Abstract
We report a pH-dependent conformational transition in short, defined homopolymeric deoxyadenosines (dA15) from a single helical structure with stacked nucleobases at neutral pH to a double-helical, parallel-stranded duplex held together by AH+-H+A base pairs at acidic pH. Using native PAGE, 2D NMR, circular dichroism (CD) and fluorescence spectroscopy, we have characterized the two different pH dependent forms of dA15. The pH-triggered transition between the two defined helical forms of dA15 is characterized by CD and fluorescence. The kinetics of this conformational switch is found to occur on a millisecond time scale. This robust, highly reversible, pH-induced transition between the two well-defined structured states of dA15 represents a new molecular building block for the construction of quick-response, pH-switchable architectures in structural DNA nanotechnology.
INTRODUCTION
Structural DNA nanotechnology is an emerging field that uses DNA to create either rigid architectures or dynamic switches (1–4). Dynamic, DNA-based nanodevices may also be described as molecular switches. They are based on structural transitions between two well-defined conformations of DNA upon the application of a stimulus. Several devices have been developed based on B-DNA assemblies employing differential hybridization of complementary strands, metal ions and indeed protons (5–11). Here we describe the poly dA helix as a new structural motif that functions as a molecular switch, which at low pH forms a parallel-stranded double helix and at neutral pH exists as a structured, single helix.
Early studies on understanding the structure, base-pairing scheme and base stacking properties of DNA and RNA duplexes used synthetic homopolymeric DNA and RNA as they were considered simplified model systems. Eventually it was found that these synthetic homopolymers actually formed different unusual conformations involving non-Watson-Crick base pairing. Poly rC and poly dC formed i-tetraplexes (12,13), while poly rG and poly dG formed G-quadruplexes (14–16). Interestingly, poly rA formed a parallel-stranded double helix, called pi-helix at acidic pH due to N1 protonation of the adenines at pH <5 (17–19). At neutral pH poly rA was found to exist as a single, right-handed helix with nine nucleotides per pitch of 25.4 Å (20,21). In fact, characteristic of the distinct nature of this helix, there are proteins called poly rA binding proteins (PABPs) that specifically bind poly rA over any random ssRNA (22,23). At neutral pH poly dA is known to exist as a structured single helix, similar to poly rA (24–27), except that the nucleobases in poly dA are more strongly stacked than in poly rA and are in the C2′-endo configuration. However, the behavior of poly dA at acidic pH is still unknown. We were encouraged by the fact that poly rA could form these structures, with no indication of any special role for the 2′OH. Further we also found a sprinkling of short DNA sequences that had an over-representation of adenines that formed parallel duplexes at acidic pH (28–30), all of which contained A–A base pairs. But, these sequences would be expected to exist as unstructured single strands at neutral pH.
We have been interested in developing alternative, non-B-DNA building blocks that rely on non-Watson-Crick base pairing, for applications in structural DNA nanotechnology (31–35). Given that poly rA exists as a right-handed, parallel-stranded, double helix at acidic pH (17) and a structured right-handed single helix at neutral pH we reasoned that poly dA may have potential as a new building block for DNA based pH-switches if is able to recapitulate poly rA behavior. In order to see whether poly dA alone could form a duplex at acidic pH and if so, could it switch reversibly between its structured single helical state at pH 7 to a structured duplex at acidic pH, we investigated a segment of poly dA. We chose a segment of poly dA 15 nucleotides long, because this is within the limits of the observed persistence length of the poly dA single helix (36). Using gel electrophoresis, circular dichroism (CD) spectroscopy and concentration dependent thermal melts we showed that poly dA15 existed in two different structural forms at acidic pH and neutral pH. 1D 1H NMR studies on a short homopolymeric deoxyadenosine sequence such as dTA6 at both pH values showed that the acidic form of short homopolymeric deoxyadenosines was a parallel duplex. The relative strand polarity in the dA15 duplex was also confirmed independently by fluorescence quenching experiments. In order to delineate the molecular basis of duplex formation by such poly dA sequences, the mode of base-pairing in dTA6 was established by 2D NMR, which revealed that the duplex was held by reverse Hoogsteen type AH+–H+A base pairs. We also present an atomistic model of the dA15 parallel duplex by molecular dynamics simulation. Importantly, we show that poly dA sequences such as dA15 undergo a pH-induced conformational transition from the single helical form to the right-handed symmetric parallel-stranded duplex form in a highly reversible manner. The kinetics of this association was found to occur on millisecond time scales. This fast association time scale makes it an ideal system for use as a molecular nanoswitch in structural DNA nanotechnology.
MATERIALS AND METHODS
Sample preparation
Desalted dA15, dTA6 and HPLC purified 5′-TAMRA as well as 3′-TMR (attached via a C3 linker) labeled dA15 were obtained from Bioserve India. HPLC purified 3′-DABCYL labeled dA15 was obtatined from Ocimum Biosolutions, India and used without further purification. Samples were prepared in buffer of desired pH by incubating them at 4°C for 12 h prior to measurement. Heating was avoided to decrease the pH-induced depurination.
Native gel electrophoresis
dA15 was phosphorylated at 5′ end with P32 by T4 PNK forward reaction and γ-P32 labeled ATP. Labeled DNA was doped with unlabeled dA15. The labeled and unlabeled dA15 mixture was incubated at different pH in 2 µM and then electrophoresed in 15% polyacrylamide gel buffered at different pH with Robinson Britton Buffer [(CH3COOH) = (H3PO4) = (H3BO3) = 0.04 M; pH adjusted with NaOH) at 10 V/cm for 3 h. The gels were dried in slab gel drier and exposed to Fujifilm BAS-IP MS 2025 imaging plate and plates were imaged in Fujifilm FLA-2000 phosphoraimager.
CD spectroscopy
All the CD experiments were done using a Jasco J-815 CD spectropolarimeter equipped with Peltier temperature controller. All the data were collected from 300 to 200 nm at a scan rate of 50 nm/min at 0.2 nm data intervals and are presented as an average of three successive scans unless specified. Samples were made at desired concentrations in phosphate buffer at pH 3 and 7 with desired ionic strength. For acidic pH, we used NaH2PO4/H3PO4 buffer and at neutral pH, Na2HPO4/NaH2PO4 buffer. Samples were annealed as described before. pH titrations were done using 0.01 N HCl or 0.01 N NaOH. Samples were used only once. Reproducibility was ensured on multiple samples prepared similarly.
Fluorescence spectroscopy
Fluorescence experiments were done on a JASCO J-815 CD Spectropolarimeter equipped with fluorescence detector or on FLUOROLOG-SPEX spectrofluorimeter using either 520 or 550 nm excitation wavelength and emission spectra were recorded from 540/560 to 700 nm. Emission spectra, presented as an average of two successive scans. Kinetics of association and dissociation of poly dA was done using a custom built single molecule tracking (Olympus IX 70) inverted microscope equipped with photon counting APD. pH jumps were performed by addition of desired strong buffer to a weakly buffered solution of 5′-TAMRA-dA15. For distance calculation experiments, samples of 1:50 3′-TMR-dA15: 3′-DABCYL-dA15 or 1:50 3′-TMR-dA15:dA15 at 5 μM were used (see Supplementary Data for details).
Molecular dynamics simulations
All the models of poly dA duplex and single strands are made using NAMOT 2 software and simulated using PMEMD (37) program of AMBER9 (38) software suite with all-atom AMBER03 force field. The equilibration protocols were followed as described previously (39,40). Structures were visualized by PyMOL and UCSF Chimera software (41,42).
NMR experiments
All NMR spectra were recorded on Bruker Avance-500 and −800 MHz spectrometer. A total of 1 mM strand concentration in 50 mM Na-acetate-d3 buffer at pH 4.0 was used to prepare samples for all 1D experiments. 10% D2O was added before taking the spectra. Whereas, for proton exchange experiments, samples in Na-acetate-d3 buffer was lyophilized overnight and reconstitute in D2O. pH of this solution was adjusted to 4 by addition of 4–5 µl of DCl and incubated at 4°C overnight. pH 8 spectra was taken after quickly elevating the pH by addition of 15 µl of 1 M NaOH to 500 μl sample. Water suppression was achieved using an excitation Sculpting solvent suppression programme (43). For 1D experiment 1024 scans were taken, the spectral width was maintained at 10 KHz, the thymine methyl chemical shift at 1.8 δppm was used as the internal standard. For NOESY experiments, (512 × 2048) complex points were collected, a 2 kHz spectral width was employed in both dimensions with acquisition times of 0.3 s in t2 and 0.3 s in t1, using a 200 ms mixing time for seeing H1′-Adenine H8 and 100 ms for H2′/H2′′-Adenine H8.
RESULTS AND DISCUSSION
Table 1 shows the poly dA sequences with the relevant modifications that were used in this study.
Table 1.
Poly dA sequences used in this study
| Name | Sequence |
|---|---|
| Poly dA15 | 5′-d(AAAAAAAAAAAAAAA)-3′ |
| dTA6 | 5′-d(TAAAAAA)-3′ |
| 3′-Dabcyl-dA15 | 5′-d(AAAAAAAAAAAAAAA)-Dabcyl-3′ |
| 3′-TMR-dA15 | 5′-d(AAAAAAAAAAAAAAA)-TMR-3′ |
| 5′-TAMRA-dA15 | 5′-TAMRA-d(AAAAAAAAAAAAAAA)-3′ |
Native PAGE evidences duplex formation
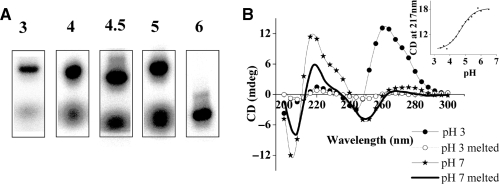
In order to see whether dA15 could self associate like its RNA analogue at acidic pH, we analyzed its electrophoretic mobility at a range of pH values from pH 3 to pH 7 by native polyacrylamide gel electrophoresis (PAGE) (Figure 1A). Samples of 2 μM 5′ P-32 labeled dA15 was equilibrated in phosphate buffer of the desired pH and electrophoresed on 15% native PAGE of the corresponding pH. At pH 3, dA15 shows a band of lower mobility, which increasingly disproportionates into a band of higher mobility with progressively increasing pH (Figure 1A). Thus, at pH 6 and above only a single band of higher mobility is observed. This clearly indicates that at acidic pH, dA15 forms a secondary structure of lower mobility and above pH 6, adopts a structure of higher mobility, with both forms being differently populated at intermediate pH values. This suggests that dA15 adopts two different forms at acidic and neutral pH values.
Figure 1.
(A) Gel electrophoresis of dA15 showing two forms with different electrophoretic mobility. P32 labeled dA15 was incubated at the indicated pH at 4°C and then electrophoresed on 15% native PAGE in Robinson–Britton buffer of corresponding pH at 20°C and visualized using PhosphorImager. pH values are indicated above each lane (Na+ = 30 mM). (B) CD spectra of 1 μM dA15 at pH 3.0 and pH 7.0 in 10 mM phosphate recorded at both 20°C and 95°C (Na+ = 10 mM). Inset: CD at 217 nm of 5 µM dA15 in 10 mM Na+ cation as a function of buffer pH.
pH-induced structural change probed by CD spectroscopy
Having established that dA15 exists in two differently migrating forms that are pH dependent, we analyzed these forms further using CD spectroscopy (Figure 1B). Samples of 1 μM dA15 were prepared at pH 3.0 and 7.0 as described in the ‘Materials and Methods’ section. At 20°C, dA15 at pH 7.0 showed a characteristic CD trace with a strong positive maximum at 217 nm with a shoulder at 232 nm, a weak positive band at 275 nm and negative bands centered at 250 nm and 206 nm. This spectrum is characteristic of single-stranded poly dA which is well documented (44). Upon heating to 95°C, this trace changed to one where the maximum at 275 nm was abolished and the minimum at 206 nm shifted to 210 nm. The CD spectrum of 1 μM dA15 at pH 3.0, on the other hand, was completely different from that at pH 7.0. At 20°C, the 217 nm positive band characteristic of the single helix was absent. Instead, only an intense, positive band maximum at 262 nm with a shoulder at 275 nm and a weak minimum at 245 nm was observed. On heating to 95°C, these bands completely disappear, flattening out to comparatively negligible CD characteristic of ssDNA. The structure of poly dA15 at acidic pH evidenced a thermal transition by CD as well as UV, where the stability of the structure was concentration dependent further supporting its intermolecular nature (see Supplementary Data). Poly dA15 at acidic pH, thus assumes a structure entirely different from the single-stranded helix, as seen clearly from their completely different CD signatures and melting behavior.
1D and 2D NMR establish structure of the duplex in solution
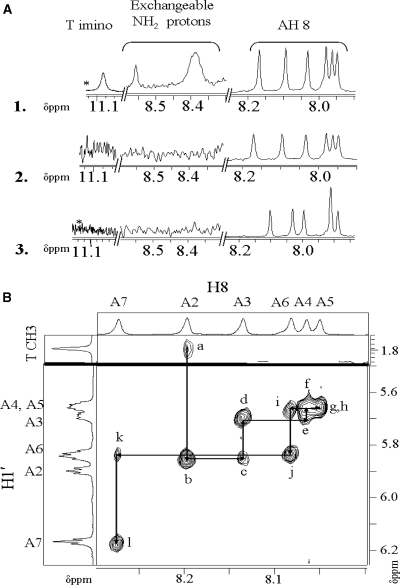
In order to get more structural detail on such short, homo A-tracts in DNA at acidic pH, high resolution NMR studies were performed on a truncated form of dA15, desymmetrized by a thymine at the 5′ end to enable complete assignment by NMR. We chose dTA6 based on literature evidence that affirmed six adenines to be the minimum length that structurally and functionally represented the poly rA helix (45). One millimolar dTA6 in 10% D2O/H2O at 10°C on a Bruker 800 MHz NMR spectrometer showed exactly six Adenine H8 protons and only one type of Thymine CH3 and H6 protons (see Supplementary Data and Figure 3A) confirming that this sequence forms a single population of dimer in bulk, precluding any slipped structures for at least six contiguous adenine tracts. Importantly, the 1D spectrum of dTA6 showed hydrogen-bonded N6 aminos that were downfield shifted to 8.4–9 δppm from the usual 6–7 δppm for these protons (see Figure 3A), characteristic of hydrogen bonding seen in A–A base pairing (Figure 2B) (30). These were not seen in either the D2O exchanged spectrum at pH 4 or the single helical, monomeric structure at pH 8 in 5% D2O [Figure 3A (2 and 3)]. Furthermore, these H-bonded amino protons also showed the characteristic dramatically reduced intensity observed for A–A base pairs bonded on their Hoogsteen faces (28–30) as indicated in Scheme 1B. Furthermore, 2D NOESY of dTA6 showed a set of eleven H8–H1′ NOEs (Figure 3B) characteristic of six A–A base pairs found in A-containing duplexes that form a parallel-stranded Π-DNA helix (29). Importantly the absence of NOEs between Adenine NH2 protons and the Adenine H2 protons are consistent with the reverse Hoogsteen base-pairing scheme seen in the dA containing parallel duplex (30).
Figure 3.
(A) 1D NMR spectrum 1 mM dTA6 at 5°C establishing NH2 involved in H bonding at pH 3. (1) Spectra taken in 50 mM Na-acetate-d3 buffer pH 4.0 in 10% D2O. (2) Spectra taken in Na-acetate-d3 buffer pH 4.0 in D2O. (3) Spectra taken in pH 8 water (Na+ = 50 mM) (B) Partial NOESY spectrum showing sugar H1′-Adenine H8 contacts of dTA6 at pH 4 Na-acetate-d3 buffer. All Spectra were recorded in Avence-500 Bruker NMR spectrometer. The NOE cross peaks a–l are assigned as follows. (a) A2(H8)-T1(CH3); (b) A2(H8)-A2(H1′); (c) A3(H8)-A2(H1′); (d) A3(H8)-A3(H1′); (e) A4(H8)-A3(H1′); (f) A4(H8)-A4(H1′); (g) A5(H8)-A4(H1′); (h) A5(H8)-A5(H1′); (i) A6(H8)-A5(H1′); (j) A6(H8)-A6(H1′); (k) A7(H8)-A6(H1′); (l) A7(H8)-A7(H1′). *Spectra acquired on a Bruker 800 MHz spectrometer.
Figure 2.
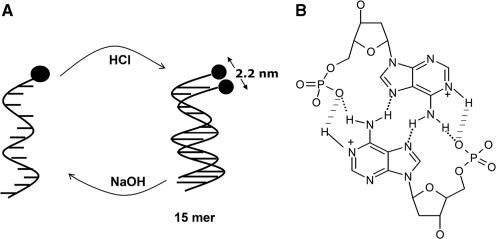
(A) Schematic showing poly dA15 changing between single helix to duplex conformations induced by alternate addition of acid and base respectively. (B) Shown in black is the base pairing scheme in AH+–H+A base pairs comprising protonated adenosines.
Salt dependence studies
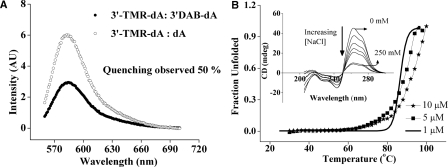
In order to investigate the effect of salt on the stability of the duplex, samples were made at 5 µM strand concentration in unbuffered solution, pH 3 and CD measured with incremental additions of NaCl. As evident from the CD profile in inset Figure 5B, with increasing salt, the intensity at 262 nm which is a clear diagnostic of the duplex, decreases (see also Supplementary Data Figure 2). It can be seen that the signal decreases sharply and tapers off at ∼250 mM NaCl. Such dependence has been observed also for the poly rA duplex which is stabilized by electrostatic interaction between N1–H+ of adenosine and phosphate oxygen (17). That such dependence is recapitulated in the poly dA duplex indicates that such an electrostatic interaction is also present here, further supporting the base pairing mode observed by NMR.
Figure 5.
(A) Fluorescence quenching experiments on the dual labeled poly dA15 duplex of 1:50 3′-TMR-dA15:3′-Dabcyl-dA15 (filled circles) and 1:50 3′-TMR-dA15: 3′-unlabeled dA15 (open circles) at 100 nM TMR- dA15 in 30 mM Na–phosphate buffer, pH 3 (Na+ = 30 mM). (B) UV thermal melting of dA15 duplex at 10 mM buffer, pH 3 (Na+ = 10 mM). Inset: CD spectra of 5 μM dA15 at 0 mM, 15 mM, 30 mM, 75 mM, 150 mM, 200 mM and 250 mM NaCl solution, pH 3.
Molecular dynamics (MD) study
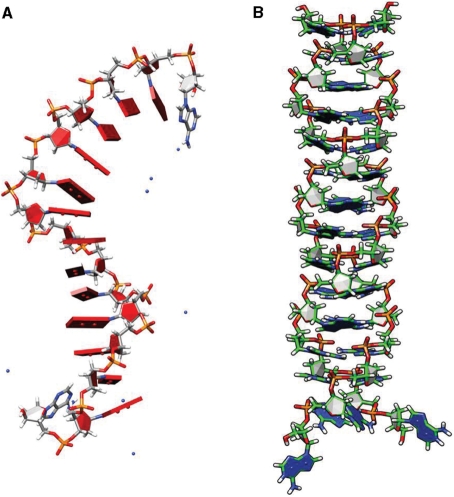
MD simulations (see Supplementary Data for details) of the single-stranded unprotonated dA15 indeed revealed a robust helical structure primarily driven by efficient stacking of the adenine nucleobases (Figure 4A) (19). Importantly, MD simulations on the parallel-stranded N1 protonated poly dA15 duplex yielded a structure which is similar to the Π-DNA helix (Figure 4B) (29). The AH+–H+A base pairs in this duplex adopted a 12° tilt from the horizontal to the helical axis. This tilting is characteristic of the AH+–H+A base pairs previously described (17). Interestingly, MD also reveals an extra strong interaction resembling an H-bond of ∼2.9 Å distance between the phosphate and the N1 protonated site on adenines shown in dashed line in Figure 2B (also see Supplementary Data). If this is true it would imply almost six hydrogen bonds per AH+–H+A base pair which is in line with UV melting studies and kinetics that evidence unusually high stability of the poly dA15 duplex (see Supplementary Data and next section).
Figure 4.
(A) Equilibrium snapshot of the single-stranded dA15 after 20 ns long MD simulation using AMBER revealing highly stacked adenine nucleobases. (B) Instantaneous snapshot of N1-protonated adenosine mediated parallel duplex of dA15 after 20 ns long MD simulation revealing a Π-helical structure with tilted base.
Thermal stability studies
To investigate the thermal stability of dA15, at both acidic and neutral pH, both samples were thermally denatured following the UV absorbance at 260 nm or CD at 262 nm. Two micromolar dA15 at pH 7.0 evidences a weakly structured form as seen from the broad and noncooperative melt centered at 46°C (see Supplementary Data for details). This is in line with previous findings on single helices of poly dA that suggest that stacking interactions are probably the only stabilizing forces in the poly dA single helix (25). For the duplex melting, freshly prepared samples of dA15 at pH 3.0 were used which evidenced a cooperative dissociation centered at ∼80°C at 1 μM dA15 (see Figure 5B). Melting temperature was found to vary with strand concentration indicating intermolecular nature of the dA15-duplex (see Supplementary Data). In all cases, regardless of strand concentration, the transitions were sharp, taking place over <12°C as seen in well-formed B-DNA duplexes indicating that the dA15 duplex is also likely to be as homogenous. Importantly, thermodynamic parameters cannot be extracted from these thermal melting profiles at acidic pH, as they could be complicated by depurination that prevents reversibility of the melts. For this reason, in this case, thermal denaturation cannot be used to establish a two-state transition. Thus, this is not a ‘melting’ experiment characteristic of a two-state transition, but the characterization of the thermal response of such dA15 duplexes. However, in order to establish whether this duplex denaturation is two state, we carried out a pH denaturation of the dA15 duplexes (see pH-induced structural transition probed by CD section).
The sharpness of the thermal melting transitions observed for the dA15 duplexes is indicative of negligible slipped intermediates (45,46). Furthermore, literature studies on the poly rA duplexes of varying lengths have shown that slipped structures and intermediates occur only when the A-tracts approach lengths greater than rA30 (45). However, in order to confirm that this is indeed the case, we performed fluorescence quenching experiments to measure the distance between two 3′ termini in the dA15 duplex, by a previously described method (31,33). Samples were prepared by mixing 1:50 3′-TMR-dA15:3′-Dabcyl-dA15 (100 nm:5 μM), 30 mM phosphate buffer, pH 3 such that every TMR-labeled dA15 strand is incorporated into a duplex containing Dabcyl-labeled dA15 strand. Any change in TMR fluorescence intensity will be due to quenching by Dabcyl-dA15 strand present in the duplex. The quenching efficiency in these dually labeled complexes was found to be 50% as compared to similarly prepared 1:50 3′-TMR-dA15:unlabeled dA15 (100 nm:5 μM) complexes (see Figure 5A). Readings were normalized to the fluorescence value of each of the samples, when they were taken to pH 7. This accounts for fluorescence changes due to both environmental effects of structure formation as well as pH effects. This quenching efficiency translates to an interfluorophore distance of 26 ± 5 Å, incorporating the distance resolution due to fluorophore linker lengths (31,33,47). Given that the diameter of the pi-helix is ∼22 Å, this translates to a maximum slippage of not more than one base in the dA15 duplex. This is consistent with the melting studies that show at these segment lengths, the dA15 duplex does not undergo any significant slipped structure formation. An equivalent of one-base slippage is seen even in the 5′E and 3′E intercalation topologies in i-motifs.
pH-induced structural transition probed by CD
The existence of two differently structured forms of dA15 as a function of pH prompted us to investigate the potential of dA15 as a nanoscale transducer, converting a proton input, into a conformational change of the poly dA single helix. For this it was essential to determine whether dA15 showed a pH induced structural transition in solution as well. Five micromolar dA15 was incubated in buffers of different pH ranging from pH 3 to 7 with a ∼0.2 pH unit increment and the CD value at 217 nm was plotted as a function of pH (inset: Figure 1B). A well-defined sharp transition centred at pH 4.8 was observed, indicating that the transition was two-state.
Reversible pH-induced structural transition in poly dA
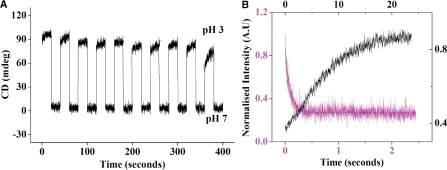
Next we investigated whether poly dA15 was capable of undergoing a reversible pH induced conformational switch from structured single helix to parallel duplex at pH 3.0. To an unbuffered solution of 5 µM dA15 at pH 7, we added acid (HCl) and base (NaOH) alternately to accordingly switch the pH of the solution from 7 to 3 reversibly. Molecular switching was visualized by monitoring CD at 262 nm where signals were very different for the single and double helical forms. As evident from Figure 6A, dA15 can switch efficiently and reversibly between the two different states with change in pH without any significant loss in efficiency. This demonstrated that poly dA15 was able to respond to a proton input, by changing its structure as evident from the changes in its CD properties.
Figure 6.
(A) CD of dA15 at 262 nm demonstrating switching between single helix and duplex upon alternately cycling between pH 7 and pH 3 (Na+ concentration at the end of 10th cycle ∼1.5 mM) (B) Kinetics of transition of dA15 from single helical to double helical form (shown in magenta) and vice versa (shown in black) probed by fluorescence from TAMRA.
Dimerization may also be followed by fluorescence self quenching
In a parallel-stranded Π-helical configuration we would expect like termini in [5′-TAMRA-dA15]2 to have an interfluorophore distance of ∼22 Å. Given that TAMRA has been shown to self-quench with a Ro of 44 Å due to exciton coupling, that has been used to determine strand polarities in unusual nucleic acid motifs at low pH (47,48), we wanted to see if this change in fluorescence property could report on dA15 duplexation. 5′-TAMRA labeled dA15 was allowed to dimerize at pH 3 and the extent of quenching, relative to 5′-TAMRA-dA15 at pH 7.0, determined. We found that the self quenching efficiency is greater than 80% consistent with the predicted strand polarity, and revealing that self-quenching could be used to follow dimerization (see Supplementary Figure 3A and Supplementary Data for details). In order to measure the response times of dA15 to this pH stimulus, kinetics experiments were performed using the fluorescence of 5′-TAMRA-dA15 which self-quenches due to duplex formation. To 20 μl solution of 0.5 µM 5′-TAMRA-dA15 in 100 μM phosphate buffer at pH 7, 5 µl of 50 mM pH 3 phosphate buffer was added to cause a pH jump to 3. Fluorescence of TAMRA-dA15 quenches due to duplex formation as shown in Figure 6B. The time scales of duplex formation at this concentration was found to be τ = 90 ms demonstrating very fast duplexation. Association time scale was found to depend on concentration of the poly dA15 strand used (see Supplementary Figure 14 and associated discussion), emphasizing the intermolecular nature of the duplex formation. Similarly dissociation of duplex to single helix was also followed in a similar way where addition of 1 M phosphate buffer, from pH 7 to 0.5 µM dA15 in 5 mM phosphate buffer to cause a pH jump to 7. This relieved the fluorescence of TAMRA from quenching which is manifested by increase in fluorescence (Figure 6A). The time scale of duplex dissociation was found to be slower (∼7 s) compared to its association. This is consistent with the compactness of the duplex as revealed by MD and high stability because of its electrically neutral character and high number of H-bonds per base pair.
CONCLUSIONS
Poly dA15 exists as a structured single helix at neutral pH (24–27). We have shown that at acidic pH, poly dA forms a right-handed parallel-stranded double helix which we term the A-motif. As evidenced by NMR, the poly dA15 duplex is held together by reverse Hoogsteen base-pairing between protonated adenosines, with molecular dynamics studies also suggesting electrostatic interactions between the phosphate backbone and N1-H+ of the base. We have delineated the structure of the poly dA15 duplex and from MD simulations, also present an atomistic model of such right-handed, parallel-stranded duplexes previously referred to as Π-DNA (29). The thermal stability of the dA15 A-motif was found to be ∼80°C as probed by both CD spectroscopy and UV spectrophotometry. The melting temperature, T1/2 was found to be dependent on concentration indicating the intermolecular nature of the A-motif. Fluorescence quenching experiments on the parallel dA15 duplex indicated that at these segment lengths, slipped hybridizations were insignificant.
Importantly we have demonstrated that dA15 undergoes a pH-induced molecular transition from its single helical to duplex form efficiently and reversibly. The kinetics of association to form the A-motif is complete within millisecond time scale at sub-micromolar concentrations. We have also shown that dA15 can be used as a proton driven molecular switch that switches reproducibly between its single helical and duplex forms with negligible loss of efficiency. The switching is two-state and is highly processive. As a switch, the A-motif has properties which would make it a valuable addition to the structural DNA nanotechnology toolkit. It has all the advantages of proton driven switches, being ‘clean’, generating only water and salt as by-products for each cycle of switching. Although slipped hybridizations could occur, these happen only in longer dA tracts, and may be avoided by employing shorter A-tracts that include a CGA motif at the 5′ end (29) to keep the strands in register. Apart from its high stability, it is simple to construct, composed of just one type of DNA base, thus minimizing interference upon its incorporation as part of a larger DNA assembly. Because it is a non-Watson–Crick-based building block, it can be integrated into Watson–Crick base-paired assemblies to realize switches with more complex functionalities.
Thus we have outlined the molecular basis of a new pH-sensitive DNA structural motif and shown its successful working as a high-performance pH-triggered molecular switch, undergoing a transition between two well-defined states triggered by a change in pH. This also represents a new mechanism by which two DNA strands may hybridize and dissociate triggered by pH, finding application as a unique method to site-specifically glue DNA assemblies together on providing a pH cue. It can thus be used to replace a critically positioned Watson–Crick base-pairing site on a given DNA assembly transforming it into a sticky or nonsticky state on the application of an external pH stimulus. Thus, with the A-motif, we can build pH responsive 1D, 2D and 3D architectures because (i) the base-pairing here requires only two strands, (ii) directionality is conferred by the parallel-stranded nature of the A-motif (as opposed to antiparallel B-DNA) and (iii) this mechanism is compatible with and does not interfere with Watson–Crick base-pairing in an assembly. The observation of millisecond association timescales for the A-motif illustrates the immense potential of non-B-DNA-based modules in structural DNA nanotechnology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NanoScience and Technology Initiative of the Department of Science and Technology, Govt of India; Fellowship from CSIR, Govt of India (to S.C. and S.S.); Innovative Young Biotechnologist Award from DBT, Govt of India (to Y.K.). The Open Access charges were partially waived by Oxford University Press. The rest of the funding was provided by National Centre for Biological Sciences, TIFR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Souvik Modi and the National facility for high-field NMR, TIFR, for NMR, D Usharani and Tod Pascal for modeling.
REFERENCES
- 1.Bath J, Turberfield AJ. DNA nanomachine. Nat. Nanotech. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 2.Pitchiaya S, Krishnan Y. First blueprint, now bricks: DNA as construction material on the nanoscale. Chem. Soc. Rev. 2006;35:1111–1121. doi: 10.1039/b602886c. [DOI] [PubMed] [Google Scholar]
- 3.Samori B, Zuccheri G. DNA codes for nanoscience. Angew. Chem. Int. Ed. Eng. 2005;44:1166–1181. doi: 10.1002/anie.200400652. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. Biochemistry and structural DNA nanotechnology: an evolving symbiotic relationship. Biochemistry. 2003;42:7259–7269. doi: 10.1021/bi030079v. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Xu Y, Li F, Yang Y, Wang W, Song Y, Liu D. Light-driven conformational switch of i-motif DNA. Angew. Chem. Int. Ed. 2007;46:2515–2517. doi: 10.1002/anie.200604589. [DOI] [PubMed] [Google Scholar]
- 6.Liedl T, Simmel FC. Switching the conformation of a DNA molecule with a chemical oscillator. Nano Lett. 2005;5:1894–1898. doi: 10.1021/nl051180j. [DOI] [PubMed] [Google Scholar]
- 7.Chan Y, Lee SH, Mao C. A DNA nanomachine based on a duplex-triplex transition. Angew. Chem. Int. Ed. 2004;43:5335–5338. doi: 10.1002/anie.200460789. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 9.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 11.Monchaud D, Yang P, Lacroix L, Teulade-Fichou MP, Mergny JL. A metal-mediated conformational switch controls G-quadruplex binding affinity. Angew. Chem. Int. Ed. 2008;47:4858–4861. doi: 10.1002/anie.200800468. [DOI] [PubMed] [Google Scholar]
- 12.Gehring K, Leroy JL, Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 13.Snoussi K, Nonon-Lacomte S, Leroy JL. The RNA i-motif. J. Mol. Biol. 2001;309:139–153. doi: 10.1006/jmbi.2001.4618. [DOI] [PubMed] [Google Scholar]
- 14.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–364. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerman SB, Cohen GH, Davies DR. X-ray fibre diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 16.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rich A, Davies DR, Crick FHC, Watson JD. The molecular structure of polyadenylic acid. J. Mol. Biol. 1961;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- 18.Fresco JR. Polynucleotides. II. The x-ray diffraction patterns of solutions of the randomly coiled and helical forms of polyriboadenylic acid. J. Mol. Biol. 1959;1:106–110. [Google Scholar]
- 19.Ts'o POP, Helmkamp GK, Sander C. Interaction of nucleosides and related compounds with nucleic acids as indicated by the change of helix-coil transition temperature. Proc. Natl Acad. Sci. USA. 1962;48:686–697. doi: 10.1073/pnas.48.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman SB, Davies DR, Navia MA. An ordered single-stranded structure for polyadenylic acid in denaturing solvents. An X-ray fibre diffraction and model building study. J. Mol. Biol. 1977;116:317–330. doi: 10.1016/0022-2836(77)90219-4. [DOI] [PubMed] [Google Scholar]
- 21.Saenger W, Riecke J, Suck D. A structural model for the polyadenylic acid single helix. J. Mol. Biol. 1975;93:529–534. doi: 10.1016/0022-2836(75)90244-2. [DOI] [PubMed] [Google Scholar]
- 22.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 23.Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J. Biol. Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- 24.Bush CA, Scheraga HA. Optical activity of single-stranded polydeoxyadenylic and polyriboadenylic acids; dependence of adenine chromophore cotton effects on polymer conformation. Biopolymers. 1969;7:395–409. [Google Scholar]
- 25.Alderfer JL, Smith SL. A proton magnetic resonance study of polydeoxyriboadenylic acid. J. Am. Chem. Soc. 1971;93:7305–7314. doi: 10.1021/ja00755a031. [DOI] [PubMed] [Google Scholar]
- 26.Olsthoorn CSM, Bostelaar LJ, vanBoom H, Altona C. Conformational characteristics of the trinucleoside diphosphate dApdApdA and its constituents from nuclear magnetic resonance and circular dichroism studies. Extrapolation to the stacked conformers. Eur. J. Biochem. 1980;112:95–110. doi: 10.1111/j.1432-1033.1980.tb04991.x. [DOI] [PubMed] [Google Scholar]
- 27.Ke C, Humeniuk M, S-Gracz H, Marszalek PE. Direct measurements of base stacking interactions in DNA by single-molecule atomic-force spectroscopy. Phys. Rev. Lett. 2007;99:018302–018305. doi: 10.1103/PhysRevLett.99.018302. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, Sarma MH, Yuan RD, Sarma RH. R study of self-paired parallel duplex of d(AAAAACCCCC) in solution. FEBS Lett. 1992;306:223–228. doi: 10.1016/0014-5793(92)81005-7. [DOI] [PubMed] [Google Scholar]
- 29.Robinson H, Wang AH-J. 5′-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc. Natl Acad. Sci. USA. 1993;90:5224–5228. doi: 10.1073/pnas.90.11.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Patel DJ. Solution structure of the d(T-C-G-A) duplex at acidic pH. A parallel-stranded helix containing C+.C, G.G and A.A pairs. J. Mol. Biol. 1994;242:508–526. [PubMed] [Google Scholar]
- 31.Chakraborty S, Modi S, Krishnan Y. The RNA2-PNA2 hybrid i-motif-a novel RNA-based building block. Chem. Commun. 2008:70–72. doi: 10.1039/b713525d. [DOI] [PubMed] [Google Scholar]
- 32.Ghodke HB, Krishnan R, Vignesh K, Kumar GVP, Narayana C, Krishnan Y. The I-tetraplex building block: rational design and controlled fabrication of robust 1D DNA scaffolds through non-Watson-Crick interactions. Angew. Chem. Int. Ed. 2007;46:2646–2649. doi: 10.1002/anie.200604461. [DOI] [PubMed] [Google Scholar]
- 33.Modi S, Wani AH, Krishnan Y. The PNA-DNA hybrid I-motif: implications for sugar-sugar contacts in i-motif tetramerization. Nucleic Acids Res. 2006;34:4354–4363. doi: 10.1093/nar/gkl443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan-Ghosh Y, Stephens E, Balasubramanian S. PNA forms an i-motif. Chem. Commun. 2005:5278–5280. doi: 10.1039/b510405j. [DOI] [PubMed] [Google Scholar]
- 35.Krishnan-Ghosh Y, Liu D, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 36.Mills JB, Vacano E, Hagerman PJ. Flexibility of single-stranded DNA: use of gapped duplex helices to determine the persistence lengths of poly(dT) and poly(dA) J. Mol. Biol. 1999;285:245–257. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 37.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, Wang J, Ross WS, Simmerling C, Darden T, Merz KM, Stanton RV, et al. AMBER 7 edit. San Francisco: University of California; 1999. [Google Scholar]
- 38.Duke RE, Pedersen LG. PMEMD 3. University of North Carolina-Chapel Hill; 2003. [PubMed] [Google Scholar]
- 39.Maiti PK, Pascal TA, Vaidehi N, Goddard WA. The stability of Seeman JX DNA topoisomers of paranemic crossover (PX) molecules as a function of crossover number. Nucleic Acids Research. 2004;32:6047–6056. doi: 10.1093/nar/gkh931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiti PK, Bagchi B. Structure and dynamics of DNA-dendrimer complexation: role of counterions, water, and base pair sequence. Nano. Lett. 2006;6:2478–2485. doi: 10.1021/nl061609m. [DOI] [PubMed] [Google Scholar]
- 41.Delano WL. San Carlos, CA, USA: DeLano Scientific; 2002. The PyMOL molecular graphics system. [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magnetic Res. Ser. A. 1995;112:139–282. [Google Scholar]
- 44.Adler AJ, Grossman L, Fasman GD. Polyriboadenylic and polydeoxyriboadenylic acids. Optical rotatory studies of pH-dependent conformations and their relative stability. Biochemistry. 1969;8:3846–3859. doi: 10.1021/bi00837a051. [DOI] [PubMed] [Google Scholar]
- 45.Brahms J, Michelson AM, van Holde KE. Adenylate oligomers in single- and double-strand conformation. J. Mol. Biol. 1966;15:467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- 46.Janik B, Sommer RG, Bobst AM. Polarography of polynucleotides. II. Conformations of poly(adenylic acid) at acidic pH. Biochim. Biophys. Acta. 1972;281:152–168. [PubMed] [Google Scholar]
- 47.Bernacchi S, Mély Y. Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Res. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty S, Krishnan Y. Kinetic hybrid i-motifs: intercepting DNA with RNA to form a DNA2-RNA2 i-motif. Biochimie. 2008;90:1088–1095. doi: 10.1016/j.biochi.2008.02.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.