Summary
ZBP1 (zipcode-binding protein 1, also known as IMP-1) is an mRNA regulator, functioning in mRNA localization, stability and translational control. ZBP1 is actively expressed during embryogenesis and tumorigenesis, but its expression is repressed in metastatic breast-cancer cell lines and tumors. In this article, we show that downregulation of ZBP1 expression results from its promoter methylation, an epigenetic process that remodels the chromatin structure and frequently represses gene activity. Demethylation of the ZBP1 promoter in metastatic cells reactivated ZBP1 expression, owing to restoration of the interaction of the ZBP1 promoter with β-catenin. Loss of ZBP1 function not only increased growth ability of metastatic cells, but also promoted cell migration. We identified a number of mRNAs that were selectively associated with ZBP1 in breast-cancer cells. Many of these are involved in cell motility and in cell-cycle regulation, and displayed altered expression patterns in the absence of ZBP1. These data suggest that repression of ZBP1 deregulates its associated mRNAs, leading to the phenotypic changes of breast cancers.
Keywords: ZBP1 repression, Promoter methylation, Wnt–β-catenin signaling, Cell proliferation, Metastasis
Introduction
When cancer cells advance to metastasis, there is an intense change in the pattern of gene expression. This change includes both the gain and loss of gene function, as well as changes in the level of expression for crucial genes. The expression of ZBP1 (zipcode-binding protein 1, also known as IMP-1; official gene name is IGF2BP1) is significantly altered during the malignant process (Wang et al., 2002).
ZBP1 belongs to a conserved family of RNA-binding proteins. It was originally isolated from chicken embryonic fibroblasts by its affinity to the `zipcode' of β-actin mRNA (Ross et al., 1997). Proteins orthologous or homologous to the chicken ZBP1 have been subsequently identified in mammalian organisms and implicated in many aspects of RNA regulation (Ioannidis et al., 2003b; Nielsen et al., 2001; Yisraeli, 2005). In the different species, the ZBP1 protein is mainly cytoplasmic, forming messenger-ribonucleoprotein (mRNP) complexes or granules with its mRNA targets (Farina et al., 2003; Huttelmaier et al., 2005; Stohr et al., 2006). In motile fibroblasts and primary neurons, ZBP1 mediates localized translation of β-actin mRNA, by which β-actin, which is required for filament growth, can be locally made to establish cellular asymmetry. In addition to the control of the localization and translation of β-actin mRNA, orthologs of ZBP1 were also identified to regulate the translation of insulin-like growth factor II (IGF2) mRNA (Liao et al., 2004; Nielsen et al., 1999) and stabilize Myc, β-catenin, CD44 and βTrCP1 mRNAs (Gu et al., 2008; Leeds et al., 1997; Noubissi et al., 2006; Vikesaa et al., 2006). This regulation has linked ZBP1 to important cellular processes, including actin dynamics and cellular polarity, cell proliferation, and metastasis (Kislauskis et al., 1997; Liao et al., 2004; Shestakova et al., 2001; Tessier et al., 2004; Wang et al., 2002).
The ZBP1-family protein is actively expressed through embryonic development but silenced or repressed in normal adult tissues. Re-activation of the gene has been detected in human primary tumors, including breast (60%), colon (81%) and non-small-cell lung carcinomas (50%) (Ioannidis et al., 2003a; Ioannidis et al., 2001; Ross et al., 2001). Recent studies have shown that in both colorectal- and breast-cancer cells, activation of the ZBP1 gene is mediated by β-catenin, which binds to the ZBP1 promoter and stimulates transcription (Gu et al., 2008; Noubissi et al., 2006). These findings suggest that the oncofetal pattern of ZBP1 expression could be a consequence of the cellular response to Wnt–β-catenin signaling. ZBP1 activation could inhibit chemotaxis and metastasis of breast-cancer cells (Lapidus et al., 2007; Wang et al., 2004) by maintaining cell polarity and directional movement through regulating the localization of β-actin mRNA (Condeelis and Singer, 2005). Hence, repression of ZBP1 expression results in behavioral changes of breast-cancer cells.
The involvement of ZBP1 repression with metastasis prompted us to investigate the underlying mechanism responsible for ZBP1 gene silencing in metastatic breast-cancer cells. We postulated that, in breast cancer cells, ZBP1 has the ability to regulate the post-transcriptional fate of a plethora of mRNA targets. When ZBP1 expression is repressed, the mRNAs that are normally associated with the protein could be misregulated, which leads to changes in cell behavior or phenotype. In this study, we show that ZBP1 repression in mammalian metastatic breast-cancer cells is a consequence of its promoter methylation. Epigenetic methylation of the ZBP1 promoter prevented β-catenin binding and resulted in the transcriptional inactivation of the gene. Repression of ZBP1 expression not only increased cell-migration potential, but also promoted the proliferation of metastatic cells. We have identified mRNAs that are selectively associated with ZBP1 in breast-cancer cells; many of these mRNAs were important for cell migration and cell proliferation, and displayed a different expression pattern in the absence of ZBP1 expression. Moreover, many ZBP1-associated mRNAs contain a conserved `zipcode' motif, which has been shown to be a structural binding target of ZBP1 (Farina et al., 2003).
Results
Downregulation of ZBP1 expression in metastatic breast-cancer cell lines and tumors
Previous studies reported ZBP1 repression in rat metastatic MTLn3 cells (Wang et al., 2002). Here, we examined ZBP1 expression in other mammalian breast-cancer cell lines: a rat non-metastatic MTC line, a human non-metastatic T47D cell line; two human MDA435 and MDA231 cell lines, both of which have been identified as highly invasive and metastatic (Zajchowski et al., 2001); and a mouse metastatic 4T1 cell line (Lelekakis et al., 1999). ZBP1 was actively expressed in non-metastatic MTC and T47D cells, whereas it was silenced in metastatic MTLn3, MDA231 and 4T1 cells (Fig. 1A). Although ZBP1 protein could be detected in human metastatic MDA435 cells, the expression was lower compared with MTC and T47D cells. A more sensitive reverse transcriptase (RT)-PCR method confirmed the western-blot results (Fig. 1B), suggesting that the ZBP1 gene in the metastatic cell lines could be transcriptionally repressed. Activation of ZBP1 expression has also been observed in breast tumors (Ioannidis et al., 2003a; Oberman et al., 2007; Yisraeli, 2005). To determine whether downregulation of ZBP1 expression occurred specifically in human breast metastatic tumors, we used a Cancer Profiling cDNA Array (BD Bioscience, catalog no. 131761) that contained four pairs of normalized human cDNA samples of breast tumor with corresponding metastasized tissues from the same individual patients (Fig. 1C). In three pairs of samples, we detected a significant reduction of ZBP1 expression in the metastatic tissues in comparison with their corresponding primary carcinoma tissues (samples 1-3). One sample showed little change in ZBP1 expression between the primary-tumor tissue and its corresponding metastatic tumor (sample 4). We extended these observations by performing in situ hybridization to examine ZBP1 transcription in a breast-tissue microarray (TMA, US Biomax, catalog no. BR1001) that contained 100 tissue biopsy sections from 50 breast-cancer patients: paired sections from two different cores of a patient, one primary tumor and the other a secondary metastatic tumor. The presence of transcription sites indicates whether the gene encoding ZBP1 is actively making RNA at the time of fixation (Capodieci et al., 2005). After hybridizing the TMA with probes for ZBP1, transcription sites were visualized by microscopy (Fig. 1D, upper). Transcription of the ZBP1 gene was detected in 48% of the primary breast tumors (24/50), in which 18% of the cells on average showed distinct transcription sites for ZBP1. However, ZBP1 expression was only detected in 20% (10/50) of the corresponding metastatic tissues, in which only 7% of cells on average showed transcription sites (Table 1). These data indicate a significant decrease in ZBP1 transcription in metastatic tissues.
Fig. 1.
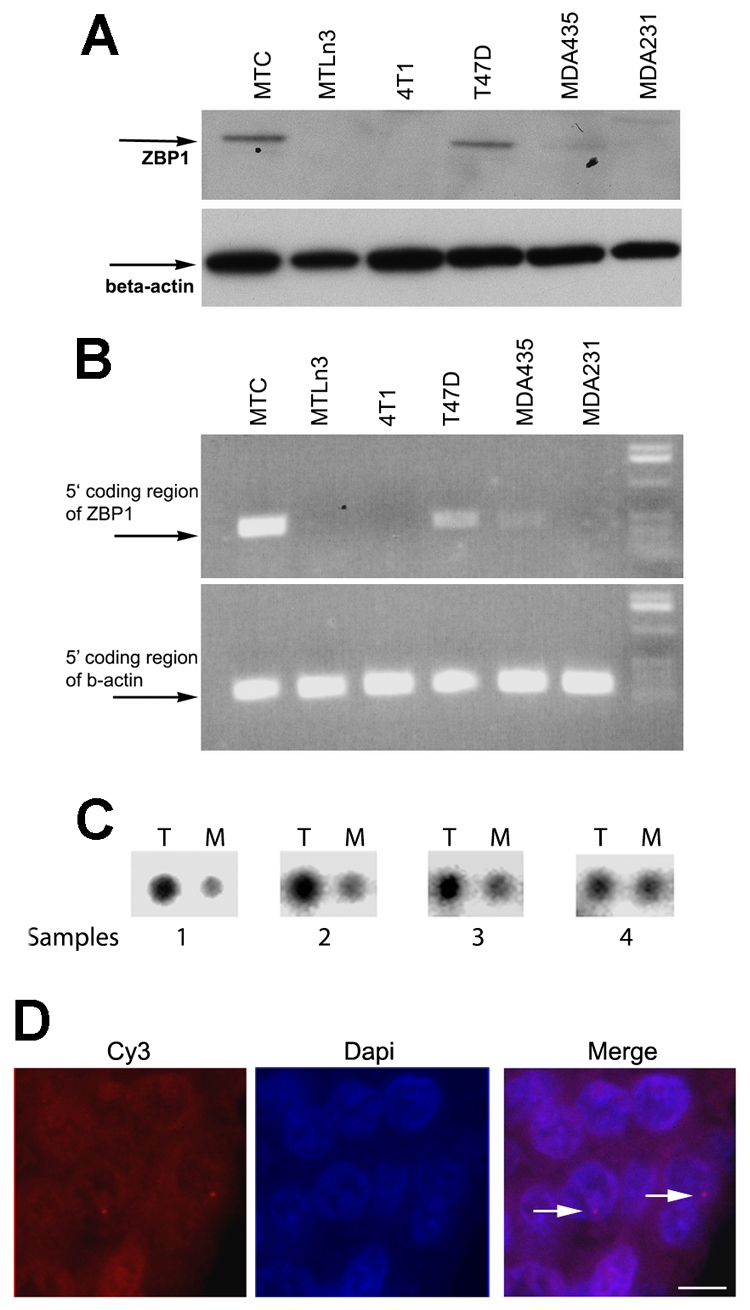
Repression of ZBP1 expression in mammalian breast-cancer cells and tumors. (A) Western blotting of ZBP1 and β-actin expression in breast-cancer cell lines. The arrows indicate the positions of the detected proteins. (B) RT-PCR analyses of ZBP1 and β-actin transcripts in breast-cancer cell lines. (C) A P32-labeled DNA probe encoding ZBP1 was hybridized to a cDNA array containing immobilized cDNA prepared from tumor patients. The array contained four pairs of breast-tumor samples corresponding to malignant tissues and metastatic tissues from the same individual patient (samples 1-4). T, tumor tissues; M, metastatic tissues. (D) Detection of active transcription sites for ZBP1 in paraffin-embedded human breast tumors by in situ hybridization. The image shows pseudo-colored transcription sites for ZBP1 (left, red spots) and DAPI-stained nuclei (middle). Also see Table 1. Arrows point to nascent mRNAs (transcription sites). Scale bar: 10 μm.
Table 1.
Quantification of ZBP1 transcription in human breast and metastatic tissues
| Primary breast tissues | Metastatic tissues | |
|---|---|---|
| % of samples with ZBP1 transcription sites | 48 (24/50) | 20 (10/50) |
|
% of cells with ZBP1 transcription sites in the samples
|
18* |
7* |
P<0.05 (two-tailed Student's t-test).
ZBP1-promoter reporters are effectively expressed in metastatic cells
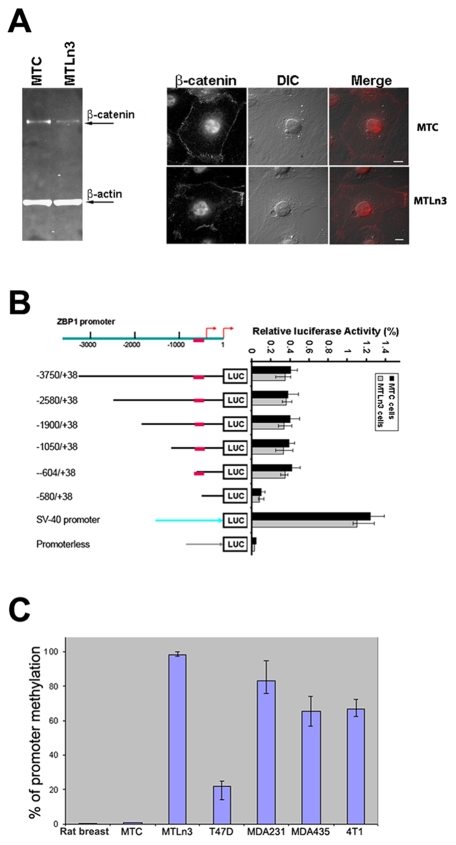
Both in non-metastatic breast-cancer cells and human 293T cells, β-catenin is responsible for activating ZBP1 gene transcription (Gu et al., 2008). To investigate whether repression of ZBP1 expression in metastatic cells was due to the absence of β-catenin signaling, we analyzed β-catenin expression and distribution in cultured MTLn3 cells. Western blots detected considerable levels of β-catenin in nuclear extracts of cultured MTLn3 cells (Fig. 2A, left). Immunofluorescence assays also demonstrated the accumulation of β-catenin within the nucleus of MTLn3 cells (Fig. 2A, right). Thus, ZBP1 repression was not a result of the absence of Wnt–β-catenin signaling.
Fig. 2.
Methylation analyses of the ZBP1 promoter. (A, left) Western blots showing the levels of β-catenin and β-actin in nuclear extracts of MTC and MTLn3 cells. (A, right) Immunofluorescence showing the cellular localization of β-catenin in MTC and MTLn3 cells. An average of 100 cells of each type were examined and representative localization is presented. β-catenin is concentrated in cell nuclei. Scale bars: 10 μm. (B) Analyses of ZBP1 promoter activities in MTLn3 and MTC cells. Left: schematic representation of the luciferase reporter constructs being driven by 5′ nested deletions of the ZBP1 promoter. Relative positions of the promoter region in each construct are marked. The red arrows indicate the two putative transcription initiation sites for the ZBP1 gene. The red block indicates the putative binding element for β-catenin (Gu et al., 2008). Right: ZBP1-promoter constructs and an internal control Renilla luciferase plasmid were co-transfected into cultured MTLn3 cells. The firefly luciferase activity from each construct was normalized to the Renilla luciferase activity and the relative luciferase activity is represented as a percentage of the pSV-40 construct. The results are the average of a total of three independent experiments each carried out in triplicate, ± s.e.m. (C) Methylation status of the ZBP1 promoter in breast-cancer cell lines and normal adult tissues. Genomic DNA was prepared from adult rat breast or cultured MTC, MTLn3, MDA231, MDA435, T47D and 4T1 cell lines, and was subjected to bisulfite-PCR. PCR products were cloned and sequenced. Relative levels of CpG site methylation within the 2-kb ZBP1 promoter is represented as a percentage of total CpG sites. The ZBP1 promoter is highly methylated in metastatic breast-cancer cell lines.
To determine whether metastatic cells have the ability to transcribe luciferase reporters driven by the ZBP1 promoter, we carried out in vivo luciferase assays using various ZBP1-promoter reporters as well as control reporters (Fig. 2B, left) (Gu et al., 2008). The reporter constructs were transiently co-transfected with an internal control, a Renilla luciferase plasmid, into cultured MTC and MTLn3 cells, and the firefly luciferase activity was determined for each construct and normalized with respect to corresponding Renilla luciferase activity. As shown in Fig. 2B, the ectopic ZBP1 promoter was active in both MTC and MTLn3 cells, and the activity of the promoter required the β-catenin-binding site. These data were consistent with our previous results (Gu et al., 2008) and suggested that, although transcription of the endogenous ZBP1 gene in metastatic cells was repressed, the ectopic ZBP1 promoter could transcribe transfected reporters and this transcription was β-catenin dependent, as is the endogenous promoter. Thus, we concluded that the silencing of the endogenous ZBP1 gene was through an epigenetic process.
The ZBP1 promoter is highly methylated in metastatic cells
Promoter methylation frequently plays a role in silencing gene expression during cell development, differentiation and tumorigenesis (Jones and Takai, 2001). To address whether methylation of the ZBP1 promoter could control ZBP1 transcription in metastatic cells, we isolated genomic DNA from cultured MTLn3 and MTC cells and treated it with bisulfite, which converts normal cytosine, but not 5-methylcytosine, to uracil (Clark et al., 1994). We then amplified the 2-kb DNA fragments of the ZBP1 promoter using specific primers designed to avoid potential CpG methylation sites and cloned the fragments. Sequencing of the clones containing the modified ZBP1 promoter from MTLn3 cells revealed that the CpG islands within the promoter were almost entirely methylated. A representative DNA fragment (–1936 to –1300) of the ZBP1 promoter that was completely methylated in the cytosine of CpG islands is shown in supplementary material Fig. S1. By contrast, this fragment was not methylated in MTC cells. These data suggest that repression of expression of the ZBP1 gene correlates with its promoter methylation and that this occurs preferentially in metastatic cells.
Because downregulation of ZBP1 expression was observed in the metastatic cell lines tested (Fig. 1A,B), we isolated genomic DNA from cultured T47D, MDA435, MDA231 and 4T1 cell lines and examined methylation patterns of their ZBP1 promoters. As expected, the CpG islands within the ZBP1 promoter were also heavily methylated in all metastatic cell lines that were tested (Fig. 2D). In addition to the MTLn3 cell line (98%), ratios of methylation of the CpG islands in the 2-kb ZBP1 promoter were 84.4% for MDA231, 65.7% for MDA435 and 71.4% for 4T1. By comparison, the ZBP1 promoter was only slightly methylated in non-metastatic cells, with a ratio of 3% in MTC and 24% in T47D cells. These data strongly suggest that promoter methylation could play a role in silencing the ZBP1 gene in mammary metastatic cells.
ZBP1 is not expressed in mammalian adult tissues or in a non-tumorigenic human breast MCF10A cell line (Gu et al., 2008). To investigate whether this was also correlated with promoter methylation, we examined the methylation pattern of the ZBP1 promoter in rat adult tissues, including breast, liver and brain (Fig. 2C and not shown). Methylation at CpG sites of the ZBP1 promoter was not detected. Immunostaining assays revealed that, in MCF10A cells, which do not express ZBP1, β-catenin was not located in the nucleus (supplementary material Fig. S2). These results suggest that, in contrast to metastatic breast-cancer cells, ZBP1 silencing in adult tissues and differentiated normal cells could be due to the absence of Wnt–β-catenin signaling.
Demethylation of the ZBP1 promoter induces endogenous ZBP1 gene expression in metastatic MTLn3 cells
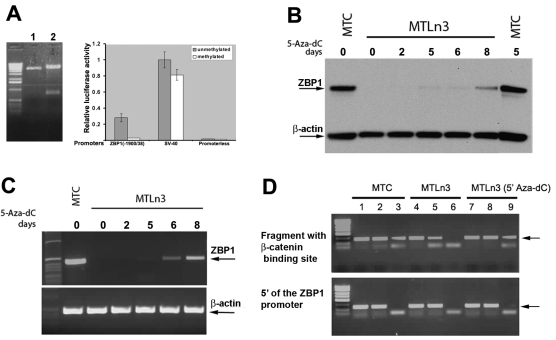
In order to assess whether a causal relationship exists between methylation of the ZBP1 promoter and repression of ZBP1 transcription, we treated two luciferase reporters, one driven by a ZBP1 promoter (–1900/+38, refer to Fig. 2B) and the other by a SV-40 promoter, with a site-specific methylase (SssI), which led to methylation of all CpG sites on the plasmid. In-vitro-methylated or mock constructs were then transiently transfected into MTC cells and luciferase activities were measured. As shown in Fig. 3A, luciferase activity was significantly reduced in MTC cells when the reporter was methylated by SssI. By contrast, in vitro methylation of the SV-40 control vector only decreased the luciferase activity by 20% compared with mock methylation. Thus, the observed repression was due to ZBP1 promoter methylation but not general methylation of the plasmid backbone. This indicates that transactivation of the in-vitro-methylated ZBP1 promoter is repressed in cells in which the endogenous promoter is active.
Fig. 3.
Repression of ZBP1 expression in metastatic cells results from promoter methylation, which prevents the binding of the promoter to β-catenin. (A) In vitro methylation of the ZBP1 promoter leads to repression of its transcriptional activity. Three luciferase constructs were in vitro methylated by SssI methyltransferase. Left: completion of in vitro methylation was evaluated by restriction digestion of methylated (lane 1) or unmethylated (lane 2) constructs with EagI, which recognizes unmethylated CGGCCG sequences. Right: after transfecting the reporters into mTC cells, luciferase activity was measured and normalized. Relative luciferase activity is represented as a percentage of the unmethylated pSV-40 construct. Values are the average of two independent assays ± s.e.m. (B) Western blot analysis of ZBP1 and β-actin protein expression in MTC and MTLn3 cells treated or non-treated with 5-Aza-dC. The arrows indicate the positions of the detected proteins. (C) Analyses of ZBP1 and β-actin mRNA expression in the same cell cultures as shown in B. The arrows indicate the positions of the ZBP1 and β-actin transcripts. (D) MTC cells and MTLn3 cells incubated with or without 5-Aza-dC were subjected to chIP using antibodies against acetylated histone and β-catenin. Total nuclear lysates were used as an input control to purified chromatin DNA. Two primer pairs, one amplifying a region of the ZBP1 promoter containing a putative CTTTG-TC binding site for β-catenin (upper) and the other amplifying a region 2-kb upstream of the ZBP1 transcription site (lower), were used in the experiments. Lanes 1, 4 and 7: inputs of MTC cells, MTLn3 cells and MTLn3 cells treated with 5-Aza-dC. Lanes 2, 5 and 8: precipitates with antibodies against acetylated histone. Lanes 3, 6 and 9: precipitates with antibodies against β-catenin. Anti-β-catenin antibody specifically pulled down the ZBP1 promoter in MTC and 5-Aza-dC-treated MTLn3 cells, but not in parental MTLn3 cells because of the promoter methylation.
To further determine whether demethylation of the ZBP1 promoter was required for gene activation, we treated cultured MTLn3 cells with 5-aza-2-deoxycytidine (5-Aza-dC), which acts as a DNA methyltransferase inhibitor and brings about genome-wide demethylation in dividing cells (Jones and Taylor, 1980). Cells treated with 5-Aza-dC have been shown to reverse expression of the genes that were repressed by DNA methylation (Abe et al., 2005; Moreau et al., 2002; Zhu et al., 2004). Treatment of cultured MTLn3 cells with 5-Aza-dC (5-8 days) activated ZBP1 expression without affecting the expression of β-actin (Fig. 3B). Transactivation of the ZBP1 gene after 5-Aza-dC treatment was also assessed by RT-PCR, which confirmed the protein results (Fig. 3C). Incubation with 5-Aza-dC did not significantly change the expression of ZBP1 mRNA in MTC cells (Fig. 3B). Evaluation of the effectiveness of 5-Aza-dC treatment on demethylation of the ZBP1 promoter showed that methylation levels of the promoter were reduced to 71% on day 2, and reached a maximal reduction of 20% on day 8, when compared with the 98% levels of promoter methylation in untreated cells. At later times the cells stopped growing, presumably owing to toxicity of the compound (Bender et al., 1998). The in vivo demethylation leading to ZBP1 activation indicated that promoter methylation is responsible for repression of endogenous ZBP1 expression in metastatic cells.
β-catenin does not associate with the methylated ZBP1 promoter in metastatic cells
In breast-cancer cells, β-catenin binds to the conserved element CTTTG-TC within the ZBP1 promoter and this interaction is required for promoter activity (the proximal binding site) (Gu et al., 2008). We postulated that, in metastatic cells, CpG methylation of the ZBP1 promoter could alter its structure and block recognition by β-catenin. To address this, we performed chromatin immunoprecipitation (ChIP) analyses (Iyer et al., 2001). Cultured MTC cells and MTLn3 cells with or without treatment of 5-Aza-dC were subjected to ChIP with equal amounts of antibodies against acetylated histone or β-catenin. One primer pair was used to amplify the ZBP1 promoter region containing the putative CTTTG-TC β-catenin–TCF4-binding site, which has an expected size of 260 bp (Fig. 3D, upper). As a negative control, an additional primer pair was targeted to a 280-bp chromatin fragment that lies 2-kb upstream of the transcription initiation site of the ZBP1 gene (Fig. 3D, lower). Anti-histone antibody was able to effectively precipitate both chromatin fragments (Fig. 3D, lanes 2, 5 and 8). β-catenin antibody could pull down the ZBP1 promoter containing the CTTTG-TC element in MTC cells but could not immunoprecipitate the identical fragment in MTLn3 cells, presumably owing to the promoter methylation that blocked β-catenin binding (Fig. 3D, lanes 3 and 6 of upper panel). However, anti-β-catenin antibody could precipitate the CTTTG-TC-containing fragment of the ZBP1 promoter in MTLn3 cells after treatment with 5-Aza-dC (Fig. 3D, lane 9 of upper panel). By contrast, the anti-β-catenin antibody could not precipitate the chromatin fragment upstream of the putative binding element (Fig. 3D, lanes 3, 6 and 9 of lower panel). In addition, DNA was not detected in control ChIP experiments using preimmune serum (not shown). These results strongly suggest that methylation prevented the physical association of β-catenin with the ZBP1 promoter in metastatic cells and thus repressed Wnt–β-catenin-mediated ZBP1 activation.
Knockdown of ZBP1 expression increased cell migration
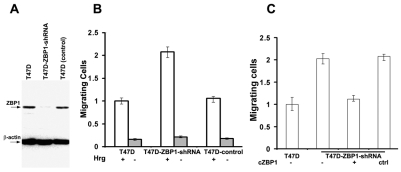
ZBP1 is able to suppress invasion and metastasis of rat breast-carcinoma cells (Lapidus et al., 2007; Wang et al., 2004). We hypothesized that ZBP1 might also function in migration and invasion of human breast cells. To address this issue, we used a lentiviral-based short hairpin RNA (shRNA) expression vector (Stove et al., 2005) to degrade ZBP1 mRNA in T47D cells (T47D-ZBP1-shRNA). shRNA-mediated ZBP1 knockdown was assessed using western blots with antibodies against ZBP1 and β-actin (Fig. 4A). Migration assays were performed using Transwell chambers in the presence of a heregulin gradient (Coniglio et al., 2008). In a control lentivirus-infected T47D cell clone (T47D-control), in which endogenous ZBP1 expression was not affected, the migration ability was not changed compared with T47D parental cells. Cells depleted of ZBP1 had a significant increase of migration through collagen (Fig. 4B). However, the observed migration was inhibited after ZBP1 function was regained by transfecting a chicken ZBP1-expressing plasmid (cZBP1) into T47D-ZBP1-shRNA cells (Fig. 4C), suggesting a role for ZBP1 in repressing migration of human breast-cancer cells. These results were consistent with previous data that ectopic ZBP1 expression reduced migration and metastatic abilities of a rat carcinoma cell line (Lapidus et al., 2007).
Fig. 4.
Loss of ZBP1 promotes cell migration. (A) Western blots showing ZBP1 knockdown in T47D-ZBP1-shRNA cells. (B) Migration assays were performed in T47D, T47D-ZBP1-shRNA and T47D lentivirus control cell clones. Migration was quantified by visual counting of the total cells on the underside of the filter. The relative numbers of migrating cells from each assay are normalized to the T47D clone and are represented as a fold change to the T47D clone. Data shown in the figure represent the means ± s.e.m. of data from six experiments. (C) Gain of ZBP1 function inhibited cell migration. An expression plasmid for chicken ZBP1 was transfected into T47D-ZBP1-shRNA cells and migration assays were performed. Data shown in the figure represent the means ± s.e.m. of data from four experiments.
Gain of ZBP1 function in metastatic MTLn3 cells reduces cell proliferation
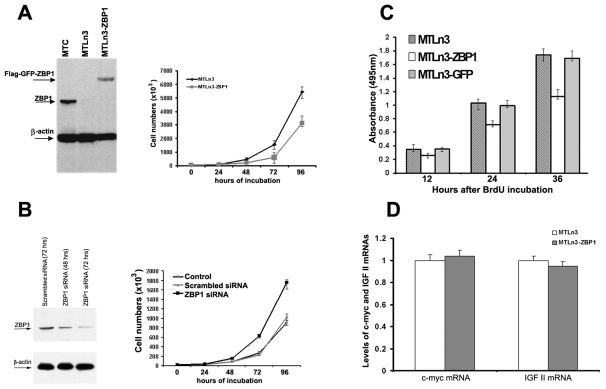
ZBP1 knockdown promotes proliferation of human leukemia cells (Liao et al., 2004). To address whether ZBP1 also affects growth of breast-cancer cells in addition to inhibiting migration and metastasis, we compared the effect of ZBP1 expression on cell proliferation in parental MTLn3 cells and MTLn3 cells constitutively expressing a FLAG-tagged ZBP1 fusion protein (MTLn3-ZBP1) (Lapidus et al., 2007; Wang et al., 2004). Western blotting confirmed expression of ZBP1 in the MTLn3-ZBP1 line (Fig. 5A, left). Accordingly, cell numbers were examined in cells cultured for 4 days. As shown in Fig. 5A (right), expression of ZBP1 markedly reduced cell numbers as assessed by trypan-blue exclusion. Cell numbers decreased by 31.5% at 24 hours, by 55.4% at 48 hours and by 36.6% at 96 hours of incubation, compared with the parental MTLn3 cells lacking ZBP1 expression. To ascertain whether the reduced proliferation was due to ZBP1 re-expression, we used siRNA to induce degradation of ZBP1 mRNA in MTLn3-ZBP1 cells. Western blotting revealed that, upon transfection with ZBP1 siRNA, relative levels of ZBP1 were decreased by approximately 75% in 72 hours when normalizing to β-actin (Fig. 5B). As a result, loss of ZBP1 function caused increased proliferation in MTLn3-ZBP1 cells (Fig. 5B, right). We also measured cell proliferation using a BrdU-incorporation assay (Calbiochem). BrdU is a substitute of [3H]-thymidine, which is incorporated into newly synthesized DNA strands during cell proliferation (Cory et al., 1991). Detection of BrdU incorporation allows the assessment of cell populations that are actively synthesizing DNA. Consistently, gain of ZBP1 activity decreased BrdU incorporation by 42% compared with MTLn3 parental cells and a control MTLn3 clone expressing GFP (Fig. 5C). These experiments indicate that ZBP1 gene silencing promotes MTLn3 cell proliferation.
Fig. 5.
Expression of ZBP1 in metastatic cells suppresses cell proliferation. (A, left) Expression of ZBP1 in MTC, MTLn3 and MTLn3-ZBP1 cells. FLAG-ZBP1 is constantly expressed in the stable MTLn3-ZBP1 cell line. The arrows indicate the positions of the detected proteins. (A, right) Growth curve of MTLn3 and MTLn3-ZBP1 cells. Expression of ZBP1 in metastatic MTLn3 cells markedly reduced their proliferation rate. Data shown in the figure represent the means ± s.e.m. of data from three experiments, each with duplicates. (B, left) Western blot analysis was performed to examine ZBP1 and β-actin expression in MTLn3-ZBP1 cells after transfection with scrambled or ZBP1 siRNAs. (B, right) MTLn3-ZBP1 cell numbers were determined at the indicated time points after transfection with scrambled or ZBP1 siRNAs. Knockdown of ZBP1 function in MTLn3-ZBP1 cells increased the number of viable cells. Data shown in the figure represent the means ± s.e.m. of data from four independent experiments. (C) Effect of ZBP1 on proliferation of MTLn3, MTLn3-ZBP1 and MTLn3-GFP cells, assessed by the BrdU-incorporation assay. The absorbance at 495 nm is proportional to the BrdU incorporation and to the number of viable cells in the culture. The experiments were repeated twice, and five wells were used for each individual cell clone. (D) Cytoplasmic levels of Myc, IGF2 and β-actin mRNAs were detected in MTLn3 and MTLn3-ZBP1 cells by RT-PCR analyses. Relative levels of Myc and IGF2 mRNAs are normalized and averaged to β-actin mRNA from three independent experiments.
ZBP1 is also capable of regulating Myc mRNA and IGF2 mRNA expression, both of which are directly related to cell growth (Bernstein et al., 1992; Ioannidis et al., 2005; Kobel et al., 2007; Lemm and Ross, 2002). To determine whether the observed effect of ZBP1 expression on proliferation of metastatic cells was a result of controlling Myc mRNA or IGF2 mRNA expression, we examined the steady-state levels of Myc and IGF2 mRNAs in parental MTLn3 cells, and transformed MTLn3-ZBP1 cells by quantitative RT-PCR. No significant changes of Myc or IGF2 mRNA expression were observed in the cells with or without ZBP1 expression (Fig. 5D). We postulated that ZBP1 function in repressing cell migration and proliferation could involve the regulation of mRNAs other than Myc and IGF2.
Identification of ZBP1-bound mRNPs in breast-cancer cells
To identify mRNAs that associate with ZBP1 mRNP in breast-cancer cells, we affinity purified ZBP1 mRNP particles from MTLn3-ZBP1 cells and performed microarray analyses that allowed us to identify previously uncharacterized ZBP1 targets (Gu et al., 2008). Among the identified transcripts, many play roles in aberrant cancer-cell proliferation and metastasis. Some of these transcripts, including β-catenin, cyclin-D1, E-cadherin and ARP2/3-complex mRNAs, were selected and are listed in Table 2. In a previous study, using SELEX assays Farina et al. identified a degenerate `ACACC' conserved motif for ZBP1 binding (Farina et al., 2003). This conserved motif, known as the `zipcode' (Kislauskis and Singer, 1992), was necessary for β-actin-mRNA targeting, and for its localized translation in motile cells and neurons (Huttelmaier et al., 2005). We found that, within the 17 selected transcripts from the microarray, 12 contained the ACACC conserved elements either in their 3′ UTR or in the coding region (Table 2).
Table 2.
Selected mRNAs associated with ZBP1 in breast-cancer cells
| Rank | Transcripts | Gene bank ID | Location of conserved `zipcode' sequences | Scald fold enrichment MTLn3-ZBP1/MTLn3 | Biological functions |
|---|---|---|---|---|---|
| A1 | β-actin | NP_112406 | 3′ UTR | 223.1 | Cytoskeleton; cell motility |
| A2 | P21-ARC (subunit 3) | NM_001105933 | Coding region | 215.0 | Actin cytoskeleton regulation |
| A3 | Galectin-3 | NP_114020 | Absent | 194.6 | Heterophilic cell adhesion |
| A4 | Endolyn | NM_031812 | Coding region | 83.9 | Cell adhesion; cell proliferation |
| A5 | P16-ARC (subunit 5) | NM_001037767 | Absent | 60.5 | Actin cytoskeleton regulation |
| A6 | Evl (RNB6) | NM_024147 | Coding region | 42.2 | Actin-filament-based movement; cell motility |
| A7 | Actinin α1 | NP_112267 | Coding region | 33.6 | Actin cross-linking; actin organization |
| A8 | E-cadherin | NP_112624 | 3′ UTR | 33.1 | Cell adhesion |
| A9 | β-catenin | NP_445809 | Coding region | 26.5 | Cell-cell contacts; Wnt signaling |
| B1 | Cyclin D1 | NP_741989 | Coding region | 241.4 | Cell proliferation; Wnt signaling |
| B2 | Hepatoma-derived growth factor | NP_446159 | 3′ UTR | 60 | Cell proliferation |
| B3 | Cyclin A2 | NM_053702 | Absent | 28.7 | Cell proliferation |
| B4 | Anxa1 | NP_037036 | Absent | 48.4 | Regulation of cell proliferation |
| B5 | Erv1, growth factor homolog | NP_037354 | Coding region | 32.7 | Cell proliferation |
| B6 | Rcl | NP_598209 | Absent | 21.9 | Cell proliferation; putative Myc-responsive |
| B7 | Cyclin G1 | NP_037055 | Coding region | 21.8 | Cytokinesis; mitosis; cell-cycle regulation |
| B8 | Frk (Src-related tyrosine kinase) | NM_024368 | Coding region | 19.4 | Cell differentiation |
Microarray experiments and data analyses were performed in the Center for Functional Genomics at the University of Albany. Two categories of 17 transcripts were selected from the microarray results. One category of mRNAs contained transcripts important for cell motility and adhesion (rank A1-A9). The second category of ZBP1-bound mRNAs included those necessary for cell mitosis and growth (rank B1-B8). Location of the conserved `zipcode' sequences within the transcripts is indicated.
ZBP1 selectively regulates its associated mRNAs in breast cancer cells
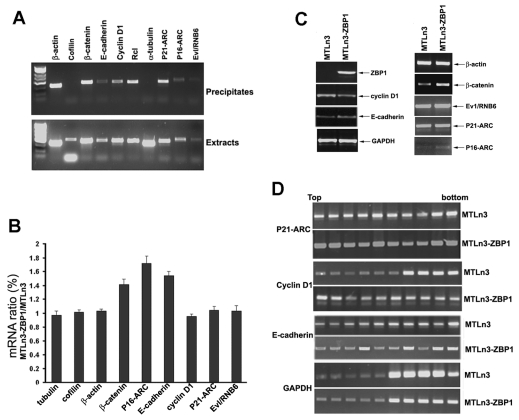
To confirm that the microarray-identified transcripts were bound to endogenous ZBP1 in vivo, we used anti-ZBP1 antibodies to immunoprecipitate extracts of MTC cells and isolated ZBP1-associated RNA. We selected eight microarray-enriched RNAs, which have been implicated as being encoded by important genes involved in the induction of breast carcinoma and in promoting cell proliferation, and two control transcripts, cofilin and α-tubulin, and used RT-PCR to determine whether the immunoprecipitates contained these transcripts (Fig. 6A). All selected transcripts except the cofilin and α-tubulin mRNAs were significantly precipitated by the anti-ZBP1 antibodies. A preimmune serum did not demonstrate any PCR-amplified DNA fragments (data not shown). These experiments indicated the physical association of the selected transcripts with ZBP1 in cancer cell extracts. We speculated that binding of ZBP1 to these transcripts could affect their cytoplasmic stability or translation. To determine whether ZBP1 could stabilize its bound mRNAs, we treated MTLn3-ZBP1 and parental MTLn3 cells with actinomycin D for 60 minutes and analyzed the changes in ZBP1-associated and non-associated (α-tubulin) mRNAs. Ratios of mRNA decay were normalized to GAPDH mRNA. As shown in Fig. 6B, cytoplasmic levels of β-catenin, P16-ARC and E-cadherin mRNAs were 41%, 76% and 54% higher in the cells expressing ZBP1, respectively, compared with cells not expressing ZBP1. The levels of β-actin, cyclin-D1, P21-ARC (ARPC3) and Evl (RNB6) mRNAs remained unchanged. We also identified the binding of ZBP1 with these mRNAs in human T47D cells and observed that the levels of E-cadherin and β-catenin mRNAs were reduced when ZBP1 was depleted (data not shown).
Fig. 6.
ZBP1 selectively regulates the turnover of its associated mRNAs in breast-cancer cells. (A) Anti-ZBP1 antibody was used to immunoprecipitate (IP) an MTC extract (upper panel). Total RNA, isolated from the precipitates and the cell extracts, was subjected to RT-PCR to detect the presence of ZBP1-associated mRNAs. Cofilin and α-tubulin mRNAs were used as negative controls. (B) ZBP1 selectively stabilizes its associated mRNAs in breast-cancer cells. Total RNAs were extracted from parental MTLn3 and MTLn3-ZBP1 cells after treatment with actinomycin D for 60 minutes. The relative change of indicated mRNA levels was analyzed by RT-PCR and was normalized to the corresponding α-tubulin mRNA. (C) Western blots showing the protein expression of ZBP1-bound mRNAs in MTLn3 and MTLn3-ZBP1 cells. Levels of β-catenin, P16-ARC and E-cadherin were increased, whereas the level of cyclin D1 was decreased in the presence of ZBP1. (D) Extracts of MTLn3 and MTLn3-ZBP1 cells were fractionated in 10-50% linear sucrose gradients. An equal volume of RNA from each fraction was subjected to RT-PCR for P21-ARE, cyclin-D1 and GAPDH mRNAs. Polysomal distribution of the mRNA in fractions of the sucrose gradient is shown in 1.5% agarose gels.
We then examined protein expression of the mRNAs (Fig. 6C). Western analysis showed that the levels of E-cadherin, β-catenin and P16-ARC in MTLn3-ZBP1 cells were higher than those in MTLn3 parental cells. Apparently, these changes resulted from their mRNA stability. An interesting observation was that the levels of cyclin D1 were significantly decreased in MTLn3 cells when ZBP1 was expressed (Fig. 6C). To address the possibility that ZBP1 could also regulate the translation of associated mRNAs in breast-cancer cells, we performed sucrose-gradient fractionations from MTLn3-ZBP1 and MTLn3 cells. Western blots showed that ZBP1 was distributed in both non-polysomal and polyribosomal fractions. Treatment of the cytoplasmic extracts with EDTA shifted ZBP1 from polyribosomal fractions into non-polysomal fractions, suggesting the disassociation of ZBP1 from polysomes (supplementary material Fig. S3). We isolated total RNA from the sucrose fractions and analyzed the translational profiles of P21-ARC, cyclin D1, E-cadherin and control GAPDH mRNAs. Levels of cyclin-D1 mRNA were significantly reduced in polyribosomal fractions of MTLn3-ZBP1 cells in comparison with parental MTLn3 cells, indicating that ZBP1 could inhibit polysomal loading of cyclin-D1 mRNA (Fig. 6D). By comparison, the levels of polysomal-bound fractions of P21-ARC and E-cadherin mRNAs were not changed (Fig. 6D). Taken together, these results suggest that ZBP1 can selectively regulate either the stability or the translation of associated mRNAs.
Discussion
ZBP1 is a multifunctional RNA-binding protein that specifically recognizes RNA targets containing the zipcode and affects their post-transcriptional fate by determining their subcellular localization, modulating their stability or translatability. The ability of ZBP1 as a RNA regulator has been associated with many cellular processes, including cell polarity, cell proliferation, tumor induction and metastasis (Condeelis and Singer, 2005; Lapidus et al., 2007; Liao et al., 2004; Tessier et al., 2004; Wang et al., 2004). We have previously shown that, in breast cancers, ZBP1 activation is a cellular response to Wnt–β-catenin signaling, which is frequently active in embryogenesis and tumorigenesis. β-catenin specifically binds to the conserved cis-element of the ZBP1 promoter and this binding transactivates ZBP1 expression (Gu et al., 2008). In the current study, we identified the molecular mechanism of ZBP1 repression in metastatic breast-cancer cells. We demonstrated that downregulation of the ZBP1 gene is a result of its promoter methylation, an epigenetic process that modifies chromatin structure and silences gene expression. Epigenetic methylation of the ZBP1 promoter disrupted its interaction with β-catenin, leading to inactivation of the gene.
It is becoming apparent that epigenetic abnormalities result in activation of oncogenes and in silencing of tumor suppressor genes in cancer cells (El-Osta, 2004; Jones and Baylin, 2002; Momparler, 2003). Similarly to ZBP1, a number of breast-cancer-related genes, including those encoding E-cadherin, TIMP3, human MLH1 and VHL, are silenced by DNA methylation (Momparler, 2003; Tsou et al., 2002). However, unlike those genes, the expressions of which are suppressed during carcinogenesis, ZBP1 inactivation by epigenetic methylation occurs in metastasis. In contrast to other metastatic cell lines that were tested, a weak ZBP1 expression is observed in MDA435 cells, although the promoter is highly methylated (65%). It is possible that the cell lines derived from different origins have altered responses to promoter methylation or that there is the presence of leakage from the methylated ZBP1 promoter in MDA435 cells. Repression of ZBP1 expression is also observed in human clinical samples, suggesting that the de novo repression of ZBP1 might serve as a useful prognostic marker for metastasis of breast cancers.
The mechanism responsible for repressing ZBP1 expression in metastatic breast-cancer cells is obviously different from that in normal breast cells, in which the ZBP1 promoter is not methylated. We hypothesize that ZBP1 silencing in normal cells might result from the absence of Wnt–β-catenin signaling. Supportive evidence is that non-tumorigenic MCF10A cells do not localize β-catenin in their nucleus nor do they express ZBP1 (supplementary material Fig. S2) (Gu et al., 2008). Because β-catenin plays roles in both transcriptional activation and cell adhesion (Gavert and Ben-Ze'ev, 2007), it is likely that normal adult cells use the protein preferentially as a cell-adhesion component and have insufficient protein in the nucleus to activate ZBP1 expression (Harris and Peifer, 2005; Nelson and Nusse, 2004).
Our results reveal that loss of ZBP1 function increases migration of human breast-cancer cells. This is consistent with previous data that showed that ZBP1 repression facilitated the invasive potential of rat carcinoma cells (Lapidus et al., 2007; Wang et al., 2004). Interestingly, we also observed that ZBP1 repression promotes proliferation of metastatic cells. It is not likely that the increased proliferation results in migration or invasion of metastatic cells. Vikesaa et al. have reported that expression of the human homologs of ZBP1 (IMP1 and IMP3) are positively correlated with the invasive capacity of cancer cells, but are not involved in cell proliferation (Vikesaa et al., 2006). Another example is that members of the Snail protein family trigger epithelial-mesenchymal transitions during tumor progression but repress cell proliferation (Vega et al., 2004).
The observation that ZBP1 is capable of reducing proliferation is in agreement with a report that showed that targeted ZBP1 knockdown increased the proliferation rates of leukemia K562 cells (Liao et al., 2004), but is contradictory to the results that showed that downregulation of ZBP1 function in MCF7 and ES-2 cells is accompanied by decreased cell proliferation rates (Ioannidis et al., 2005; Kobel et al., 2007). One way to reconcile these contradictory results is to recognize that cancer cells arising from different origins behave differently in response to changes in ZBP1 expression. Cells have distinct RNA targets that are post-transcriptionally regulated by ZBP1. These cells might be differentially affected depending on the complexity of the mRNA targets of ZBP1. ZBP1 knockdown in a particular cell line can cause a noticeable change in the levels of mRNA it regulates, leading to changes in cell polarity, migration and proliferation. For instance, ZBP1 could promote cancer progression by controlling the expression of cell-adhesion molecules, or inhibit cell invasion and chemotaxis-like movement by regulating localized translation of mRNAs that are involved in motility (Condeelis and Singer, 2005; Huttelmaier et al., 2005).
We therefore hypothesize that the plethora of mRNA targets present in the cells could have a variety of effects on cell proliferation and metastasis when bound to ZBP1. On the basis of this hypothesis, we employed microarrays and identified mRNAs that are associated with ZBP1 in breast-cancer cells. The results suggest that roughly 3% of the transcripts in breast-cancer cells are possible targets for ZBP1 (Gu et al., 2008). However, it might be only a few mRNAs, perhaps those with the greatest binding affinity to ZBP1, that are mostly affected by ZBP1 expression (Table 2). One category of mRNAs contains transcripts that are important for cell motility and adhesion (Table 2, A1-A9). Among these transcripts, cytoplasmic expression of β-actin and β-catenin mRNAs has previously been shown to be regulated by ZBP1 (Huttelmaier et al., 2005). Others, such as ARP-21 and ARP-16 (two components of the Arp2/3 complex), E-cadherin (a transmembrane protein involved in establishing cell polarity and maintaining normal tissue morphology) and Evl (an actin-associated protein), are all required to be properly targeted for function (Biliran et al., 2005; Gumbiner, 2000; Mingle et al., 2005). In the group of transcripts that are important for cell motility and adhesion, three – β-catenin, P16-ARC and E-cadherin – have been shown to be selectively stabilized by ZBP1, indicating that ZBP1 regulation of these mRNAs could strengthen cell-cell contacts and maintain an intrinsic polarity, thus inhibiting tumor-cell invasion. A second category of ZBP1-associated mRNAs includes those that are necessary for cell mitosis and growth (Table 2, B1-B8); within this group, cyclin D1 (Ccnd1) has been implicated as an important gene involved in induction of breast carcinoma by promoting cell-cycle progression (Yu et al., 2001). In MTLn3 cells, ZBP1 re-expression effectively suppressed cell proliferation; gain of ZBP1 function also reduced the polyribosomal loading of Ccnd1 mRNA. These results indicate that the role of ZBP1 on metastatic-cell proliferation could be through regulation of growth-related genes, such as Ccnd1 expression. Noticeably, most of the transcripts identified (Table 2) contain a conserved zipcode (Farina et al., 2003), which is consistent with their binding to ZBP1. Supportive evidence is that ZBP1 binds to a zipcode-containing region of β-catenin mRNA (Gu et al., 2008). We expect that microarray identification of the ZBP1 targets in this study will permit further investigation of the role of ZBP1 and the physiological relevance of its activation to cancer-cell invasion and proliferation.
On the basis of these data, we propose the following mechanism. In some types of breast-cancer cells, the ZBP1 gene is activated as a result of a cellular response to Wnt–β-catenin signaling. ZBP1 activation represses metastasis and proliferation through controlling the post-transcriptional expression of mRNAs to which it binds. However, when cancer cells metastasize, the chromatin-remodeling pathway methylates the ZBP1 promoter, altering the promoter structure and inactivating ZBP1 gene transcription by preventing the binding of the β-catenin–TCF4 complex. ZBP1 repression results in misregulation of its target transcripts, leading to enhanced invasive potential and proliferation of cancer cells. This mechanism is supported by our previous experiments (Lapidus et al., 2007; Wang et al., 2004) and by the data in this study.
Materials and Methods
Cell culture, transfection and luciferase assay
MTLn3 and MTC cells were cultured in α-MEM supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) (Wang et al., 2002). MDA231, MDA435, T47D and 4T1 cell lines were grown in DMEM supplemented with 10% FBS and 1% P/S in a humidified incubator at 37°C with 5% CO2. For transfection, cells were harvested and transiently co-transfected with a mixture containing 1.0 μg of the individual luciferase reporter plasmids and 0.1 μg of an internal control plasmid by using a nuclearfector (Lonza). After transfection, the cells were placed into wells of the 96-multiwell plastic plate and cultured for 16-24 hours. Where indicated, cells transfected with a promoterless pG2 vector were used as a negative control. Luciferase assays were performed as previously described (Gu et al., 2008).
Western blotting
Primary antibodies used in western blotting were mouse anti-β-actin, mouse anti-β-catenin, mouse anti-cyclin-D1, mouse anti-GPDH and polyclonal anti-ZBP1 antibodies (laboratory-generated). The western blots were developed using the ECL method.
Northern blot analyses and RT-PCR
Total RNA was isolated from cultured cell lines and from adult rat tissues using a RAeasy Mini Kit (Qiagen). Northern blots and RT-PCR were performed as previously described (Gu et al., 2008).
Immunofluorescence and imaging
Cells cultured on glass coverslips were permeabilized with 0.5% Triton X-100 and fixed with 3% PFA in PBS. The coverslips were incubated with monoclonal antibody against β-catenin, which was purchased from Abcam (Cambridge, MA). The secondary antibodies were Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories).
RNA FISH to detect ZBP1 transcription sites in paraffin-embedded tissues
Paraffin-embedded tissue FISH was performed as described (Capodieci et al., 2005). Fluorescent signals were detected with an Olympus BX61 microscope and analyzed using IPLab software version 3.61 (BD Biosciences). Only nuclei located entirely within the imaged field were scored for transcription sites.
In vitro methylation of the constructs
Luciferase reporters driven by the ZBP1 promoter (–1900/+38) or SV-40 promoter were incubated with SssI DNA methyltransferase (New England Biolab), which led to in vitro methylation of all cytosine residues at the CpG sites of the constructs. Completion of in vitro methylation was ascertained by cleavage of the constructs with EagI and subsequent analysis by electrophoresis on a 1% agarose gel and ethydinebromide staining.
Bisulfite genomic modification and DNA methylation assays
Genomic DNAs were prepared from cultured cells and rat adult spleen and liver tissues with a DNeasy Kit from Qiagen. Bisulfite modification of genomic DNA was performed using an `EZ DNA Methylation Kit' (Zymo Research) following the manufacturer's instructions. For PCR amplification of the ZBP1 promoter after bisulfite modification independent of methylation status, primers were designed that anneal specifically to converted (C-to-T) sequences. Amplificates were then purified and ligated into a Topo vector (Invitrogen). Plasmid containing the PCR products was prepared (Qiagen Mini Plasmid Kit) and sequenced in AECOM DNA sequencing facility. An average of five to ten clones from each individual ligation were sequenced and analyzed. The percentage of CpG methylation sites in the ZBP1 promoter was determined.
5-Aza-dC treatment
Various amounts of 5-Aza-dC (2-10 μM) were added into MTLn3 and MTC cultures, and the cells were incubated for 2-8 days. Culture medium was changed every 2 days. Genomic DNA, total RNA and cell lysates were prepared form 5-Aza-dC-treated or untreated cell cultures and were subjected to bisulfite-PCR for methylation assays, and RT-PCR and western blotting for protein and RNA analyses.
RNA interference
ZBP1 siRNA was transfected into MTLn3 cells with Lipofectamine 2000 (Invitrogen) as previously described (Huttelmaier et al., 2005). Cells were analyzed 24-96 hours after transfection by either western blotting or RT-PCR.
ChIP assays
ChIP experiments were performed as mentioned previously (Iyer et al., 2001; Pan et al., 2007). For PCR after ChIP experiments, 1-2 μl of a 50 μl DNA extraction and 20-30 cycles of amplification were used.
Isolation of ZBP1 mRNP complexes, RNA extraction and microarray analysis
Isolation of ZBP1 mRNP complexes in MTLn3-ZBP1 cells and RNA extraction were carried out as previously described (Gu et al., 2008). Briefly, MTLn3 and MTLn3-ZBP1 (a MTLn3 stable cell line constitutively expressing FLAG-tagged ZBP1) cells were lysed in an ice-cold lysis buffer containing 10 mM HEPES, pH 7.8, 40 mM NaCl, 10 mM KCl, 0.5% NP-40, 0.5 μg/ml PMSF, and 1× protease inhibitor mixture (Roche). Supernatants were obtained after high-speed centrifugation (21,000 g for 60 minutes at 4°C) and were incubated with FLAG-specific monoclonal antibody M2 covalently coupled agarose beads (Sigma) at 4°C in the presence of RNase inhibitor (250 units/ml). After overnight incubation, the supernatant was removed by a short centrifugation and the beads were extensively washed in lysis buffer followed by adding 1 ml of TRIzol. RNAs were extracted in TRIzol as described by the manufacturer (Invitrogen).
RNA microarray analyses and data assays were performed in the Center for Functional Genomics at University of Albany using a Rat Genome 230 Plus 2.0 Array. The data was analyzed using the Affymetrix software and filtered for probe sets `present' and `increased' in MTLn3-ZBP1 RNA with a 10.0-fold change or greater.
Lentivirus generation and infection
A pGIPZ lentiviral vector encoding an shRNA for ZBP1 was purchased from Open Biosystems (ID: V2LMM_64131). The sequence for ZBP1 shRNA was 5′-CCTGAAGATCCTGGCTCATAA-3′. Lentiviruses were generated by co-transfecting 20 μg of lentiviral vector and 1 μg of five individual packaging vectors (coding for Gag, Pol, Tat, Rev and VSVG) in HEK 293T cells by using Fugene 6 Transfection Reagent (Roche). Supernatants were collected 48 hours after transfection and filtered through a 0.4-μm membrane. Viral particles were pelleted by centrifugation at 100,000 g for 2 hours and resuspended in 200 μl serum-free medium. 50 μl of the viral suspension was mixed with 0.5 ml culture medium (serum-free) and used to infect T47D cells that were seeded in a six-well dish with 20% confluence in the presence of 3 μg polybrene (Sigma). Stably transfected cell clones were selected by puromycin.
Migration assay
Cell migration was tested using 8-μm-pore polycarbonate membrane Transwell chambers (Corning Costar Products, Acton, MA) as previously described (Coniglio et al., 2008; Marone et al., 2004).
Cell-proliferation assays
Equal amounts of MTLn3 and MTLn3-ZBP1 cells were seeded and cultured for 24, 48, 72 and 96 hours. Cell counts were obtained by standard trypan blue (Sigma) exclusion according to the manufacturer's protocol. The BrdU-incorporation assay was performed using the BrdU Cell Proliferation Assay Kit following the manufacturer's instruction (Calbiochem). A total of 20 μl of BrdU-labeling reagent was added to each well of a 96-well culture plate containing tested cells in 100 μl (4×105/ml) of culture medium. Cells were incubated for periods of 12, 24 and 36 hours at 37°C. The absorbance at 495 nm was measured using a 96-well plate reader (Tecan). The 495-nm absorbance was proportional to the number of proliferating cells in the culture.
Sucrose-gradient fractionation
MTLn3 and transformed MTLn3 (MTLn3-ZBP1) cells were cultured to 50-70% confluency. After incubating with cycloheximide (100 μg/ml) to arrest polyribosome migration, cells were harvested and lysed. Cell lysates were centrifuged for 30 minutes at 21,000 g and the supernatants were loaded onto 10 ml, 10-50% linear sucrose gradients and fractionated at 35,000 r.p.m. for 2 hours in an SW41 rotor (Beckman). Fractions (1.0 ml each) were collected from top to bottom and the OD254 profile was monitored.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/11/1895/DC1
We thank Jeffrey Segall and John Condeelis for providing the metastatic breast-cancer cell lines and Kyle Lapidus for providing the MTLn3-ZBP1 stable cell line. We also thank Daniel Medrano for help in methylation assays and members of the Singer Lab for valuable discussions. This work was supported by NIH grants CA83208 and AR41480 to R.H.S. Deposited in PMC for release after 12 months.
References
- Abe, T., Toyota, M., Suzuki, H., Murai, M., Akino, K., Ueno, M., Nojima, M., Yawata, A., Miyakawa, H., Suga, T. et al. (2005). Upregulation of BNIP3 by 5-aza-2′-deoxycytidine sensitizes pancreatic cancer cells to hypoxia-mediated cell death. J. Gastroenterol. 40, 504-510. [DOI] [PubMed] [Google Scholar]
- Bender, C. M., Pao, M. M. and Jones, P. A. (1998). Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 58, 95-101. [PubMed] [Google Scholar]
- Bernstein, P. L., Herrick, D. J., Prokipcak, R. D. and Ross, J. (1992). Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 6, 642-654. [DOI] [PubMed] [Google Scholar]
- Biliran, H., Jr, Wang, Y., Banerjee, S., Xu, H., Heng, H., Thakur, A., Bollig, A., Sarkar, F. H. and Liao, J. D. (2005). Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin. Cancer Res. 11, 6075-6086. [DOI] [PubMed] [Google Scholar]
- Capodieci, P., Donovan, M., Buchinsky, H., Jeffers, Y., Cordon-Cardo, C., Gerald, W., Edelson, J., Shenoy, S. M. and Singer, R. H. (2005). Gene expression profiling in single cells within tissue. Nat. Methods 2, 663-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. J., Harrison, J., Paul, C. L. and Frommer, M. (1994). High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis, J. and Singer, R. H. (2005). How and why does beta-actin mRNA target? Biol. Cell 97, 97-110. [DOI] [PubMed] [Google Scholar]
- Coniglio, S. J., Zavarella, S. and Symons, M. H. (2008). Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanisms. Mol. Cell. Biol. 28, 4162-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory, A. H., Owen, T. C., Barltrop, J. A. and Cory, J. G. (1991). Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3, 207-212. [DOI] [PubMed] [Google Scholar]
- El-Osta, A. (2004). The rise and fall of genomic methylation in cancer. Leukemia 18, 233-237. [DOI] [PubMed] [Google Scholar]
- Farina, K. L., Huttelmaier, S., Musunuru, K., Darnell, R. and Singer, R. H. (2003). Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160, 77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert, N. and Ben-Ze'ev, A. (2007). beta-Catenin signaling in biological control and cancer. J. Cell Biochem. 102, 820-828. [DOI] [PubMed] [Google Scholar]
- Gu, W., Wells, A. L., Pan, F. and Singer, R. H. (2008). Feedback regulation between ZBP1 and {beta}-catenin mRNAs in breast cancer cells. Mol. Cell. Biol. 28, 4963-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner, B. M. (2000). Regulation of cadherin adhesive activity. J. Cell Biol. 148, 399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T. J. and Peifer, M. (2005). Decisions, decisions: beta-catenin chooses between adhesion and transcription. Trends Cell Biol. 15, 234-237. [DOI] [PubMed] [Google Scholar]
- Huttelmaier, S., Zenklusen, D., Lederer, M., Dictenberg, J., Lorenz, M., Meng, X., Bassell, G. J., Condeelis, J. and Singer, R. H. (2005). Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512-515. [DOI] [PubMed] [Google Scholar]
- Ioannidis, P., Trangas, T., Dimitriadis, E., Samiotaki, M., Kyriazoglou, I., Tsiapalis, C. M., Kittas, C., Agnantis, N., Nielsen, F. C., Nielsen, J. et al. (2001). C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int. J. Cancer 94, 480-484. [DOI] [PubMed] [Google Scholar]
- Ioannidis, P., Mahaira, L., Papadopoulou, A., Teixeira, M. R., Heim, S., Andersen, J. A., Evangelou, E., Dafni, U., Pandis, N. and Trangas, T. (2003a). 8q24 Copy number gains and expression of the c-myc mRNA stabilizing protein CRD-BP in primary breast carcinomas. Int. J. Cancer 104, 54-59. [DOI] [PubMed] [Google Scholar]
- Ioannidis, P., Mahaira, L., Papadopoulou, A., Teixeira, M. R., Heim, S., Andersen, J. A., Evangelou, E., Dafni, U., Pandis, N. and Trangas, T. (2003b). CRD-BP: a c-Myc mRNA stabilizing protein with an oncofetal pattern of expression. Anticancer Res. 23, 2179-2183. [PubMed] [Google Scholar]
- Ioannidis, P., Mahaira, L. G., Perez, S. A., Gritzapis, A. D., Sotiropoulou, P. A., Kavalakis, G. J., Antsaklis, A. I., Baxevanis, C. N. and Papamichail, M. (2005). CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J. Biol. Chem. 280, 20086-20093. [DOI] [PubMed] [Google Scholar]
- Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. and Brown, P. O. (2001). Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533-538. [DOI] [PubMed] [Google Scholar]
- Jones, P. A. and Taylor, S. M. (1980). Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85-93. [DOI] [PubMed] [Google Scholar]
- Jones, P. A. and Takai, D. (2001). The role of DNA methylation in mammalian epigenetics. Science 293, 1068-1070. [DOI] [PubMed] [Google Scholar]
- Jones, P. A. and Baylin, S. B. (2002). The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415-428. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E. H. and Singer, R. H. (1992) Determinants of mRNA localization. Curr. Opin. Cell Biol. 4, 975-978. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E. H., Zhu, X. and Singer, R. H. (1997). beta-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136, 1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel, M., Weidensdorfer, D., Reinke, C., Lederer, M., Schmitt, W. D., Zeng, K., Thomssen, C., Hauptmann, S. and Huttelmaier, S. (2007). Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene 26, 7584-7589. [DOI] [PubMed] [Google Scholar]
- Lapidus, K., Wyckoff, J., Mouneimne, G., Lorenz, M., Soon, L., Condeelis, J. S. and Singer, R. H. (2007). ZBP1 enhances cell polarity and reduces chemotaxis. J. Cell Sci. 120, 3173-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds, P., Kren, B. T., Boylan, J. M., Betz, N. A., Steer, C. J., Gruppuso, P. A. and Ross, J. (1997). Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14, 1279-1286. [DOI] [PubMed] [Google Scholar]
- Lelekakis, M., Moseley, J. M., Martin, T. J., Hards, D., Williams, E., Ho, P., Lowen, D., Javni, J., Miller, F. R., Slavin, J. et al. (1999). A novel orthotopic model of breast cancer metastasis to bone. Clin. Exp. Metastasis 17, 163-170. [DOI] [PubMed] [Google Scholar]
- Lemm, I. and Ross, J. (2002). Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 22, 3959-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, B., Patel, M., Hu, Y., Charles, S., Herrick, D. J. and Brewer, G. (2004). Targeted knockdown of the RNA-binding protein CRD-BP promotes cell proliferation via an insulin-like growth factor II-dependent pathway in human K562 leukemia cells. J. Biol. Chem. 279, 48716-48724. [DOI] [PubMed] [Google Scholar]
- Marone, R., Hess, D., Dankort, D., Muller, W. J., Hynes, N. E. and Badache, A. (2004). Memo mediates ErbB2-driven cell motility. Nat. Cell Biol. 6, 515-522. [DOI] [PubMed] [Google Scholar]
- Mingle, L. A., Okuhama, N. N., Shi, J., Singer, R. H., Condeelis, J. and Liu, G. (2005). Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118, 2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momparler, R. L. (2003). Cancer epigenetics. Oncogene 22, 6479-6483. [DOI] [PubMed] [Google Scholar]
- Moreau, P., Rousseau, P., Rouas-Freiss, N., Le Discorde, M., Dausset, J. and Carosella, E. D. (2002). HLA-G protein processing and transport to the cell surface. Cell Mol. Life Sci. 59, 1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W. J. and Nusse, R. (2004). Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303, 1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, F. C., Nielsen, J. and Christiansen, J. (2001). A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand. J. Clin. Lab. Invest. Suppl. 234, 93-99. [PubMed]
- Nielsen, J., Christiansen, J., Lykke-Andersen, J., Johnsen, A. H., Wewer, U. M. and Nielsen, F. C. (1999). A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19, 1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi, F. K., Elcheva, I., Bhatia, N., Shakoori, A., Ougolkov, A., Liu, J., Minamoto, T., Ross, J., Fuchs, S. Y. and Spiegelman, V. S. (2006). CRD-BP mediates stabilization of beta TrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature 441, 898-901. [DOI] [PubMed] [Google Scholar]
- Oberman, F., Rand, K., Maizels, Y., Rubinstein, A. M. and Yisraeli, J. K. (2007). VICKZ proteins mediate cell migration via their RNA binding activity. RNA 13, 1558-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, F., Huttelmaier, S., Singer, R. H. and Gu, W. (2007). ZBP2 Facilitates Binding of ZBP1 to {beta}-Actin mRNA during Transcription. Mol. Cell. Biol. 27, 8340-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L. and Singer, R. H. (1997). Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J., Lemm, I. and Berberet, B. (2001). Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene 20, 6544-6550. [DOI] [PubMed] [Google Scholar]
- Shestakova, E. A., Singer, R. H. and Condeelis, J. (2001). The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA 98, 7045-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr, N., Lederer, M., Reinke, C., Meyer, S., Hatzfeld, M., Singer, R. H. and Huttelmaier, S. (2006). ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175, 527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stove, V., Van de Walle, I., Naessens, E., Coene, E., Stove, C., Plum, J. and Verhasselt, B. (2005). Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8alphabeta. J. Virol. 79, 11422-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier, C. R., Doyle, G. A., Clark, B. A., Pitot, H. C. and Ross, J. (2004). Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 64, 209-214. [DOI] [PubMed] [Google Scholar]
- Tsou, J. A., Hagen, J. A., Carpenter, C. L. and Laird-Offringa, I. A. (2002). DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene 21, 5450-5461. [DOI] [PubMed] [Google Scholar]
- Vega, S., Morales, A. V., Ocana, O. H., Valdes, F., Fabregat, I. and Nieto, M. A. (2004). Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18, 1131-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikesaa, J., Hansen, T. V., Jonson, L., Borup, R., Wewer, U. M., Christiansen, J. and Nielsen, F. C. (2006). RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 25, 1456-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Wyckoff, J. B., Frohlich, V. C., Oleynikov, Y., Huttelmaier, S., Zavadil, J., Cermak, L., Bottinger, E. P., Singer, R. H., White, J. G. et al. (2002). Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 62, 6278-6288. [PubMed] [Google Scholar]
- Wang, W., Goswami, S., Lapidus, K., Wells, A. L., Wyckoff, J. B., Sahai, E., Singer, R. H., Segall, J. E. and Condeelis, J. S. (2004). Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64, 8585-8594. [DOI] [PubMed] [Google Scholar]
- Yisraeli, J. K. (2005). VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97, 87-96. [DOI] [PubMed] [Google Scholar]
- Yu, Q., Geng, Y. and Sicinski, P. (2001). Specific protection against breast cancers by cyclin D1 ablation. Nature 411, 1017-1021. [DOI] [PubMed] [Google Scholar]
- Zajchowski, D. A., Bartholdi, M. F., Gong, Y., Webster, L., Liu, H. L., Munishkin, A., Beauheim, C., Harvey, S., Ethier, S. P. and Johnson, P. H. (2001). Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 61, 5168-5178. [PubMed] [Google Scholar]
- Zhu, W. G., Hileman, T., Ke, Y., Wang, P., Lu, S., Duan, W., Dai, Z., Tong, T., Villalona-Calero, M. A., Plass, C. et al. (2004). 5-aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J. Biol. Chem. 279, 15161-15166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.