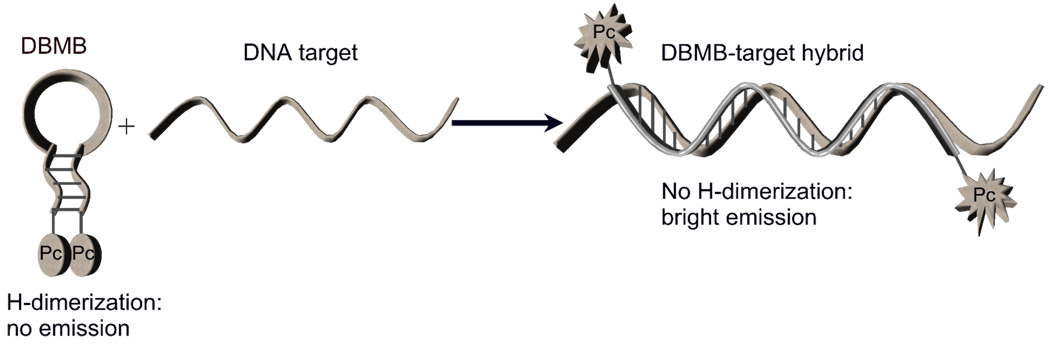
An H-dimer-based MB system utilizing tetramethylrhodamine has been reported, but the quenching efficiencies were only ~80 %.12 In another report, a DCDHF dimerization-based MB was reported with a quenching efficiency of ~97%.13 However, the use of this MB system depended intimately on the viscosity and polarity of the solvent environment.14 Thus, we believe that the use of the self-dimerization behavior of Pc’s will be particularly compelling for MB applications due to the nature of the quenching mechanism potentially expanding the applications of these beacon probes.11, 15 The Pc dimerization-based MB (DBMB-Pc) systems we have designed are presented in Figure 1. Two identical Pc molecules (Pc1 or Pc2, Scheme 1) in the MB’s closed state form a non-fluorescent H-dimer. In the presence of a complementary DNA, the MB’s loop hybridizes to its target forcing the MB to open and disrupting the Pc dimer and restoring fluorescence emission.
Figure 1.
DBMB assay using Pc dyes.
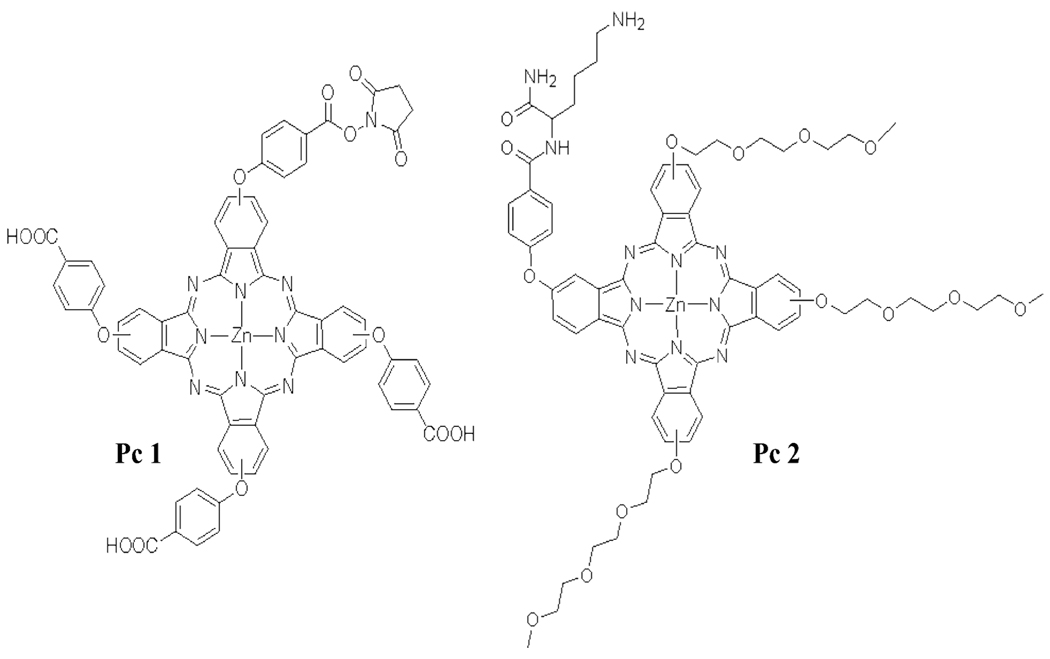
Scheme 1.
The MB’s were synthesized using either Pc succinimidyl-ester amine conjugation10 (for Pc1) or reductive amination of an aldehyde (for Pc2) with both purified via HPLC. Details on the synthesis, characterization and purification are provided in the Supporting Information.
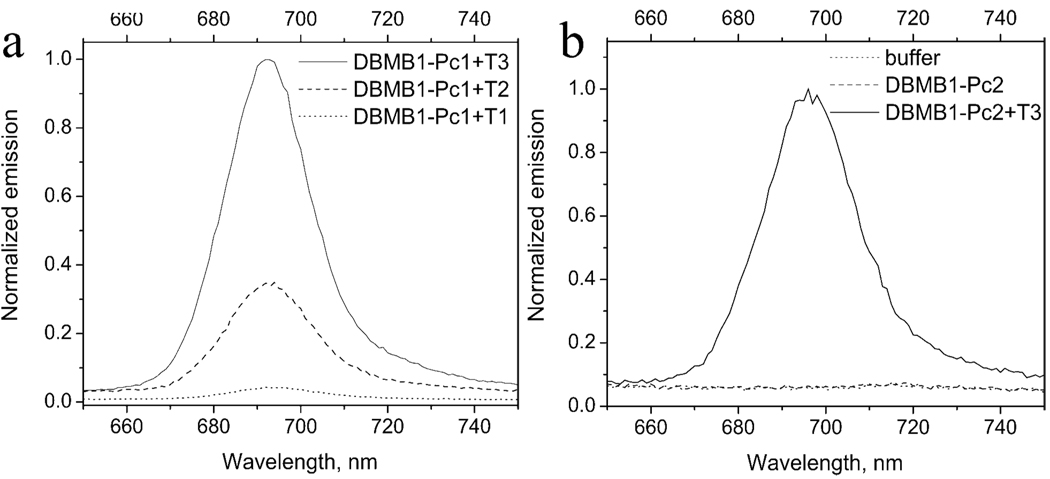
Preliminary evaluations of hybridization of DBMB1-Pc1 to its complementary target did not produce significant fluorescence signals in the open form. However, after careful examination, it was found that those systems had a high propensity for the so called, “sticky-end” base paring,16 leading to substantial emission quenching even in the open form. Also, Pc-Pc interactions contributed significantly to the overall stability of the “sticky-end” paired structures depicted in Figure S4. The use of longer template strands (T2, Table 1) and “stem inclusive” templates (T3, Table 1) lead to restoration of the monomeric Pc-fluorescence as noted in Figure 2a. Thus, the propensity of dyes towards dimer-based interactions should be an important consideration when designing DBMB’s.
Table 1.
Sequences for oligonucleotides used to obtain DBMB’s and single strand target DNAs used in hybridization experiments (MB stem and its complements are underlined).
| Name | 5' – 3’ sequence |
|---|---|
| DBMB1 | *CTGACGAGTCCTTCCACGATACCAGTCAG* |
| DBMB2 | **CTGACGAGTCCTTCCACGATACCAGTCAG** |
| T1 | TGGTATCGTGGAAGGACTC |
| T2 | (T)20TGGTATCGTGGAAGGACTC(T)20 |
| T3 | (T)20TGGTATCGTGGAAGGACTCGTCAG(T)20 |
Modified with NH2- or aldehyde groups attached via C6H12. The conjugation of this sequence with Pc1 or Pc2 produces DBMB1-Pc1 or ABMB1-Pc2 (see Scheme 1 for Pc1 and Pc2 structures).
Modified with NH2-group attached via C3H6.
Figure 2.
Emission from (a) DBMB1-Pc1 (200 nM) hybridized to various types of complementary DNA templates (2 µM) and; (b) DBMB1-Pc2 (200 nM) and DBMB1-Pc2 (200 nM) hybridized to T3 (2 µM).
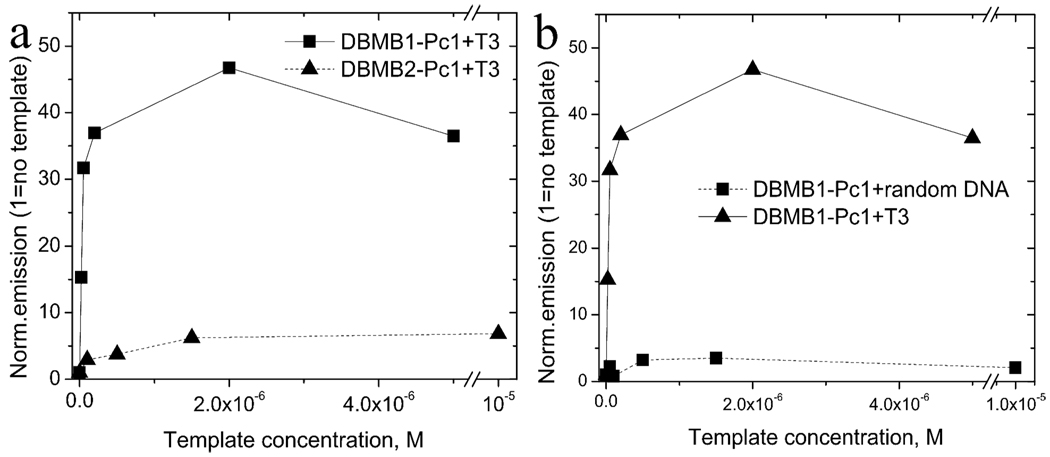
An important element in the DBMB’s performance was the length of the aliphatic spacer used between the stem and fluorophore, because the linker length can affect both signal (shorter linkers cause less fluorescence upon hybridization17) and background (shorter linkers result in better dimerization in the closed form of the MB, see Figure S8). Two identical DBMB’s (Table 1) based on either C6H12 (DBMB1-Pc1) or C3H6 (DBMB2-Pc1) linkers were obtained and evaluated when hybridized to their complementary templates. Results (Figure 3a) indicated that the beacon modified with a longer linker (C6H12, DBMB1-Pc1) provide better SB ratios compared to one modified with a shorter linker (C3H6, DBMB2-Pc1). Results indicated that dimerization was more efficient for DBMB2-Pc1 based on entropic effects, but the fluorescence yield in the open form of DBMB1-Pc1 was higher due to competition between Pc1/Pc1 dimerization and hybridization between the MB loop and its complement (see Supporting Information for discussion).
Figure 3.
Changes in emission of (a) DBMB1-Pc1 and DBMB2-Pc1 (200 nM) upon addition of various amounts of complementary target, T3. (b) DBMB1-Pc1 (200 nM) emission upon addition of various DNA targets; complementary target T3 or non-complementary random DNA sequence.
One of the attractive properties of many MBs is their ability to specifically detect complementary target DNA from non-complementary DNAs, even when the non-targets contain a single base mismatch (SNP detection). Thus, experiments targeting evaluation of DBMB1-Pc1’s ability to discriminate between matched DNA and random non-matched DNA were conducted (Figure 3b). The perfectly matched template produced significantly higher signal enhancement when compared to the random DNA template. The ability of DBMB1-Pc1 to detect SNPs was also evaluated (see Figure S7) and demonstrated an ~5-fold increase in the SB for the perfect match target compared to the single base mismatch.
Another Pc was evaluated as the reporter for the DBMB, which contained a triethylene glycol modification, Pc2 (Scheme 1). When this Pc dye was used as the reporter for the DBMB, it produced practically complete quenching (Figure 2b, quenching efficiency = 98%) and a SB ratio as high as 59.
In conclusion, we have, for the first time, designed, prepared and evaluated the use of a double-labeled dimerization-based MB with near-IR emission. The DBMB produced quenching efficiencies and SB ratios exceeding or comparable to currently available fluorophore-quencher systems and thus, could be very beneficial for live cell imaging or other applications requiring monitoring in complex biological matrices due to lower light scattering and autofluorescence obtained in the near-IR compared to other regions of the electromagnetic spectrum. An especially attractive feature of our design is the doubling of the extinction coefficient and fluorescent photon yield in the open form due to the presence of two fluorescent reporter moieties attached to the DBMB instead of one as is typically used. In addition, this format provides an easier one-step synthesis/purification protocol compared to the tedious and costly synthesis of regular MB’s using a fluorescent dye and quenching moiety, which requires a two-step conjugation process with purification at each step. Another important aspect of our Pc-based DBMB system is the non-susceptibility to increase its emission upon probe degradation due to the lower emission from shorter DNA fragments10 such as those produced via nuclease degradation that is often an issue in live cell imaging using MBs.18 Finally, we have shown that the Pc-based dyes are considerably more photochemically stable compared to other molecular dye systems, making them particularly attractive for imaging applications.11
Supplementary Material
Description of DBMB identification, characterization, sticky-end pairing, SNP detection and discussion of fluorescence quenching in the closed form is included in the Supporting Information that is available free of charge over the internet.
ACKNOWLEDGMENT
NIH grant R01 EB-006639
References
- 1.Tyagi S, Kramer F. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Yang C, Lin H, Tan W. J. Am. Chem. Soc. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon P, Yang C, Wu Y, Chen Y, Martinez K, Kim Y, Stevens N, Marti A, Jockusch S, Turro N, Tan W. J. Am. Chem. Soc. 2008;130:336–342. doi: 10.1021/ja076411y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimoto K, Shimizu H, Inouye M. J. Org. Chem. 2004;69:3271–3275. doi: 10.1021/jo049824f. [DOI] [PubMed] [Google Scholar]
- 5.Zhang P, Beck T, Tan W. Angew. Chem., Int. Ed. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 6.Yang C, Pinto M, Schanze K, Tan W. Angew. Chem., Int. Ed. 2005;44:2572–2576. doi: 10.1002/anie.200462431. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Yun W, Dong P, Zhou J, He PG, Fang Y. Langmuir. 2008;24:2200–2205. doi: 10.1021/la702345p. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Morikis D, Ozkan M. Sens. Actuators, B. 2004;102:315–319. [Google Scholar]
- 9.Michalet X, Pinaud F, Bentolila L, Tsay J, Doose S, Li J, Sundaresan G, Wu A, Gambhir S, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesterova I, Verdree V, Pakhomov S, Strickler K, Allen M, Hammer R, Soper S. Bioconjugate Chem. 2007;18:2159–2168. doi: 10.1021/bc700233w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdree V, Pakhomov S, Su G, Allen M, Countryman A, Hammer R, Soper S. J. Fluoresc. 2007;17:547–563. doi: 10.1007/s10895-007-0210-4. [DOI] [PubMed] [Google Scholar]
- 12.Bernacchi S, Mely Y. Nucleic Acids Res. 2001;29:e62/1–e62/8. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conley N, Pomerantz A, Wang H, Twieg R, Moerner W. J. Phys. Chem. B. 2007;111:7929–7931. doi: 10.1021/jp073310d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura S, Lord S, Klein L, Willets K, He M, Lu Z, Twieg R, Moerner W. J. Phys. Chem. B. 2006;110:8151–8157. doi: 10.1021/jp0574145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lord S, Lu Z, Wang H, Willetst K, Schuck P, Lee H, Nishimura SY, Twieg R, Moerner W. J. Phys. Chem. A. 2007;111:8934–8941. doi: 10.1021/jp0712598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Tan W. Anal. Biochem. 2003;312:251–254. doi: 10.1016/s0003-2697(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 17.Randolph J, Waggoner A. Nucleic Acids Res. 1997;25:2923–2929. doi: 10.1093/nar/25.14.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee W, Santangelo P, Jo H, Bao G. Nucleic Acids Res. 2008;36:e30. doi: 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of DBMB identification, characterization, sticky-end pairing, SNP detection and discussion of fluorescence quenching in the closed form is included in the Supporting Information that is available free of charge over the internet.