Abstract
The Sec1/Munc18 (SM) protein Munc18-1 and the SNAREs syntaxin-1, SNAP-25 and synaptobrevin form the core of the membrane fusion machinery that triggers neurotransmitter release. Munc18-1 binds to syntaxin-1 folded into a closed conformation and to the SNARE complex formed by the three SNAREs, which involves an open syntaxin-1 conformation. The former interaction is likely specialized for neurotransmitter release, whereas SM protein/SNARE complex interactions are likely key for all types of intracellular membrane fusion. It is currently unclear whether the closed conformation is highly or only marginally populated in isolated syntaxin-1, and whether Munc18-1 stabilizes the close conformation or helps to open it to facilitate SNARE complex formation. A detailed NMR analysis now suggests that the closed conformation is almost quantitatively populated in isolated syntaxin-1 in the absence of oligomerization, and indicates that its structure is very similar to that observed previously in the crystal structure of the Munc18-1/syntaxin-1 complex. Moreover, we demonstrate that Munc18-1 binding prevents opening of the syntaxin-1 closed conformation. These results support a model whereby the closed conformation constitutes a key intrinsic property of isolated syntaxin-1 and Munc18-1 binding stabilizes this conformation; in this model, Munc18-1 plays in addition an active role in downstream events after another factor(s) helps to open the syntaxin-1 conformation.
Keywords: conformational exchange, membrane traffic, munc18, neurotransmitter release, syntaxin, TROSY
Introduction
The exquisite spatial and temporal regulation of neurotransmitter release is critical for brain function. To achieve such regulation, release is controlled by a complex protein machinery and occurs in a series of steps that include docking of synaptic vesicles to the plasma membrane, a priming step(s) that leaves the vesicles in a release-ready state, and Ca2+-triggered release of neurotransmitters by vesicle exocytosis (Sudhof, 2004). Central components of the release apparatus are the member of the Sec1/Munc18 (SM) family Munc18-1 and the SNARE proteins syntaxin-1, SNAP-25 and synaptobrevin/VAMP (Jahn and Scheller, 2006; Brunger, 2005; Rizo et al., 2006; Toonen and Verhage, 2007). These proteins have homologues in most types of intracellular membrane traffic and are thus thought to form the core of a conserved membrane fusion machinery (Rizo and Sudhof, 2002). Among these proteins, syntaxin-1 plays a particularly central role in the regulation and execution of neurotransmitter release.
Syntaxin-1 contains a single C-terminal transmembrane sequence and a cytoplasmic region of 265 residues that includes an autonomously folded three-helix bundle domain (called the Habc domain, residues 27–146), a linker sequence, and a SNARE motif (which is the signature of SNARE proteins; residues 190–259) (Fernandez et al., 1998). The syntaxin-1 SNARE motif forms a tight four-helix bundle with the SNARE motifs of synaptobrevin and SNAP-25 that is known as the SNARE complex (Poirier et al., 1998; Sutton et al., 1998). Formation of this complex brings the synaptic vesicle and plasma membranes together and is key for membrane fusion (Hanson et al., 1997; Jahn and Scheller, 2006). Syntaxin-1 also binds tightly to Munc18-1 (Hata et al., 1993; Pevsner et al., 1994b; Garcia et al., 1994), an interaction that is incompatible with the SNARE complex and requires a ‘closed conformation’ of syntaxin-1 involving intramolecular binding of its SNARE motif to the Habc domain (Dulubova et al., 1999).
All SNAREs from the syntaxin family share the domain structure of syntaxin-1 and form analogous SNARE complexes. However, while the closed conformation is also adopted by Sso1p, the syntaxin from the yeast plasma membrane (Nicholson et al., 1998; Fiebig et al., 1999), this feature is not generally found in syntaxins from other membrane compartments (Dulubova et al., 2001; Dulubova et al., 2002; Yamaguchi et al., 2002). Moreover, the SM protein that functions in the yeast plasma membrane, Sec1p, binds to the SNARE complex containing Sso1p rather than to isolated Sso1p (Carr et al., 1999), and syntaxins from diverse membrane compartments of yeast and vertebrates bind to their cognate SM proteins through an N-terminal motif preceding the Habc domain (Dulubova et al., 2002; Yamaguchi et al., 2002; Dulubova et al., 2003; Bracher and Weissenhorn, 2002). The apparently confusing picture that emerged from the observation of such diverse interactions between highly conserved proteins has been recently clarified by increasing evidence suggesting that SM proteins generally bind to SNARE complexes and that such interactions involve in many cases the N-terminal motifs of the corresponding syntaxins (Collins et al., 2005; Peng and Gallwitz, 2002; Carpp et al., 2006; Latham et al., 2006). Indeed, a potential mechanism for how these SM protein/SNARE complex macromolecular assemblies may act as the core of the fusion machinery has been proposed (Rizo et al., 2006), and Munc18-1 was shown recently to bind to the neuronal SNARE complex (Dulubova et al., 2007) and to enhance SNARE-mediated liposome fusion (Shen et al., 2007).
These findings suggest that the binary interaction between Munc18-1 and the closed conformation of syntaxin-1 is not universal but rather represents a specific feature that evolved to meet the tight regulatory requirements of Ca2+-evoked exocytosis. Thus, a popular model envisages that the transition from the closed conformation of syntaxin-1 to its open conformation is key for priming vesicles into the release-ready state and that the priming factor Munc13 plays a crucial role in this transition (Richmond et al., 2001; Rizo and Sudhof, 2002). Based on the crystal structures of the Munc18-1/syntaxin-1 complex and the closed conformation of isolated Sso1p, it has also been proposed that Munc18-1 binding could play an active role in helping to open the syntaxin-1 conformation (Misura et al., 2000; Munson et al., 2000). Conversely, single molecule FRET studies suggested that Munc18-1 binding stabilizes the closed conformation and that this conformation is only populated 15–30% of the time in isolated syntaxin-1 (Margittai et al., 2003). Assessing the merit of these different proposals is critical to understand the mechanism of neurotransmitter release, but has been hindered by the lack of high-resolution structural information on the conformational state of the cytoplasmic region of syntaxin-1.
Here we describe an NMR study of a fragment encompassing the minimal sequence required to form the closed conformation of syntaxin-1. Our data indicate that much of the closed conformation of isolated syntaxin-1 is very similar to that of syntaxin-1 bound to Munc18-1, but is less stable. In combination with 1H-15N HSQC spectra of diverse syntaxin-1 fragments and a 1H-15N TROSY-HSQC spectrum of one fragment bound to Munc18-1, our data show that the stability of the closed conformation depends on the ionic strength and on the syntaxin-1 fragment used, and that Munc18-1 prevents opening of syntaxin-1 at physiological salt concentrations. Overall, our results strongly support the notion that Munc18-1 binding to syntaxin-1 stabilizes the closed conformation, preventing rather than facilitating SNARE complex assembly, even though Munc18-1 likely assists in SNARE complex formation and membrane fusion after other components of the release machinery help to open the syntaxin-1 conformation.
Materials and methods
Vectors for bacterial expression of rat syntaxin-1A fragments spanning residues 27–146, 2–243 and 2–253, as well as full-length rat Munc18-1 were described previously (Fernandez et al., 1998; Dulubova et al., 1999; Dulubova et al., 2007). The vector to express the Sxcl fragment (residues 26–230 of rat syntaxin-1A) was generated by standard PCR-based procedures and subcloned into pGEX-KT (Hakes and Dixon, 1992). All proteins were expressed in bacteria as GST-fusion proteins and purified after cleavage of the GST moiety as described (Dulubova et al., 1999; Dulubova et al., 2007). Uniform 2H, 15N and 13C labeling in different combinations was accomplished by growing the bacteria in media made with D2O as the solvent, using 15NH4Cl and 13C6-labeled glucose as the sole nitrogen and carbon sources, respectively, as needed. The complex between 2H,15N-labeled syntaxin-1(2–253) and unlabeled Munc18-1 was prepared by mixing equimolar amounts of the two purified proteins and purification of the complex by gel filtration.
All NMR experiments were acquired at 28 °C on a Varian INOVA600 spectrometer equipped with a triple resonance probe. All samples were prepared in 20 mM phosphate buffer (pH. 7.4) containing no NaCl, unless otherwise indicated, using H2O:D2O 95:5 (v/v) or D2O as the solvent. The protein concentration was 100–200 µM for 1H-15N HSQC experiments of syntaxin-1 fragments, 200 µM for the 1H-15N TROSY-HSQC spectrum of the Munc18-1/2H,15N-syntaxin-1(2–253) complex, and 700 µM in all experiments used for resonance assignment of the syntaxin-1(26–230) fragment and determination of the structure of the Habc domain within this fragment. These experiments were performed using standard sensitivity-enhanced, pulsed-field gradient-based pulse sequences for double and triple resonance spectra (Zhang et al., 1994; Muhandiram and Kay, 1994; Kay et al., 1993; Yamazaki et al., 1994), and included: i) 2D DQF-COSY, 2D NOESY, 2D TOCSY, 3D 1H-15N TOCSY-HSQC and 3D 1H-15N NOESY-HSQC acquired on a uniformly 15N-labeled sample; ii) 3D 1H-15N TOCSY-HSQC, 3D 1H-15N NOESY-HSQC, 3D 1H-13C NOESY-HSQC, HNCO, HNCACB, CBCA(CO)NH, (H)C(CO)NH-TOCSY, H(C)(CO)NH-TOCSY, and HCCH-TOCSY acquired on a uniformly 15N, 13C-labeled sample; iii) HNCO, HNCA, HN(CO)CA, HN(CA)CB and HN(COCA)CB acquired on constant time and non-constant time modes in the 13C dimension on a uniformly 2H,15N,13C-labeled sample; and iv) 3D 1H-15N NOESY-HSQC and 3D 1H-15N,1H-15N HSQC-NOESY-HSQC (only one 1H dimension developed) acquired on a uniformly 2H,15N-labeled sample. All data were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRView (Johnson and Blevins, 1994). The assignments for the Sxcl fragment have been deposited in BMRB (accession code 15646). Determination of the structure of the Habc domain within the Sxcl fragment was performed based on interproton distance restraints derived from NOEs, hydrogen bond restraints derived from NH/H2O exchange cross-peaks observed in the 3D 1H-15N TOCSY-HSQC and NOESY-HSQC spectra, and torsion angle restraints derived by analysis of chemical shifts using Talos (Cornilescu et al., 1999). The general methodology used for resonance assignment and structure determination was described earlier (Shao et al., 1998; Ubach et al., 1999; Dulubova et al., 2001; Chen et al., 2002).
Results
Resonance assignments
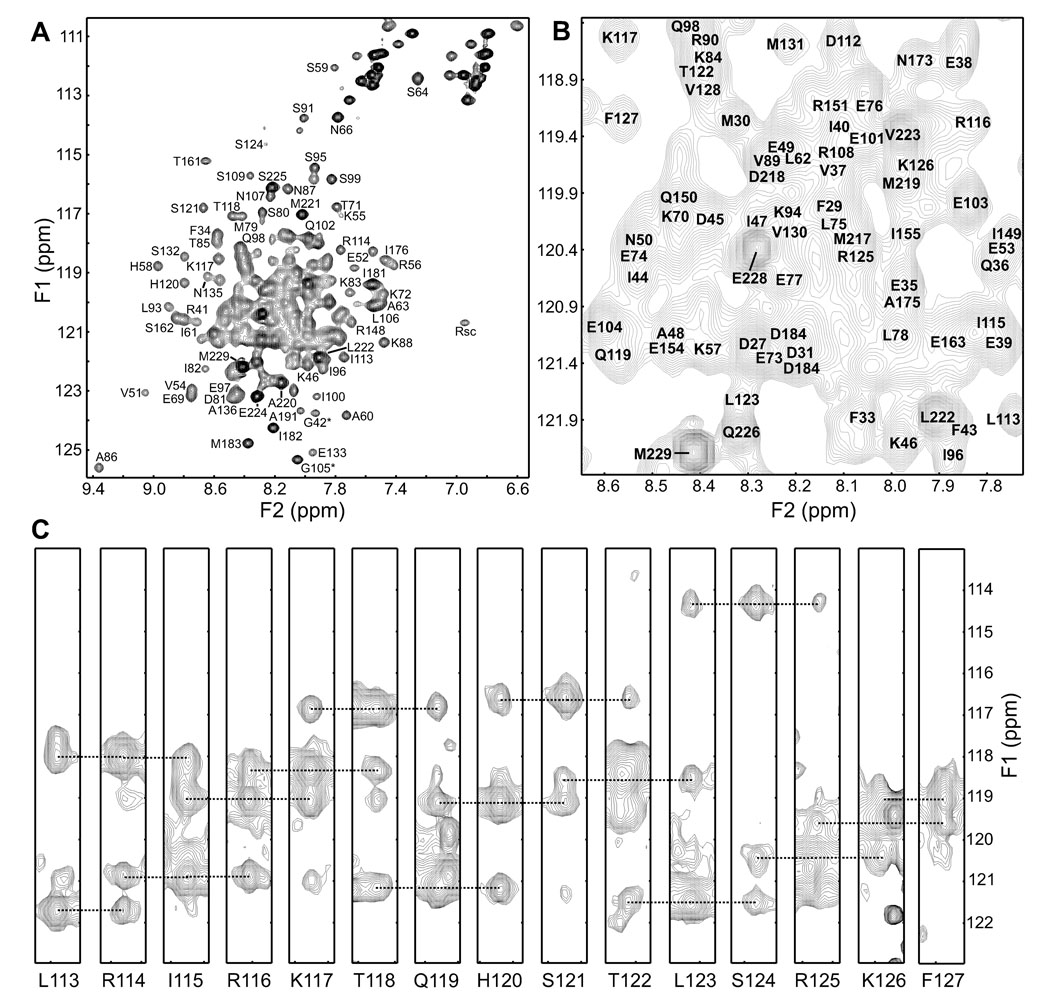
Previous studies of syntaxin-1 fragments spanning most of its cytoplasmic region (up to residue 253) using 1H-15N HSQC spectra showed that residues 2–26 and 227–253 are flexible and largely unstructured, thus implicating residues 27–226 in formation of the closed conformation (Dulubova et al., 1999; Fernandez et al., 1998). Thus, to study in detail the closed conformation by NMR spectroscopy, we prepared a minimal fragment encompassing these residues (the fragment contained residues 26–230 and will be hereafter referred to as Sxcl fragment). Analysis of the fragment was performed in 20 mM phosphate (pH 7.4) without addition of salt for optimal solubility. As shown in Figures 1A,B, 1H-15N HSQC spectra of the Sxcl fragment exhibited some well dispersed cross-peaks, but also severe overlap in the central region. Such overlap likely arises in part from the highly helical nature of the fragment and the paucity of aromatic residues in its sequence, but also suggests that some regions of the fragment might be flexible. Although homogeneous intensities were observed for many cross-peaks, we also observed low intensities and severe broadening of other cross-peaks, which did not depend on the protein concentration in a range from 20 µM to 700 µM. Hence, the observed broadening originates from conformational exchange rather than from aggregation.
Figure 1.
Assignment of amide groups of the Sxcl fragment. (A,B) 1H-15N HSQC spectrum of uniformly 2H,15N-labeled Sxcl fragment (A) and expansion of the central region of the spectrum (B) showing the assignment of backbone cross-peaks. Note that a few cross-peaks could not be assigned. (C) (F3,F1) stripes of a 3D 1H-15N,1H-15N HSQC-NOESY-HSQC spectrum of uniformly 2H,15N-labeled Sxcl fragment where both 15N dimensions were developed, taken at the F2 plane corresponding to the 15N chemical shift of the residues indicated below each stripe. The lines illustrate the connectivities from diagonal to sequential NH/NH cross-peaks that yield the sequential assignment for residues 113–127 of the Sxcl fragment.
A battery of experiments that included standard homonuclear 2D and heteronuclear 3D spectra acquired on four samples with different labeling schemes was used to assign the resonances of the Sxcl fragment, although complete assignment was hindered by the problems with resonance overlap and broadening. To obtain as many assignments as possible, we acquired HNCA, HN(CO)CA, HN(CA)CB and HN(COCA)CB spectra on a uniformly 2H,15N,13C-labeled sample both in constant time mode, to improve the resolution in the 13Cα and 13Cβ dimensions and thus help correlating unambiguously the corresponding chemical shifts, and in non-constant time mode to improve the sensitivity for cross-peaks broadened by conformational exchange. Because of the highly helical nature of the Sxcl fragment, assignment of backbone amide groups was facilitated by the observation of NH/NH NOEs, which were particularly well resolved through a 1H-15N,1H-15N HSQC-NOESY-HSQC experiment acquired in three dimensional mode whereby the two 15N dimensions were developed during t1 and t2, and one 1H dimension was acquired during t3. Sample stripes illustrating the assignment of sequential NOEs for residues 113–127 of the Sxcl fragment are shown in Figure 1C. Altogether, we were able to obtain full or partial assignments for 80% of the residues in the Sxcl fragment (assignments of selected 1H-15N HSQC cross-peaks are shown in Figures 1A,B)
Structural analysis
To interpret our NMR data on the Sxcl fragment in solution, we made extensive use of the crystal structure of syntaxin-1 bound to Munc18-1, where syntaxin-1 adopts a closed conformation (Misura et al., 2000). A ribbon diagram of residues 27–230 within this structure (Figure 2A) shows that the SNARE motif (helix Hf) packs against helices Hb and Hc of the Habc domain, forming a four-helix bundle, and that the linker region forms two short helices (Hd and He) and three loops at one end of the bundle. The diagram is color coded to illustrate the residues for which assignments were obtained, showing that practically the entire Habc domain was assigned, whereas only limited information could be obtained for the rest of the Sxcl fragment. This observation can be attributed to the marginal stability of the closed conformation. Thus, even though the population of the closed conformation appears to be close to 100% (see below), the Sxcl fragment likely samples a significant amount of open conformations where the Habc domain is structured but the remaining sequences (the linker and the SNARE motif) become flexible.
Figure 2.
Secondary structure of the Sxcl fragment. (A) Ribbon diagram of residues 27–230 within the crystal structure of the syntaxin-1/Munc18-1 complex [PDB code 1DN1; (Misura et al., 2000)]. Residues for which full or partial resonance assignments were obtained are indicated in blue for the Habc domain and in orange for the linker region and SNARE motif; other residues are in gray. The helices are labeled Ha-Hf. The numbers indicate the positions of selected residues in the structure. (B) Differences between the Cα chemical shifts observed for the Sxcl fragment and those expected for a random coil. The positions of the helices are indicated in red at the bottom. The short extended segment between helices Hc and Hd is indicated in blue.
Even with the limited information available for part of the Sxcl fragment, the available NMR data strongly suggest that the closed conformation of isolated syntaxin-1 is very similar to that of syntaxin-1 bound to Munc18-1. Thus, the differences between the observed Cα chemical shifts and those of a random coil (ΔδCα; see Figure 2B) correspond well with those expected according to the secondary structure of syntaxin-1 bound to Munc18-1, including the patterns observed for the loops and helices of the linker region. For instance, negative values of ΔδCα were observed for a short stretch between helices Hc and Hd that adopts an extended structure in the complex, whereas all helical residues exhibited positive values of ΔδCα. These positive values were generally of lower magnitude in the linker region than for the helices of the Habc domain, which can be attributed to some flexibility in the linker and correlates with the high structure factors observed in this region of the crystal structure of the syntaxin-1/Munc18-1 complex (Misura et al., 2000). It is also noteworthy that a gradient of ΔδCα values is observed at the C-terminus of the Sxcl fragment, which illustrates how a decreasing population of helical conformation is adopted from the N- to the C-terminus of the segment spanning residues 216 to 230. This observation correlates with the fact that, in the crystal structure of the complex, the helix formed by residues 216–230 is oriented slightly away from the Habc domain and does not pack well against this domain. Hence, the helical structure of this segment is mostly likely stabilized in the complex by its tight packing against Munc18-1, but the helix frays toward the C-terminus in isolated syntaxin-1 because it is not stabilized by tertiary or quaternary contacts. Indeed, the resonances of residues 220–230 were much sharper than those of the rest of the Sxcl fragment, showing its high flexibility.
The resonance assignments obtained for the linker region and SNARE motif were largely limited to backbone nuclei and we were thus unable to perform a full structure determination for the Sxcl fragment. However, all the backbone NOE patterns observed were consistent with the secondary structure of syntaxin-1 bound to Munc18-1, further reinforcing the notion that Munc18-1 binding does not induce major alterations in the syntaxin-1 closed conformation (although it does induce structure in the flexible C-terminal sequences that do not form part of the closed conformation). We did assign sufficient NOEs for the Habc domain to determine its structure within the closed conformation. A backbone superposition of the ten conformations with the lowest energies shows that the structure is well defined throughout the domain (Figure 3A), and the structural statistics revealed low deviations from the experimental restraints as well as good geometries (see legend of Figure 3A). A ribbon diagram of the lowest energy structure is displayed in Figure 3B, and is shown superimposed with ribbon diagrams of the crystal structure of the isolated Habc domain (Lerman et al., 2000) and of the Habc domain within the crystal structure of the syntaxin-1/Munc18-1 complex (Misura et al., 2000) in Figure 3C. The similarity between the three structures shows that neither the transition from the open to the closed conformation nor binding of Munc18-1 to the closed conformation induce large structural changes in the Habc domain. The former conclusion is further supported by comparison of the chemical shifts of the Habc domain within the closed conformation obtained here with those of the isolated Habc domain obtained previously (Fernandez et al., 1998). This analysis revealed generally small changes in aliphatic 1H and 13C chemical shifts (data not shown). The changes in amide 1H and 15N chemical shifts (Figure 4A) are larger but still of moderate magnitude and, considering the high sensitivity of amide chemical shifts to the chemical environment, they can be attributed to slight rearrangements induced by the intramolecular interactions of the SNARE motif with the Habc domain. Indeed, the most substantial chemical shift changes were observed in the regions of the Habc domain that pack against the SNARE motif in the closed conformation (Figure 4B).
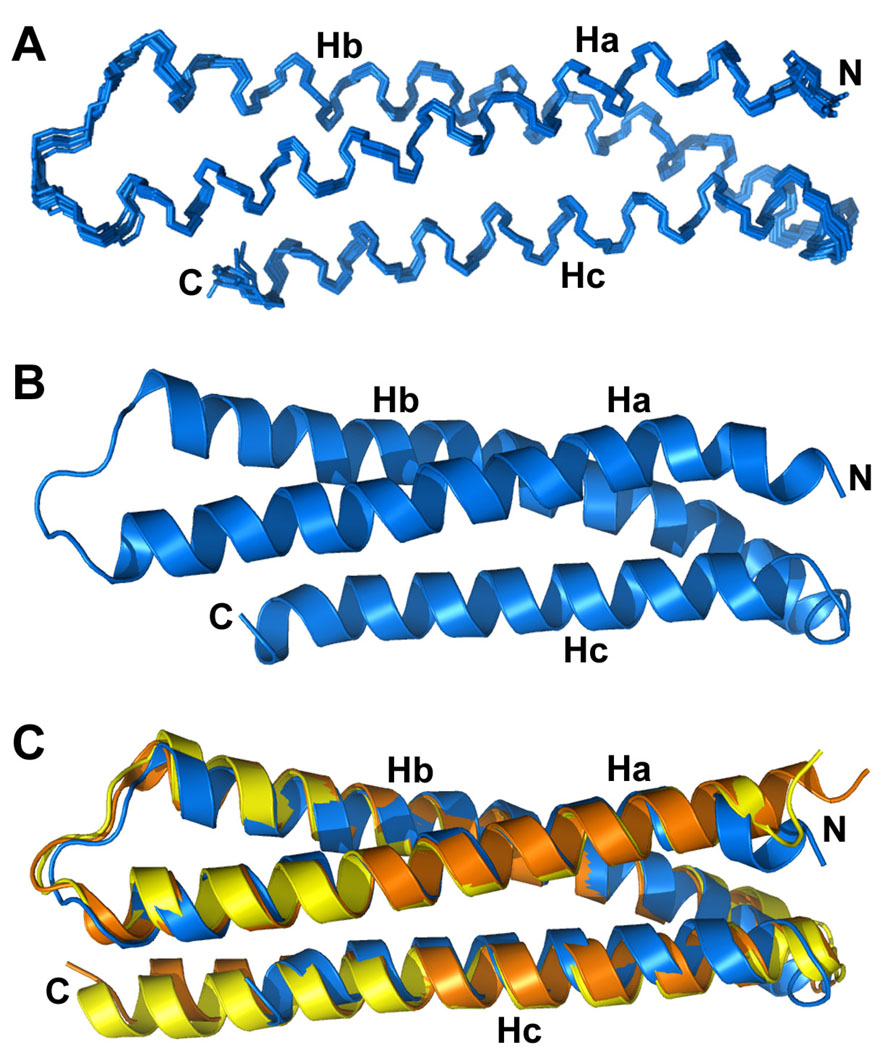
Figure 3.
Structure of the Habc domain within the syntaxin-1 closed conformation. (A) Backbone superposition of the ten lowest energy structures. The structures (residues 27–141) were calculated using 1675 NOE-derived interproton distance restraints (which include 433 long-range restraints), 186 H bond restraints and 195 backbone torsion angle restraints, and there were no violations larger than 0.3 Å or 5°. The structures exhibited low deviations from idealized covalent geometry (averages of 0.0027 Å for bonds, 0.47° for angles and 0.37° for impropers) and good Ramachandran map statistics (94.9% of residues in the most favored regions and none in disallowed regions). The average rms deviation among the ten structure is 0.38 Å and 0.90 Å for the backbone and all heavy atoms, respectively. (B) Ribbon diagram of the lowest energy structure of the Habc domain within the syntaxin-1 closed conformation. (C) Superposition of ribbon diagrams of the structure shown in (B) with the crystal structure of the isolated Habc domain [orange; PDB code 1EZ3; (Lerman et al., 2000)] and the Habc domain within crystal structure of the syntaxin-1/Munc18-1 complex [yellow; PDB code 1DN1; (Misura et al., 2000)].
Figure 4.
Differences between the amide chemical shifts of the Habc domain within the Sxcl fragment and those of the isolated Habc domain (Fernandez et al., 1998). (A) The combined differences in 1H and 15N backbone chemical shifts were calculated as ΔδN,HN = [(Δδ15N/5.5)2 + (Δδ1H)2]1/2, where Δδ15N and Δδ1H are the differences in backbone 15N and 1H chemical shifts, respectively. The positions of helices Ha-Hc are indicated at the bottom. (B) Ribbon diagram of residues 27–230 within the crystal structure of the syntaxin-1/Munc18-1 complex [PDB code 1DN1; (Misura et al., 2000)] illustrating the residues with the largest amide chemical shift changes (ΔδN,HN > 0.2 yellow; ΔδN,HN > 0.1 orange). The positions of the helices and of selected residues are indicated.
Munc18-1 stabilizes the closed conformation
The differences in the 1H-15N HSQC cross-peaks of the isolated Habc domain (residues 27–146), which represents the open conformation, and the Sxcl fragment (residues 26–230), which represents the closed conformation, are also illustrated in Figure 5A, where a few diagnostic cross-peaks have been labeled. These differences are analogous to those observed previously with a longer syntaxin-1 fragment including the entire SNARE motif, which led to the conclusion that syntaxin-1 adopts a closed conformation involving intramolecular interactions between the Habc domain and the SNARE motif (Dulubova et al., 1999). As stated above, the picture that emerged from the NMR analysis of the Sxcl fragment suggests that there is exchange between the closed conformation and a significant albeit small population of open conformations where the linker and SNARE motif become flexible while the Habc domain remains structured. Although we cannot completely rule out the potential existence of multiple states, which would hinder determination of their populations, the fact that two distinct cross-peaks can be assigned to the open and closed states for a longer syntaxin-1 fragment (see below and Fig. 5C) suggests that a two-state model can be assumed to a first approximation; based on this assumption, the sensitivity of our 1H-15N HSQC spectra of the Sxcl fragment and the absence of cross-peaks corresponding to the open conformation, our data suggest that the closed conformation is populated more than 98% in the Sxcl fragment. This conclusions contrasts with single molecule FRET studies that suggested that the population of the closed conformation in isolated syntaxin-1 is only 15–30% populated (Margittai et al., 2003). To try to resolve this apparent paradox and gain further insights into the equilibrium between open and closed conformations, we analyzed perturbations in 1H-15N HSQC spectra of the Sxcl fragment under a variety of conditions. However, changes in the NaCl concentration from 0 to 200 mM, or addition of agents that stabilize protein structure such as glycerol or TMAO, did not induce substantial changes in chemical shifts or linewidths that would suggest destabilization or stabilization of the closed conformation of the Sxcl fragment, and the alterations observed upon gradually increasing the temperature from 12 to 35 °C did not suggest a substantial opening of the conformation (data not shown).
Figure 5.
Munc18-1 binding stabilizes the closed conformation. (A) Superposition of 1H-15N HSQC spectra of the Sxcl fragment (residues 26–230; black contours) and the isolated Habc domain (residues 27–146; red contours) in 20 mM phosphate (pH 7.4). The cross-peaks corresponding to residues 58–60 and 64 are labeled. (B) Structure of the syntaxin-1/Munc18-1 complex [PDB code 1DN1; (Misura et al., 2000)] with the surface of Munc18-1 shown in gray and syntaxin-1 shown in ribbon diagram. The linker and SNARE motif are colored in orange, and the Habc domain is colored in blue except for residues 58–60 and 64, which are colored in yellow. (C) Expansions of the regions containing the cross-peaks of S64 (above) and S59 (below) in 1H-15N HSQC spectra of syntaxin-1(2–253) in the presence of 0 mM, 100 mM and 150 mM NaCl, and of syntaxin-1(2–243) in the presence of 150 mM NaCl. (D) 1H-15N TROSY-HSQC spectrum of 2H,15N-labeled syntaxin-1(2–253) bound to unlabeled Munc18-1 in 20 mM phosphate (pH 7.4) containing 200 mM NaCl. (E) Superposition of the 1H-15N TROSY-HSQC spectrum shown in (D) with a 1H,15N HSQC spectrum of 2H,15N-labeled syntaxin-1(2–253) in 20 mM phosphate (pH 7.4) without NaCl.
Interestingly, the closed conformation is much more sensitive to the ionic strength in the context of a longer syntaxin-1 fragment (residues 2–253) that we used to analyze the effects of Munc18-1 binding (see below). This conclusion is illustrated by monitoring the cross-peaks of S59 and S64, which exhibit marked shifts upon closing of the syntaxin-1 conformation (Figure 5A) because these residues are located in a region of the Habc domain that packs against the SNARE motif (Figure 5B). In the absence of NaCl, strong S59 and S64 cross-peaks are observed at the positions corresponding to the closed conformation; a small cross-peak is also observed in the position corresponding to the open conformation for S64, although this cross-peak is almost at noise level (Figure 5C). In 100 mM NaCl, cross-peaks can be observed at the positions of both the open and the closed conformations, which some substantial broadening for the S59 cross-peaks, and no cross-peaks corresponding to the closed conformation are observable in 150 mM NaCl (Figure 5C). These results show that increasing the ionic strength gradually shifts the equilibrium from the closed to the open conformation for this fragment. The different sensitivity of the closed conformation in the context of syntaxin-1(2–253) and the Sxcl fragment (residues 26–230) can be attributed to the tendency of the longer fragment to oligomerize, which was shown previously by analytical ultracentrifugation (Lerman et al., 2000) and requires an open conformation. This tendency arises from the known propensity of the syntaxin-1 SNARE motif to form coiled coils and promiscuously associate with itself or with a wide variety of proteins through hydrophobic interactions (Jahn and Scheller, 2006), which are favored by increasing the ionic strength. This notion explains the higher stability of the closed conformation of the Sxcl fragment to NaCl, as this fragment contains only half of the SNARE motif and hence has a much lower tendency to self-associate through hydrophobic interactions. Indeed, only the cross-peaks corresponding to the closed conformation are observed for a syntaxin-1 fragment containing residues 2–243 in 150 mM NaCl (illustrated for S59 and S64 HN in Figure 5C), showing that just deleting the ten C-terminal residues of syntaxin-1(2–253) is sufficient to strongly impair its oligomerization.
To investigate the effects of Munc18-1 binding on the syntaxin-1 closed conformation, we acquired a 1H-15N TROSY-HSQC spectrum of uniformly 2H,15N-labeled syntaxin-1(2–253) bound to unlabeled Munc18-1 in the presence of 200 mM NaCl to help with complex solubility (Figure 5D). The strong cross-peak overlap observed in the spectrum prevented a thorough analysis of the changes in the chemical shifts of the syntaxin-1 fragment induced by Munc18-1 binding. However, the spectrum had good sensitivity, considering the relatively large size of this complex (95 kDa), and allowed comparison of the locations of diagnostic, well-resolved cross-peaks with those of the 1H-15N HSQC spectrum of isolated syntaxin-1(2–253) (acquired at 0 mM NaCl to preserve the closed conformation; Figure 5E). Particularly informative in this respect are the cross-peaks of residues 58–60 and 64, which are located at the C-terminus of helix A and do not contact Munc18-1 (Figure 5B) but exhibit substantial changes in the transition from the open to the closed conformation [Figure 5A]. All these cross-peaks appear in positions close to those corresponding to the closed conformation in the 1H-15N TROSY-HSQC spectrum of syntaxin-1(2–253) bound to Munc18-1 (Figure 5E), showing that Munc18-1 keeps the syntaxin-1 fragment in the closed conformation in 200 mM NaCl, as expected from the high affinity of Munc18-1 for the syntaxin-1 closed conformation. Note that this NaCl concentration is above that required to completely open isolated syntaxin-1(2–253) at the protein concentrations used (Figure 5C). Furthermore, the similarity of the chemical shifts of the diagnostic cross-peaks in the syntaxin-1/Munc18-1 complex with those of the closed conformation of isolated syntaxin-1 further supports the conclusion from our structural analysis that Munc18-1 binding does not induce large changes in the closed conformation. These results demonstrate that Munc18-1 binding stabilizes the syntaxin-1 closed conformation and strongly suggest that the binding does not induce substantial rearrangements to help opening the conformation.
Discussion
Initial studies of SM protein/SNARE interactions revealed a surprising diversity of binding modes (Dulubova et al., 1999; Yamaguchi et al., 2002; Dulubova et al., 2002; Carr et al., 1999), but it now seems clear that SM proteins generally bind to assembled SNARE complexes (Carr et al., 1999; Peng and Gallwitz, 2002; Carpp et al., 2006; Latham et al., 2006; Dulubova et al., 2007; Shen et al., 2007; Collins et al., 2005), in interactions that may be key for membrane fusion (Rizo et al., 2006) and most often involve syntaxin N-terminal sequences (Khvotchev et al., 2007). Thus, the binary association between Munc18-1 and the closed conformation of syntaxin-1 now appears to constitute a specialization that may have evolved to meet unique regulatory requirements of Ca2+-evoked exocytosis. Previous studies led to contradictory conclusions about the stability of the closed conformation (Dulubova et al., 1999; Margittai et al., 2003) and to opposing models for the role of Munc18-1 binding to syntaxin-1, which proposed that Munc18-1 stabilizes the closed conformation of syntaxin-1 (Margittai et al., 2003) or alters this conformation to facilitate its opening (Munson et al., 2000). Our data now suggest that the closed conformation is almost quantitatively formed in isolated syntaxin-1 in the absence of oligomerization, and strongly support the notion that the binary interaction of Munc18-1 with syntaxin-1 stabilizes the closed conformation, hindering SNARE complex formation, even though it is clear that Munc18-1 must have an additional, active role in downstream events.
The idea that Munc18-1 prevents SNARE complex assembly emerged from the early observation that this protein competes with SNAP-25 and synaptobrevin for syntaxin-1 binding (Pevsner et al., 1994a). This conclusion was reinforced in part by the finding that the so-called ‘LE mutation’ in syntaxin-1, which destabilizes the closed conformation and impairs Munc18-1 binding, enhances SNARE complex formation (Dulubova et al., 1999). However, these conclusions seemed counterintuitive given the crucial role of Munc18-1 for neurotransmitter release (Verhage et al., 2000), leading to the proposal that, even if there is competition between Munc18-1 and SNAP-25/synaptobrevin for syntaxin-1 binding, Munc18-1 could still somehow assist in SNARE complex formation with the help of other factors (Dulubova et al., 1999; Misura et al., 2000). Moreover, the crystal structure of the closed conformation of Sso1p, the syntaxin-1 homologue from the yeast plasma membrane, revealed significant differences with respect to the closed conformation of syntaxin-1 bound to Munc18-1, particularly in the linker region (Munson et al., 2000). This observation suggested that Munc18-1 binding might modify the syntaxin-1 closed conformation, partially releasing the inhibition of the SNARE motif by the Habc domain and thus helping to promote binding to SNAP-25. However, our results are clearly inconsistent with this proposal. First, all our NMR data suggest that the closed conformation of isolated syntaxin-1 is very similar to that adopted in the syntaxin-1/Munc18-1 complex; note that, although these data are not definitive because a full structure determination could not be achieved, the fact that Munc18-1 binding does not significantly shift the cross-peaks of residues 58–60 and 64 strongly suggests that the binding does not substantially alter the linker region, given its proximity to these residues (see Figures 5B,E). Second, and perhaps more important, our analysis by 1H-15N HSQC spectra clearly demonstrate that Munc18-1 binding stabilizes the closed conformation.
Previous single molecule FRET studies led to a similar conclusion but, surprisingly, also suggested that only 15–30% of isolated syntaxin-1 molecules adopt the closed conformation (Margittai et al., 2003), which raises the question as to whether isolated syntaxin-1 truly has an intrinsic tendency to adopt a closed conformation with any biological relevance. However, the low population of closed conformation was largely derived from data obtained with FRET pair labels placed at residues 91 and 225 of syntaxin-1, and measurements with FRET pairs in other positions did not appear to unambiguously support such low population. Since residue 225 is at the C-terminal region that is highly flexible according to our NMR data, the FRET results could be attributed to this flexibility rather than to the opening-closing model assumed to conclude that syntaxin-1 is largely open. Note also that it seems unlikely that oligomerization might have induced opening of the syntaxin-1 conformation in the FRET experiments, since the protein concentrations used for these experiments [10–100 pM) were much lower than those required for oligomerization [in the low µM range; see (Lerman et al., 2000)]. Regardless of whether the interpretation of the FRET study of Margittai et al. was correct, our NMR data conclusively demonstrate that the closed conformation is highly populated in the absence of oligomerization, and hence constitutes an intrinsic property of isolated syntaxin-1 that is expected to hinder intermolecular interactions of its promiscuous SNARE motif. This property may be crucial to prevent immediate SNARE complex reassembly after the complex is disassembled by the ATPase activity of NSF (Hanson et al., 1995), which would lead to a futile expense of ATP.
Our NMR data also show that, even though the closed conformation of syntaxin-1 is highly populated, this conformation is marginally stable and can easily open. Furthermore, our data leave no doubt that Munc18-1 binding stabilizes the closed conformation. Hence, from a purely thermodynamic point of view, Munc18-1 binding prevents opening of the syntaxin-1 closed conformation and cannot directly facilitate SNARE complex formation. This conclusion does not rule out the possibility that Munc18-1 binding to syntaxin-1 might have an indirect, beneficial effect for SNARE complex assembly by preventing the promiscuous SNARE motif of syntaxin-1 from engaging in non-specific interactions such as those involved in formation of non-productive 2:1 syntaxin-1/SNAP-25 heterodimers, which should also hinder formation of the SNARE complex. In addition, Munc18-1 binding likely plays a role in the overall stabilization of syntaxin-1 in vivo, since syntaxin-1 levels are dramatically decreased in Munc18-1 knockout mice (Verhage et al., 2000), and there is little doubt that Munc18-1 participates in downstream events that help to form the SNARE complex, since Munc18-1 binds to the SNARE complex and facilitates it assembly in vitro when starting from co-expressed syntaxin-1/SNAP-25 heterodimers (Dulubova et al., 2007; Shen et al., 2007). Nevertheless, the stabilization of the syntaxin-1 closed conformation by Munc18-1 binding implies that the binary Munc18-1/syntaxin-1 interaction necessarily imposes an energy barrier to SNARE complex assembly and that additional factors should help to overcome this barrier for assembly to occur during the fast time scales of synaptic vesicle priming and fusion. Primary candidates to perform this role are the members of the Munc13 family, which has been recently supported by the finding that the Munc13-1 MUN domain binds to membrane anchored SNARE complexes and syntaxin-1/SNAP-25 heterodimers (Guan et al., 2008; Weninger et al., 2008). Importantly, Munc13s contain additional domains that likely mediate diverse forms of regulation of the efficiency of release during presynaptic plasticity processes involved in some forms of information processing in the brain, which has led to the proposal that regulation of MUN domain activity by these various domains underlies these processes (Basu et al., 2005).
In an exquisitely regulated biological process such as neurotransmitter release, inhibition of the process is as critical as its activation. Hence, the picture that emerges from all these observations is that the binary interaction of Munc18-1 with the syntaxin-1 closed conformation, which may not be generally conserved in other forms of membrane traffic, might have evolved as a roadblock that provides a key point for diverse forms of regulation of neurotransmitter release. The insights into the nature of this block offered by our NMR data provide a nice illustration of how NMR spectroscopy can yield a wealth of information on biological processes even when a full structure determination cannot be achieved.
Acknowledgements
We thank Josep Ubach, Imma Fernandez and Iryna Huryeva for preparation of recombinant proteins, Lewis Kay for providing all pulse sequences used in this study, and Thomas Südhof for fruitful discussions. This work was supported by grant I-1304 from the Welch Foundation and NIH grant NS37200 (to J.R.).
Abbreviations
- COSY
correlation spectroscopy
- FRET
fluorescence resonance energy transfer
- HSQC
heteronuclear single quantum coherence
- NOESY
nuclear Overhauser effect spectroscopy
- NSF
N-ethylmaleimide sensitive factor
- SM protein
Sec1/Munc18 protein
- SNAP
soluble NSF attachment protein
- SNAP-25
synaptosomal associated protein of 25 kDa
- SNARE
SNAP receptor
- TMAO
trimethylamine N-oxide
- TOCSY
total correlation spectroscopy
- TROSY
transverse relaxation optimized spectroscopy
- VAMP
vesicle associated membrane protein
References
- Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, Grishin NV, Rosenmund C, Rizo J. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- Bracher A, Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- Collins KM, Thorngren NL, Fratti RA, Wickner WT. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - A Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dulubova I, Khvotchev M, Liu S, Huryeva I, Sudhof TC, Rizo J. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Arac D, Li H, Huryeva I, Min SW, Rizo J, Sudhof TC. Convergence and divergence in the mechanism of SNARE binding by Sec1/Munc18-like proteins. Proc Natl Acad Sci U S A. 2003;100:32–37. doi: 10.1073/pnas.232701299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/Syntaxin 16 “Snares” Vps45. EMBO J. 2002 doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Garcia EP, Gatti E, Butler M, Burton J, De Camilli P. A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci U S A. 1994;91:2003–2007. doi: 10.1073/pnas.91.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN Domain to Membrane-Anchored SNARE Complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- Hakes DJ, Dixon JE. New Vectors for High-Level Expression of Recombinant Proteins in Bacteria. Analytical Biochemistry. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Otto H, Barton N, Jahn R. The N-ethylmaleimide-sensitive fusion protein and alpha-SNAP induce a conformational change in syntaxin. J Biol Chem. 1995;270:16955–16961. doi: 10.1074/jbc.270.28.16955. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs-engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. Nmr View - A Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kay LE, Xu GY, Singer AU, Muhandiram DR, Formankay JD. A Gradient-Enhanced Hcch Tocsy Experiment for Recording Side-Chain H-1 and C-13 Correlations in H2O Samples of Proteins. Journal of Magnetic Resonance Series B. 1993;101:333–337. [Google Scholar]
- Khvotchev M, Dulubova I, Sun J, Dai H, Rizo J, Sudhof TC. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham CF, Lopez JA, Hu SH, Gee CL, Westbury E, Blair DH, Armishaw CJ, Alewood PF, Bryant NJ, James DE, Martin JL. Molecular dissection of the Munc18c/syntaxin4 interaction: implications for regulation of membrane trafficking. Traffic. 2006;7:1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- Lerman JC, Robblee J, Fairman R, Hughson FM. Structural analysis of the neuronal SNARE protein syntaxin-1A. Biochemistry. 2000;39:8470–8479. doi: 10.1021/bi0003994. [DOI] [PubMed] [Google Scholar]
- Margittai M, Widengren J, Schweinberger E, Schroder GF, Felekyan S, Haustein E, Konig M, Fasshauer D, Grubmuller H, Jahn R, Seidel CA. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc Natl Acad Sci U S A. 2003;100:15516–15521. doi: 10.1073/pnas.2331232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-Enhanced Triple-Resonance 3-Dimensional Nmr Experiments with Improved Sensitivity. Journal of Magnetic Resonance Series B. 1994;103:203–216. [Google Scholar]
- Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- Peng R, Gallwitz D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994a;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Scheller RH. n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci U S A. 1994b;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. Snares and munc18 in synaptic vesicle fusion. Nat Rev Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Toonen RF, Verhage M. Munc18-1 in secretion: lonely Munc joins SNARE team and takes control. Trends Neurosci. 2007;30:564–572. doi: 10.1016/j.tins.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Ubach J, Garcia J, Nittler MP, Sudhof TC, Rizo J. Structure of the Janus-faced C2B domain of rabphilin. Nat Cell Biol. 1999;1:106–112. doi: 10.1038/10076. [DOI] [PubMed] [Google Scholar]
- Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, Geuze HJ, Sudhof TC. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory Proteins Stabilize the Acceptor Complex for Synaptobrevin, the 1:1 Syntaxin/SNAP-25 Complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Sudhof TC. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A Suite of Triple-Resonance Nmr Experiments for the Backbone Assignment of N-15, C-13, H-2 Labeled Proteins with High-Sensitivity. Journal of the American Chemical Society. 1994;116:11655–11666. [Google Scholar]
- Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]