Abstract
In mammalian cells, microRNAs (miRNAs) are incorporated into miRNA-induced silencing complexes (miRISCs), which regulate protein expression post-transcriptionally through binding to 3′-untranslated regions of target mRNAs. Argonaute2 (Ago2), a key component of the miRISC, recruits GW182, a component of the processing body (GW/P-body), to the target mRNAs. To elucidate the function of GW182 in an miRNA-mediated translational repression, we analyzed Argonaute-binding sites in GW182. We found that human GW182 contains three binding sites for Ago2, within the amino-terminal glycine tryptophan (GW/WG)-repeated region that is characteristic of the GW182 family proteins. We also found that the first and second Ago2-binding site is conserved within the amino-terminal half of TNRC6B, which is a paralog of GW182. Each of the Ago-binding sites is alone sufficient to bind Ago2. Furthermore, we demonstrated that multiple Argonaute proteins were connected via the GW182 protein. A GW182 fragment containing the Ago2-binding region partially relieved let-7-mediated repression of protein synthesis in a mammalian cell-free system. Coincidentally, let-7-directed target mRNA deadenylation was delayed. Together, these results strongly suggested that the interactions of GW182 with Argonautes may induce the formation of large complexes containing miRNA target mRNAs, and may be critical for miRNA-mediated translational repression.
Keywords: microRNA, translational repression, miRISC, GW182, Argonaute, cell-free system
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) small RNAs that regulate gene expression at the post-transcriptional level (Ambros 2004; Bartel 2004). A large number of miRNAs influence the expression of genes that control cell growth and differentiation, as well as viral pathogenesis (Kloosterman and Plasterk 2006; Sarnow et al. 2006). In mammalian cells, precursor miRNAs are processed by a protein complex containing Dicer, and the mature miRNAs are incorporated into miRNA-induced silencing complexes (miRISCs). Recruitment of miRISCs to target sites located in the 3′-untranslated region (3′-UTR) of mRNA, which is partially complementary to the miRNA, result in translational repression of the mRNA. The mechanisms of miRNA-mediated translational repression remain controversial. Several models have been proposed based on experiments using different systems, including Drosophila and mammalian cell-free systems (Wang et al. 2006; Mathonnet et al. 2007; Standart and Jackson 2007; Thermann and Hentze 2007; Wakiyama et al. 2007; Eulalio et al. 2008a; Filipowicz et al. 2008)
Argonaute and GW182 are the key components of the miRISC. GW182 is a member of a conserved protein family localized to processing bodies (P bodies or GW bodies), which are cytoplasmic foci related to translational repression and degradation of mRNA (Eystathioy et al. 2003; Ding and Han 2007; Eulalio et al. 2007a; Parker and Sheth 2007). GW182 was first discovered using an autoimmune serum from a patient with motor and sensory polyneuropathy (Eystathioy et al. 2002). Two GW182 orthologs, called AIN-1 (ALG-1 interacting protein 1) and AIN-2, were identified in Caenorhabditis elegans, and one ortholog was identified in Drosophila (Ding et al. 2005; Behm-Ansmant et al. 2006). In mammals, there are two GW182 paralogs, TNRC6B and TNRC6C. TNRC6B was identified through a biochemical search for Argonaute interacting proteins (Meister et al. 2005). The GW182 family proteins contain many glycine-tryptophan or tryptophan-glycine (GW/WG) repeats (Ding and Han 2007). With the exception of AIN-1 and AIN-2, the GW182 family proteins contain an RNA recognition motif in their carboxy-terminal regions (Ding and Han 2007).
AIN-1 and AIN-2 are associated with a large number of miRNAs and mRNAs, and they also interact with the miRNA-specific Argonautes, ALG-1 and ALG-2, which are involved in translational inhibition (Ding et al. 2005; Zhang et al. 2007). In Drosophila cells, Ago1 functions in miRNA-mediated translational repression, and the N terminus of GW182 interacts with Ago1 (Okamura et al. 2004; Behm-Ansmant et al. 2006). Depletion of Drosophila GW182 and overexpression of the N terminus of Drosophila GW182 inhibit miRNA-mediated gene silencing (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006; Eulalio et al. 2008b). GW182 interacts with Ago2 and functions in miRNA-mediated gene silencing in mammalian cells (Jakymiw et al. 2005; Liu et al. 2005). Previously, in a HEK293F cell-free system, we demonstrated that let-7 miRNA-mediated translational repression is enhanced by the addition of GW182, and that GW182 was recruited to the target mRNA by Ago2, depending on the let-7 miRNA (Wakiyama et al. 2007). These results suggested that GW182 is involved in miRNA-mediated translational repression, and that the interaction between Ago2 and GW182 may be important.
To elucidate the function of GW182 and the importance of its interactions with Argonaute proteins, we investigated the Ago2-binding site of GW182. Recently, it was reported that an Ago2-binding site, termed the Ago hook, existed in the N terminus of human TNRC6B isoform2, and one quite similar region existed in GW182 (Till et al. 2007). (Note that the GW182 and TNRC6B clones used in the study by Till et al. [2007] are the longer N terminus GW182 isoform and the shorter N terminus TNRC6B isoform, as compared with those used in this study.) Moreover, they showed that the addition of the Ago hook peptide relieved the miR-2-mediated repression in a Drosophila cell-free system (Till et al. 2007).
In this study, we show that three distinct Ago2-binding sites, including the aforementioned Ago hook motif, exist in the amino-terminal half of GW182, and that GW182 can bind multiple Ago2. Furthermore, we show that TNRC6B, a GW182 paralog, also contains two distinct Ago2-binding sites, which are conserved in GW182. Amino acid substitution experiments revealed that highly conserved tryptophan residues are crucial for Ago2 binding. Using a cell-free system that recapitulates let-7-mediated translational repression, we show that the interaction between Ago2 and GW182 is important for miRNA-mediated repression.
RESULTS
Human GW182 contains multiple Argonaute-binding sites
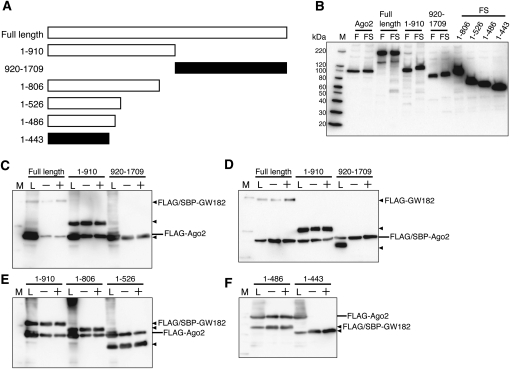
In Drosophila, the amino-terminal region of GW182, containing the GW/WG repeats, is important for the interaction with Ago1, the Drosophila Argonaute protein that functions in the miRNA pathway (Okamura et al. 2004; Behm-Ansmant et al. 2006). To confirm the ability of Ago to bind to the amino-terminal domain of human GW182, we first carried out pull-down assays of human Ago2 with amino-terminal (residues 1–910) and carboxy-terminal (residues 920–1709) fragments of human GW182 (Fig. 1A,B). These fragments were fused with a tandem tag containing the FLAG sequence and the streptavidin-binding peptide (SBP) sequence (Wilson et al. 2001). Each FLAG/SBP-tagged GW182 fragment was coexpressed with FLAG-tagged Ago2 in HEK293F cells, and the GW182 fragment was purified with streptavidin-agarose. These assays revealed that Ago2 was precipitated with the amino-terminal half, but not the carboxy-terminal half of GW182 (Fig. 1C). In addition, FLAG/SBP-tagged Ago2 was able to bind the FLAG-GW182 (1–910) fragment, but not the FLAG-GW182 (920–1709) fragment (Fig. 1D). As for the Ago paralogs, FLAG-tagged Ago1, Ago3, and Ago4 were precipitated with the FLAG/SBP-GW182 (1–910) fragment (data not shown).
FIGURE 1.
The amino-terminal half of GW182, containing GW/WG repeats, binds Ago2. (A) Schematic representation of the GW182 fragments used in the SBP pull-down assay. The polypeptides that bound Ago2 are represented by the white bars, while the black bars represent fragments that did not bind Ago2. (B) Expression of Ago2 and the GW182 fragments in HEK293F cells. The expressed proteins were detected by Western blotting with an anti-FLAG M2 antibody. The sizes of the molecular markers are indicated on the left (lane M). (Lanes F,FS) FLAG- and FLAG/SBP-tagged proteins, respectively. (C–F) SBP pull-down assay of Ago2 and GW182. HEK293F cells were cotransfected with FLAG/SBP or FLAG-tagged Ago2 and the GW182 expression vectors. After 48 h, the cells were harvested and the prepared cell lysates were subjected to SBP pull-down assays with (+) or without (−) RNaseA. The precipitates were analyzed by Western blotting using an anti-FLAG M2 antibody. The positions of the GW182 fragments are indicated with triangles on the right. (Lane L) The cell lysate sample.
To clarify the Ago2-binding site within GW182, we performed SBP pull-down assays with several fragments of the amino-terminal region of GW182 (Fig. 1A,B). In these assays, the GW182 fragments 1–806, 1–526, and 1–486 were able to bind to Ago2, but 1–443 was not (Fig. 1E,F). These results suggested that the GW182 fragment including residues 444–486 contained an Ago2-binding site.
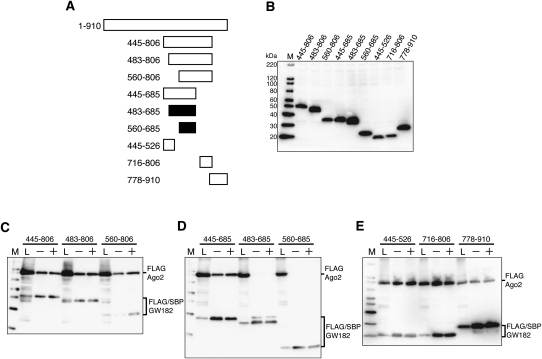
Next, to investigate whether another region is responsible for the interaction with Ago2, we used different amino-terminal and/or carboxy-terminal truncated GW182 fragments in SBP pull-down assays (Fig. 2A,B). In agreement with the results shown in Figure 1F, GW182 fragments 445–806 and 445–685 containing amino acids 445–486, bound to Ago2, whereas 483–685 and 560–685 did not (Fig. 2C,D). Interestingly, despite the absence of the aforementioned binding site, GW182 fragments 483–806 and 560–806 also interacted with Ago2 (Fig. 2C). These results indicated that another binding site exists within amino acids 686–806.
FIGURE 2.
Multiple Ago2-binding sites are present in the amino-terminal half of GW182. (A) Schematic representation of the GW182 fragments used in the SBP pull-down assay. The polypeptides that can bind Ago2 are represented by the white bars, while the black bars represent those that cannot bind Ago2. (B) Expression of Ago2 and each GW182 fragment in HEK293F cells. The expressed proteins were detected by Western blotting with an anti-FLAG M2 antibody. (C–E) SBP pull-down assays of Ago2 and GW182. HEK293F cells were cotransfected with FLAG-Ago2 and FLAG/SBP-GW182 expression vectors. SBP-pull down assays and Western blotting were performed as shown in Figure 1C.
To examine whether the minimal polypeptides containing the predicted Ago2 interaction site are sufficient to bind Ago2, we produced two GW182 fragments, 445–526 and 716–806, which are highly conserved regions between GW182 and TNRC6B (see Supplemental Fig. 1). SBP pull-down assays revealed that these two fragments were associated with Ago2 (Fig. 2E). It was reported that the Ago hook motif of TNRC6B isoform2, which can associate with Argonaute, is also conserved in the GW182 isoform, within residues 1075–1093. (Again, note that the GW182 used by Till et al. [2007] was the longer N-terminus isoform. Thus, the Ago2 hook motif exists within residues 822–840 of the GW182 used in this study.) Therefore, we performed a pull-down assay using the GW182 778–910 polypeptide. As shown in Figure 2E, this polypeptide was able to bind Ago2. Since GW182 fragments 716–806 and 778–910 partially overlap, we confirmed the binding of GW182 (amino acids 806–910) to Ago2, ruling out the possibility that the Ago2-binding site is located within amino acids 778–806 (data not shown).
Collectively, there are three Ago2-binding sites in GW182, and each site is independently sufficient to interact with Ago2.
A conserved tryptophan residue is necessary for Ago2 binding
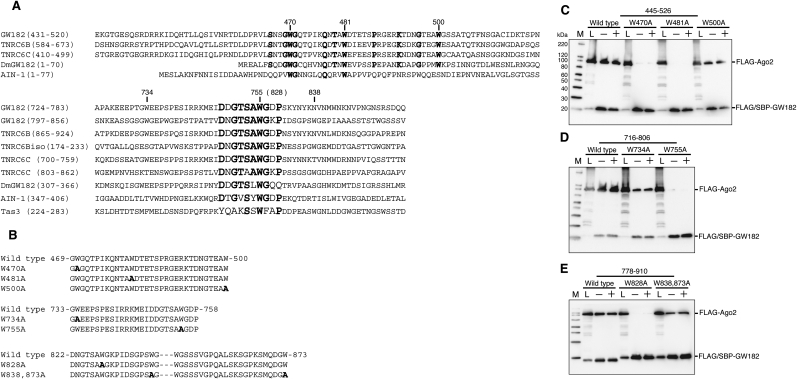
To further investigate the three Ago-binding sequences in human GW182, we compared the corresponding amino acid sequences with those of human TNRC6B, TNRC6B isoform2, TNRC6C, Drosophila GW182, C. elegans AIN-1, and Schizosaccharomyces pombe Tas3 by an amino acid alignment and a BLAST search. The multiple alignment of the sequences containing the first binding site (residues 445–526) revealed that several tryptophan residues are well conserved among human GW182, TNRC6B, TNRC6C, Drosophila GW182, and AIN-1 (Fig. 3A). The second and third binding sites contain almost identical sequences, located in residues 749–758 and 822–831. Identical or similar sequences are present in the GW182 paralogs and orthologs (Fig. 3A). (Note that the two conserved regions shown in Figure 3A correspond to the highly conserved motifs I and II in the N-terminal GW repeat of Drosophila GW182 described by Behm-Ansmant et al. [2006].) These results revealed that tryptophan residues are especially highly conserved in each individual binding domain (i.e., residues 470, 481, 755, and 828). It was previously reported that the tryptophan residues in the Ago hook motif are important for Argonaute binding (Till et al. 2007). Together, these results suggest that these tryptophan residues are important for the interaction with Ago2.
FIGURE 3.
A conserved tryptophan residue is important for Ago2 binding. (A) Alignment of the highly conserved regions of human GW182 (AY035864), TNRC6B (AB029016), TNRC6C (NM_018996), TNRC6Bisoform2 (NM_001024843), Drosophila melanogaster (Dm) GW182 (NP_726596), Caenorhabditis elegans AIN-1 (NM_078286), and Schizosaccharomyces pombe Tas3 (O94687). Highly conserved amino acids are indicated in bold. (B) The partial amino acid sequences with the point mutations in GW182 445-526 (top), 716-806 (middle), and 778-910 (bottom). Mutated amino acid residues are indicated in bold. (C–E) SBP pull-down assays of Ago2 and GW182 mutants. HEK293F cells were cotransfected with FLAG-Ago2 and FLAG/SBP-GW182 mutant expression vectors. SBP pull-down assays and Western blotting were performed as shown in Figure 1C.
To examine the role of these tryptophan residues, each of the residues was substituted with alanine (Fig. 3B). The FLAG/SBP (FS)-tagged GW182 variants and FLAG-tagged Ago2 proteins were coexpressed in HEK293F cells, and then SBP pull-down assays were performed. The W470A and W481A mutations in the GW182 fragment 445–526 abolished the interaction with Ago2. These two tryptophans are also conserved in AIN-1. On the contrary, the mutant of Trp500, which is not conserved in AIN-1, was able to bind to Ago2 (Fig. 3C). Analyses of the GW182 fragments 716–806 and 778–910 revealed that Trp755 and Trp828 are also important for Ago2 binding (Fig. 3D,E). The interaction with Ago2 was also affected by the W734A mutant, but was not abolished (Fig. 3D). The W838A and W873A mutants had little effect on the binding of Ago2 (Fig. 3E).
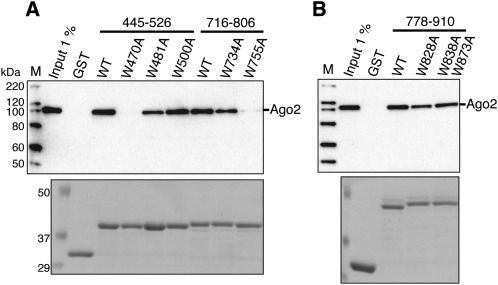
The importance of these tryptophan residues was also ascertained by a GST pull-down assay. Although the wild-type and W500A proteins bound Ago2, the W470A mutant did not in this assay (Fig. 4A). These results are consistent with the data obtained by the SBP pull-down assay. In contrast, the W481A GST fusion protein was able to interact with Ago2, although the amount of bound Ago2 was reduced, as compared with the wild type (Fig. 4A). In the GST pull-down assays, the W755A mutant did not bind Ago2 and the interaction with Ago2 was weakened by W734A, in agreement with the SBP pull-down results (Fig. 4A). On the other hand, Ago2 coprecipitated with the W828A mutant, which was inconsistent with the SBP pull-down results (Fig. 4B).
FIGURE 4.
GST pull down assay of Ago2 and GST-fused GW182 mutants. (A) The wild-type and mutated GST-GW182 (445–526 and 716–806) fragments were each expressed in E. coli. (B) The wild-type and mutated GST-GW182 (778–910) fragments were each expressed in E. coli. HEK293F cell extracts overexpressing FLAG-Ago2 were incubated with each MagneGST particle-immobilized GST-GW182 fragment. The resulting complexes were resolved by SDS-PAGE, followed by Coomassie Brilliant Blue (CBB) staining (bottom), and were analyzed by Western blotting using an anti-FLAG M2 antibody (top).
Collectively, these data indicate that Trp470, in the first binding site, and Trp755, in the second binding site, are crucial for the interaction with Ago2. Although Trp734 in the second binding site is important for the interaction, it is probably not essential. The importance of Trp481 and Trp828 for Ago2 binding remains elusive.
The amino-terminal GW/WG-repeated region of TNRC6B contains two Ago2-binding sites
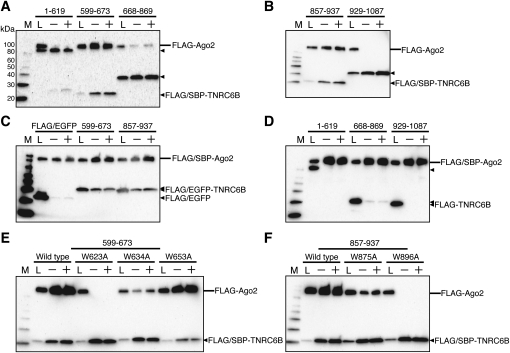
It has been reported that TNRC6B is involved in RNA silencing and also associates with Ago2 (Meister et al. 2005). As shown in the alignment of the GW182 and TNRC6B amino acid sequences, the first and second Ago2-binding sites are also conserved in the amino terminus of TNRC6B (Fig. 3A; Supplemental Fig. 1). To examine whether these binding sites are sufficient for the interaction, we generated a number of TNRC6B fragments, in which 599–673 and 857–937 contain the first and the second binding site, respectively. Due to degradation or their low molecular weight, the expression of FLAG-tagged 599–673 and 857–937 fragments was not detected. Hence, we used the FLAG-EGFP-fused 599–673 and 857–937 fragments in the SBP pull-down assay. The pull-down assays revealed that TNRC6B 599–673 and 857–937, which contain predicted Ago2-binding sites, could bind Ago2 (Fig. 5A–C). In contrast, the fragments 1–619, 668–869, and 929–1087 did not interact with Ago2, or exhibited only weak binding, as compared with the 599–673 and 857–937 fragments (Fig. 5A,B,D). These results indicated that at least two distinct Ago2-binding sites, which have high similarity to the first and second Ago2-binding sites of GW182, exist in TNRC6B.
FIGURE 5.
Two Ago2-binding sites exist in the amino-terminal half of TNRC6B. (A,B,D) SBP pull-down assays of Ago2 and TNRC6B. HEK293F cells were cotransfected with FLAG/SBP or FLAG-tagged Ago2 and the TNRC6B expression vectors. (C) HEK293F cells were cotransfected with FLAG-Ago2 and FLAG-EGFP-TNRC6B or FLAG-EGFP expression vectors as a negative control. (E,F) HEK293F cells were cotransfected with FLAG-Ago2 and wild-type or mutant FLAG/SBP-TNRC6B expression vectors. SBP-pull down assays and Western blotting were performed as shown in Figure 1C.
Next, we carried out SBP pull-down assays using amino acid substitution variants of 599–673 and 857–937. The substituted tryptophan residues in TNRC6B fragments correspond to the conserved amino acids substituted in GW182 (Fig. 3A; Supplemental Fig. 1). In the fragment 599–673, W623A abolished the Ago2 binding and W634A scarcely interacted with Ago2, whereas W653A had no effect (Fig. 5E). In the fragment 857–937, Trp896 was essential for the Ago2 binding (Fig. 5F, W896A). W875A had some effect on the binding, but was not crucial (Fig. 5F, W875A).
These results indicate that the amino-terminal halves of the GW182 family proteins contain multiple Ago2-binding sites, and in these regions, the highly conserved tryptophan residues are important for the interaction with Ago2.
Each binding site cannot simultaneously bind Ago2
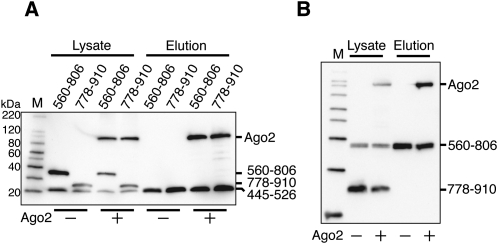
To investigate whether the three binding sites simultaneously bind to Ago2, fragments of GW182 polypeptides including the individual binding sites were coexpressed in HEK293F cells with or without Ago2, and SBP pull-down assays were performed. As a result, FLAG/SBP-GW182 (445–526) precipitated FLAG-Ago2, but neither FLAG-GW182 (560–806) nor FLAG-GW182 (778–910) (Fig. 6A). Likewise, FLAG/SBP-GW182 (560–806) precipitated FLAG-Ago2, but not FLAG-GW182 (778–910) (Fig. 6B). These data suggested that Ago2 binds to one of the three binding sites in GW182 and cannot associate with multiple GW182 molecules.
FIGURE 6.
Each Ago2-binding domain of GW182 binds to an overlapping region of Ago2. (A) HEK293F cells were cotransfected with the FLAG/SBP-GW182 445–526, FLAG-GW182 560–806, or 778–910 expression vector, with or without the FLAG–Ago2 expression vector. (B) HEK293F cells were cotransfected with the FLAG/SBP-GW182 560–806 and FLAG-GW182 778–910 expression vectors, with or without the FLAG-Ago2 expression vector. SBP pull-down assays and Western blotting were performed as described in Figure 1C, in the presence of RNaseA.
GW182 can associate with multiple Argonaute proteins
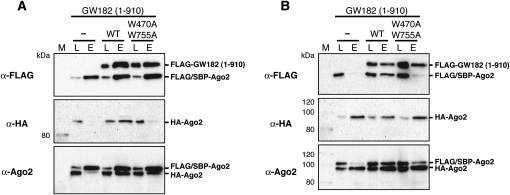
The presence of three Ago-binding sites in GW182 raises the possibility that GW182 could associate with multiple Ago proteins. To verify this, FLAG/SBP-Ago2 and HA-tagged Ago2 (HA-Ago2) were coexpressed in HEK293F cells in the presence or absence of FLAG-GW182 (1–910) and SBP pull-down and coimmunoprecipitation assays with anti-HA agarose were performed. As shown in Figure 7A, HA-Ago2 was recovered with FLAG/SBP-Ago2 in the presence of wild-type (WT) GW182 (1–910), which contains all three binding sites for Ago2 (Fig. 7A, WT). In the absence of GW182 (1–910), HA-Ago2 was slightly recovered (Fig. 7A, “−”). This was probably because of the effect of endogenous TNRC6 proteins. Unexpectedly, HA-Ago2 was recovered even with the W470A/W755A double mutant GW182 (1–910), which contains only one Ago2-binding site, though the amount was smaller than that of the wild type (Fig. 7A, W470A W755A). The recovery of HA-Ago2 may be due to the effect of endogenous TNRC6 proteins, and the weak binding activity of the mutant protein. Results in accord with the SBP pull-down assays were obtained by anti-HA immunoprecipitation. FLAG/SBP-Ago2 was detected in the presence of the wild-type GW182 fragment (Fig. 7B, WT). A weaker signal was detected in the presence of the mutant form of the GW182 fragment (Fig. 7B, W470A W755A). Also, a slight band was detected in the absence of GW182 fragments (Fig. 7B, “−”). Thus, these results strongly suggest that GW182 could associate with multiple Argonaute proteins.
FIGURE 7.
GW182 can associate with multiple Ago2 proteins. FLAG/SBP-Ago2 and HA-Ago2 were coexpressed with or without FLAG-GW182 1–910 or FLAG-GW182 1–910 (W470A and W755A) in HEK293F cells. (A) SBP pull-down assays were performed, and the precipitates were analyzed by Western blotting using an anti-FLAG M2 antibody (top), an anti-HA antibody (middle), and an anti-hAgo2 antibody (bottom). (Lane L) Cell lysate (1.25 μL equivalent); (lane E) fraction eluted with biotin (300 μL of cell lysate was subjected to the assay and 14% of the eluted fraction was loaded). (B) HA-Ago2 protein was precipitated with anti-HA agarose beads, and the precipitates were analyzed by an anti-FLAG M2 (top), an anti-HA (middle), and an anti-hAgo2 (bottom) Western blotting. (Lane L) Cell lysate (1.25 μL equivalent); (lane E) fraction eluted with 100 μg/mL of HA peptide solution. (300 μL of cell lysate was subjected to the assay and 14% of the eluted fraction was loaded).
The interaction between GW182 and Argonaute is important in the miRNA-mediated repression pathway
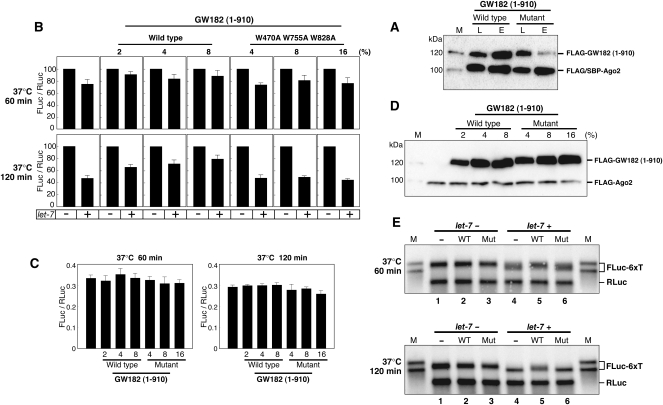
We previously reported a mammalian cell-free system that recapitulates let-7-miRNA-mediated translational repression (Wakiyama et al. 2007). This system is composed of cell extracts prepared from HEK293F cells and HEK293F cells overexpressing Ago2 and GW182. In this system, Ago2 overexpression was required for the efficient processing of externally added let-7 pre-miRNAs. The cell extracts containing overexpressed Ago2 exhibited ∼50% miRNA-mediated repression of the target mRNA, depending on the addition of let-7 pre-miRNA. Furthermore, the addition of cell extracts containing overexpressed GW182 enhanced miRNA-mediated repression up to 70%. In the present work, we used a modified cell-free system composed of diluted cell extracts prepared from HEK293F cells and HEK293F cells overexpressing Ago2. This system exhibited ∼60% repression with let-7. In order to elucidate whether GW182 binding to Ago2 is required for the miRNA-mediated translational repression in this cell-free system, we tested the effect of the addition of the wild-type GW182 (1–910) expressed extract and a triple-mutant GW182 (1–910) extract (W470A W755A W828A), on the synthesis of firefly luciferase. The affinity of the GW182 (1–910) fragment for Ago2 was greatly reduced by introducing triple mutations (Fig. 8A). The translation of capped and polyadenylated FLuc-6xT mRNA (firefly luciferase mRNA containing six let-7 target sites) was examined in the absence (−) and the presence (+) of let-7 miRNA. Capped and polyadenylated RLuc (Renilla luciferase) mRNA was also translated to normalize the activities of FLuc in different experiments.
FIGURE 8.
Let-7-mediated translational repression is inhibited by a GW182 fragment containing an Ago2-binding site. (A) FLAG/SBP-Ago2 was coexpressed with FLAG-GW182 (1–910) or FLAG-GW182 (1–910, W470A, W755A, W828A) in HEK293F cells. SBP pull-down assays were performed, as described in the Figure 7A legend, and the precipitates were analyzed by Western blotting using an anti-FLAG antibody. Lane L, cell lysate; lane E, fraction eluted with biotin. (B) In vitro translation experiments were performed with HEK293F extracts (containing 10% [v/v] Ago2 extract) along with 2%, 4%, and 8% (v/v) of the wild-type, or 4%, 8%, and 16% (v/v) of the mutant FLAG-GW182 (1–910) extract. Capped FLuc-6xT with a poly(A) tail mRNA was translated in the absence (−) or presence (+) of let-7. The FLuc and RLuc activities were measured, and the FLuc-to-RLuc activity ratio in the reaction without let-7 was set at 100. The data shown constitute an average of three independent experiments, with standard deviations. (C) Comparison of the translation efficiencies of the extracts with different ratios of FLAG-GW182 (1–910) extract in the absence of let-7. The average number of light units produced by the translation of FLuc mRNAs in the absence of FLAG-GW182 (1–910) lysate was ∼2,200,000. (D) Expression of FLAG-Ago2 and wild-type or mutant FLAG-GW182 (1–910) in the cell extract mixture using the translation experiment. The expressed proteins were detected by Western blotting using an anti-FLAG M2 antibody. (E) Analysis of capped and polyadenylated FLuc-6xT and RLuc mRNAs by Northern blotting with an FLuc probe and an RLuc probe. Translation reactions of mRNAs were performed with the cell extract mixture containing 8% (v/v) of the wild-type GW182 (1–910) and 16% (v/v) of the mutant GW182 (1–910). As molecular markers, nonadenylated (shorter) and polyadenylated (longer) transcripts were loaded in lane M. After 60 min (upper panel) and 120 min (lower panel) of translation, the RNAs were extracted from the reaction mixture, and similar amounts were loaded in each lane.
In the absence of the GW182 (1–910) extract, the repression was ∼25% and ∼60% after 60 min and 120 min incubations, respectively, in the cell extract composed of the HEK293F and Ago2 extracts (Fig. 8A). Since the expression level of the GW182 (1–910) mutant was lower than that of the wild type (Fig. 8C), we added twice the volume of the mutated GW182 (1–910) extract to the reaction mixtures. At the 60-min time point, we observed ∼25% repression in the absence of the GW182 fragment, or in the presence of the GW182 (1–910) mutant (W470A W755A W828A) extract, and ∼15% repression in the presence of the wild-type GW182 (1–910) extract (Fig. 8A). The addition of the wild-type extract to the translation reaction mixture relieved the translational repression to ∼25% after a 120-min incubation (Fig. 8A, wild type). On the other hand, the GW182 mutant had no apparent effect on the repression (Fig. 8A, W470A W755A W828A). As shown in Figure 8B, the addition of the GW182 (1–910) extract had minimal influence on the translational efficiencies in the absence of let-7. These data suggested that the interaction between GW182 and Ago2 is important in the miRNA-mediated repression of protein synthesis.
Target mRNA deadenylation is coincident with miRNA-mediated translational repression
It has been shown that miRNAs direct the deadenylation of their target mRNAs (Behm-Ansmant et al. 2006; Giraldez et al. 2006; Mishima et al. 2006; Wu et al. 2006; Eulalio et al. 2007b). We previously reported the correlation between mRNA deadenylation and translational repression in a mammalian cell-free system (Wakiyama et al. 2007). However, the mechanisms of miRNA-mediated translational repression remain controversial. Hence, we investigated whether the target mRNAs were deadenylated under conditions where miRNA-mediated repression was inhibited.
The mRNAs were isolated after 60- and 120-min incubations in the absence or presence of the wild-type or mutant GW182 (1–910) extract, and were analyzed by Northern blotting. In order to detect a change in the poly(A) length, we used a different lot of FLuc6xT mRNAs, which have a longer poly(A) tail as compared with that of the mRNAs used in the experiments shown in Figure 8, A and B. The repression with this target mRNA after a 120-min incubation, at the time point when the mRNAs were extracted, was 47% in the absence of GW182 (1–910), 27% in the presence of the wild type, and 55% in the presence of the variant (data not shown).
Northern blotting revealed that mRNA deadenylation was not completed, even after a 120-min incubation in the presence of GW182 (1–910) wild type (Fig. 8D, 120 min, lane 5), while the target mRNAs were deadenylated to nearly the same length as the nonpolyadenylated mRNAs in the absence of the GW182 fragment or the presence of the mutant form of the GW182 fragment (Fig. 8D, 120 min, lanes 4,6). Although the deadenylation of Luc6xT mRNAs proceeded in each sample in the presence of let-7 miRNAs at 60 min, it seemed to be delayed by the addition of the wild-type GW182 (1–910) extract (Fig. 8D, 60 min, lanes 4–6). The translational repression was 21%, 5%, and 31% with GW182 (1–910), the wild-type GW182 (1–910), and the mutant GW182 (1–910), respectively. Although the mRNA levels in the presence of let-7 were reduced, the reduction was not as dramatic as compared with the control RLuc mRNAs. Therefore, these results suggest that miRNA-mediated mRNA deadenylation is very closely correlated with translational repression. Furthermore, the inhibition of miRNA-mediated deadenylation by the GW182 fragment suggests that GW182 is involved in this step and may have an important role.
DISCUSSION
Two recent studies proposed that the conserved amino-terminal GW/WG repeated domain is crucial for Ago2 binding. El-Shami et al. (2007) reported that the reiterated WG/GW motifs, which also exist in Arabidopsis NRPD1b, of GW182 are the conserved Argonaute-binding platform. Till et al. (2007) reported that the Ago hook of TNRC6B isoform2 is sufficient for the interaction with Ago2, and that the conserved tryptophan residues in this motif are important.
In the present work, we demonstrated for the first time that GW182 contains multiple Ago-binding sites. The sequence of the first binding site, which is located at the extreme amino-terminal region, is novel, and it is conserved in TNRC6B, TNRC6C, and Drosophila GW182, and possibly in C.elegans AIN-1. The second and third binding sites contain almost identical sequences. We found identical or similar sequences in TNRC6B, TNRC6B isoform2, TNRC6C, Drosophila GW182, and C. elegans AIN-1. Although the number of potential binding sites differs among the paralogs and orthologs, it seems that all of the GW182 family members analyzed thus far contain two to three Ago-binding sites. Each of the Ago-binding sites is alone sufficient to bind Ago2. Site-directed mutagenesis revealed that the tryptophan residues conserved among GW182, TNRC6B, TNRC6C, Drosophila GW182, and AIN-1 are important for Ago2 binding, at least in the case of the GW182 fragment used in this study.
We demonstrated that a fragment containing one of the three Ago-binding sites can bind Ago2, but multiple fragments cannot simultaneously bind to Ago2. Most importantly, we demonstrated that GW182 is able to associate with multiple Ago2 proteins. MiRNA-mediated translational repression is generally detected with target mRNAs containing multiple miRNA target sites in their 3′-UTRs. Thus, the binding of multiple Argonaute proteins to the target mRNAs may facilitate the recruitment of GW182, which may cause strong repression of the target mRNA. Alternatively, it is possible that GW182 connects mRNA molecules by associating with multiple Argonaute proteins bound to different mRNA molecules, to form large complexes of mRNA and proteins. This is consistent with the observation reported by Thermann and Hentze (2007), who observed the miRNA-induced formation of a pseudo-polysome, heavy mRNP, in a Drosophila cell-free system.
In this study, we showed that the addition of the GW182 (1–910) fragment containing Ago2-binding sites into a mammalian cell-free system partially relieved the miRNA-mediated translational repression. In agreement with our results, it was previously reported that in HeLa cells, the expression of the amino-terminal fragment of GW182 (1–498), containing the first Ago-binding site we found, interfered with the assembly of P-bodies and the small RNA-mediated gene-silencing pathway (Jakymiw et al. 2005). In addition, it was recently shown that the overexpression of the amino-terminal domain of Drosophila GW182 (1–592), and the addition of the Ago hook peptide inhibited the miRNA pathway in Drosophila cells and in a Drosophila cell-free system, respectively (Till et al. 2007; Eulalio et al. 2008b).
Several models of the mechanisms of miRNA-mediated translational repression have been proposed, including repression at translation initiation (Humphreys et al. 2005; Pillai et al. 2005; Chendrimada et al. 2007; Mathonnet et al. 2007; Thermann and Hentze 2007), repression at post-translation initiation (Maroney et al. 2006; Nottrott et al. 2006; Petersen et al. 2006), and target mRNA deadenylation and degradation (Behm-Ansmant et al. 2006; Giraldez et al. 2006; Wu et al. 2006; Eulalio et al. 2007b; Wakiyama et al. 2007). We demonstrated here that the reduction of protein synthesis is coincident with mRNA deadenylation. However, the deadenylation of the target mRNAs was not fully inhibited in this study. Therefore, it is not clear whether miRNA-mediated translational repression occurs, when mRNA deadenylation is completely inhibited.
In conclusion, we propose that the interactions of GW182 with Argonautes may provide a structural platform for the formation of large complexes of proteins and miRNA target mRNAs, and may have a crucial role in the miRNA-mediated translational repression.
MATERIALS AND METHODS
Plasmid construction
The plasmids pCMV-SPORT/FLAG-Ago2, pCMV-SPORT/FLAG-GW182, pSP36T7-Luc-6xT, and pSP36T7-RLuc were described previously (Wakiyama et al. 2007). The pOripCAG vector was created by replacing the CMV promoter in the pOriP vector (Mukai et al. 2008) with the CAG promoter derived from pAxCAwtit (TaKaRa). To create pOripCAG-FLAG/SBP-GW182 and pOripCAG-FLAG-GW182, the DNA fragment encoding full-length GW182 was inserted into the BamHI-XhoI sites of the pOripCAG vector with a FLAG-tag and a TEV protease digestion site or a FLAG-tag, a streptavidin-binding peptide tag (SBP-tag), and a TEV protease digestion site fused to the N terminus. The GW182 fragments, 1–910, 1–806, 1–526, 1–486, 1–443, 445–806, 483–806, 560–806, 445–685, 483–685, 560–685, 445–526, 716–806, 778–910, 806–910, and 920–1709, were amplified from pCMV-SPORT/FLAG-GW182 or pOripCAG-FLAG/SBP-GW182 fragments using PrimeStar HS DNA Polymerase (TaKaRa), and were cloned into the BamHI-XhoI sites of pOripCAG-FLAG/SBP. pOripCAG-FLAG/SBP-GW182 1–910, 560–806, 778–910, and 920–1709 were excised and cloned into the BamHI-XhoI sites of pOripCAG-FLAG vector, to create pOripCAG-FLAG-GW182 1–910, 560–806, 778–910, and 920–1709, respectively. Point mutants in GW182 were generated from pOripCAG-FLAG/SBP-GW182 445–526, 716–806, 778–910, and 1–910, respectively, using PCR. The wild-type and mutated GW182 445–526, 716–806, and 778–910 fragments were excised from pOripCAG-FLAG/SBP and inserted into the BamHI-XhoI sites of the pGEX-6P-2 vector (GE Healthcare) to create plasmids expressing GW182 fragments with a glutathione-S-transferase tag (GST-tag) fused to the N terminus. The clone for the full-length TNRC6B (KIAA1093) was obtained from the Kazusa DNA Research Institute. The cDNAs were cloned into the pCMV-SPORT/FLAG vector (Wakiyama et al. 2007). The TNRC6B fragments 1–619, 599–673, 668–869, 857–937, and 929–1087 were amplified from pCMV-SPORT/FLAG-TNRC6B and were cloned into the BamHI-XhoI or BamHI-NotI sites of pOripCAG-FLAG/SBP and pOripCAG-FLAG. To create pOripCAG-FLAG/EGFP, the DNA fragment encoding EGFP was inserted into the BamHI-XhoI sites of the pOripCAG-FLAG vector. The pOripCAG-FLAG-TNRC6B 599–673 and 857–937 vectors were digested at the KpnI-BamHI sites, and the DNA fragment encoding FLAG/EGFP was inserted into these sites. The resulting plasmids are designated as pOripCAG-FLAG/EGFP-TNRC6B 599–673 and 857–937. Point mutants in TNRC6B were generated from pOripCAG-FLAG/SBP-TNRC6B 599–673 and 857–937, respectively, using PCR. To create pOripCAG-FLAG/SBP-Ago2 and pCMV-SPORT/HA-Ago2, the DNA fragment encoding full-length Ago2 was inserted into the BamHI-NotI sites of the pOripCAG-FLAG/SBP or the BamHI-XbaI sites of the pCMV-SPORT vector with a hemaglutinin tag (HA-tag).
RNA experiments
Chemically synthesized let-7 pre-miRNA was obtained from DHARMACON. The sequence of the let-7 pre-miRNA used in this work is as follows: 5′-p-UGAGGUAGUAGGUUGUAUAGUAGUAAUUACACAUCAUACUAUACAAUGUGCUAGCUUUCU-3′. The mRNAs encoding Fluc and RLuc were transcribed in vitro from the DNA template, as described previously (Wakiyama et al. 2007). Purification of RNAs from reaction mixtures and Northern blotting were performed as described previously (Wakiyama et al. 2007).
Cell culture, transfection, and cell extract preparation
HEK293F cells (FreeStyle 293F cell, Invitrogen) were grown in suspension as described previously (Wakiyama et al. 2007). The plasmids (1 μg) were transfected into 1 × 106 cells/mL of FreeStyle 293 cells by the use of 293fectin (Invitrogen). Cell extracts from HEK293F cells and HEK293F cells overexpressing FLAG-Ago2 were prepared as previously described (Wakiyama et al. 2007).
SBP pull-down and coimmunoprecipiation
HEK293F cells were harvested 48 h after transfection, lysed in binding buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% [w/v] Nonidet-P40, and protease inhibitor cocktail [Roche]), and centrifuged at 16,000g for 5 min at 4°C. Figures 1–3 and 5: The cell lysates were incubated with streptavidin agarose beads (Novagen) for 1 h at 4°C. After this incubation, the lysates containing the streptavidin agarose beads were partitioned into two equal volumes, and then were incubated for 1.5 h at 4°C in the presence or absence of 100 μg/mL RNaseA (Nakalai Tesque). Figures 6, 7, and 8A: The cell lysates were incubated with streptavidin agarose beads for 2 h at 4°C with 100 μg/mL RNaseA. After this incubation, the beads were washed three times with wash buffer (50 mM Tris-HCl at pH 7.4, 300 mM NaCl, 5 mM MgCl2), and then the binding proteins were eluted by 2 mM D-biotin and subjected to Western blotting analysis. As for the coimmunoprecipiation with the anti-HA antibody, the cell lysates were incubated with anti-HA agarose beads (Sigma) for 2 h at 4°C with 100 μg/mL RNaseA. After this incubation, the beads were washed three times with wash buffer (50 mM Tris-HCl at pH 7.4, 300 mM NaCl, 5 mM MgCl2), and then the binding proteins were eluted by 100 μg/mL of HA peptide (Sigma) and subjected to Western blotting analysis. Western blotting was performed with an anti-FLAG M2 monoclonal antibody (1:4000) (Sigma), an anti-HA monoclonal antibody (1:1000) (Roche), or an anti-human Ago2 monoclonal antibody (1:1000) (Wako). Primary antibodies were detected with anti-mouse IgG Horseradish Peroxidase-linked whole antibody (GE Healthcare).
GST pull-down assay
GST pull-down assays were performed using a MagneGST pull-down system (Promega). GST fusion proteins were expressed in Escherichia coli BL21 (TakaRa), induced with 1 mM IPTG at 37°C for 3 h. The cells were harvested and suspended in PBS (Nacalai Tesque) containing protease inhibitor cocktail (Roche). The suspension was sonicated and centrifuged. The supernatant was incubated with the MagneGST particles at 4°C for 0.5 h. The particles were washed three times with PBS containing protease inhibitor. The MagneGST particles were suspended in 295 μL of binding buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10% [w/v] glycerol, 1% (w/v) NP-40, protease inhibitor cocktail), mixed with 5 μL of FLAG-tagged Ago2-overexpressing HEK293F cell extracts, and incubated at 4°C for 2 h. After this incubation, the particles were captured with a magnet and were washed three times with binding buffer. The binding fraction was eluted in elution buffer (Promega). The FLAG-Ago2 protein in the eluted fraction was detected by Western blotting analysis, using an anti-FLAG M2 antibody (Sigma).
In vitro translation assay
HEK293F cell extracts, FLAG-Ago2, and wild-type or mutant FLAG-GW182 (1–910)-overexpressing cell extracts were mixed. The mixed cell extracts were preincubated with or without let-7 pre-miRNA (0.2 pmol/μL) for 60 min at 37°C, and then cooled on ice. The cell extracts were diluted 1/4 (v/v) with lysis solution containing 4 mg/mL creatine kinase (Roche). Translation reactions were carried out in a final volume of 20–40 μL containing a 15% volume of the preincubated cell extracts, 30 mM Hepes-KOH (pH 7.4), 8 mM creatine phosphate, 0.5 mM spermidine, 1 mM ATP, 0.2 mM GTP, 20 μM amino acids, 1.5 mM magnesium acetate, 80 mM potassium acetate, 1 ng/μL FLuc mRNA, and 1 ng/μL RLuc mRNA, for 60 or 120 min at 37°C. After the translation reaction, the reaction mixture was cooled on ice, and then diluted fivefold with ice-cold Passive Lysis Buffer (Promega). The luciferase activities were analyzed in a MiniLumat LB9506 (Berthold) with the Dual-Luciferase Reporter Assay System (Promega).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Dr. Osamu Ohara (Kazusa DNA Research Institute, Japan) for the TNRC6B clone (KIAA1093), Dr. Nobumasa Hino for the pOriP vector, and Seisuke Yamashita for creating the pOripCAG vector. This work was supported by the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1363109.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Ding L., Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S.F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007b;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., 3rd, Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney P.A., Yu Y., Fisher J., Nilsen T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A.J., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Kobayashi T., Hino N., Yanagisawa T., Sakamoto K., Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P., Jopling C.L., Norman K.L., Schutz S., Wehner K.A. MicroRNAs: Expression, avoidance, and subversion by vertebrate viruses. Nat. Rev. Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- Standart N., Jackson R.J. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes & Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- Thermann R., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Takimoto K., Ohara O., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Love T.M., Call M.E., Doench J.G., Novina C.D. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wilson D.S., Keefe A.D., Szostak J.W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., Liu X., Yates J.R., 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]