Abstract
Junctophilins (JPHs) are members of a junctional membrane complex protein family important for the physical approximation of plasmalemmal and sarcoplasmic/endoplasmic reticulum membranes. As such, JPHs facilitate signal transduction in excitable cells between plasmalemmal voltage-gated calcium channels and intracellular calcium release channels. To determine the molecular evolution of the JPH gene family, we performed a phylogenetic analysis of over 60 JPH genes from over 40 species and compared conservation across species and different isoforms. We found that JPHs are evolutionary highly conserved, in particular the membrane occupation and recognition nexus motifs found in all species. Our data suggest that an ancestral form of JPH arose at the latest in a common metazoan ancestor and that in vertebrates four isoforms arose, probably following two rounds of whole genome duplications. By combining multiple prediction techniques with sequence alignments, we also postulate the presence of new important functional regions and candidate sites for posttranslational modifications. The increasing number of available sequences yields significant insight into the molecular evolution of JPHs. Our analysis is consistent with the emerging concept that JPHs serve dual important functions in excitable cells: structural assembly of junctional membrane complexes and regulation of intracellular calcium signaling pathways.
Keywords: calcium signaling, junctional membrane complex, excitation-contraction coupling, MORN motif, excitable cells
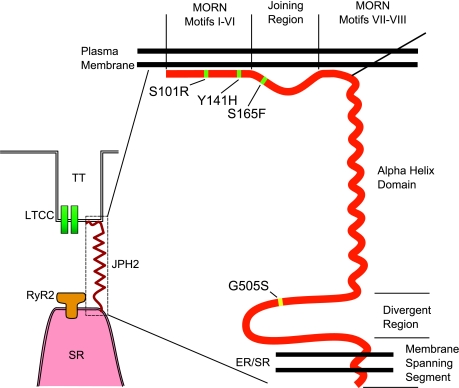
junctional complexes between the plasma membrane (PM) and endo/sarcoplasmic reticulum (ER/SR) are found in excitable cell types (46). Close apposition of the PM and ER/SR membranes is a requirement for efficient cross talk between ion channels that localize to the PM and ER/SR (Fig. 1) and is mediated by junctophilins (JPHs) (3, 46). Consistently, members of the JPH family are expressed in excitable cells (3, 46). Spatial coupling of the PM and ER/SR by JPHs ensures optimal transmission of the transmembrane (TM) Ca2+ influx signal to Ca2+ release from within the ER/SR, a process that underlies contraction of muscle, insulin release from pancreatic β-cells, and synaptic plasticity in neurons (Fig. 1) (14, 42).
Fig. 1.
Schematic representation of junctophilin (JPH). Model figure showing a JPH2 protein (red) spanning the junctional membrane domain between the plasma membrane (PM) and endo/sarcoplasmic reticulum (ER/SR). Functional domains within JPH are indicated. MORN, Membrane Occupation and Recognition Nexus; TT, transverse tubule; LTTC, L-type Ca2+ channel; RyR2, ryanodine receptor type 2. Mutations identified in patients with hypertrophic cardiomyopathy are indicated in green (7) and yellow (13).
In skeletal muscle, junctophilin-1 (JPH1) is believed to facilitate direct protein-protein interactions between voltage-dependent L-type Ca2+ channels (LTCC) on the PM and intracellular Ca2+ release channels (CRCs), specifically the type 1 ryanodine receptor (RyR1), on the SR (36, 40). In cardiac muscle, JPH2 determines the distance that the flux of Ca2+ entering through LTCC must travel, before reaching the activating Ca2+ binding sites on type 2 ryanodine receptors (RyR2) (19). As such, JPH2 might indirectly determine the gain of Ca2+-induced Ca2+ release (CICR) in cardiac myocytes (16). In neurons, JPH3 and JPH4 contribute to the formation of similar junctional membrane structures in subsurface cisternae, although their functions are not as well understood as those in skeletal/cardiac muscle (37).
Despite the important functional role for JPHs in muscle and neurons, little is known about the canonical JPH protein structure as X-ray crystallography data are lacking at present. Based on primary sequence analysis, it has been proposed that all JPH isoforms contain eight “membrane occupation and recognition nexus” (MORN) motifs clustered near the amino terminus, followed by a putative α-helical domain, and a carboxyl-terminal TM segment (Fig. 1) (36). The MORN motifs likely mediate association with the PM, either directly by interacting with lipids or possibly indirectly through membrane-bound adapter proteins (23). The α-helical domain might determine the distance between the PM and the ER/SR, which is estimated to be ∼12 nm based on electron microscopy measurements (24, 25, 46). Several in vivo studies support this notion that JPHs determine the spacing between the PM and ER/SR membrane (26). Genetic deletion of JPH2 in mice leads to the formation of fewer and improperly aligned junctional membrane complexes (JMCs) in cardiac muscle, as evidenced by an increased PM-SR distance (46). On the other hand, overexpression of JPH1 in amphibian embryonic cells is sufficient to generate de novo 12 nm junctions between the ER/SR and PM (26).
To date, most cloned JPH genes are from mammalian origin. However, the recent availability of nucleotide sequencing data of JPH genes from lower vertebrates and invertebrates provides the opportunity to compare JPH amino acid sequences from a variety of different organisms (Table 1). In this study, we performed a comprehensive phylogenetic study of all known JPH sequences currently available in genomic databases. By analyzing multisequence alignments, we demonstrate a high level of conservation among vertebrate JPH isoforms, suggesting the four mammalian JPH genes that encode JPH1–JPH4 arose from an ancestral gene via two whole genome duplications. Our data also reveal excellent evolutionary conservation of the joining region between the first six and the last two MORN motifs, suggesting this region might serve an accessory protein-binding region. These studies provide novel insights into the molecular evolution of JPHs and highlight the potential role of JPHs in excitable cells.
Table 1.
Results from BLASTP search showing orthologous sequences and accession numbers from NCBI database
| Isoform | Species | Common Name | Accession No. | Length | HSP Value |
|---|---|---|---|---|---|
| JPH1 | Homo sapiens | human | NP_065698.1 | 661 | 1,756 |
| JPH1 | Macaca mulatta | Rhesus monkey | XP_001086528.1 | 661 | 1,757 |
| JPH1 | Oryctolagus cuniculus | rabbit | NP_001075465.1 | 662 | 3,375 |
| JPH1 | Bos taurus | cattle | XP_616955.2 | 658 | 1,777 |
| JPH1 | Canis lupus familiaris | dog | XP_849971.1 | 662 | 3,410 |
| JPH1 | Rattus norvegicus | Norway rat | NP_001100100.1 | 660 | 1,747 |
| JPH1 | Mus musculus | house mouse | NP_065629.1 | 660 | 1,763 |
| JPH1 | Monodelphis domestica | gray short-tailed opossum | XP_001367740.1 | 659 | 1,708 |
| JPH1 | Gallus gallus | red jungle fowl | XP_418302.2* | 666* | 1,798 |
| JPH1 | Xenopus (Silurana) tropicalis | Western clawed frog | NP_001120224.1 | 560 | 2,334 |
| JPH1 | Tetraodon nigroviridis | pufferfish | CAF98018.1 | 694 | 2,285 |
| JPH1 | Danio rerio | zebrafish | NP_001037813.1 | 673 | 2,345 |
| JPH2 | Homo sapiens | human | NP_065166.2 | 696 | 3,657 |
| JPH2 | Macaca mulatta | Rhesus monkey | XP_001083346.1 | 696 | 3,590 |
| JPH2 | Rattus norvegicus | Norway rat | NP_001033063.1 | 692 | 3,166 |
| JPH2 | Mus musculus | house mouse | NP_067541.1 | 696 | 3,159 |
| JPH2 | Oryctolagus cuniculus | rabbit | NP_001075467.1 | 694 | 3,352 |
| JPH2 | Monodelphis domestica | gray short-tailed opossum | XP_001369593.1 | 726 | 2,706 |
| JPH2 | Danio rerio | zebrafish | NP_001082833.1 | 781 | 1,981 |
| JPH2 | Tetraodon nigroviridis | pufferfish | CAG03835.1 | 476 | 1,037 |
| JPH3 | Homo sapiens | human | NP_065706.2 | 748 | 3,927 |
| JPH3 | Rattus norvegicus | Norway rat | NP_001100907.1 | 749 | 3,587 |
| JPH3 | Mus musculus | house mouse | NP_065630.1 | 744 | 3,551 |
| JPH3 | Equus caballus | horse | XP_001502768.2 | 753 | 1,487 |
| JPH3 | Bos taurus | cattle | XP_584374.3 | 754 | 3,576 |
| JPH3 | Canis lupus familiaris | dog | XP_546789.2 | 754 | 3,579 |
| JPH3 | Monodelphis domestica | gray short-tailed opossum | XP_001366198.1 | 755 | 1,509 |
| JPH3 | Gallus gallus | red jungle fowl | XP_414192.1 | 758 | 3,446 |
| JPH3 | Danio rerio | zebrafish | NP_001116174.1 | 791 | 1,147 |
| JPH4 | Homo sapiens | human | Q96JJ6.2 | 628 | 1,157 |
| JPH4 | Rattus norvegicus | Norway rat | EDM14220.1 | 628 | 3,207 |
| JPH4 | Mus musculus | house mouse | NP_796023.2 | 628 | 1,144 |
| JPH4 | Bos taurus | cattle | XP_001249446.2 | 628 | 3,203 |
| JPH4 | Sus scrofa | pig | XP_001928705.1 | 628 | 1,154 |
| JPH4 | Canis lupus familiaris | dog | XP_547737.2 | 599 | 1,078 |
| JPHa | Ciona intestinalis | sea squirt | XP_002122697.1 | 914 | 1,309 |
| JPHa | Branchiostoma floridae A | Florida lancelet (draft) | XP_002241825.1 | 441 | 1,217 |
| JPHa | Branchiostoma floridae B | Florida lancelet (draft) | XP_002213020.1 | 377 | 1,134 |
| JPHa | Nematostella vectensis | starlet sea anemone | XP_001633625.1 | 391 | 847 |
| JPHa | Hydra magnipapillata | hydra | XP_002168568.1 | 741 | 490 |
| JPHa | Strongylocentrotus purpuratus | purple sea urchin | XP_781706.2 | 842 | 1,192 |
| JPHa | Drosophila erecta | fruit fly | XP_001969316.1 | 1,067 | 807 |
| JPHa | Drosophila yakuba | fruit fly | XP_002088942.1 | 1,075 | 806 |
| JPHa | Drosophila sechellia | fruit fly | XP_002036346.1 | 984 | 806 |
| JPHa | Drosophila simulans | fruit fly | XP_002078784.1 | 594 | 1,078 |
| JPHa | Drosophila melanogaster | fruit fly | NP_523525.2 | 1,054 | 1,003 |
| JPHa | Drosophila ananassae | fruit fly | XP_001964889.1 | 1,077 | 1,082 |
| JPHa | Drosophila pseudoobscura | fruit fly | XP_001356378.2 | 1,114 | 994 |
| JPHa | Drosophila persimilis | fruit fly | XP_002014505.1 | 958 | 1,078 |
| JPHa | Drosophila willistoni | fruit fly | XP_002065678.1 | 1,087 | 818 |
| JPHa | Drosophila mojavensis | fruit fly | XP_002002795.1 | 1,074 | 808 |
| JPHa | Drosophila virilis | fruit fly | XP_002057766.1 | 1,129 | 1,088 |
| JPHa | Drosophila grimshawi | fruit fly | XP_001988245.1 | 1,083 | 1,093 |
| JPHa | Aedes aegypti (Stegomyia aegypti) | yellow fever mosquito | XP_001661427.1 | 925 | 915 |
| JPHa | Anopheles gambiae str. PEST | African malaria mosquito | XP_565673.3 | 910 | 916 |
| JPHa | Pediculus humanus corporis | human body louse | EEB13577.1 | 706 | 1,149 |
| JPHa | Acyrthosiphon pisum | pea aphid | XP_001947644.1 | 464 | 816 |
| JPHa | Nasonia vitripennis (jewel wasp) | jewel wasp | XP_001599701.1 | 1,014 | 916 |
| JPHa | Apis mellifera | honey bee | XP_624956.1 | 1,027 | 936 |
| JPHa | Tribolium castaneum | red flour beetle | XP_973598.1 | 871 | 1,234 |
| JPHa | Brugia malayi | (agent of lymphatic filariasis) | XP_001901447.1 | 810 | 1,120 |
| JPHa | Caenorhabditis elegans | nematode | NP_492193.2 | 747 | 1,109 |
| JPHa | Caenorhabditis briggsae AF16 | nematode | XP_001668645.1 | 755 | 1,089 |
JPH, junctophilin; HSP, high scoring pair.
The first 150 amino acids of XP_418302.2 were removed (see methods).
MATERIALS AND METHODS
Sequence Data
All sequences used in this study were obtained from either the Ensembl Genome Browser (Wellcome Trust Genome Campus, Hinxton, Cambridge, UK) or the National Center for Biotechnology Information (NCBI, Bethesda, MD) (4, 22). A custom program was written using the BioPython tools package (http://biopython.org) to catalogue results obtained from a protein basic local alignment search tool (BLASTP) with human JPH1–4 isoforms as queries (accession numbers NP_065698.1, NP_065166.2, NP_065706.2, and Q96JJ6.2, respectively). Full protein sequences from the top 200 hits of each BLASTP search were entered into a database, and any duplicate entries were removed. This yielded 252 unique database entries. The database was then filtered using a minimum high scoring pair (HSP) score of 300 (1), because most E values were very small (typically smaller than e−100). The resulting data showed a biphasic distribution; most full-length sequences had an HSP value >300, whereas most small fragments with high homology attained <300. In case multiple entries were identified for a given isoform of the same species, all entries were analyzed in detail, and the most representative or complete sequence was chosen to be representative of that isoform.
The results were then manually curated to eliminate possible artifacts due to sequencing or prediction errors. In particular, we created a modified Gallus gallus JPH1 sequence by removing the first 150 amino acids from the original sequence (XP_418302.2). By examining the sequence and conservation across species, we find it probable that the extra segment was due to an incorrect prediction of the start codon in the 5′ region upstream of the consensus start codon. During the process of verifying each sequence manually, we eliminated several protein sequences from the phylogeny analysis even though they satisfied our HSP threshold of 300. The Macaca mulatta JPH3 (XP_001092976.1) had >200 additional amino acids in the sequence, suggesting an error in intron prediction, whereas all other M. mulatta isoforms exhibited >98% homology to the human isoforms. Two sequences, Sus scrofa JPH1 (XP_001925343.1) and Bos taurus JPH2 (XP_001788032.1), were eliminated from the analysis, as they, despite high conservation, ended prematurely. Equus caballus JPH4 (XP_001918361.1) had a 123-amino acid gap in the middle of the sequence that matches a gap in the sequence (UCSC Genome Browser, Horse Assembly January 2007, chrUn:181082763-181083762), suggesting that the missing amino acids are due to incomplete sequencing. Similarly, Canis lupus familiaris JPH2 (XP_853529.1) had a gap in the sequence (May 2005 assembly, chr24:34678376-34680451) and was excluded from further analysis. Although Drosophila silvestris sequences ABY55761.1 and AAC06033.1 had an HSP score >300, both fragments (345 and 147 amino acids long, respectively) were short compared with the other Drosophila JPHs that were ∼1,000 residues long.
The complete list of sequences used in the analysis is shown in Table 1. After screening with a minimum HSP score and discarding sequences that where incomplete or otherwise truncated, we analyzed 64 isoforms from 41 species (27 invertebrates and 14 vertebrates). Only three species had sequences for all four isoforms: Homo sapiens, Mus musculus, and Rattus norvegicus.
Alignments and Phylogenetic Analysis
The protein sequences for the resulting 64 isoforms were aligned using ClustalW2 (version 2.0.10) (28) and verified using MUSCLE. The phylogeny tree was generated from the ClustalW2 alignment using PhyML Online (version 3.0.1) (17, 18). The tree topology search was obtained using Nearest Neighbor Interchange, with the LG (29), JTT, WAG, and Dayhoff substitution models and BIONJ initial tree. Branch support was calculated using the approximate likelihood-ratio test [aLRT(2)]. Using the UPGMA and NJ methods and using CLC Sequence Viewer version 5.0.1 and a bootstrap value of 1,000, we also generated additional phylogeny trees. Sequence alignment figures and phylogeny tree depictions were generated using CLC Sequence Viewer version 5.0.1. Conservation scores were calculated by the same method as by ClustalW2 (by dividing the number of identities in the best alignment by the number of residues compared). Sequence logos (44) were created with the online program WebLogo version 2.8.2 (8) interface. Inkscape (v. 0.46) was used to annotate protein domains and other information in alignment figures.
In Silico Models
Secondary protein structure predictions were performed on the human JPH isoforms, using the consensus of the SOPM, HNN, DPM, DSC, GOR IV, PHD, PREDATOR, and SIMPA96 models provided by NPS@ (Network Protein Sequence @nalysis, http://www.bioinf.manchester.ac.uk/dbbrowser/bioactivity/NPS2.html). Phosphorylation sites were predicted using the KinasePhos online website with a prediction specificity of 100% (http://kinasephos.mbc.nctu.edu.tw/index.php), which is based on profile hidden Markov models (HMM) (21).
RESULTS
Phylogeny of the JPHs
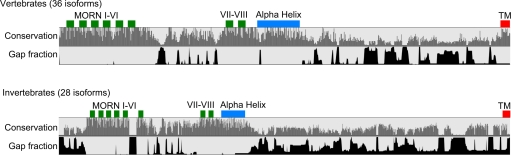
JPHs were found to be present and highly conserved across phyla of the animal kingdom. By comparing JPH protein sequences from different species, we can derive the evolutionary history of the JPH family. Therefore, we decided to examine the molecular evolution of JPH in detail. After an extensive search and review of the NCBI database for all proteins similar to JPHs, 64 full-length nonredundant isoforms from 41 species were included in the phylogenetic analysis (27 invertebrates and 14 vertebrates). There were 19 mammalian JPH sequences and 45 nonmammalian sequences, of which 17 were from lower vertebrates and 28 from invertebrates. All JPH sequences identified and used for phylogenic analysis are listed in Table 1. The JPH proteins range in size from 377 to 1,129, with the vast majority between 600 and 900 amino acids. Only five sequences are <560 amino acids; their short length is apparently an artifact of incomplete sequencing of the 3′ end, but the NH2 terminus of the protein still yields important conservation information in the MORN and joining regions. The only sequences that exceed 925 amino acids are those of the honey bee (Apis mellifera), wasp (Nasonia vitripennis), and Drosophila spp. The increased length is mostly due to extra segments in the divergent region and may be a result of an arthropod-specific divergence. The α-helical region, thought to account for most of the PM-SR distance, is not significantly affected. This correlates with the reported PM-SR distance in Drosophila junctional regions, which are reported to range from 10 to 25 nm (53). The sequence alignment using ClustalW2 (Supplemental Fig. S1)1 was compared with results of MUSCLE alignment (Supplemental Fig. S2) and yielded highly similar results.
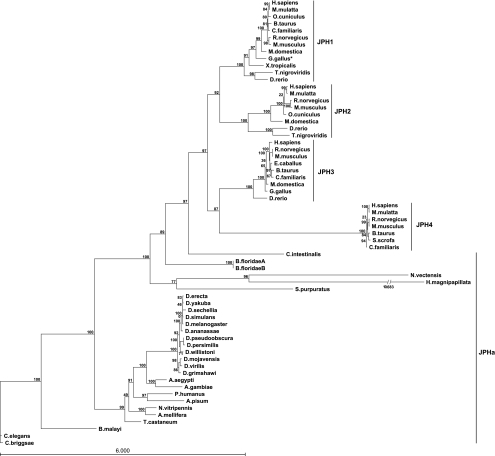
Figure 2 shows the unrooted maximum-likelihood tree for all 64 JPH sequences. The resulting tree has a log-likelihood of −41800.47734 and a tree size of 48.60002 (17). Trees generated using additional three different ML substitution methods (Dayhoff, JTT, and WAG) showed almost identical results and are included in the supplementary material (Supplemental Figs. S3–S5). UPGMA and NJ tress are also shown (Supplementary Figs. S6–S7). The tree shows a clear separation of the ancestral invertebrate JPH isoforms (JPHa), with further branching into phylum-based groups containing Nematodas, Arthropodas, Cnidarias, Echinodermata, and Chordata sequences. Although the tree follows the commonly accepted evolutionary path, the Cnidarians (Hydra magnipapillata and Nematostella vectensis) are paradoxically shown among the bilateria, between the echinoderms and chordates. They are also shown with particularly long branch lengths. This is most likely a combination of artifacts due to sequence gaps and the short (possibly truncated) sequence of the species included, combined with the rapid divergence and gene loss of the arthropod and nematode genomes compared with the vertebrates and cnidarians (41).
Fig. 2.
JPH phylogeny tree. Phylogeny tree of 64 JPH isoforms representing 41 species generated using the maximum likelihood method, with the LG substitution model and BIONJ initial tree. Branch support is calculated using the approximate likelihood ratio test and shown for each branch out of 100. Isoforms are listed by the Latin name. The branch length for Hydra magnipapillata was truncated for clarity; the actual length is shown. *The first 150 amino acids Gallus gallus JPH1 were removed (see methods).
The divergence of JPH into four genes took place after chordates branched from other deuterostomes but before the vertebrates arose, as indicated by the relative positions of the amphioxus (this genome is in draft form, so two candidate sequences are included) and sea squirt genomes, leading to separation from the invertebrate JPHa clade. The ancestor to the vertebrate JPH family underwent two separate gene duplication events, most likely first giving rise to a muscle and a neuronal isoform. The JPH1–2 clades then separated from the common muscle ancestor, whereas the JPH3–4 clades diverged from the neuronal ancestor. The actual timing of these serial gene duplications could not be established due to the lack of JPH sequences from organisms spanning the evolutionary gap between invertebrates and lower vertebrate fish species. The vertebrate JPH sequences are distinctly grouped into four clades, including the four JPH1, JPH2, JPH3, and JPH4 protein groups. Within each of these four groups, mammalian and nonmammalian vertebrate proteins are grouped in a manner consistent with the evolution of the organisms. Overall, these vertebrate JPH isoforms remained tightly conserved across a variety of species, suggesting that they have been subject to significant pressure to remain relatively unchanged, even though four distinct isoforms were generated very early among vertebrates. JPH4 exhibits a longer branch length relative to the other isoforms. This implies that JPH4 has undergone the most sequence changes, which suggests it has been under less evolutionary pressure than the other JPHs. This may indicate that whereas JPH1–3 all have very specific roles in skeletal and cardiac muscle, and in the brain respectively, JPH4 may only have an accessory or complementary role in the brain.
Sequence Alignments
Following the identification of the JPH protein family, it has been postulated that each protein contains eight MORN motifs in the NH2-terminal domain, an α-helix, and a COOH-terminal transmembrane motif (46). The conservation of these protein domains in JPH isoforms throughout evolution is evidenced by the high homology in the full amino acid sequence alignment (Fig. 3). Indeed, the highest degree of conservation was observed in the first group of MORN motifs (I–VI), the second series of MORN motifs (VII–VIII), the α-helical domain, and the transmembrane segment. The joining region was not thought to play a particular role in JPH function. However, the alignment in Fig. 3 shows significant conservation islands in the region, for example amino acids 190–210 in human JPH2. This suggests these sites may have a specific role in JPH function.
Fig. 3.
Conservation and secondary structure of JPH domains. Alignment of all 64 isoforms of JPH, showing location of reported domains, conservation, and gaps in alignment. Note the conserved region between the MORN motifs. TM, transmembrane.
Among the four human JPH isoforms (JPH1–4), amino acid conservation of the full-length sequences ranges from 42 to 59% (Table 2). Conservation percentages for the α-helical domain and the TM segment across the human JPH isoforms range from 40 to 68% and 35 to 75%, respectively. Interspecies sequence homology was determined for 8 JPH2 isoforms included in this study (Table 3). Among the six mammalian JPH2 isoforms, amino acid sequence conservation of the entire proteins was ≥73%. When the Tetraodon nigroviridis (green spotted puffer fish) and Danio rerio (zebrafish) were included in the alignments, the minimal conservation percentage was 55%. The overall high degree of conservation suggests that the various domains all serve specific purposes, and mutations in any of them could have significant impact on the protein structure/function.
Table 2.
Percent amino acid conservation for human JPH
| H. sapiens | JPH1 | JPH2 | JPH3 | JPH4 |
|---|---|---|---|---|
| JPH1 | ||||
| JPH2 | 53 | |||
| JPH3 | 53 | 45 | ||
| JPH4 | 59 | 43 | 42 |
Table 3.
Percent amino acid conservation for JPH2 across species
| JPH2 | H. sapiens | M. mulatta | O. cuniculus | R. norvegicus | M. musculus | M. domestica | T. nigroviridis | D. rerio |
|---|---|---|---|---|---|---|---|---|
| H. sapiens | ||||||||
| M. mulatta | 98 | |||||||
| O. cuniculus | 91 | 92 | ||||||
| R. norvegicus | 87 | 87 | 88 | |||||
| M. musculus | 86 | 87 | 88 | 97 | ||||
| M. domestica | 76 | 76 | 75 | 74 | 73 | |||
| T. nigroviridis | 66 | 67 | 64 | 65 | 65 | 67 | ||
| D. rerio | 57 | 57 | 57 | 55 | 55 | 57 | 80 |
MORN motifs.
JPHs contain multiple repeats of MORN motifs. Prior studies have demonstrated that MORN motifs are able to target proteins to the cell's PM (30). In addition to JPHs, the MORN motif is also found in multiple copies in several other classes of proteins including histone-lysine N-methyltransferase [SETD7 (UniProt Q8WTS6)] and phosphatidylinositol-4-phosphate 5-kinase (23).
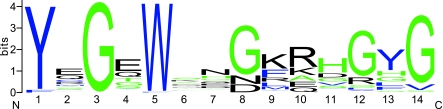
In JPHs, the MORN motifs are highly conserved, with amino acid homology ranging from 76% (MORN II) to 90% (MORN I) across all species. When analyzing the conservation of individual amino acids within the 14 amino acid-containing MORN motifs, we accounted for most of the variation (both within isoforms and across species) by the sixth and seventh amino acid. Whereas most amino acids were between 80 and 100% conserved, these two amino acid positions were conserved at an average of 51 and 52%, respectively. Interestingly, this variability was so pronounced that the MORN IV motif in Drosophilia spp. actually contains an additional lysine, and H. magnipapillata an asparagine between the sixth and seventh position.
Comparison of the different MORN motifs has resulted in a consensus MORN motif that can be described as YxGxWxxGKRH GYG, with x representing ambiguous amino acids (Fig. 4). Considering that the MORN motifs are hypothesized to bind the PM, in particular PIP2(30), these amino acids could be critical in forming the binding site. So far, human JPH2 is the only JPH isoform for which genetic mutations have been associated with human disease (27). Interestingly, one of the reported mutations, Y141H, occurs within a MORN motif, (amino acid 13 of MORN motif IV). Thus, this particular mutation might alter membrane binding of JPH2, thereby affecting JPH function in the heart, leading to cardiomyopathy in patients.
Fig. 4.
Conservation of MORN motifs. Sequence logo from all 8 MORN motifs of all 64 isoforms combined, resulting in a total of 512 MORN motifs averaged. The extra amino acid (at position 7) in Drosophila and Hydra MORN motif 4 was excluded (see text). Hydrophobic residues are shown in black, hydrophilic in blue, and neutral in green (8).
Joining region.
The joining region is defined as the sequence linking the first group of MORN motifs (I–VI) to the second group containing MORN motifs VII–VIII. To date, the joining region is not known to have a particular function. Here, however, we demonstrate that the joining region exhibits several areas of high conservation. To compare the amino acid conservation across various joining regions of JPH1–4 isoforms, we aligned only those sequences for which all four JPH isoforms were available. This limited the analysis to H. sapiens, M. musculus, and R. norvegicus. The results revealed a higher level of conservation among the neuronal JPHs, with the joining region in JPH3 being 99.3% conserved (only 1 of 144 amino acids is different among the three species), and JPH4 being 98.7% conserved (2 of 139 amino acids being different). Conservation is slightly lower among muscle JPHs, with 97.0 and 87.3% conservation of the joining region for skeletal JPH1 and cardiac JPH2, respectively. The overall high level of conservation suggests an important functional role for the joining region in JPHs, in particular for the first segment.
α-Helix domain.
Previous studies have reported an α-helical domain spanning ∼100 amino acids halfway along the protein (46). We confirmed that the α-helical domain, spanning approximately from amino acids 350 to 420 in human JPH2, is the main region within JPH proteins with an extensive predicted secondary structure. The α-helix is thought to bridge the gap between the PM and the SR (36). The domain is conserved across isoforms, ranging from 40 to 68%. The average length of this domain is ∼70 amino acids, which equates to an α-helix spanning ∼10.5 nm (assuming that each amino acid contributes 1.5 Å along the helix axis). This prediction correlates well with electron microscopy data showing a distance between the PM and SR in cardiac muscle of 12 nm (46). Since this distance decreases significantly in cardiac muscle lacking JPH2, JPHs might actively determine the distance between the cell surface and intracellular (SR/ER) membranes.
Divergent region.
The role of the divergent region is less clear at present. Despite its name, the region exhibits significant conservation across species. Comparison of the divergent regions of human, mouse, and rat sequences (as described above for the joining region) revealed high degrees of conservation for the four JPH isoforms: JPH1 (85.7%), JPH2 (83.7%), JPH3 (86.4%), and JPH4 (91.4%). Comparing the different JPH isoforms (JPH1–4), the divergent regions exhibit very low conservation percentages of 17.2% in humans, 15.5% in mice, and 17.5% in rats. This significant divergence between isoform and interspecies conservation suggests that the divergent region might play an important role in isoform-specific JPH functions. One hypothesis is that the divergent region mediates interactions with tissue-specific binding partners within the JMC. Another possibility is that the divergent region hosts sites for posttranslational modifications that serve to regulate protein function.
TM segment.
The TM segment is conserved almost completely across species, ranging from 95 to 100% conservation and is also highly conserved across isoforms, from 35 to 75%. Secondary structure analysis also consistently predicts the formation of a TM segment, anchoring the carboxy terminus of the protein into the ER/SR membrane.
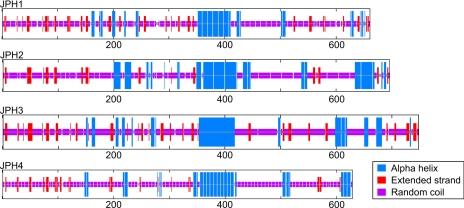
Secondary Structure Analysis
To further refine the structure of JPH, a consensus of secondary structure models was used to predict the most likely configuration of each functional region. Figure 5 shows the predicted structure for each of the human JPH proteins. The previously reported α-helical region is immediately apparent, constituting an ∼70-amino acid α-helix near the middle of the sequence. Additional smaller, interspersed α-helices are predicted throughout the protein sequence, comprising on average 20% of total JPH sequence. In particular, a small triplet of helices is predicted in the joining region. The remainder of the protein is predicted to consist of random coil regions (65%), extended strands (10%), or be ambiguous at present (5%).
Fig. 5.
Predicted secondary structure of human JPH isoforms. Blue, alpha helix; purple, random coil; red, extended strand.
Posttranslational Modifications
Posttranslational modifications are used extensively to modulate protein functions. Protein kinases A and C (PKA, PKC) are just two examples of many protein kinases that play a critical role in calcium signaling (50). In the heart, phosphorylation by these kinases significantly affects the dynamics of the heart, impacting contractility, relaxation, and cardiac output (51). In the case of JPH, a phosphorylation site could be key to regulating protein binding, or act as a structural switch, altering the distance between the PM and SR membranes. An unbiased kinase site search using HMM of the four human JPHs predicted up to 24 possible phosphorylation sites in JPH1, 35 in JPH2, 33 in JPH3, and 29 in JPH4, including targets of PKA and PKC (Table 4 and Supplemental Table S1). Large-scale mass spectrometry screens for phosphorylation sites in enriched phosphopeptide preparations have postulated a variety of candidates in mouse JPH1 [serine-448 and 452 (47)], human JPH1 [threonine-448 and serine-452 (38)], human JPH2 [serine-469 (aligns with JPH1 S452), serine-486 and threonine-490 (38)], and mouse JPH3 [serine-420 (35) and serine-506 (47)]. Three of these five human phosphorylation sites overlapped with the ones predicted by the HMM analysis. Cysteines can also play a key role in protein-protein interactions. No disulfide bonds have been found in JPHs, but there are three hyperreactive cysteine sites in JPH1 that are not conserved in JPH2 (40). These available cysteines were found to play a key role in protein-protein interactions with the skeletal muscle RyR1 (40). These findings suggest that JPHs have a role beyond serving as a structural foundation for the JMCs; the phosphorylation and protein-protein interaction sites could regulate calcium signaling even after the JMC is established.
Table 4.
Phosphorylation kinases and residues
| JPH1 | JPH2 | JPH3 | JPH4 | |||||
|---|---|---|---|---|---|---|---|---|
| Number of Residues | ||||||||
| Serines | 17 | 26 | 22 | 21 | ||||
| Threonine | 4 | 6 | 7 | 5 | ||||
| Tyrosine | 3 | 3 | 4 | 3 | ||||
| Kinase | ||||||||
| Ca2+/calmodulin-dependent protein kinase II (CaMKII) | 2 | 2 | ||||||
| Casein kinase I | 1 | 7 | 2 | 2 | ||||
| Casein kinase II (CKII) | 2 | 1 | 1 | |||||
| I-κB kinase (IKK) | 5 | 7 | 8 | 6 | ||||
| Mitogen-activated protein kinase (MAPK) | 5 | 8 | 4 | 4 | ||||
| cAMP-dependent protein kinase A (PKA) | 3 | 1 | 6 | 4 | ||||
| Protein kinase B (PKB) | 2 | 2 | ||||||
| Protein kinase C (PKC) | 2 | 1 | 1 | 3 | ||||
| cGMP-dependent protein kinase (PKG) | 4 | 5 | 7 | 5 | ||||
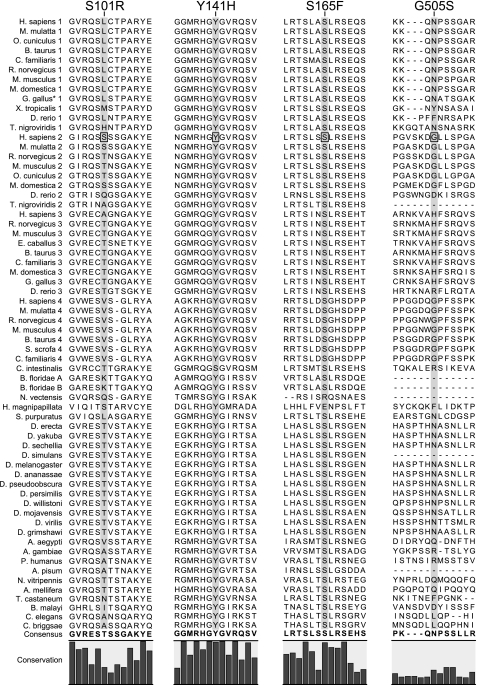
Human Disease-associated Mutations in JPH2
Recently, several missense mutations in JPH2 have been associated with hypertrophic cardiomyopathy (HCM) in patients in the US (27) and Japan (31). Conservation of the residues affected by these disease-associated mutations (S101R, Y141H, S165F, and G505S) is shown in Fig. 6. The alignment shows that residues Y141H and S165F are highly conserved across all isoforms, whereas G505S was only conserved within JPH2 isoforms, and S101R was poorly conserved overall. Interestingly, the Y141H and S165F are near the start of the joining region, where we propose a functional domain may be present. This hypothesis is supported by in vitro functional analysis of these mutations in immortalized cardiac cell lines in which overexpression of mutated JPH2 demonstrated vacuolated SR, attenuation of calcium-induced calcium release, and induction of cellular hypertrophy (26). Importantly, Y141H and S165F, which localize to residues highly conserved across multiple species, demonstrated the most impressive in vitro phenotype. These findings represent the first human disease associated with mutations in JPH2.
Fig. 6.
Alignment of published JPH2 mutations associated with cardiomyopathy in patients. Four mutations in JPH2 associated with heart disease have been published. The mutation is highlighted by a black box, and the alignment of the mutated site is shown by the gray shading. S101R, Y141H, and S165F from Ref. 27; G505S from Ref. 31.
DISCUSSION
In this study, we report the first comprehensive phylogenetic analysis of the JPH family. JPHs are a class of structural proteins that play an important role in the structural organization of JMCs, important subcellular structures in excitable cells involved in intracellular calcium signaling. We report the first extensive use of in silico techniques fully leveraging the data published in genomic databases. By aligning multiple isoforms and different species, we determine conservation at the single amino acid level and predict regions with structural importance and possible sites of posttranslational modifications. We postulate conserved regions that may play a role in protein structure and function, as well as a likely evolutionary history of the protein family. These findings are important to guide future research into the role(s) played by JPHs. Our study was limited by the incomplete sequencing or draft nature of several genomes, which yielded truncation artifacts. In particular, the early chordate sequences have incomplete COOH-terminal sequences, which slightly decreased the fidelity of our analysis. Nevertheless, the inclusion of sequences that met our inclusion criteria despite truncations was critical to explore the vertebrate/invertebrate split and the subsequent gene duplications that took place.
Evolution of JPH Isoforms
The JPH family is highly conserved throughout many species. Our studies reveal that JPHs have been predicted to be part of cnidarian, nematode, arthropod, and chordate genomes. The cnidarians (H. magnipapillata and N. vectensis) appear paradoxically among the deuterostomes, whereas they would be expected to be the first branch to split. This is most likely an artifact of the alignment (the N. vectensis sequence is likely truncated at the 3′ end) coupled with the rapid divergence of the arthropod and nematode genomes from deuterostomes that artificially brings the cnidarians closer to chordates and echinoderms. This artificial grouping probably causes the models to assume a high rate of mutation, which explains the particularly long branch lengths that are generated. The amphioxus and sea squirt genomes highlight the split between early chordates and the vertebrates, which are theorized to have undergone whole genome duplications (10). Although two amphioxus JPH sequences met our criteria and are shown, it is probably an artifact of sequencing or assembly (the genome used is the first draft assembly). In fact, both A and B isoforms are probably NH2-terminal fragments of the full-length Branchiostoma floridae JPH.
Our phylogenetic analysis suggests that gene duplications in the vertebrate lineage produced multiple copies. In the human, JPH1 is found on 8q21, JPH2 on 20q13.12, JPH3 on 16q24.3, and JPH4 on 14q11. The duplication and subsequent divergence allow each different isoform to specialize in a specific function and tissue. JPH1 and JPH2 became critical to the formation of triads in skeletal muscle and dyads in cardiac myocytes, respectively, while JPH3 and JPH4 are associated with subsurface cisternae in neurons. The presence of these four isoforms in mammals, as well as in the bony fish, and their close conservation with isoforms from other species (see Fig. 2) indicate that these different roles were established around the fish-tetrapod split (at least as far back as the divergence of gnathostomes, or jawed vertebrates, approximately 450 million years ago). This is in accord with the 2R hypothesis, which postulates two rounds of whole genome duplication took place in that period (10). These findings are similar to the evolution of other proteins in the junctional membrane complex, including the intracellular CRC, NCX, and the Ca2+ release-activated Ca2+ channel subunit Orai. The whole genome duplication theory also suggests that an additional round of replication (3R) took place in the teleosts (32). The genomes of D. rerio and T. nigroviridis harbor six and seven possible JPH genes (or pseudogenes), respectively (data not shown). Although the fragments are too short to pass our inclusion criteria or be included in the phylogeny tree, they could be the traces of the additional paralogs predicted by the 3R hypothesis.
Another interesting finding is the peculiar position of JPH4. The longer branch length in JPH4 implies that it has undergone the most sequence changes, which suggests it has been under less evolutionary pressure than the other JPHs. This may indicate that whereas JPH 1–3 all have very specific roles in skeletal and cardiac muscle and in the brain, respectively, JPH4 may only have an accessory or complementary role in the brain. The expression of JPH3 and 4 in almost identical regions of the brain supports this hypothesis: both are most highly expressed in the hippocampus, granule cells of the cerebellum, caudate putamen, nucleus accumbens, and the olfactory bulb and anterior olfactory nuclei. Additionally, JPH3 has a low level of expression in the ventrolateral, ventroposterior, and posterior thalamic nuclei and spinal gray matter, whereas JPH4 is undetectable in these regions (37). Furthermore, in knockout mouse models a phenotype was only apparent when both JPH3 and 4 where knocked out simultaneously (34).
Interestingly, other junctional membrane proteins seem to have lost the fourth isoforms predicted by the 2R hypothesis altogether [e.g., NCX (39) and RYR (48) only have three isoforms], suggesting that JPH4 may have a redundant function (as suggested by the knockout data). Conversely, the evolutionary conservation of JPH4 in vertebrates suggests that it may have evolved to carry out a particular function (probably, but not necessarily similar to JPH3 function) that was not detected in the knockout model but was sufficient to maintain JPH4 conserved in the genome. Additional studies of the function of JPH4 and indeed JPHs in general will help to answer the question of whether JPH4 is a gene on its way out or has developed a particular function yet to be discovered.
Our search also found a low but consistent similarity between JPH and phosphatidylinositol-4-phosphate-5-kinases [PIP(4,5)K] in protists and some plants (data not shown). This is probably the result of the presence of multiple MORN motifs with these sequences, which have been found to bind the negatively charged membrane lipids, in particular PtdIns(4,5)P2 and PtdOH (23). This suggests that the PM-associated region of JPH could play a role in signaling regulation through PIP2, although no data have been published on whether it acts upstream or downstream in the pathway. The Drosophila gene Undertaker, which encodes two small transcripts, also contains MORN motifs, suggesting they may be generic membrane binding domains. Although Undertaker was reported recently as a novel JPH (9), Undertaker actually has very little similarity to the prototypic structure that is highly conserved among all JPHs examined in this study, including the Drosophila JPHs. However, its role in calcium signaling and forming of junctional complexes between the PM and ER/SR membranes suggests that there may be a larger family of JPH-like membrane proteins, and those examined here (8 MORN domains, an α-helical structure and a TM domain) may constitute a specific subgroup.
Functional Roles of JPHs
In excitable cells, intracellular signaling processes occur locally within periplasmic subspaces often referred to as microdomains. The assembly of these JMCs enables communication between the PM and intracellular compartments such as the SR/ER. JPHs are believed to play an essential role in the proper assembly of these JMCs in muscle and neuronal cell types (37, 46). In cardiac myocytes, a complex network of PM invaginations called T-tubules propagate the depolarization signal, which originates at the cell surface, throughout the cell interior. JMCs are found at regular intervals along the T-tubules, where the PM comes into close proximity to the SR membrane. Here, the cellular depolarization signal triggers the opening of voltage-sensitive L-type Ca2+ channels, allowing the influx of extracellular Ca2+ into the cell. After entering the JMC, Ca2+ diffuses until it reaches cardiac CRCs (RyR2) on the SR membrane to trigger channel opening. This in turn allows the release of large amounts of Ca2+ from the SR, increasing the cytoplasmic Ca2+ concentration several-fold (50). This amplification of Ca2+ is termed CICR (12) and plays a critical role in the initiation of contraction of cardiac myocytes. The short distance between the LTCC and RyR2 is thought to be critical for efficient triggering of RyR2-mediated SR-stored Ca2+ release by extracellular Ca2+. Conversely, improper approximation or alignment of LTCC and RyR2 channels in the JMC has been observed in hypertrophic and failing hearts (45, 54). Suboptimal coupling between cytoplasmic and SR Ca2+ channels results in a decreased excitation-contraction coupling efficiency and impaired contractility of the heart. Other studies have shown that disruption of JPH expression in cardiac muscle by genetic deletion of JPH2 in mice leads to the formation of fewer and improperly aligned JMCs as evidenced by the lower incidence of 12 nm junctions between the PM and SR (46). At the whole organism level, absence of JPH2 in mouse heart muscle causes abnormal, stochastic heartbeats and death by embryonic day 10.5 (46). Loss of JPH1 in mice results in poor skeletal muscle function, resulting in early postnatal death of JPH1 knockout mice as a result of an inability to feed (24). These findings attest to the critical role of JPHs in the formation and functionality of JMCs.
JPH in Disease
In humans, mutations in JPH2 are associated with HCM (27, 31). Indeed, several distinct missense mutations identified in a cohort of unrelated patients with HCM were found to impair the ability of JPH2 to maintain the critical dyadic geometry necessary for proper CICR through in vitro analyses. In rodent genetic models of HCM and dilated cardiomyopathy (DCM), reduced JPH2 expression has been associated with pathogenesis of these diseases (33). Furthermore, in rodent models of “compensated” and “decompensated” pressure-induced hypertrophy, this downregulation of JPH2 occurred early during a period of intermolecular remodeling, ultimately resulting in uncoupling of CICR and disease pathogenesis (54). Whether this downregulation is a maladaptive physiological response or directly contributory to the failing myocyte remains unanswered.
Trinucleotide repeat expansion in the JPH3 gene has been associated with Huntington disease-like 2 (HDL2), a disease characterized by progressive dementia, ataxia and movement disorders (including chorea) (20). The expanded CAG/CTG repeats may result in the formation of RNA foci associated with cellular toxicity (43). Therefore, it appears that JPH3 defects associated with HDL2 may not involve JPH3 expression abnormalities although this remains to be determined.
Posttranslational Modifications
One advantage of a large alignment of protein sequences is that it allows conservation to be assessed at the single amino acid level. This degree of resolution is important to identify key residues involved in posttranslational modifications and protein binding. Phosphorylation is a common mechanism to modify protein function, and is known to regulate calcium signaling (6, 51). For example, PKA phosphorylation of phospholamban increases Ca2+ entry into the SR, improving cardiac relaxation (5). We identified several possible phosphorylation sites on human JPH isoforms, some of which correlate with previously proposed sites. In vivo confirmation of even some of these sites could reveal regulatory pathways that directly affect intracellular calcium signaling.
The variable nature of regulation site predictions from these different approaches confirms that they all suffer from intrinsic biases and limitations. However, it is interesting to note that some predictions cluster in highly conserved regions across isoforms, suggesting generic JPH regulation sites. Some sites are consistently predicted in regions that are highly conserved in one specific isoform. These could serve as isoform-specific regulatory sites. For example, the two postulated phosphorylation sites in mouse JPH3 are highly conserved in all species but are only found in JPH3 isoforms, suggesting an isoform-specific function or regulatory mechanism. These findings are the first step in determining how and where each JPH isoform is regulated. It is also interesting to note that most of these sites are in the joining and divergent regions, supporting the hypothesis that these two domains are responsible for protein interaction and regulation.
Protein-protein Interactions
Little information is available on JPH regulation and interactions with other proteins. Interactions are difficult to explore, since they span from transient (such as a phosphorylation event) to prolonged (binding to another structural element to form a complex, such as TM segments in the SR membrane), with highly variable effects. The two most likely sites for protein-protein interactions would be the joining region and the divergent region, since these are both highly conserved but do not seem to play a role in forming JPH structure.
Two protein-protein interactions have been specifically reported in the literature: JPH1 was shown to coimmunoprecipitate with RyR1 (40) and JPH2 coimmunoprecipitated with caveolin-3 (33). Interaction with RyR1 supports the hypothesis of JPH being a key structural component of the JMC and opens an avenue of research into how CICR is regulated. Caveolin-3 forms caveolae in muscle and is involved in the proper formation of T-tubules and the scaffolding of the PM; mutations in CAV3-encoded caveolin-3 have been associated with myopathies (52), long QT syndrome (49), and sudden infant death syndrome (7). Binding of caveolin-3 to JPH could thus be an important component for the formation of JMCs in myocytes. Large-scale analyses of protein interactions have shown three additional binding candidates: SMAD3 coimmunoprecipitated with JPH1(15), slc2a4 (the glut-4 glucose transporter) in rats coimmunoprecipitated with JPH2 (13), and a coimmunoprecipitation showed an interaction between JPH3 and STK23, a serine/threonine kinase (11). Although these large-scale approaches using tagged proteins are less precise, they do point to a variety of possible interactions and regulatory pathways that JPH could influence.
Summary
JPHs are proteins found in JMC in excitable cells, where they play a critical role in approximating the plasmalemmal and SR/ER membranes. A phylogenetic analysis of over 60 JPH proteins from over 40 species revealed that JPHs are highly conserved in evolution, in particular the MORN motifs found in all species. Our data suggest that an ancestral form of JPH arose at the latest in a common metazoan ancestor and that in vertebrates four isoforms arose, probably following two rounds of whole genome duplications. The increasing number of available JPH sequences yields significant insight into the molecular evolution and the functional roles of JPHs in excitable cells. Our analysis is consistent with the emerging concept that JPHs serve dual important functions in excitable cells: structural assembly of JMC and regulation of intracellular calcium signaling pathways.
GRANTS
X. H. T. Wehrens is a W. M. Keck Foundation Distinguished Young Scholar in Medical Research and is also supported by Muscular Dystrophy Association Grant 69238, National Institutes of Health (NIH) Grant R01-HL-089598, March of Dimes Grant MOD24172, and by the Hankamer Foundation. A. Garbino is supported in part by NIH Grant T32 GM-07330. R. J. van Oort is a recipient of the 2008–2009 American Physiological Society Postdoctoral Fellowship in Physiological Genomics. S. S. Dixit is supported by NIH Grant T32 HL-07676-20. A. P. Landstrom is supported by an American Heart Association (AHA) Predoctoral Fellowship. M. J. Ackerman is an Established Investigator of the AHA and is also supported by NIH Grant HD-42569, the CJ Foundation for SIDS, the Dr. Scholl Foundation, and the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death research program.
Address for reprint requests and other correspondence: X. H. T. Wehrens, Dept. of Molecular Physiology and Biophysics, Baylor College of Medicine, 1 Baylor Plaza, BCM335, Houston, TX 77030 (e-mail wehrens@bcm.edu).
Footnotes
The online version of this manuscript contains supplemental material.
REFERENCES
- 1.Altschul SF, Gish W. Local alignment statistics. Methods Enzymol 266: 460–480, 1996. [DOI] [PubMed] [Google Scholar]
- 2.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Bers DM Macromolecular complexes regulating cardiac ryanodine receptor function. J Mol Cell Cardiol 37: 417–429, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res 14: 988–995, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brittsan AG, Kranias EG. Phospholamban and cardiac contractile function. J Mol Cell Cardiol 32: 2131–2139, 2000. [DOI] [PubMed] [Google Scholar]
- 6.Chen-Izu Y, Ward CW, Stark W Jr, Banyasz T, Sumandea MP, Balke CW, Izu LT, Wehrens XH. Phosphorylation of RyR2 and shortening of RyR2 cluster spacing in spontaneously hypertensive rat with heart failure. Am J Physiol Heart Circ Physiol 293: H2409–H2417, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Cronk LB, Ye B, Kaku T, Tester DJ, Vatta M, Makielski JC, Ackerman MJ. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm 4: 161–166, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, Eid JP, Quirin M, Franc NC. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135: 524–534, 2008. [DOI] [PubMed] [Google Scholar]
- 10.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3: 89, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabiato A Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Physiol Cell Physiol 245: C1–C14, 1983. [DOI] [PubMed] [Google Scholar]
- 13.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC). J Proteome Res 5: 64–75, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Graves TK, Hinkle PM. Ca2+-induced Ca2+ release in the pancreatic beta-cell: direct evidence of endoplasmic reticulum Ca2+ release. Endocrinology 144: 3565–3574, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Grimsby S, Jaensson H, Dubrovska A, Lomnytska M, Hellman U, Souchelnytskyi S. Proteomics-based identification of proteins interacting with Smad3: SREBP-2 forms a complex with Smad3 and inhibits its transcriptional activity. FEBS Lett 577: 93–100, 2004. [DOI] [PubMed] [Google Scholar]
- 16.Guatimosim S, Amaya MJ, Guerra MT, Aguiar CJ, Goes AM, Gomez-Viquez NL, Rodrigues MA, Gomes DA, Martins-Cruz J, Lederer WJ, Leite MF. Nuclear Ca2+ regulates cardiomyocyte function. Cell Calcium 44: 230–242, 2008. [DOI] [PubMed] [Google Scholar]
- 17.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–W559, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez-Cruz A, Escobar AL, Jimenez N. Ca2+-induced Ca2+ release phenomena in mammalian sympathetic neurons are critically dependent on the rate of rise of trigger Ca2+. J Gen Physiol 109: 147–167, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes SE, O'Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, Fleisher A, Stevanin G, Brice A, Potter NT, Ross CA, Margolis RL. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet 29: 377–378, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Huang HD, Lee TY, Tzeng SW, Wu LC, Horng JT, Tsou AP, Huang KT. Incorporating hidden Markov models for identifying protein kinase-specific phosphorylation sites. J Comput Chem 26: 1032–1041, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res 30: 38–41, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im YJ, Davis AJ, Perera IY, Johannes E, Allen NS, Boss WF. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J Biol Chem 282: 5443–5452, 2007. [DOI] [PubMed] [Google Scholar]
- 24.Ito K, Komazaki S, Sasamoto K, Yoshida M, Nishi M, Kitamura K, Takeshima H. Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J Cell Biol 154: 1059–1067, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komazaki S, Ito K, Takeshima H, Nakamura H. Deficiency of triad formation in developing skeletal muscle cells lacking junctophilin type 1. FEBS Lett 524: 225–229, 2002. [DOI] [PubMed] [Google Scholar]
- 26.Komazaki S, Nakamura H. Functional analysis of mammalian genes using amphibian embryonic cells. J Electron Microsc (Tokyo) 53: 87–92, 2004. [DOI] [PubMed] [Google Scholar]
- 27.Landstrom AP, Weisleder N, Batalden KB, Bos JM, Tester DJ, Ommen SR, Wehrens XH, Claycomb WC, Ko JK, Hwang M, Pan Z, Ma J, Ackerman MJ. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol 42: 1026–1035, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320, 2008. [DOI] [PubMed] [Google Scholar]
- 30.Ma H, Lou Y, Lin WH, Xue HW. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res 16: 466–478, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita Y, Furukawa T, Kasanuki H, Nishibatake M, Kurihara Y, Ikeda A, Kamatani N, Takeshima H, Matsuoka R. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet 52: 543–548, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27: 937–945, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Minamisawa S, Oshikawa J, Takeshima H, Hoshijima M, Wang Y, Chien KR, Ishikawa Y, Matsuoka R. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem Biophys Res Commun 325: 852–856, 2004. [DOI] [PubMed] [Google Scholar]
- 34.Moriguchi S, Nishi M, Komazaki S, Sakagami H, Miyazaki T, Masumiya H, Saito SY, Watanabe M, Kondo H, Yawo H, Fukunaga K, Takeshima H. Functional uncoupling between Ca2+ release and afterhyperpolarization in mutant hippocampal neurons lacking junctophilins. Proc Natl Acad Sci USA 103: 10811–10816, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, Panse C, Schlapbach R, Mansuy IM. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics 6: 283–293, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Nishi M, Mizushima A, Nakagawara K, Takeshima H. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun 273: 920–927, 2000. [DOI] [PubMed] [Google Scholar]
- 37.Nishi M, Sakagami H, Komazaki S, Kondo H, Takeshima H. Coexpression of junctophilin type 3 and type 4 in brain. Brain Res Mol Brain Res 118: 102–110, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648, 2006. [DOI] [PubMed] [Google Scholar]
- 39.On C, Marshall CR, Chen N, Moyes CD, Tibbits GF. Gene structure evolution of the Na+-Ca2+ exchanger (NCX) family. BMC Evol Biol 8: 127, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phimister AJ, Lango J, Lee EH, Ernst-Russell MA, Takeshima H, Ma J, Allen PD, Pessah IN. Conformation-dependent stability of junctophilin 1 (JP1) and ryanodine receptor type 1 (RyR1) channel complex is mediated by their hyper-reactive thiols. J Biol Chem 282: 8667–8677, 2007. [DOI] [PubMed] [Google Scholar]
- 41.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94, 2007. [DOI] [PubMed] [Google Scholar]
- 42.Rose CR, Konnerth A. Stores not just for storage. Intracellular calcium release and synaptic plasticity. Neuron 31: 519–522, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington's disease-like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol 61: 272–282, 2007. [DOI] [PubMed] [Google Scholar]
- 44.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song LS, Sobie EA, McCulle S, Lederer WJ, Balke CW, Cheng H. Orphaned ryanodine receptors in the failing heart. Proc Natl Acad Sci USA 103: 4305–4310, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell 6: 11–22, 2000. [DOI] [PubMed] [Google Scholar]
- 47.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics 5: 914–922, 2006. [DOI] [PubMed] [Google Scholar]
- 48.Tunwell RE, Wickenden C, Bertrand BM, Shevchenko VI, Walsh MB, Allen PD, Lai FA. The human cardiac muscle ryanodine receptor-calcium release channel: identification, primary structure and topological analysis. Biochem J 318: 477–487, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 114: 2104–2112, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Wehrens XH, Lehnart SE, Marks AR. Intracellular calcium release and cardiac disease. Annu Rev Physiol 67: 69–98, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Wehrens XH, Lehnart SE, Reiken S, Vest JA, Wronska A, Marks AR. Ryanodine receptor/calcium release channel PKA phosphorylation: a critical mediator of heart failure progression. Proc Natl Acad Sci USA 103: 511–518, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodman SE, Sotgia F, Galbiati F, Minetti C, Lisanti MP. Caveolinopathies: mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases. Neurology 62: 538–543, 2004. [DOI] [PubMed] [Google Scholar]
- 53.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol 174: 803–813, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu M, Zhou P, Xu SM, Liu Y, Feng X, Bai SH, Bai Y, Hao XM, Han Q, Zhang Y, Wang SQ. Intermolecular failure of L-type Ca2+ channel and ryanodine receptor signaling in hypertrophy. PLoS Biol 5: e21, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]