Abstract
A major outcome of the canonical Wnt/β-catenin-signalling pathway is the transcriptional activation of a specific set of target genes. A typical feature of the transcriptional response induced by Wnt signalling is the involvement of Tcf/Lef factors that function in the nucleus as the principal mediators of signalling. Vertebrate Tcf/Lef proteins perform two well-characterized functions: in association with β-catenin they activate gene expression, and in the absence of Wnt ligands they bind TLE/Groucho proteins to act as transcriptional repressors. Although the general characteristics of Tcf/Lef factors are well understood, the mechanisms that control their specific roles in various cellular backgrounds are much less defined. In this report we reveal that the evolutionary conserved Dazap2 protein functions as a TCF-4 interacting partner. We demonstrate that a short region proximal to the TCF-4 HMG box mediates the interaction and that all Tcf/Lef family members associate with Dazap2. Interestingly, knockdown of Dazap2 not only reduced the activity of Wnt signalling as measured by Tcf/β-catenin reporters but additionally altered the expression of Wnt-signalling target genes. Finally, chromatin immunoprecipitation studies indicate that Dazap2 modulates the affinity of TCF-4 for its DNA-recognition motif.
INTRODUCTION
The Wnt-signalling pathway is essential during different developmental processes for determining cell fate. In addition, aberrant activation of this pathway has been implicated in cellular transformation and cancer [see some recent reviews (1–3)]. Transcription factors of the Tcf/Lef family are important downstream effectors of the so-called canonical Wnt/β-catenin-signalling pathway. In vertebrates the family consists of four members: Tcf-1, Tcf-3, Tcf-4 and Lef-1 (4). All vertebrate Tcf/Lef proteins (further referred to as Tcfs) contain virtually identical DNA-binding domains, a high mobility group (HMG) box, and a highly conserved β-catenin-interacting region. In the absence of the Wnt signal, Tcf/Lef factors interact with Transducin-like enhancer of split (TLE)/Groucho co-repressors to mediate the transcriptional repression of Tcf-bound genes (5–7). Alternatively, upon initiation of Wnt signalling the constitutive degradation of β-catenin is inhibited allowing this protein to accumulate both in the cytoplasm and nucleus, with the nuclear form able to displace TLE/Groucho co-repressors from Tcfs (8). Since β-catenin contains a strong transactivation domain, Tcf/β-catenin heterocomplexes function as transcriptional activators of specific Wnt-responsive genes such as c-myc (9), Cyclin D1 (10,11), Axin2 (12) and CD44 (13). For a more comprehensive survey on Wnt signalling, please refer to the Wnt signalling home page at http://www.stanford.edu/%7ernusse/wntwindow.html.
Although the general function of Tcfs as transcriptional repressors or co-activators is well understood, their specific roles in Wnt signalling or cell physiology are much less defined. Besides β-catenin and TLE/Groucho co-repressors several other proteins associate with the HMG box of Tcfs. Such factors include proteins containing the I-mfa domain that mask the DNA-interacting region of Tcf-3, thereby preventing Tcf-3/β-catenin heterodimers from activating transcription (14). Likewise, RUNX3 forms a ternary complex with β-catenin and Tcfs to attenuate the transactivation potential of Tcf/β-catenin complexes by decreasing their DNA-binding activity (15).
Expression of mouse Tcf/Lef genes during embryogenesis and in adult tissues often overlaps. Nevertheless, gene-targeting experiments have demonstrated that individual Tcf members control their own cell biological programs (16–19). This observation implies that throughout evolution the functions originally executed by a single Tcf polypeptide have been distributed in more complex organisms among several family members. A plausible explanation for the functional diversity among Tcfs would be their selective interaction with distinct partners as the amino-acid sequences outside the highly conserved DNA- and β-catenin-binding domains are less homologous. Indeed, it has been reported that LEF-1 activates some promoters together with ALY, a nuclear protein that specifically binds LEF-1 and AML-1 (20). Additionally, LEF-1 cooperates with the Microphthalmia-associated transcription factor (MITF) to activate the expression of melanocyte-specific genes (21). Interestingly, although the activity of LEF-1 is suppressed by association with PIASy (a nuclear matrix-associated SUMO E3 ligase), this interaction results in increased TCF-4-regulated transcription (22,23). Two Tcf/Lef family members, Tcf-3 and Tcf-4, contain binding motifs for C-terminal-binding proteins (CtBPs) at their C-termini (24–26). As CtBPs operate as short-distance transcriptional repressors, interaction with such factors strengthens the repressive potential of these Tcfs in the absence of Wnt signalling (27). Besides CtBP, TCF-4 also binds the Hypermethylated in cancer 1 (HIC1) tumour suppressor. This interaction leads to the recruitment of TCF-4 into nuclear ‘speckles’ called HIC1 bodies. Upon association with HIC1, TCF-4 is unable to bind Wnt-responsive gene promoters. Thus, HIC1 functions as a nuclear TCF-4-specific Wnt pathway inhibitor (27). Finally, to add another layer of complexity to the regulation of Wnt target genes it has also been demonstrated that alternative promoters and/or alternative splicing of Tcf/Lef mRNAs occurs (28,29). A mechanism by which distinct Lef/Tcf isoforms may acquire individual properties is illustrated by their interaction with Hic-5 (hydrogen peroxide-induced clone 5). Hic-5 has been shown to bind a highly conserved and alternatively spliced exon of Lef/Tcf proteins and this results in the formation of a Lef/Tcf subtype-specific repressive complex that prevents target gene activation (30).
Mammalian Dazap2, also known as Proline codon-rich transcript, brain expressed (Prtb), was originally isolated in a mouse gene trap screen as a transcript expressed in the inner ear (31). This gene encodes a small 17 kDa protein that is highly conserved throughout evolution. The protein does not share significant sequence homology with any protein family and its most notable feature is a high content of prolines (17%) and several potential Src homology 2 (SH2)- and SH3-binding motifs (32). The Dazap2 gene is broadly expressed during mouse embryonic development and in adult mouse and human tissues (31,33–35). Interestingly, Dazap2 mRNA and protein are frequently down-regulated in multiple myeloma patients (36) whilst Dazap2 mRNA is known to increase in adhering mouse osteoblasts or in rat astrocytes grown in high ammonia or hypo-osmotic conditions (37). In humans, Dazap2 interacts with RNA-binding testes-specific proteins DAZ and DAZL1 (35). In addition, Dazap2 also binds the Sox6 transcription factor to regulate L-type Ca++ channel α1c expression during cardiac myocyte development (33). Recently, Kim and colleagues (38) described the interaction of Dazap2 with the Eukaryotic initiation factor 4G (eIF4G) which is essential for the formation of discrete cytoplasmic foci, named stress granules (SGs). SGs are formed upon translation inhibition and contain translation initiation factors and 40S ribosomal subunits (34,38). Finally, the protein level of Dazap2 is regulated by its interaction with NEDD4, an E3 ubiquitin ligase (39). Taken together, the aforementioned data indicate that Dazap2 functions in diverse roles in cell biology and physiology.
In this study we have used a yeast two-hybrid screen to identify Dazap2 as a TCF-4 interacting partner. Furthermore, we show that a short region proximal to the TCF-4 HMG box mediates this interaction. Interestingly, although this region is only partially conserved among Tcfs, all Tcf/Lef family members associate with Dazap2 in mammalian cells. Upon interaction with TCF-4 the subcellular distribution of Dazap2 is dramatically shifted from the cytoplasm into the nucleus. Upon knockdown of Dazap2 a reduction in the activity of Tcf/β-catenin reporters was observed along with the expression of several Wnt-signalling target genes. Chromatin immunoprecipitation experiments performed in cells with down-regulated Dazap2 expression revealed a remarkable decrease in TCF-4 binding to Tcf-responsive elements in the promoters of genes tested. We propose that Dazap2 modulates the affinity of Tcfs to their recognition motifs.
MATERIALS AND METHODS
Plasmid constructs
Constructs encoding proteins fused at the N-terminus to EGFP were prepared using the pEGFP-C vector (Clontech); plasmids encoding Myc-tagged proteins were generated using the pK-Myc vector (26) and plasmids expressing Flag-tagged polypeptides were constructed in the vector pFlag-CMV-5a (Sigma). cDNAs encoding human TCF-4 (GenBank accession number NM_030756), human TCF-1 (NM_003202), and mouse Lef-1 (NM_010703) were described previously (26,40,41). Full-length cDNA encoding human TCF-3 (NM_031283) was purchased from Open Biosystems whilst the cDNA encoding human β-catenin (NM_205081) was kindly provided by B. Vogelstein (The Johns Hopkins Kimmel Cancer Center); cDNA encoding the full-length human DAZAP2 protein (NP_055579) was cloned by RT–PCR using template mRNA isolated from DLD-1 cells. PCR amplification steps were performed with Phusion High-Fidelity DNA Polymerase (Finnzymes). Mouse cDNA encoding Wnt-1 [a gift from M.van Dijk (University Hospital Utrecht)] was subcloned into the mammalian expression vector pXJ41 (kindly provided by L. Andera, IMG, Prague, Czech Republic). Mouse cDNA encoding Grg4 [a TLE/Groucho repressor (NM_011600)] was kindly provided by Z. Kozmik (IMG). Human TAB1 (U49928), mouse Tak1 (D76446) and mouse Nlk (NM_008702) cDNAs were obtained from T. Ishitani (Nagoya University). PCR-derived constructs were verified by sequencing; details of plasmid constructs are available on request.
Yeast two-hybrid screen
The cDNA encoding the N-terminal part of TCF-4 lacking the β-catenin-interacting domain [amino acids (aa) 31–333] was subcloned into the vector pGBKT7 (Clontech) and introduced into yeast AH109 cells by the standard lithium acetate transformation protocol. Expression of the TCF-4 fragment and GAL-4 DNA-binding domain (DBD) bait protein was tested in cell lysates by immunoblotting using anti-Myc and anti-TCF-4 monoclonal antibodies. A pre-transformed mouse 17-day embryo Matchmaker cDNA library amplified in the yeast strain Y187 (Clontech) was used for the screen according to the manufacturer's instructions. The ‘library’ and ‘bait’ cells were first mated in liquid cultures before subsequent plating on selective agar plates. After incubation at 30°C for 7–10 days, clones of growing cells were picked and streaked onto fresh selective plates and subjected to β-galactosidase filter lift assays. Plasmids isolated from positive clones were transformed into the yeast strain Y187 and their specificity tested by mini-mating with AH109 yeast cells that expressed the GAL-4 DBD or GAL-4 DBD-Lamin fusion proteins as bait. Clones that specifically interacted with the GAL-4 DBD-TCF-4 bait were sequenced. For the interaction domain mapping experiment cDNAs encoding corresponding fragments of TCF-4 (see legend to Figure 1 for details) were subcloned into the pGBKT7 vector and the resulting constructs were transformed into AH109 yeast cells. Individual yeast clones were mated with Y187 cells containing the Dazap2 prey and the growth of diploid yeast was tested on agar plates under selective conditions.
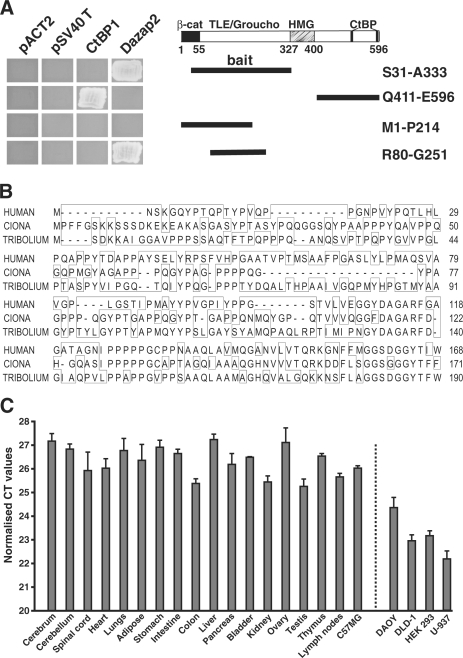
Figure 1.
Interaction between Dazap2 and TCF-4 in a yeast two-hybrid screen. (A) Deletion mutants of human TCF-4 (schematically represented on the right as thick black lines) were tested in an Y2H mini-mating assay for interaction with full-length mouse Dazap2. The left panel shows the growth of clones of yeast cells on selective agar plates. The yeast cells contain plasmids as indicated above and express TCF-4 deletion mutants that are depicted on the right. None of the TCF-4 proteins binds to the separate GAL4 activation domain (AD) encoded by ‘empty’ library vector pACT2 or to the fusion protein GAL4 AD-SV40 T large antigen (pSV40 T). β-cat, β-catenin interaction domain; TLE/Groucho, TLE/Groucho-binding domain; CtBP, CtBP-binding sites; HMG, DNA-binding domain. (B) Amino-acid comparison of human, sea squirt (Ciona) and red flour beetle (Tribolium) Dazap2. Protein sequences were aligned by the ClustalV program. The amino-acid differences are boxed. GenBank accession numbers: Homo sapiens, NP_055579; Tribolium castaneum, XP_973572; Ciona intestinalis, NM_001032667. (C) The Dazap2 gene is broadly expressed in tissues and cell lines. Results of qRT–PCR analyses performed with Dazap2-specific primers on cDNA generated from adult mouse tissues, mouse mammary epithelium C57MG cells, human medulloblastoma DAOY, human embryonic kidney HEK 293, human adenocarcinoma DLD-1 and human lymphoma U-937 cells. The reactions were performed in triplicate. The results shown are from one representative experiment from a total of two. The expression levels of Dazap2 mRNA in the indicated tissues or cell lines are presented as average CT values and the corresponding standard deviations (SD) after normalization to the levels of β-actin cDNA.
Cell lines, transfections, retrovirus production and infection
Human embryonic kidney (HEK) 293, human HeLa, DLD-1 and U2OS cells and mouse Wnt3a-producing L cells were purchased from ATCC. Mouse C57MG cells were kindly provided by R. Nusse and K. Willert (Stanford University). HEK 293 FT cells used for production of retroviral stocks were obtained from Invitrogen. pSuperTOPFLASH HEK 293 (STF 293) cells containing the integrated variant of the Wnt/β-catenin-responsive luciferase reporter pSuperTOPFLASH (42) were a kind gift from Q. Xu and J. Nathans (Johns Hopkins University). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone) and antibiotics. Cells in cultures were regularly checked for the presence of mycoplasma and only mycoplasma-free cells were used in experiments. Transfections of human cells were performed using the Lipofectamine RNAiMAX [small inhibitory RNAs (siRNA) transfections] or Lipofectamine 2000 reagent (plasmid or combined plasmid and siRNAs transfections). Both reagents were purchased from Invitrogen. C57MG cells were transfected using Fugene HD (Roche). Retroviral stocks were produced as described previously (26). Lentiviral stocks were prepared using the Trans-Lentiviral Packaging System (Open Biosystems) according to the manufacturer’s protocol. Retroviral (or lentiviral) infections have been described previously (26); puromycin (Alexis; final concentration 5 µg/ml) resistant cells were selected without subcloning for 10 days and used in subsequent experiments.
GST interaction assays
Constructs expressing Glutathione S-transferase (GST)-TCF-4 fusion proteins were prepared using the pET-42b vector (Novagen). GST-TCF-4 (full-length), GST-TCF-4-N-term (aa 1–333), GST-TCF-C-term (aa 333–596), GST-TCF-4 (aa 1–214), GST-TCF-4 (aa 1–228), GST-TCF-4 (aa 214–310), GST-TCF-4 (aa 228–310) and GST-β-catenin (full-length) fusion proteins were expressed in the BL21 (DE3) strain of Escherichia coli. The mouse Dazap2 and human TCF-4 proteins were produced in vitro using the Quick TNT Coupled Reticulocyte System (Promega). A detailed protocol describing GST pull-down assays was reported previously (26).
Antibodies, co-immunoprecipitation and western blotting
Antiserum to Dazap2 was produced by immunization of rabbits or hens with a bacterially expressed mouse full-length polypeptide. An anti-β-catenin rabbit polyclonal antibody was produced by immunization with a bacterially expressed C-terminal fragment (aa 585–781) derived from the human polypeptide; an anti-β-catenin mouse monoclonal antibody was prepared in collaboration with Exbio Praha (Czech Republic) using standard techniques from the splenocytes of mice immunized with a bacterially produced β-catenin fragment. The anti-TCF-4 monoclonal and anti-TCF-4 and anti-EGFP rabbit polyclonal antibodies were reported previously (27). The following commercially available mouse monoclonal antibodies were used: anti-Myc 9E10 (Roche), anti-Flag M2 (Sigma), anti-Flag (Exbio), anti-α-tubulin TU-01 (Exbio). A detailed protocol describing the immunoblotting procedure can be found in the Supplementary Data.
Immunofluorescent microscopy
For immunofluorescence studies, Dazap2 polyclonal antibodies were purified by affinity chromatography using the GST-Dazap2 antigen coupled to glutathione Sepharose 4B (Amersham Pharmacia Biotech) (43). The purified antibodies were subsequently stored at 4°C in PBS supplemented with 1% BSA [(w/v), Sigma, molecular biology grade]. Monoclonal antibodies were used as hybridoma cell culture supernatants without dilution. Cells grown on coverslips were fixed (when cells were transfected prior to staining, the fixation was performed 18 h after transfection) in cold methanol (−20°C, 5 min) followed by a brief incubation in acetone (−20°C, 30 s). Alternatively, fixation was performed for 10 min at room temperature using a 4% (v/v) solution of paraformaldehyde (Electron Microscopy Sciences) diluted in PBS before cells were subsequently permeabilized with 0.2% (v/v) Triton X-100 (Sigma; room temperature, 15 min) diluted in PBS. After washing with PBS, cells were pre-incubated in 1% BSA (Fraction V; Sigma) for 20 min at room temperature. The cells were then stained with primary GST-Dazap2-purified polyclonal antibodies (5 µg/ml in PBS) or with an anti-TCF-4 monoclonal antibody (undiluted hybridoma culture supernatant; room temperature, 60 min). The samples were washed three times with PBS, and consecutively incubated with a relevant fluorescently labelled secondary antibody. The ALEXA 488 dye conjugated to goat anti-chicken or goat anti-rabbit antibodies and ALEXA 594 conjugated to a goat anti-mouse antibody (dilution 1:500; Molecular Probes) were used. To pre-block the Dazap2 antibody, antigen-purified antibodies (20 µg/ml) were incubated with the bacterially expressed Dazap2 or EGFP (negative control) proteins (overnight at 4°C; the concentration of recombinant protein in each sample was 50 µg/ml), before being diluted in PBS (final concentration 5 µg of the antibody per ml) and used for staining. Finally, the samples were washed three times in PBS, incubated with DAPI nuclear stain (Molecular Probes; 1 min, room temperature, final concentration 1 µM), washed and mounted in MOWIOL (Calbiochem). Immunofluorescence was visualized using a confocal laser scanning microscope (TCS SP5; Leica). The system was carefully tested for overlaps between individual optical channels and the microscopic images were taken separately for each fluorescence channel using the sequential scanning mode.
RNA purification and real-time quantitative RT–PCR (qRT–PCR)
Standard procedures were used for RNA purification and reverse transcription. Briefly, total RNAs were isolated from cells using the Trizol reagent (Invitrogen); random or oligo dT-primed cDNA was prepared in a 20 µl reaction from 1 µg of total RNA using Superscript II RNAseH− reverse transcriptase (Invitrogen). cDNAs were produced from at least two independent RNA isolations and the PCR reactions were performed in triplicate for each primer set. Two percent of the resulting cDNA was used for one quantitative PCR reaction. Control reactions (containing corresponding aliquots from cDNA synthesis reactions that were performed without reverse transcriptase; minus RT controls) were run in parallel duplicates. PCR reactions were run using the LightCycler 480 Real-Time PCR System (Roche). Typically, a 5 µl reaction mixture contained 2.5 µl of LightCycler 480 SYBR Green I Master mix (Roche), 0.5 µl of primers (final concentration 0.5 µM) and cDNA diluted in 2 µl of deionized water. Crossing-threshold (CT) values were calculated by LightCycler® 480 Software (Roche) using the second-derivative maximum algorithm. The specificity of each PCR product was analysed using the in-built melting curve analysis tool for each DNA product identified; additionally, some selected PCR products were verified by sequencing. All primers were calculated using Primer 3 computer services at http://frodo.wi.mit.edu/. Two housekeeping genes, β-actin and ubiquitin C (Ubc) were used as internal control genes to standardize the quality of different cDNA preparations (44). Primer sequences are listed in Supplementary Data.
Wnt3a purification
Recombinant mouse Wnt3a ligand was isolated from the culture medium of Wnt3a-producing L cells according to a detailed protocol published on the Internet (http://www.stanford.edu/%7ernusse/assays/W3aPurif.htm). The activity of individual batches of purified Wnt3a protein were tested using Wnt3a-stimulated and control (Wnt3a storage buffer added only) STF 293 cells and luciferase assays.
Knockdown of Dazap2
For gene knockdowns of human DAZAP2, four duplex siRNAs were purchased from Dharmacon. The target sequences (on the plus DNA strand) were as follows: #1 5′-GGA GCC AAC GUC CUC GUA A-3′, #2 5′-CAC CAU GUC AGC CGC AUU U-3′, #3 5′-UCA GAG CUC UAU CGU CCG A-3′, #4 5′-CUU CAU GGG UGG UUC AGA U-3′. Control EGFP siRNA (Dharmacon) target sequence was: 5′-GCG ACG TAA ACG GCC ACA AGT TC-3′. Cells transfected with duplex siRNAs at a concentration of 30 nM were grown for 24–72 h before further analysis. To generate a stable knockdown of Dazap2 in mouse C57MG cells, the cells were transduced with retroviruses (purchased from Open Biosystems) that express Dazap2 shRNA (shRNA #1 code: TRCN0000085966, shRNA #2 code: TRCN0000085965). Non-silencing lentiviral shRNAmir (pGIPZ; Open Biosystems) were used as a control. The constructs were packaged and transduced into the target cells as described by the manufacturer. The cells were selected without subcloning using appropriate antibiotics before they were used for further analysis.
Reporter gene assays and Wnt stimulation
To assay TCF-mediated transcription, firefly luciferase pTOPFLASH and pFOPFLASH reporters containing either three copies of the optimal Tcf motif GATCAAAGG or three copies of a mutant motif GGCCAAAGG, respectively were used (45). Additionally, Cyclin D1 reporter constructs containing one copy of the Tcf-interacting motif (designated 163CD1LUC) or its mutated variant (163mtLefCD1LUC) (10) (a gift from A. Ben-Ze'e;v, The Weizmann Institute of Science) and the Axin2 promoter reporter (46) (a gift from F. Costantini, Columbia University Medical Center) were used. Reporter gene assays were performed as described previously (26). Briefly, cells were seeded into 24-well plates (∼105 cells per well, depending on the cell type) and transfected 2 h later with a Lipofectamine mixture containing 100 ng Renilla pRL-SV40 plasmid (Promega) as an internal control, 500 ng luciferase reporter plasmid, and up to 1 µg of the particular expression vector. The total amount of DNA was kept constant by adding empty expression vector where necessary. For transfection into STF 293 cells (containing an integrated TCF-dependent reporter pSuperTOPFLASH) (42,47), a mixture that included 50 ng Renilla plasmid and up to 1.5 µg of a specific expression or stuffer vector was prepared. Two independent systems were utilized to activate Wnt signalling: (i) co-transfection of cells with a Wnt1-expressing plasmid. Cells were analysed 24 h post-transfection; (ii) stimulation of cells with purified recombinant Wnt3a ligand. Cells were transfected with corresponding constructs and 15 h post-transfection recombinant Wnt3a (only the vehicle was used in control experiments) was added and the cells cultured for additional 16 h before their harvest and lysis. The activity of firefly and Renilla luciferase in cell lysates were determined using the Dual luciferase system (Promega) and a single tube luminometer Sirius (Berthold). All reporter gene assays were done in triplicate. Reporter gene activities shown are average values plus standard deviations calculated from at least three independent experiments after normalization against the activity of Renilla luciferase.
Chromatin immunoprecipitation (ChIP)
For ChIP experiments, STF 293 cells were grown in 10-cm culture dishes and transfected either with siRNA #2 targeting Dazap2 or control siRNA (non-silencing siRNA set; Dharmacon). Two days later the cells were transferred into 15-cm culture dishes and subsequently stimulated with Wnt3a or Wnt3a storage buffer alone for 16 h. The cells were fixed directly in the dishes using formaldehyde [1% (v/v); Sigma] then harvested and subjected to ChIP analysis according to Kirmizis (48). Usually, chromatin isolated from cells grown in one 15-cm culture dish was used for immunoprecipitation with one specific antibody. Rabbit anti-Dazap2, TCF-4 and β-catenin polyclonal antibodies were used for ChIP; the negative control experiments were performed with a rabbit anti-EGFP polyclonal antibody. The amount of precipitated DNA was analysed using the LightCycler 480 Real-Time PCR System (Roche) in an analogous manner to real-time qRT–PCR. Half a percent of decrosslinked and purified (by phenol extraction) input chromatin (chromatin that was not subjected to ChIP) was analysed in control PCR reactions. The primers used for the PCR amplification are listed in the Supplementary Data.
RESULTS
Dazap2, a small evolutionary conserved protein, interacts with the TCF-4 N-terminus
It is well known that the function of the nuclear effectors of Wnt signalling, the Tcf/Lef transcription factors, can be modulated by various interacting partners. As such, we decided to perform a yeast two-hybrid screen (Y2H) to search for novel TCF-interacting proteins. An N-terminal part of human TCF-4 protein (aa 31–333) was used as bait. This truncated protein lacks its very N-terminus that encodes the main β-catenin-interacting domain (8,49) as well as the C-terminal DNA-binding HMG box (Figure 1A). From a cDNA library collected from a Day 17 mouse embryo we obtained seven yeast colonies (out of ∼5 × 107 diploid yeast cells) growing in selective broths. Of these clones only one encoded a protein that specifically interacted with the TCF-4 bait and not the Gal-4 DNA-binding domain (DBD) alone, Gal-4 DBD-Lamin and Gal-4 DBD-p53 fusion proteins or with the C-terminal part of TCF-4 used as bait in our previous study (26) (Figure 1A and data not shown). The resulting plasmid DNA isolated from the yeast cells encoded a full-length 168 aa polypeptide described previously as Proline codon-rich transcript, brain expressed (Prtb) or DAZ-associated protein 2 (Dazap2) (31,35). Although Dazap2 does not share significant homology with any known protein family, Dazap2 orthologues isolated from various species display remarkable sequence similarity (there is only one amino acid change between human and mouse proteins), especially within the C-terminal half (Figure 1B and Supplementary Figure 1). To determine the expression pattern of Dazap2 mRNA in adult mouse tissues and several different cell lines we performed quantitative real-time RT–PCR analysis (qRT–PCR), which revealed virtually ubiquitous expression (Figure 1C). We further delineated the minimal region in TCF-4 required for association with Dazap2. We generated several deletions in the TCF-4 bait used for the primary screen and tested their ability to interact with full-length Dazap2 via Y2H. As shown in Figure 1A, truncated bait containing aa 80–251 was able to bind Dazap2, however, a protein spanning aa 1–214 could not. Taken together, these results reveal that the relatively short sequence in TCF-4 spanning aa 214–251 is essential for the interaction.
All Tcf/Lef family members associate with Dazap2 in mammalian cells
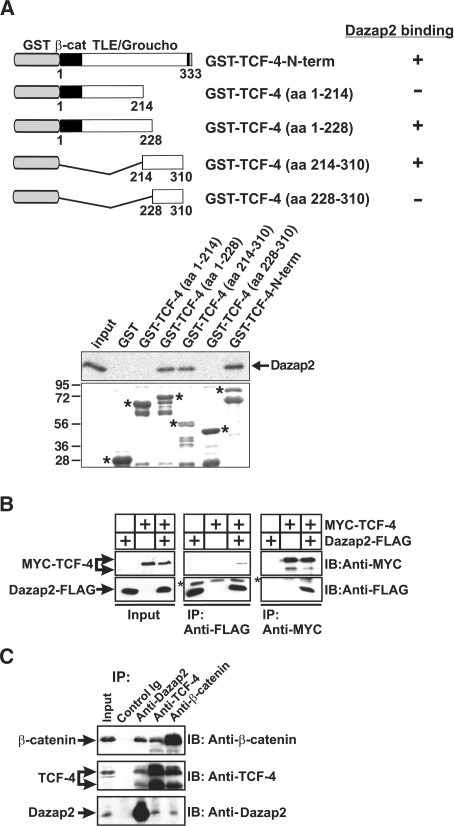
Direct binding between Dazap2 and TCF-4 was evaluated in vitro by pull-down assays that utilized bacterially expressed GST-tagged TCF-4 and in vitro translated Dazap2. Dazap2 associated both with full-length GST-TCF-4 and with the truncated TCF-4 N-terminal fragment. No interaction was detected between Dazap2 and GST alone or with the C-terminal part of TCF-4 (TCF-4-C-term) that encompassed the DNA-binding HMG box domain (Figure 2A and data not shown). Furthermore, we performed a detailed mapping of the putative interaction domains in both proteins using pull-down assays. In agreement with Y2H the Dazap2-binding region was mapped to aa 214–228 in the TCF-4 N-terminus (Figure 2A). On the other hand, any truncation of Dazap2 abolished its association with TCF-4 (Supplementary Figure 2 and data not shown), indicating that non-adjacent parts of the Dazap2 polypeptide participate in the interaction interface. The interaction of Dazap2 with TCF-4 was further confirmed in mammalian cells using co-immunoprecipitations. Experiments involving HEK 293 cells double-transfected with constructs expressing Dazap2-Flag and TCF-4-Myc demonstrated that Flag-tagged Dazap2 could be co-isolated with Myc-tagged TCF-4 when an anti-Myc monoclonal antibody was used for precipitation; conversely, Myc-tagged TCF-4 was present in the anti-Flag precipitates (Figure 2B). The interaction of these two proteins is specific since parallel single-transfection assays did not reveal any binding of Dazap2 and TCF-4 to the anti-Myc- or anti-Flag-tag antibodies, respectively (Figure 2B). In addition, we analysed the ability of endogenous TCF-4 and Dazap2 to interact by performing co-immunoprecipitation assays with rabbit polyclonal antibodies raised against these polypeptides. Analysis of a variety of cell types (DLD-1, HEK 293, C57MG) by co-immunoprecipitation using the anti-Dazap2 or anti-TCF-4 antibodies confirmed our earlier observations and demonstrated that endogenous Dazap2 and TCF-4 do associate. Negative control reactions using an anti-EGFP rabbit polyclonal antibody failed to pull down any proteins again confirming that the interaction is specific (Figure 2C and data not shown). The human colon adenocarcinoma cell line, DLD-1 harbours a mutation in the tumour suppressor APC that results in the accumulation of nuclear TCF-4/β-catenin complexes. Interestingly, Dazap2 was present in the precipitates obtained by incubating DLD-1 cell lysates with an anti-β-catenin antibody and similarly, β-catenin was isolated using an anti-Dazap2 antibody (Figure 2C). Since we did not detect any association between Dazap2 and β-catenin in GST pull-down assays (Supplementary Figure 3) we conclude that these proteins do not interact directly but are instead brought to one heterocomplex via association with their common partner, the TCF-4 factor. We also noted that the endogenous TCF-4 protein extracted from various mammalian cells migrates in the denaturing gels as a double band representing polypeptides of the apparent molecular weight 65 and 85 kDa, respectively (Figures 2C, 5A, 6A and data not shown). Interestingly, both these bands showed immunoreactivity with various monoclonal antibodies recognizing different epitopes in TCF-4 (data not shown). Moreover, ectopic expression of TCF-4 generated two different protein forms similar to their endogenously produced counterparts (Figure 2B and Supplementary Figure 5). As the expression of the both putative TCF-4 proteins was specifically down-regulated by TCF-4 shRNA (Supplementary Figure 4) and the predicted Mw of TCF-4 is 65.3 kDa, we concluded that the slower migrating and mostly more prominent band represents the TFC-4 polypeptide, possibly modified by sumoylation (23).
Figure 2.
Association between Dazap2 and TCF-4 in vitro and in mammalian cells. (A) Interaction of Dazap2 with TCF-4 in GST pull-down assays. The top panel represents a schematic diagram of the TCF-4 proteins used in the in vitro pull-down assays. The bottom panel displays the pull-down assay results between the different bacterially expressed GST fusion TCF-4 proteins and in vitro translated [35S]-labelled Dazap2. Ten percent of the total reaction was loaded in the lane denoted ‘Input’. Under the autoradiograph is a Coomassie Blue-stained gel that shows the amount of GST-tagged TCF-4 proteins used in the experiment. The putative intact forms of the recombinant proteins are labelled by asterisks; the faster migrating bands result from a partial degradation of the corresponding GST fusion proteins. Molecular weight markers in kDa are indicated on the left. (B) Co-immunoprecipitation of FLAG-tagged Dazap2 and MYC-tagged TCF-4. Cell lysates prepared from HEK 293 cells transfected with constructs as indicated were precipitated using anti-MYC and anti-FLAG monoclonal antibodies. The asterisks indicate the light chains of immunoglobulins used in the experiments. (C) Endogenous complexes of Dazap2, TCF-4 and β-catenin in human cells. Lysates prepared from DLD-1 cells were subjected to immunoprecipitation with anti-Dazap2, anti-TCF-4, anti-β-catenin or anti-EGFP (Control Ig) rabbit polyclonal antibodies. Blots were probed by either anti-TCF-4, anti-β-catenin mouse monoclonal or anti-Dazap2 chicken polyclonal antibodies. In lanes denoted ‘Input’, 10% of the total lysate used for one immunoprecipitation was loaded; IP, immunoprecipitation; IB, immunoblotting.
Figure 5.
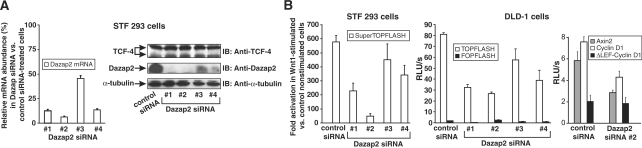
The activity of the Wnt/β-catenin-dependent reporters positively correlates with the amounts of Dazap2 in the cells. (A) HEK 293 cells containing the integrated reporter pSuperTOPFLASH (STF 293 cells) were transfected with four different Dazap2-specific siRNAs marked 1 to 4 or with control siRNA (EGFP-specific) and the changes in the levels of Dazap2 mRNA or protein were tested 24 h post transfection. The left panel shows the results obtained by qRT–PCR analysis. The relative abundance of Dazap2 mRNA in Dazap2 siRNA- versus control siRNA-transfected cells (the level of Dazap2 mRNA in control cells was set to 100%) was derived from the average CT values after normalizing to the levels of β-ACTIN cDNA. Data from two independent experiments performed in triplicate were combined in the graph. SDs (standard deviations) are shown by error bars. The right panel shows western blots of whole cell extracts prepared from STF 293 cells treated with siRNAs as indicated. Blots were probed with anti-Dazap2, anti-TCF-4 or anti-α-tubulin (as an internal control) antibodies. (B) Results of the reporter gene assays in human cells. STF 293 and DLD-1 adenocarcinoma cells were transfected with Dazap2 or control siRNAs. To stimulate the integrated pSuperTOPFLASH in STF 293 cells the siRNA transfection mixtures additionally contained a Wnt1-expression construct; mixtures including an empty vector were used in control transfections. DLD-1 cells were co-transfected with siRNA and the Tcf/β-catenin-responsive reporter pTOPFLASH or pFOPFLASH as a negative control. After 24 h, cells were harvested and luciferase (firefly) activities were determined in the lysates. The histograms represent mean values of triplicate experiments with SDs. The reporter activity in unstimulated (empty vector) STF 293 cells was arbitrarily set to 1. For DLD-1 cells the average luciferase light units per second (RLU/s) are given; the values were corrected for the efficiency of transfection using the internal control Renilla luciferase expression plasmid. The results of one representative experiment from two in total are shown.
Figure 6.
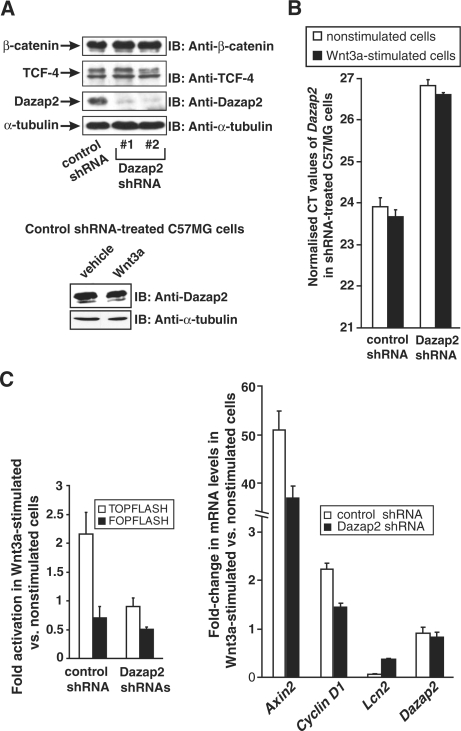
Down-regulation of Dazap2 reduces the responsiveness of C57MG cells to Wnt3a stimulation. (A) The top panel shows a series of western blots from whole cell extracts prepared from mammary gland epithelium C57MG cells transduced with retroviral vectors expressing either a non-silencing control shRNA or two different Dazap2 shRNAs. Prior to harvesting, cells were stimulated with Wnt3a ligand for 16 h. The blots were probed with antibodies as indicated. The bottom panel shows the western blot of whole cell extracts prepared from non-silencing control shRNA-treated cells incubated with Wnt3a or vehicle for 16 h. (B) The relative abundance of Dazap2 mRNA in control or Dazap2 shRNAs-expressing (a mixed culture of cells containing Dazap2 shRNA 1 and 2 was used) cells as measured by qRT–PCR. The cells were cultured without activation or stimulated with Wnt3a for 16 h. The expression of the Dazap2 gene is indicated by average CT values obtained by qRT–PCR assay on the corresponding mRNA after normalization to the levels of β-actin cDNA. (C) The left panel depicts control or Dazap2 shRNA #1 and #2-producing C57MG cells that were transfected with the indicated reporters. Twenty-four hours post-transfection, cells were either stimulated with Wnt3a ligand or grown without stimulation. After additional 16 h, the cells were harvested and luciferase activities were determined in lysates. These activities were corrected for the efficiency of transfection by determining the luciferase/Renilla ratio. Values in unstimulated cells were arbitrarily set to 1. Transfections were done in triplicates and the results from one experiment out of two in total are presented with SDs. The right panel shows cDNAs prepared from Wnt3a-treated or control cells expressing non-silencing or Dazap2 shRNAs analysed by qRT–PCR. The relative abundance of the indicated mRNAs in given cells (the levels of the tested mRNAs in unstimulated cells was set to 1) was derived from the average CT values after normalizing to the levels of β-actin cDNA. Data shown are from one representative experiment from a total of two.
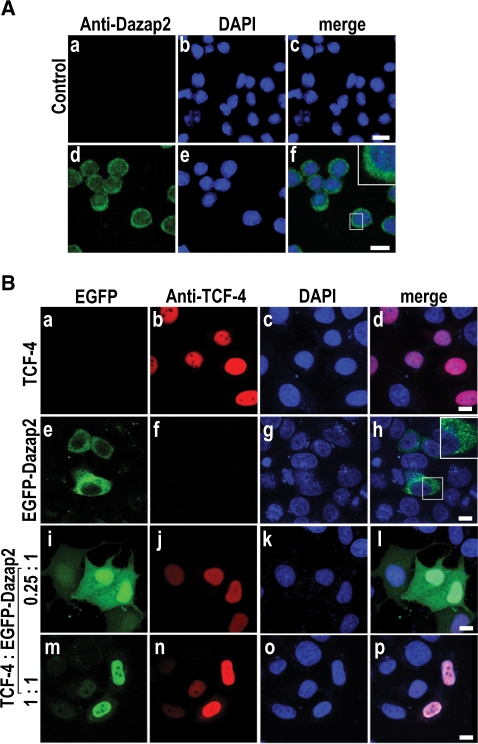
To visualize the subcellular distribution of endogenous DAZAP2 protein we selected the human lymphoma U-937 cell line as it exhibits a relatively high level of DAZAP2 mRNA expression (Figure 1C). Analysis of Dazap2-stained cells by confocal microscopy revealed a predominantly cytoplasmic distribution with some additional nuclear staining (diffuse or in distinct dots or ‘puncta’) (Figure 3A). The staining of putative Dazap2 protein was specific as both polyclonal antisera showed a virtually identical subcellular distribution. Moreover, the observed reactivity was completely abolished by pre-incubation of the primary antibodies with Dazap2 (and not with control EGFP) recombinant protein (Figure 3A and data not shown). Finally, we tested the co-localization of ectopically expressed TCF-4 and Dazap2 (either EGFP- or Myc-tagged) in HeLa cells. In single-transfected cells expressing either TCF-4 or Dazap2, TCF-4 was clearly nuclear whilst Dazap2 (visualized by either EGFP or Myc antibodies) displayed a primarily cytoplasmic and partly nuclear localization. When both proteins were co-expressed, Dazap2 was sequestered to the nucleus in a dose-dependent manner unlike the control EGFP-only protein, which remained uniformly distributed between the cytoplasm and nucleus irrespective of the presence or absence of the TCF-4 factor (Figure 3B and data not shown). Altogether, the data reported here indicate that Dazap2 interacts directly with TCF-4 and that this interaction results in its subcellular redistribution to the nucleus.
Figure 3.
TCF-4 translocates Dazap2 into the nucleus. (A) Endogenous Dazap2 protein shows a mostly cytoplasmic distribution in human cells. Laser scanning confocal micrographs of U-937 cells stained with an antigen-purified chicken anti-Dazap2 polyclonal antibody. The merged images (c) and (f) were generated by an overlay of endogenous Dazap2 captured in the green channel and the DAPI nuclear stain captured in the blue channel. The first row, labelled ‘Control’, shows the cells stained with the primary antibody pre-blocked with Dazap2 recombinant protein as described in the ‘Materials and methods’ section; inset in (f) shows a magnified image as indicated. Bar, 10 µm. (B) Nuclear co-localization of ectopically expressed EGFP-Dazap2 and TCF-4 proteins. Laser scanning confocal microscopy images of HeLa cells transfected with constructs (as indicated on the left) and subsequently stained with the mouse anti-TCF-4 monoclonal antibody. The images (i, j, k, l) show cells transfected with TCF-4 and EGFP-Dazap2 constructs at a ratio 0.25 (TCF-4) to 1 (EGFP-Dazap2); the images (m, n, o, p) were obtained at a ratio 1 to 1. The merged images (d, h, l, p) were generated by an overlay of the corresponding images gained either in the green input channel detecting EGFP-Dazap2, red input channel detecting TCF-4 or blue channel to detect the DAPI nuclear stain. Inset in (h) shows a magnified image as indicated. Bar: 10 µm.
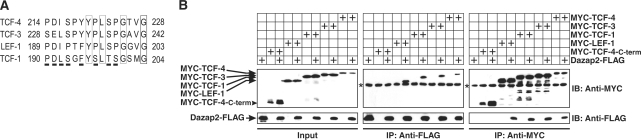
As the essential region in TCF-4 required for binding to Dazap2 displays some sequence homology with the corresponding sequences in other Tcf/Lef family members (Figure 4A), we decided to further analyse whether Dazap2 can interact with additional Tcf/Lef proteins. Constructs expressing Myc-tagged TCF-1, -3, -4, Lef-1 and Flag-tagged Dazap2 were either single- or double-transfected into HEK 293 cells and their ability to interact was tested by co-immunoprecipitation using anti-Myc or anti-Flag antibodies as described above. All TCF/Lef proteins analysed were isolated from cell lysates containing Flag-Dazap2 using the anti-Flag antibody. Reciprocally, Dazap2 was similarly pulled down from cells co-expressing Myc-tagged TCF/Lef proteins with the anti-Myc antibody (Figure 4B). In agreement with previous results Dazap2 did not co-immunoprecipitate with the C-terminal part of TCF-4 (TCF-4-C-term) lacking the Dazap2-interaction domain (Figure 4B). Since the short stretch of amino acids mediating the Dazap2 binding is in close proximity to a region which is alternatively spliced in most of Tcf/Lef family members (alternative splicing involves exon IVa in Tcf-1, exon VI in Lef-1 and exon VIII in Tcf-4) (50–52), we generated a TCF-4 variant lacking exon VIII and tested its ability to bind Dazap2 in GST pull-down and co-immunoprecipitation assays. As this variant associated with Dazap2 to the same extent as the non-spliced form (Supplementary Figure 5), the assays further confirmed the importance of the already defined interaction motif.
Figure 4.
Dazap2 interacts with all members of the Tcf/Lef family. (A) The region of TCF-4 required for interaction with Dazap2 as identified by GST pull-down assays (aa 214–228) is partly conserved in other Tcf/Lef family members. The corresponding sequences of the human proteins were aligned using the ClustalV program. Fully conserved amino-acid residues are boxed whilst amino-acid residues with similar biochemical properties are underlined. GenBank accession numbers: TCF-4, NP_110383; TCF-3, NP_112573; LEF-1, NP_057353; TCF-1, NP_003193. (B) Co-immunoprecipitation of Dazap2-FLAG and MYC-tagged TCF/LEF transcription factors in human HEK 293 cells transfected with various Tcf expression constructs as indicated. Co-immunoprecipitation of TCF/LEF proteins using an anti-FLAG antibody is also shown. In the gel denoted ‘Input’, 10% of total lysates was loaded. The asterisks indicate the heavy chains of immunoglobulins used in the experiments.
Dazap2 knockdown decreases Wnt/β-catenin-mediated transcription
To examine whether Dazap2 overexpression has any effect on Wnt-mediated transcription we co-transfected a TCF/β-catenin-responsive luciferase reporter, pTOPFLASH or its mutated variant, pFOPFLASH into HEK 293 cells in conjunction with a Dazap2 expression construct. The cells were subsequently stimulated by recombinant Wnt3a and the luciferase activities were determined from cell lysates. The ectopic expression of Dazap2 displayed neither an inhibitory nor stimulatory effect on Wnt signalling. Furthermore, analyses of Wnt-regulated transcription in DLD-1 cells that should exhibit constitutive Wnt signalling or U2OS cells that are activated by the Wnt3a ligand were not affected by Dazap2 overexpression (data not shown). Finally, RNA interference (RNAi) was used to down-regulate the expression of Dazap2 and the resulting Tcf/β-catenin-dependent transcription was analysed. First, we utilized four distinct siRNA duplexes and tested their efficiency towards Dazap2 mRNA using qRT–PCR and western blotting. As shown in Figure 5A, all four siRNAs reduced the levels of Dazap2 mRNA and protein to different extents, with siRNA #1 and #2 showing the highest efficiency (as compared to the control EGFP siRNA). Subsequently, we introduced by lipofection the siRNAs into cells containing the genome-integrated variant of the pTOPFLASH reporter, pSuperTOPFLASH [the resultant cells were termed pSuperTOPFLASH (STF) 293]. To stimulate Wnt signalling the cells were co-transfected with a Wnt1 expression construct. Twenty-four hours post-transfection the cells were harvested, cell lysates prepared and luciferase activities measured. In a parallel analogous experiment, DLD-1 cells were co-transfected with siRNA duplexes and a DNA mixture containing the pTOPFLASH or pFOPFLASH (negative control) reporter in addition to a Renilla luciferase-expressing plasmid that acted as an internal control. The next day cells were harvested and processed as described for STF 293 cells. Surprisingly, Tcf-driven transcription measured from the integrated or ectopic reporter pTOPFLASH was substantially reduced in the Dazap2 siRNA-treated cells. Of importance, the extent of signalling reduction corresponded to the efficiency of the Dazap2 knockdown as documented for the individual Dazap2-specific siRNAs (Figure 5B). Virtually identical results were obtained with Wnt-stimulated U2OS cells (data not shown). Furthermore, Dazap2 protein knockdown also negatively affected the transcription of two additional well-defined Tcf/β-catenin-dependent reporter constructs that contained either a 5-kb promoter region of the Axin2 (46) gene or a 163-nt long enhancer element proximal to the transcription start of the Cyclin D1 gene (10) (Figure 5B). Importantly, when a single Tcf/Lef-binding site in the Cyclin D1 reporter was mutated, the resulting plasmid (designated as ΔLEF-Cyclin D1) not only lost its responsiveness to Wnt signalling but its basal expression did not change in cells treated with Dazap2 or control siRNA (Figure 5B). To ensure the two Dazap2-specific siRNAs used did not induce any non-specific ‘off-target’ effects, two lentiviral constructs containing shRNA against mouse Dazap2 were purchased and introduced into mouse mammary epithelium C57MG cells. Since both shRNAs showed a similar efficiency to down-regulate Dazap2 expression (Figure 6A) and the Wnt stimulation had no effect on Dazap2 expression (Figure 6A and B), the polyclonal cell cultures were mixed and luciferase reporter assays performed. As expected, the cells with down-regulated Dazap2 displayed a decrease in Wnt-stimulated transcription as compared to control cells containing non-silencing shRNAs (generated in parallel experiments) (Figure 6C). Therefore, these experiments confirm our previous results obtained from transient transfection assays performed in human cells.
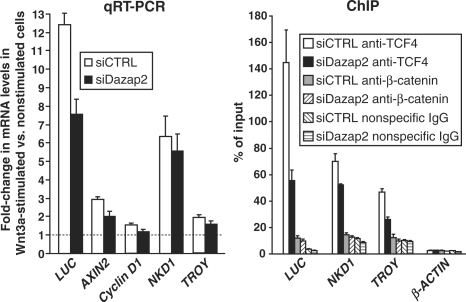
To analyse whether Dazap2 knockdown has any effect on the transcription of endogenous Wnt-signalling target genes, C57MG cells expressing Dazap2 or control shRNAs were stimulated with Wnt3a ligand and the expression of several putative Tcf/β-catenin targets was assessed by qRT–PCR. As shown in Figure 6C, Dazap2 down-regulation resulted in the lower stimulation of the Cyclin D1 and Axin2 genes by Wnt3a than observed in control shRNA-producing cells. Interestingly, the reduced level of Dazap2 protein partly relieved the Wnt-mediated repression of the lipocalin 2 (Lcn2) gene (Figure 6C). Additionally, we performed qRT–PCR analysis in STF 293 cells transiently transfected with control or Dazap2 siRNA. As described above, STF 293 cells contain a stably integrated luciferase reporter that is under the control of eight Tcf/Lef-binding sites. This endogenous reporter, which we named Luciferase (LUC), was activated at the mRNA level up to 13-fold upon stimulation with Wnt3a for 16 h (Figure 7). In agreement with the reporter gene assays, Dazap2 knockdown resulted in an approximately forty percent reduction in transcription of this gene after treatment with Wnt3a. A similar although less robust decrease in Wnt-activated expression was observed in several other Wnt-signalling target genes including AXIN2, CYCLIN D1, naked cuticle homologue 1 (NKD1; 53,54) and tumor necrosis factor receptor superfamily, member 19 [(TNFRSF19), known also as TROY; (55,56) (Figure 7)]. To clarify the possible mechanism behind Dazap2 function we wanted to perform ChIP analysis. Since we did not succeed to perform ChIP on the promoters of the Axin2 and Cyclin D1 genes in any mouse or human cells (including C57MG and STF 293 cells) we extended our analysis to the TCF-responsive enhancers of the Luciferase, NKD1 and TROY genes. We did not observe ChIP using our anti-Dazap2 antibody and furthermore, we only detected a limited pull-down of the Luciferase transgene (the most extensively up-regulated gene in the assay) by the anti-β-catenin antibody. Instead we used our anti-TCF-4 antibody to immunoprecipitate DNA elements that spanned the promoter regions of the tested genes (Figure 7). Interestingly, reduction of Dazap2 lowered the binding of TCF-4 to the promoters of these Wnt-signalling target genes. To verify the ChIP results, two control experiments were carried out. First, we performed ChIP with a non-specific antibody (rabbit anti-EGFP). Second, we used an anti-TCF-4 antibody in an attempt to immunoprecipitate irrelevant chromosomal DNA that concealed the open reading frames of the β-ACTIN or LUC genes. In these experiments we never noted any effect of the cellular levels of Dazap2 on the quantity of precipitated DNA (Figure 7 and data not shown). As Dazap2 knockdown did not alter the amount of TCF-4 and β-catenin in the cell (Figures 5A, 6A and data not shown) these results imply that the Dazap2/TCF-4 interaction might influence the efficiency of TCF-4 binding to the promoters of the genes regulated by the canonical Wnt-signalling pathway.
Figure 7.
Dazap2 knockdown decreases binding of TCF-4 to the promoters of the Wnt-signalling target genes. STF 293 cells were transfected with Dazap2 siRNA #2 (siDazap2) or control siRNA (siCTRL). After a 2-day expansion the cells were transferred to large culture dishes and stimulated with either Wnt3a ligand or Wnt3a storage buffer only. After 16 h, the cells were harvested and used for mRNA isolation and cDNA preparation. Alternatively, the cells were fixed directly in the dishes and utilized for chromatin immunoprecipitation (ChIP). The left panel shows the results from qRT–PCR analysis. The relative abundance of the indicated mRNAs in Wnt3a-stimulated versus control cells was derived from the average CT values after normalizing to the levels of β-ACTIN cDNA. The right panel shows the ChIP analysis of chromatin isolated from STF 293 cells transfected with control or Dazap2 siRNA #2 duplexes. Only results for Wnt3a-stimulated cells are shown. The diagram represents the relative amounts of the respective DNA element pulled-down by the indicated antibodies. The amount of input non-immunoprecipitated DNA (evaluated separately for each primer set) was arbitrarily set to 100%. Data from one representative experiment from two in total are given.
DISCUSSION
In this report we provide evidence for an association between the nuclear Wnt-signalling pathway effectors, Tcf/Lef proteins, and a small evolutionary conserved protein Dazap2. Dazap2 was isolated in a Y2H screen that utilized the N-terminal part of the TCF-4 factor as bait. Although the Dazap2-interacting domain in TCF-4 is only partially preserved in other Tcf/Lef proteins, co-immunoprecipitation assays carried out in mammalian cells clearly demonstrated that all Tcf/Lef family members bind to Dazap2 with similar affinities (Figure 4B). We further delineated a short region in TCF-4 spanning aa 214–228 as the interaction domain essential for Dazap2 binding (Figure 2A). Interestingly, the homologous region in all Tcfs contains amino acids that are not identical but display similar biochemical properties (Figure 4A). This might indicate a common structural basis for the association of Tcf/Lef proteins with Dazap2.
Dazap2 was originally identified as a transcript expressed in the mouse inner ear with its expression further observed in the embryonic heart and developing and adult mouse brain (31). This report is in stark contrast to our finding that illustrates ubiquitous expression of Dazap2 mRNA in various mouse tissues (Figure 1C). Nevertheless, our data are in agreement with northern blot analyses that indicate broad expression of the Dazap2 gene in different human and mouse tissues and cell lines (32–34). The most remarkable feature of Dazap2 is the conservation of its DNA and protein sequence throughout evolution. The identity between human and mouse protein orthologues is virtually 100% (there is only one aa change from 168 aa in total) (Supplementary Figure 1) and human Dazap2 aligns well, especially at the C-terminus, with the putative Dazap2 proteins identified in the invertebrates Ciona intestinalis and Tribolium castaneum (Figure 1B). Given that Dazap2 was identified as a binding partner of many cellular proteins (57), the necessity to preserve these interactions might possibly explain the low mutational rate of the Dazap2 sequence throughout evolution. With respect to the high sequence homology of Dazap2 in various species it is rather striking that Dazap2 mutant mice do not display any remarkable phenotype and are born and bred as their wild-type littermates (31). The Dazap2−/− mice were generated from ES cells via gene trap technology. Although we cannot exclude that the modified Dazap2 locus still produces an intact protein, the insertion site of the reporter gene just several nucleotides downstream of the putative translation start site and a complete absence of Dazap2 mRNA expression indicate that the mutant animals are really Dazap2 null. There are two possibilities that might explain an absence of phenotype in Dazap2−/− mice. First, the phenotype may be very subtle and/or the mice have to be challenged in some way to display any phenotype and second, there is another Dazap2 homologue in the mouse genome that can functionally replace the damaged gene. Indeed, a sequence database search in the mouse genome revealed a Dazap2 pseudogene localized on chromosome 4 and one gene similar to Dazap2 on chromosome 13. This Dazap2-like gene comprises several exons and introns and encodes a putative 168 aa polypeptide that is highly homologous to Dazap2 (Supplementary Figure 6). Therefore, we speculate that these genes are redundant and Dazap2-like can compensate for the absence of the Dazap2 gene. Interestingly, we have not been able to detect the expression of the Dazap2-like product in any of the cell lines tested (data not shown). Thus, to settle this matter definitively, the possible redundancy of these two genes should be tested directly in the Dazap2 mutant animals. Another noticeable feature of the Dazap2 protein is its subcellular localization. Several authors have used confocal or fluorescent microscopy to visualize ectopically expressed Dazap2 as a wild-type untagged protein or as a variant fused to different tags (mostly N-terminal EGFP). These authors described the subcellular distribution of Dazap2 as diffuse in the cytoplasm and nucleus (32,36), in nuclear puncta (39) or in the nucleus and cytoplasmic SG bodies (38). These experiments were predominantly performed in HeLa cells. Interestingly, we noted in HeLa (and Cos-7) cells that both tagged or untagged Dazap2 principally localizes in the cytoplasm and partly in the nucleus but this distribution can differ in a limited fraction of the cells possibly as a consequence of the cell cycle. Nevertheless, Dazap2 was efficiently translocated to the nuclei (or retained in the nuclei) in cells expressing TCF-4, thus further confirming the interaction between Dazap2 and TCF-4 (Figure 3B).
To assess the biological significance of the association between Dazap2 and Tcfs, we first ectopically expressed Dazap2 together with three different Tcf/β-catenin-responsive reporters in various cells. The cells were further stimulated with purified Wnt3a ligand and the activities of the reporters were determined in lysates. Surprisingly, we never observed changes in the transcriptional activity of the reporters depending on the increased levels of Dazap2. Subsequently, we utilized RNAi technology to test the influence of Dazap2 knockdown on Wnt-dependent transcription. In cells with reduced levels of Dazap2 mRNA and protein the activities of the Tcf/β-catenin reporters were significantly decreased and the extent of reduction correlated well with the ability of each particular siRNA to down-regulate Dazap2 (Figure 5A and B). The observed results were not related to the non-specific ‘off-target’ effects of the siRNA duplexes used since control non-silencing siRNA did not show any impact on the transcriptional activity of the tested constructs. Additionally, similar effects on Wnt signalling were observed in cells stably expressing Dazap2 shRNAs (Figure 6C). Importantly, we also demonstrated by qRT–PCR that Dazap2 knockdown also negatively influenced the transcriptional activation of endogenous Wnt-signalling target genes, although to a lesser extent than observed for the reporter genes (Figures 6C and 7). There are several possibilities that could explain the partial discrepancy between the results obtained from the reporter gene assays and the qRT–PCR analysis of endogenous genes. First, the transcriptional regulation of the reporter genes is less complex than that of the endogenous promoters and possibly more dependent on the activity of the Wnt pathway. Second, in the transient siRNA transfections we noted fast ‘exhaustion’ of Dazap2 siRNA followed by rapid return of Dazap2 mRNA and protein to the original levels (data not shown). This in fact could be responsible for the less pronounced effects of Dazap2 knockdown especially in experiments where prolonged treatment with Dazap2 siRNA was needed (Figure 7). Finally, the reporters used in the study encode a ‘standard’ luciferase protein that is quite stable and accumulates in the cells. This presumably would also explain the differences between luminometric measurements and the qRT–PCR analysis.
A possible explanation for the negative impact of Dazap2 down-regulation on Wnt-stimulated transcription is that Dazap2 is a stabilizing component of TCF-4/β-catenin heterocomplexes (Figure 2C). To evaluate this possibility we tried to examine whether reduced levels of Dazap2 could disrupt these complexes. However, co-immunoprecipitation experiments from shRNA-producing cells did not reveal any difference in the amounts of β-catenin pulled down by TCF-4 in cells with normal or decreased Dazap2 expression (data not shown). We also excluded the possibility that Dazap2 is important for the production or stability of TCF-4 or β-catenin which could have accounted for the decrease in transcriptional activation from the reporters as Dazap2 knockdown did not influence the expression of either of these Wnt-signalling effectors (Figures 5A, 6A and data not shown). The Dazap2 interacting domain in Tcfs partly overlaps with a region that in vivo is bound by TLE/Groucho proteins (5,6,8). Therefore, we tested the hypothesis that Dazap2 binding might block the association of Tcfs with these co-repressors and would in turn increase the activating function of the Tcf/Lef transcription factors. We transfected pTOPFLASH into control siRNA- or Dazap2 siRNA-treated HEK 293 cells together with Grg4, a mouse TLE/Groucho homologue. The cells were subsequently stimulated with Wnt3a, harvested and reporter activities measured in cell lysates. In a parallel experiment, control siRNA- or Dazap2 siRNA-treated cells were lipofected with pTOPFLASH and cDNA encoding either a negative regulator of Wnt signalling, nemo-like kinase (NLK) alone or in combination with its upstream activating kinases TAB1 and TAK1. The TAB/TAK/NLK cascade is a downstream component of a negative feed-back loop that is activated by Wnt signalling (58). Active NLK phosphorylates Tcfs and importantly, this phosphorylation prevents the binding of Tcf/β-catenin complexes to DNA and consequently leads to Tcf ubiquitylation and degradation (59–61). Interestingly, the NLK phosphorylation sites in Tcfs are located just proximal to the region indispensable for Dazap2 binding (59). Intriguingly, Winkel and colleagues (62) recently reported that Dazap2 (referred to as PRTB) interacts with TAK1 kinase and enhances its enzymatic activity. Nevertheless, we did not observe any of the repressive effects of Grg4 or NLK on Wnt signalling (using the ectopic or integrated reporter) and thus we did not notice any enhancement of this repression in cells with reduced Dazap2 expression. Based on these observations we excluded the possibility that Dazap2 functions as a blocker of the negative functions of TLE/Groucho and NLK. Finally, we performed a ChIP experiment utilizing chromatin isolated from STF 293 cells with DAZAP2 down-regulated. The assay revealed a remarkable decrease in the association of TCF-4 to the Tcf-responsive sites in the promoters of Wnt-signalling target genes (Figure 7). These data imply that the Tcf/Lef interacting partner Dazap2 can modulate in vivo the affinity of Tcfs for their recognition motifs. Interestingly, in mouse mammary gland epithelium C57MG cells, decreased levels of Dazap2 partly relieved the repression on the Lcn2 gene mediated by Wnt3a treatment (Figure 6C). As the repressive effect of active Wnt signalling on Lcn2 does not depend on the direct binding of Tcf/β-catenin complexes to the Lcn2 promoter (63), this result suggests that Dazap2 levels might modulate the complex output of the Wnt-signalling pathway. To prove this hypothesis, we tested the expression of genes that respond differentially to the Wnt signal in C57MG cells, Stromelysin-1 (Sl-1/Mmp3) and mesothelin (Msln) (64). However, qRT–PCR analysis revealed that the transcription of these two genes depends on the plating density of cells in culture rather than specifically on Wnt signalling (P.M. and V.K., unpublished data). Nevertheless, preliminary cDNA microarray data obtained from mRNAs isolated from Wnt-stimulated C57MG cells expressing Dazap2-specific or control shRNAs indicate that indeed, both the activating and also the inhibitory function of Tcf/Lef proteins might be influenced by the amounts of Dazap2 (P.M., R.I. and V.K., unpublished data).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant Agency of the Czech Republic [grant number 204/07/1567]; qChIP/chip06 project from the Ministry of Education, Youth and Sports of the Czech Republic [B06077]; Institutional grant from the Academy of Sciences of the Czech Republic [AV0Z50520514] to the Institute of Molecular Genetics. Funding for open access charge: Grant Agency of the Czech Republic and the Ministry of Education, Youth and Sports of the Czech Republic.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank L. Andera, A. Ben-Ze’ev, F. Costantini, T. Ishitani, Z. Kozmik, M.van Dijk and B. Vogelstein for vectors, plasmid constructs and reporters. We are grateful to J. Nathans, R. Nusse, K. Willert and Q. Xu for cell lines used in the study. We further thank A. Corlett and S. Takacova for critically reading the manuscript.
REFERENCES
- 1.Huang H, He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polakis P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 5.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 6.Brantjes H, Roose J, van de Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co- repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 9.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 10.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'A;mico M, Pestell R, Ben-Ze'e;v A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 12.Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snider L, Thirlwell H, Miller JR, Moon RT, Groudine M, Tapscott SJ. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell Biol. 2001;21:1866–1873. doi: 10.1128/MCB.21.5.1866-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 18.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial- mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 19.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 20.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 21.Yasumoto K, Takeda K, Saito H, Watanabe K, Takahashi K, Shibahara S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 2002;21:2703–2714. doi: 10.1093/emboj/21.11.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H, Ihara M, Matsuura Y, Kikuchi A. Sumoylation is involved in beta-catenin-dependent activation of Tcf-4. EMBO J. 2003;22:2047–2059. doi: 10.1093/emboj/cdg204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Flejou JF, Hamelin R, Duval A. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–4448. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]
- 26.Valenta T, Lukas J, Korinek V. HMG box transcription factor TCF-4's; interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31:2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25:2326–2337. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2003;278:3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 29.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 30.Ghogomu SM, van Venrooy S, Ritthaler M, Wedlich D, Gradl D. HIC-5 is a novel repressor of lymphoid enhancer factor/T-cell factor-driven transcription. J. Biol. Chem. 2006;281:1755–1764. doi: 10.1074/jbc.M505869200. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Mansour SL. Expression and genetic analysis of prtb, a gene that encodes a highly conserved proline-rich protein expressed in the brain. Dev. Dyn. 1999;215:108–116. doi: 10.1002/(SICI)1097-0177(199906)215:2<108::AID-DVDY3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Luo S, Peng J, Huang C, Tan D, Hu W. The structure, expression and function prediction of DAZAP2, a down-regulated gene in multiple myeloma. Genomics Proteomics Bioinformatics. 2004;2:47–54. doi: 10.1016/S1672-0229(04)02007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen-Barak O, Yi Z, Hagiwara N, Monzen K, Komuro I, Brilliant MH. Sox6 regulation of cardiac myocyte development. Nucleic Acids Res. 2003;31:5941–5948. doi: 10.1093/nar/gkg807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommerfeldt DW, Zhi J, Rubin CT, Hadjiargyrou M. Proline-rich transcript of the brain (prtb) is a serum-responsive gene in osteoblasts and upregulated during adhesion. J. Cell Biochem. 2002;84:301–308. doi: 10.1002/jcb.10018. [DOI] [PubMed] [Google Scholar]
- 35.Tsui S, Dai T, Roettger S, Schempp W, Salido EC, Yen PH. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 36.Shi YW, Shen R, Ren W, Tang LJ, Tan DR, Hu WX. Molecular features and expression of DAZAP2 in human multiple myeloma. Chin. Med. J. 2007;120:1659–1665. [PubMed] [Google Scholar]
- 37.Warskulat U, Kreuels S, Muller HW, Haussinger D. Identification of osmosensitive and ammonia-regulated genes in rat astrocytes by Northern blotting and differential display reverse transcriptase-polymerase chain reaction. J. Hepatol. 2001;35:358–366. doi: 10.1016/s0168-8278(01)00149-0. [DOI] [PubMed] [Google Scholar]
- 38.Kim JE, Ryu I, Kim WJ, Song OK, Ryu J, Kwon MY, Kim JH, Jang SK. Proline-rich transcript in brain protein induces stress granule formation. Mol. Cell Biol. 2008;28:803–813. doi: 10.1128/MCB.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton MH, Tcherepanova I, Huibregtse JM, McDonnell DP. Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrates. J. Biol. Chem. 2001;276:26324–26331. doi: 10.1074/jbc.M101205200. [DOI] [PubMed] [Google Scholar]
- 40.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 41.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 43.Bar-Peled M, Raikhel NV. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 1996;241:140–142. doi: 10.1006/abio.1996.0390. [DOI] [PubMed] [Google Scholar]
- 44.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma [see comments] Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 46.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaykas A, Moon RT. A plasmid-based system for expressing small interfering RNA libraries in mammalian cells. BMC Cell Biol. 2004;5:16. doi: 10.1186/1471-2121-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 50.Gradl D, Konig A, Wedlich D. Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements. J. Biol. Chem. 2002;277:14159–14171. doi: 10.1074/jbc.M107055200. [DOI] [PubMed] [Google Scholar]
- 51.Pukrop T, Gradl D, Henningfeld KA, Knochel W, Wedlich D, Kuhl M. Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J. Biol. Chem. 2001;276:8968–8978. doi: 10.1074/jbc.M007533200. [DOI] [PubMed] [Google Scholar]
- 52.Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Wharton KA, Jr, Fuchs SY, von Schweinitz D, Pietsch T. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res. 2005;11:4295–4304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Cagatay T, Amanai M, Zhang M, Kline J, Castrillon DH, Ashfaq R, Oz OK, Wharton KA., Jr Viable mice with compound mutations in the Wnt/Dvl pathway antagonists nkd1 and nkd2. Mol. Cell Biol. 2007;27:4454–4464. doi: 10.1128/MCB.00133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buttitta L, Tanaka TS, Chen AE, Ko MS, Fan CM. Microarray analysis of somitogenesis reveals novel targets of different WNT signaling pathways in the somitic mesoderm. Dev. Biol. 2003;258:91–104. doi: 10.1016/s0012-1606(03)00116-7. [DOI] [PubMed] [Google Scholar]
- 56.Phesse TJ, Parry L, Reed KR, Ewan KB, Dale TC, Sansom OJ, Clarke AR. Deficiency of Mbd2 attenuates Wnt signaling. Mol. Cell Biol. 2008;28:6094–6103. doi: 10.1128/MCB.00539-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 58.Smit L, Baas A, Kuipers J, Korswagen H, van de Wetering M, Clevers H. Wnt activates the Tak1/Nemo-like kinase pathway. J. Biol. Chem. 2004;279:17232–17240. doi: 10.1074/jbc.M307801200. [DOI] [PubMed] [Google Scholar]
- 59.Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol. Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta- catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 61.Yamada M, Ohnishi J, Ohkawara B, Iemura S, Satoh K, Hyodo-Miura J, Kawachi K, Natsume T, Shibuya H. NARF, an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF) J. Biol. Chem. 2006;281:20749–20760. doi: 10.1074/jbc.M602089200. [DOI] [PubMed] [Google Scholar]
- 62.Winkel A, Stricker S, Tylzanowski P, Seiffart V, Mundlos S, Gross G, Hoffmann A. Wnt-ligand-dependent interaction of TAK1 (TGF-beta-activated kinase-1) with the receptor tyrosine kinase Ror2 modulates canonical Wnt-signalling. Cell Signal. 2008;20:2134–2144. doi: 10.1016/j.cellsig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Ziegler S, Rohrs S, Tickenbrock L, Langerak A, Chu ST, Feldmann I, Jakubowski N, Muller O. Lipocalin 24p3 is regulated by the Wnt pathway independent of regulation by iron. Cancer Genet. Cytogenet. 2007;174:16–23. doi: 10.1016/j.cancergencyto.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 64.Prieve MG, Moon RT. Stromelysin-1 and mesothelin are differentially regulated by Wnt-5a and Wnt-1 in C57mg mouse mammary epithelial cells. BMC Dev. Biol. 2003;3:1–10. doi: 10.1186/1471-213X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.