Abstract
Although the interconversion between C4 and C3 compounds has an important role in overall metabolism, limited information is available on the properties and regulation of enzymes acting on these metabolites in hyperthermophilic archaea. Malic enzyme is one of the enzymes involved in this interconversion, catalyzing the oxidative decarboxylation of malate to pyruvate as well as the reductive carboxylation coupled with NAD(P)H. This study focused on the enzymatic properties and expression profile of an uncharacterized homolog of malic enzyme identified in the genome of a heterotrophic, hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 (Tk-Mae). The amino acid sequence of Tk-Mae was 52–58% identical to those of malic enzymes from bacteria, whereas the similarities to the eukaryotic homologs were lower. Several catalytically important regions and residues were conserved in the primary structure of Tk-Mae. The recombinant protein, which formed a homodimer, exhibited thermostable malic enzyme activity with strict divalent cation dependency. The enzyme preferred NADP+ rather than NAD+, but did not catalyze the decarboxylation of oxaloacetate, unlike the usual NADP-dependent malic enzymes. The apparent Michaelis constant (Km) of Tk-Mae for malate (16.9 mM) was much larger than those of known enzymes, leading to no strong preference for the reaction direction. Transcription of the gene encoding Tk-Mae and intracellular malic enzyme activity in T. kodakaraensis were constitutively weak, regardless of the growth substrates. Possible roles of Tk-Mae are discussed based on these results and the metabolic pathways of T. kodakaraensis deduced from the genome sequence.
Keywords: carbon metabolism, hyperthermophile, malate, pyruvate, tricarboxylic acid cycle
Introduction
Phosphoenolpyruvate (PEP), pyruvate and oxaloacetate (OAA) are key metabolites in central carbon and energy metabolism. In general, interconversion between C4 and C3 metabolites is important in maintaining the levels of intermediates in the tricarboxylic acid (TCA) cycle. When TCA cycle metabolites are consumed, OAA can be replenished from PEP or pyruvate, or both. When these metabolites are in excess, this link can be utilized in the opposite direction to remove and direct the carbon flux toward other pathways such as gluconeogenesis (Owen et al. 2002). The enzymes catalyzing these transformations have been well-investigated in a wide variety of organisms, but limited information is available for the enzymes in hyperthermophilic archaea.
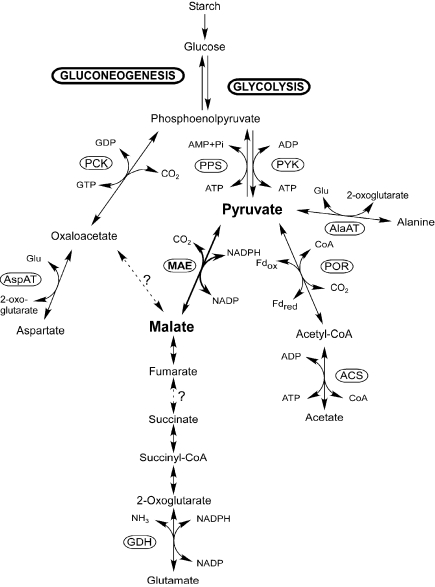
Thermococcus kodakaraensis KOD1 is a sulfur-reducing hyperthermophilic archaeon isolated from a hot spring on Kodakara Island, Kagoshima, Japan (Morikawa et al. 1994, Imanaka and Atomi 2002, Atomi et al. 2004). This archaeon prefers amino acids as carbon and energy sources in the presence of elemental sulfur; in the absence of sulfur, it can assimilate and grow on starch or pyruvate. A variant of the Embden-Meyerhof pathway is present in Thermococcus and the closely related Pyrococcus spp. (Verhees et al. 2003). In addition, recent whole genome analysis of T. kodakaraensis (Fukui et al., unpublished results) identified enzymes that may be involved in the metabolism of C4 and C3 intermediates, namely, pyruvate kinase, PEP synthase, PEP carboxykinase (PCK) and malic enzyme (Figure 1). However, obvious homologs for both PEP carboxylase and pyruvate carboxylase are missing. These enzymes are major anaplerotic enzymes in many organisms. The genome analysis also revealed that the TCA cycle is incomplete because of the absence of enzymes for many reactions. We have recently reported the first characterization and expression profile of an archaeal PCK from T. kodakaraensis (Tk-Pck) (Fukuda et al. 2004). The characteristics of the recombinant protein and expression profile of Tk-Pck were consistent with a role in providing PEP from OAA as the first step of gluconeogenesis. Additionally, it was suggested that the enzyme recycled excess PEP produced in the presence of abundant pyruvate.
Figure 1.
Predicted pyruvate metabolism in Thermococcus kodakaraensis KOD1. Abbreviations: ACS = acetyl-CoA synthetase; AlaAT = alanine aminotransferase; AspAT = aspartate aminotransferase; Fd = ferredoxin; GDH = glutamate dehydrogenase; MAE = malic enzyme; PCK = phosphoenolpyruvate carboxykinase; POR = pyruvate:ferredoxin oxidoreductase; PPS = phosphoenolpyruvate synthase; and PYK = pyruvate kinase.
Malic enzyme is another enzyme capable of interconverting C4 and C3 compounds. This enzyme catalyzes the NADP(H)-dependent oxidative decarboxylation of malate to pyruvate and the reductive carboxylation of pyruvate. It is a ubiquitous enzyme and is widely distributed in both eukaryotes and prokaryotes. Malic enzymes are highly conserved and divided into three groups based on their coenzyme requirements and their ability to decarboxylate OAA. The first class of malic enzymes (EC 1.1.1.38) utilizes NAD(H) and can decarboxylate oxaloacetate. The second class (EC 1.1.1.39) prefers NAD(H) rather than NADP(H), and is unable to decarboxylate OAA. The third class (EC 1.1.1.40) is NADP(H)-dependent and able to catalyze the decarboxylation of OAA. It has generally been assumed that NADP-dependent malic enzymes provide the reducing power necessary for biosynthesis of various cellular components, while NAD-dependent enzymes promote the entrance of carbon derived from amino acids or C4-dicarboxylates into gluconeogenesis. Although the reductive formation of malate is thermodynamically favored (–7.3 kJ mol–1) (Thauer et al. 1977, Goldberg et al. 1993), many malic enzymes kinetically prefer the oxidative direction. However, malic enzymes from different sources or organelles often show different characteristics and functions. With regard to malic enzyme from archaea, an NADP+-dependent enzyme from a thermoacidophilic crenarchaeon Sulfolobus solfataricus has been studied (Bartolucci et al. 1987), but the physiological function of this enzyme has not been documented.
The present work focused on the homolog of malic enzyme identified in the T. kodakaraensis genome. The primary structure, enzymatic characterization and expression profile were investigated to gain insight into the role of this universal enzyme in the hyperthermophilic archaeon.
Materials and methods
Microorganisms, plasmids and media
Thermococcus kodakaraensis KOD1 cells were grown anaerobically at 85 °C in MA-YT medium consisting of 4.8 and 26.4 g l–1 of Marine Art SF agents A and B, respectively (Senjyu Seiyaku, Osaka, Japan), 5.0 g l–1 of yeast extract and 5.0 g l–1 of tryptone supplemented with 5.0 g l–1 of elemental sulfur. For fermentative growth, 5.0 g l–1 of pyruvate or 5.0 g l–1 of starch was added in place of elemental sulfur. Escherichia coli DH5α and plasmid pUC118 were used for general DNA manipulation and sequencing, and E. coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) and pET21a(+) (Novagen, Madison, WI) was used for gene expression. Escherichia coli strains were cultivated in Luria-Bertani (LB) medium (10 g l–1 of tryptone, 5 g l–1 of yeast extract and 10 g l–1 of NaCl (pH 7.0)) at 37 °C. Ampicillin was added to the medium at a concentration of 100 µg ml–1 when needed.
DNA manipulation and sequencing
We performed DNA manipulations by standard methods as described by Sambrook and Russell (2001). Restriction and modification enzymes were purchased from Toyobo (Osaka, Japan) or Takara Bio (Otsu, Japan). Small-scale preparations of plasmid DNA were performed with a Qiagen Plasmid Mini Kit (Qiagen, Hilden, Germany). The polymerase for PCR was KOD Plus (Toyobo), and DNA fragments were recovered with a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Little Chalfont, U.K.) from agarose gels after electrophoresis. We performed DNA sequencing with a BigDye Terminator Cycle Sequencing Kit v.3.0 and a Model 3100 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Alignment and phylogenetic analyses of malic enzyme sequences
An unrooted phylogenetic tree was constructed with the homologous core regions (residues 9 to 382 in Tk-Mae) in malic enzymes from various sources with the neighbor-joining method using the ClustalW program provided by the DNA Data Bank of Japan (DDBJ). Calculations were performed under the following conditions: ENDGAPS (End gap separation) and NOPGAPS (Pascarella gaps off) were on, and NOHGAPS (hydrophilic gaps off) and Kimura’s correction were off. Other parameters were default values. Bootstrap resampling was performed 1000 times with the BSTRAP program. Alignment analysis between malic enzymes from T. kodakaraensis and pigeon was performed under the same conditions as phylogenetic tree construction.
Expression of the malic enzyme gene from T. kodakaraensis KOD1 (Tk-mae)
The expression plasmid for Tk-mae was constructed as follows. The gene was amplified with T. kodakaraensis genomic DNA as a template and two oligonucleotide primers (sense, 5′-AAAAAAGGATCCCATATGAATCCCCTCGAATTCCATAGGG-3′, and antisense, 5′-AAAAAAGGATCCCTAGGG GGAACTCCCTCTACCGCTGAA-3′; underlined sequences indicate NdeI and BamHI sites in the sense and antisense primers, respectively). The amplified fragment was inserted into pUC118. After confirming the absence of unintended mutations in the amplified region, the NdeI–BamHI restriction fragment was inserted into pET21a(+) at the corresponding sites, and the resulting plasmid, pET-mae, was used to transform E. coli BL21-CodonPlus(DE3)-RIL. The recombinant cells were grown in LB medium at 37 °C, and gene expression was induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside at the mid-exponential growth phase with further incubation for 4 h at 37 °C. The cells were harvested by centrifugation (5000 g, 4 °C, 20 min), resuspended in 50 mM Tris-HCl buffer (pH 8.0) and sonicated for 7 min (output:pause = 10:20 s; output power = about 80 W) with a Misonix Sonicator Ultrasonic Cell Processor XL2020 (Misonix, Farmingdale, NY). The lysate was clarified by centrifugation (17,000 g, 4 °C, 15 min). The soluble cell extract was heat-precipitated for 20 min at 80 °C to remove heat-labile proteins derived from the host. The supernatant was applied to an anion exchange column Resource Q (6 ml; Amersham Biosciences), and the recombinant protein was eluted with a linear gradient of NaCl (400 to 500 mM) in 50 mM Tris-HCl buffer (pH 8.0). The resulting solution was applied to a gel filtration column Superdex 200 HR 10/30 (Amersham Biosciences) with a mobile phase of 50 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl at a flow rate of 0.25 ml min–1. The native molecular mass was calibrated with standard proteins of thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (13.7 kDa). Protein concentration was determined with the Protein Assay Kit (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions, with bovine serum albumin as the standard. Determination of N-terminal amino acid sequences of proteins was performed by Biologica (Nagoya, Japan) after separation by SDS-PAGE and electroblotting onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA).
Malic enzyme assay
Oxidative decarboxylation of L-malate was determined by monitoring the increase in NADPH at 340 nm with a spectrophotometer (UV-1600, Shimadzu, Kyoto, Japan), equipped with an internal temperature regulator that covers the cell and maintains the temperature up to 99 °C. The standard reaction mixture was composed of 50 mM Tris-HCl buffer (pH 8.0), 5 mM MnCl2, 5 mM NADP+, 30 mM L-malate and 2.3 µg of enzyme at a final volume of 1 ml. The buffer and salts, comprising 94% of the total reaction mixture volume, were incubated at the respective assay temperature before adding the remaining components. One unit (U) of activity was defined as the amount of activity reducing 1 µmol of NADP+ per min. Reductive carboxylation of pyruvate was measured with similar procedures at 50 °C with a reaction mixture (0.5 ml) containing 50 mM MES-NaOH buffer (pH 6.0), 5 mM MnCl2, 0.05 mM NADPH, 12.5 mM NaHCO3, 1–60 mM pyruvate and 1.9 µg of enzyme. The reaction was followed by measuring the decrease in absorbance at 340 nm. Oxaloacetate decarboxylase activity was assayed by a continuous coupling reaction with lactate dehydrogenase. The reaction mixture (1 ml) was composed of 8 mM MnCl2, 0.1 mM NADH, 0.05 mM oxaloacetate, 50 U of rabbit muscle lactate dehydrogenase (Sigma, St. Louis, MO) and 0.4–1.7 µg of enzyme in 50 mM Tris-HCl buffer (pH 8.0). The reaction was initiated by addition of OAA, and the decrease in NADH accompanied by pyruvate reduction catalyzed by lactate dehydrogenase was determined spectrophotometrically at 50 °C. The thermal decomposition of OAA forming pyruvate in the absence of enzyme was subtracted from each measurement. The optimum pH for Tk-Mae was determined by measuring the oxidative decarboxylation of malate at 50 °C with 50 mM acetate-NaOH (pH 4.5 to 5.5), 50 mM MES-NaOH (pH 5.5 to 7.0), HEPES-NaOH (pH 7.0 to 8.0) or 50 mM Tris-HCl (pH 8.0 to 9.0). For measurement of malic enzyme activity in T. kodakaraensis KOD1 extract, the harvested cells were resuspended in 50 mM HEPES-NaOH buffer (pH 7.5) and sonicated under the same conditions described above for 5 min. The soluble cell extract after centrifugation (15,000 g, 4 °C, 15 min) was assayed, and measurements were performed at 60 °C. The reaction mixture (0.5 ml) was composed of 5 mM MnCl2, 30 mM L-malate, 0.25 mM thio-NADP+ (Oriental Yeast, Tokyo, Japan), 0.1 mM NADH, 50 U of rabbit muscle lactate dehydrogenase and various amounts of cell extract. The reduction of thio-NADP+ and oxidation of NADH were monitored at 398 and 340 nm, respectively. The pH values of all buffers were adjusted at each assay temperature.
RNA isolation and Northern blot analysis
Total RNA from T. kodakaraensis KOD1 was isolated from cells harvested at the early exponential growth phase (optical density at 660 nm ≈ 0.1–0.2) with RNeasy Midi Kit (Qiagen, Valencia, CA). Total RNA (15 µg) was denatured by heat treatment at 65 °C for 15 min, separated by 1% agarose gel electrophoresis and transferred onto a positively charged nylon membrane (Roche Diagnostics, Basel, Switzerland) by vacuum blotting. Labeling of a partial fragment of Tk-mae with digoxigenin (Roche Diagnostics), hybridization and detection of signals were performed according to the manufacturer’s instructions.
Nucleotide sequence accession number
The nucleotide sequence data reported here are available in the EMBL/GenBank/DDBJ nucleotide sequence databases under Accession No. AB195292.
Results
Primary structure of Tk-Mae
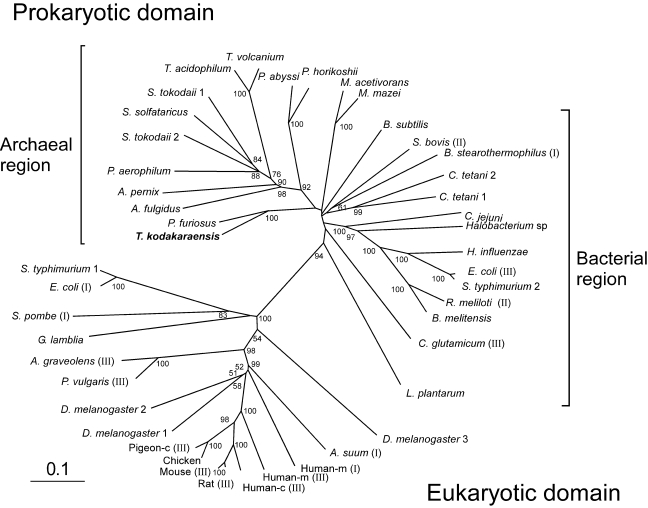
One open reading frame (ORF) on the T. kodakaraensis genome, TK1963, encoding a 425-amino-acid protein (45,380 Da), was found to share high amino acid sequence similarity to previously characterized bacterial malic enzymes from Bacillus stearothermophilus (NAD-dependent) (Kobayashi et al. 1989), Streptococcus bovis (NAD-dependent) (Kawai et al. 1996) and Corynebacterium glutamicum (NADP-dependent) (Gourdon et al. 2000). The percentages of identical residues were 58, 53 and 52%, respectively. We therefore designated the putative malic enzyme gene Tk-mae. Many archaeal genomes also contain malic enzyme homologs with high sequence similarity to Tk-Mae (45–80% identitical residues). However, homologs cannot be identified on the genomes ofmost methanogenic archaea, with the exception of two Methanosarcina species. The archaeal enzymes lack the N-terminal extension found in most eukaryotic enzymes (Boles et al. 1998, Yang et al. 2000, Coleman et al. 2002, Yang et al. 2002), as well as the C-terminal phosphotransacetylase domain present in a subset of bacterial enzymes (Mitsch et al. 1998). The Tk-Mae homolog and other archaeal homologs are more related to bacterial malic enzymes than eukaryotic malic enzymes. The percentage of identical amino acids of Tk-Mae was no greater than 27% when compared with malic enzymes from pigeon cytosol (NADP-dependent) (Yang et al. 2002), human mitochondria (NAD-dependent) (Yang et al. 2000) and Ascaris suum mitochondria (NAD-dependent) (Coleman et al. 2002). The topology of the unrooted phylogenetic tree, constructed with the homologous core regions in malic enzymes from various sources, reflects these data. A clear separation can be observed between eukaryotic enzymes and prokaryotic enzymes (Figure 2). Although bacterial and archaeal enzymes form clusters in this tree, many deep branches are present with low bootstrap values, suggesting that the distinction between the two groups is not necessarily clear-cut. The scattered distribution of the three classes of malic enzymes suggest that cofactor specificity and intracellular function do not provide a significant phylogenetic signal (Boles et al. 1998). The homologs in T. kodakaraensis and Pyrococcus furiosus comprise a branch distinct to that composed of the homologs from the closely related species P. abyssi and P. horikoshii, which may suggest a gain of the genes from a different lineage. This assumption is also supported by the observation that the malic enzyme gene in both P. abyssi and P. horikoshii is next to a probable malate dehydrogenase gene, which is lacking in T. kodakaraensis and P. furiosus.
Figure 2.
Radial phylogenetic tree of representative bacterial, eukaryotic and archaeal malic enzymes. Multiple sequence alignments and phylogenetic analysis were performed with the neighbor-joining method using the the ClustalW program provided by the DNA Data Bank of Japan (DDBJ). Scale bar indicates 10 substitutions per 100 amino acids. Bootstrap resampling was performed 1000 times and only values observed in more than 50% of the replicas are shown. The accession numbers of the sequences are as follows: Aeropyrum pernix, G72731; Apium graveolens (III), Q9SDL2; Archaeoglobus fulgidus, F69465; Ascaris suum (III), S29742; Bacillus stearothermophilus (I), P16468; Bacillus subtilis, O34962; Brucella melitensis, AI3372; Campylobacter jejuni, Q9PN12; chicken, AF408407; Clostridium tetani1, AE015937; C. tetani 2, AE015944; Corynebacterium glutamicum (III), CAF18947; Drosophila melanogaster 1, Q9U1J0; D. melanogaster 2, AAF43603; D. melanogaster 3, Q9U1J2; Escherichia coli (I), P26616; E. coli (III), P76558; Giardia lamblia, U59300; Haemophilus influenzae, P43837; Halobacterium sp. NRC-1, A84315;human cytosolic (Human-c) (III), P48163; human mitochondrial (Human-m) (I), P23368; Human-m (III), Q16798; Lactobacillus plantarum, Q88XT0; Methanosarcina acetivorans, AAM05142; Methanosarcina mazei, AAM32328; mouse (III), P06801; Phaseolus vulgaris (III), P12628; pigeon cytosolic (Pigeon-c) (III), P40927; Pyrobaculum aerophilum, AAL63657; Pyrococcus abyssi, G75131; Pyrococcus furiosus, AAL81150; Pyrococcus horikoshii, H71072; rat (III), P13697; Rhizobium meliloti (II), O30807; Salmonella typhimurium 1, AE008768; S. typhimurium 2, Q9ZFV8; Schizosaccharomyces pombe (I), P40375; Streptococcus bovis (II), U35659; Sulfolobus solfataricus, A90465; Sulfolobus tokodaii1, BAB65071; S. tokodaii2, BAB65246; Thermococcus kodakaraensis,AB195292;Thermoplasma acidophilum, Q9HKY7; Thermoplasma volcanium, BAB59923. Roman numerals in parentheses indicate the class of each malic enzyme previously characterized: I, EC 1.1.1.38; II, EC 1.1.1.39; and III, EC 1.1.1.40.
In the primary structures of malic enzymes, two highly conserved dinucleotide-binding signatures, GXGXXG/A, are present regardless of the cofactor specificities (Wierenga et al. 1986). The majority of archaeal homologs also possess the two signatures, such as 72-GLGDIG-77 and 188-GAGAAG-193 in Tk-Mae (Figure 3), whereas malic enzymes from Thermoplasma, Archaeoglobus and crenarchaeapossess GXGXXN as the second signature. Recently, the crystal structures of the NAD(P)-enzyme from human mitochondria (Yang et al. 2000) and an NADP-enzyme from pigeon cytosol (Yang et al. 2002) have been solved, and several important residues in binding to substrates and metals have been identified. Many of these residues are highly conserved in malic enzymes from a wide range of sources. For instance, Glu130, Asp131 and Asp156 in Tk-Mae appear to be conserved for binding of a divalent metal cation. The Lys88 corresponds to the Lys162 (numbered Lys183 in Yang et al. 2002) in the pigeon enzyme and may be an important residue for catalytic activity. Mutation of this residue in the pigeon enzyme resulted in a 230-fold loss in kcat (Kuo et al. 2000). In addition, the 2′-phosphate group of NADP+ interacts with Ser325 (numbered Ser346 in Yang et al. 2002) and the side chain amino group of Lys340 (numbered Lys362 in Yang et al. 2002) in the pigeon enzyme. These two residues are highly conserved among NADP-dependent malic enzymes (Yang et al. 2002, Chang and Tong 2003). In particular, a K340A mutation in the pigeon enzyme leads to a 70-fold increase in the apparent Michaelis constant (Km) for NADP, whereas other kinetic parameters were unaffected (Kuo et al. 2000). All archaeal enzymes were found to harbor a Lys (or an Arg) residue corresponding to Lys340. While Ser325 was also conserved in most archaeal enzymes, the residue was replaced by Arg in those from T. kodakaraensis (Arg213) and P. furiosus, and by Leu in those from P. abyssi and P. horikoshii.
Figure 3.
Alignment of malic enzyme sequences from T. kodakaraensis and pigeon (Columba livia). The sequences of NADP-dependent malic enzyme from T. kodakaraensis (Tk-Mae; AB195292) and pigeon (Cl-Mae; P40927) are shown. Symbols: # = amino acid residues referred to in the text; and ☆ = identical amino acid residues. Hyphens in the sequences represent gaps introduced to maximize alignment.
Purification and characterization of recombinant Tk-Mae
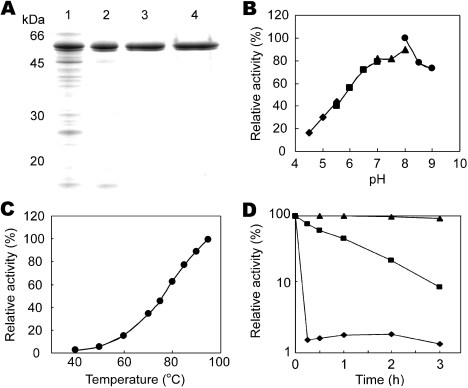
To characterize the malic enzyme from T. kodakaraensis, the Tk-mae gene was overexpressed in E. coli BL21-CodonPlus (DE3)-RIL under control of the T7 promoter. The recombinant protein, expressed in a soluble form, was purified by heat treatment followed by anion exchange chromatography. After SDS-PAGE analysis (Figure 4A), we observed two distinct bands with molecular masses of 50 and 49 kDa. The bands could not be separated by additional gel filtration chromatography. These proteins displayed identical elution profiles in both chromatographical steps. Moreover, the N-terminal amino acid sequences of both proteins were MNPLE and identical to each other as well as to that deduced from the nucleotide sequence of Tk-mae. The different mobilities of these bands probably reflect different denatured states of the same protein, as has been observed in several enzymes from T. kodakaraensis (Ezaki et al. 1999, Nakatani et al. 2000). We therefore used the fraction after gel filtration chromatography for further enzymatic characterization. The native molecular mass of Tk-Mae was calculated as 101 kDa by gel filtration chromatography. Considering the subunit molecular mass, Tk-Mae was a homodimer like the homolog from the crenarchaeaon S. solfataricus(Bartolucci et al. 1987). In contrast, most malic enzymes possess a tetrameric structure.
Figure 4.
Purification and pH and temperature dependencies of recombinant T. kodakaraensis malic enzyme (Tk-Mae). (A) SDS-PAGE of purified recombinant Tk-Mae. Lanes: 1 = soluble fraction after sonication; 2 = soluble fraction after heat treatment at 80 °C for 20 min; 3 = after anion exchange chromatography with Resource Q; and 4 = after gel filtration chromatography with Superdex 200 10/30HR. (B) Effect of pH on enzyme activity. Symbols: ◆ = 50 mM acetate-NaOH (pH 4.5–5.5); ■ = 50 mM MES-NaOH (pH 5.5–7.0); ▲ = 50 mM HEPES-NaOH (pH 7.0–8.0); and ● = 20 mM Tris-HCl (pH 8.5–9.0). (C) Effect of temperature on enzyme activity. (D) Thermostability of Tk-Mae. Incubation temperatures: ◆ = 99 °C; ■ = 95 °C; and ▲ = 90 °C.
The recombinant Tk-Mae possessed malic enzyme activity, producing pyruvate from malate in the presence of NADP+, with a specific activity of 5.4 U mg–1 at 50 °C. The stoichiometric formation of pyruvate accompanied by malate oxidation was confirmed by a coupling assay with lactate dehydrogenase. It was a thermostable enzyme with an optimal pH of 8.0 and temperature greater than 95 °C (Figures 4B and 4C). The half-life at 95 °C was determined to be 1 h (Figure 4D). The Tk-Mae preferred NADP+ as an electron acceptor. Although NAD+ could substitute NADP+, the activity was seven times lower than that with NADP+. This cofactor specificity clearly indicates that Tk-Mae is a class III malic enzyme. However, Tk-Mae did not exhibit OAA decarboxylase activity, which is usually observed in class III malic enzymes, including the enzyme from S. solfataricus (Bartolucci et al. 1987). It has been reported that the OAA decarboxylation activities of malic enzymes from pigeon liver and A. suum increased at a lower pH range (Park et al. 1986). We therefore examined the OAA decarboxylase activity of Tk-Mae at a pH range of 6 to 8, but the activity still could not be detected. The lack of OAA decarboxylase activity is one feature that distinguishes the enzyme from typical NADP-dependent class III malic enzymes. As expected from the primary structure described above, Tk-Mae absolutely required a divalent metal cation as a cofactor for its activity. When Mn2+, Co2+, Mg2+, Ni2+, Zn2+, Cu2+, Sr2+ or Ca2+ was added to the reaction mixture at a concentration of 2.5 mM, the most effective cation was Mn2+. The cations Co2+, Mg2+ and Ni2+ also supported relatively high enzyme activity (ca. 60–80% compared with that with Mn2+). The other cations, Ca2+, Cu2+ and Sr2+, did not promote the reaction.
Kinetic analysis
Kinetic analyses of the oxidative decarboxylation of malate and the reductive carboxylation of pyruvate at 50 °C were performed. The enzyme showed typical Michaelis-Menten kinetics in both directions, indicating no homotropic allosteric regulation for this enzyme. It has been clarified that the majority of malic enzymes kinetically prefer oxidative decarboxylation of malate, with a much lower apparentKm value for malate than that for pyruvate (Ziegler 1974, Bukato et al. 1995, Kawai et al. 1996, Stols and Donnelly 1997, Gourdon et al. 2000). In contrast, Tk-Mae showed no striking preference for either direction, as indicated by the similar values of kinetic parameters for both directions (Table 1). This was attributed to the much larger apparent Km of Tk-Mae for malate (16.9 mM) than those for typical malic enzymes (0.09–3.8 mM), despite the comparable apparent Km values (3–14 mM) for pyruvate among Tk-Mae and other enzymes.
Table 1.
Kinetic analysis of recombinant T. kodakaraensis malic enzyme (Tk-Mae). All cofactors and co-substrates were present at saturating concentrations. For oxidative decarboxylation, NADP+ was added at a concentration of 5 mM. For reductive carboxylation, NADPH and NaHCO3 were added at concentrations of 0.05 and 12.5 mM, respectively. For both reactions, Mn2+ was present at a concentration of 5 mM. Abbreviations: Km = Michaelis constant; Vmax = maximum velocity; and kcat/Km = catalytic efficiency.
| Substrate | Direction | Apparent | Vmax | kcat/Km |
| Km (mM) | (U mg–1) | (mM–1 s–1) | ||
| Malate | Oxidative | 16.9 | 7.1 | 0.32 |
| decarboxylation | ||||
| Pyruvate | Reductive | 7.3 | 4.5 | 0.46 |
| carboxylation | ||||
Expression profile of Tk-mae
Thermococcus kodakaraensis cells were grown on different carbon sources (amino acids, pyruvate, starch or pyruvate + starch). To obtain information concerning the physiological function, the effects of growth conditions on the expression profile of Tk-mae were examined by Northern blot analysis and enzyme assay. In the Northern blot analysis, the signals detected with the Tk-mae probe corresponded to transcripts with a length of about 1.3 kb, consistent with a monocistronic transcription of Tk-mae gene. There was no obvious difference in the signal intensities under all conditions examined, indicating constitutive transcription without regulation depending on the carbon source. This profile is significantly different from that of Tk-pck, of which transcription was induced under gluconeogenic conditions (Fukuda et al. 2004). We further noticed that the transcription of Tk-mae appeared to be much weaker than that of Tk-pck. Indeed, quantitative reverse transcription-PCR analysis indicated that the signal intensities for Tk-mae were lower than those for Tk-pck by one order of magnitude. We further performed enzyme assays using cell extracts of T. kodakaraensis grown on the different carbon sources, by determining the reduction of thio-NADP+ and simultaneous formation of pyruvate. Only trace levels of activity (< 0.005 U mg–1) could be observed, if at all, under the conditions examined, probably reflecting the low transcription described above.
Discussion
In this study, we set out to determine the enzymatic properties and expression profile of malic enzyme from the hyperthermophilic archaeon T. kodakaraensis. This enzyme could be involved in C4 and C3 interconversions in this archaeon, following the previous report that PEP carboxykinase links OAA and PEP (Fukuda et al. 2004). The malic enzyme examined here was a homodimeric enzyme preferentially utilizing NADP(H) as a cofactor. Unlike the homolog from the thermoacidophilic crenarchaeon S. solfataricus, Tk-Mae did not possess oxaloacetate decarboxylase activity, and therefore can be presumed to interconvert only malate and pyruvate. When compared with known malic enzymes, notable properties of Tk-Mae were its extreme thermostability and high apparent Km value towards malate (16.9 mM). The apparent Km and Vmax values of Tk-Mae were comparable for both the oxidative and reductive directions, leading to similar catalytic efficiencies.
Malic enzymes are widely distributed in a variety of organisms and possess highly conserved primary structures, suggesting the physiological importance of this enzyme. Generally, it has been proposed that NAD-dependent malic enzyme has a role in the entry of carbons from C4-dicarboxylates or amino acids to gluconeogenesis, while the NADP-dependent enzyme acts as a provider of the reducing equivalent, NADPH, for biosynthesis of cellular components. Nevertheless, the presence of isozymes and alternative pathways that can compensate for the absence of malic enzyme has hampered studies to obtain functional evidence through simple mutation analysis. For example, in E. coli, an ATP-dependent PCK can also participate in the carbon flux between C3 compounds and C4-dicarboxylates, and a mutation in the NAD-dependent malic enzyme gene had no significant phenotype (Hansen and Juni 1975). A double mutant lacking both ATP-dependent PCK and NAD-dependent malic enzyme showed a severe growth defect only when grown on C4-dicarboxylates. This defect could be corrected with the overexpression of the NADP-dependent enzyme in the double mutant (Hansen and Juni 1975). When NAD-dependent malic enzyme was induced in a strain blocked in the fermentative metabolism of pyruvate, the enzyme reductively carboxylated pyruvate, conferring the ability to form succinate as a major fermentation product from glucose (Stols and Donnelly 1997, Hong and Lee 2001). These facts suggested that the in vivo function of malic enzymes can vary depending on the expression levels in and the metabolic demands of the host cells.
In order to gain insight into the role of malic enzyme in T. kodakaraensis and possibly distinguish its role from that of PCK, we examined the transcription of the gene and activity of the enzyme in cells grown on various carbon sources. It was surprising to find that expression was constitutive and extremely low, both at the transcript and protein levels. These results make it difficult to draw a definite conclusion about the physiological role of the enzyme. A majority of NADP-dependent malic enzymes are presumed to function in the provision of intracellular NADPH. However, in Thermococcus and the related genus Pyrococcus, abundant NADPH can be produced through the oxidative deamination of amino acids by the concerted action of glutamate dehydrogenase (Rahman et al. 1998) and aminotransferases (Adams et al. 2001). Excess NADPH is proposed to be recycled through reduction of sulfur to hydrogen sulfide or reductive amination of pyruvate to alanine in the presence or absence of elemental sulfur, respectively. In this metabolic environment, it is hard to image that Tk-Mae, with its low expression levels, plays a major role in NADPH metabolism.
Another possible role of malic enzyme is involvement in carbon metabolism. In the presently understood metabolism of T. kodakaraensis, malate and fumarate are metabolically isolated, because linkages between malate and OAA (malate oxidoreductase) as well as fumarate and succinate (succinate oxidoreductase) are missing in the metabolic pathways deduced from the whole genome analysis (Figure 1). However, we have observed that glutamate or glutamine, or both, were consumed during growth of T. kodakaraensis cells in a nutrient-rich medium supplemented with pyruvate in the absence of sulfur. Although glutamate and glutamine are expected to be catabolized to succinate via 2-oxoglutarate, the total glutamate plus glutamine consumption (0.5 mM) was inconsistent with the amount of succinate accumulation (0.1 mM). The lack of C6 acids-processing enzymes in this archaeon suggests the presence of a metabolic pathway for further succinate utilization. With a pathway from succinate to malate, Tk-Mae may have a role in the entry of carbons derived from amino acids via 2-oxoglutarate into central metabolism by catalyzing oxidative decarboxylation of malate to pyruvate. However, the significance of this role depends critically on whether there is an additional oxidoreductase that can link malate with oxaloacetate. If present, the role proposed above for Tk-Mae could be replaced by Tk-Pck catalyzing PEP formation from OAA. This would be most probable because of the much higher expression levels of Tk-pck than those of Tk-mae. If a metabolic linkage between malate and OAA was absent, Tk-Mae might be essential to catabolize carbons from amino acids such as glutamate and glutamine, which are metabolically converted via 2-oxoglutarate.
In order to address the physiological roles of Tk-Mae and other enzymes for pyruvate and PEP metabolism, further detailed studies will be needed. Recently, we have successfully developed a targeted gene disruption system for T. kodakaraensis (Sato et al. 2003). Analyses of knockout strains with defined medium containing a sole carbon source such as glutamate, and a search for the presently unidentified malate oxidoreductase will help elucidate the unique metabolic features of hyperthermophilic archaea.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research to T.I. (No. 14103011) from the Ministry of Education, Science, Sports, Culture and Technology.
References
- R1.Adams M.W., Holden J.F., Menon A.L., et al. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus . J. Bacteriol. 2001;183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea. 2004;1:263–267. doi: 10.1155/2004/204953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Bartolucci S., Rella R., Guagliardi A., Raia C.A., Gambacorta A., De Rosa M., Rossi M. Malic enzyme from archaebacterium Sulfolobus solfataricus. Purification, structure, and kinetic properties. J. Biol. Chem. 1987;262:7725–7731. [PubMed] [Google Scholar]
- R4.Boles E., de Jong-Gubbels P., Pronk J.T. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5.Bukato G., Kochan Z., Œwierczyñski J. Purification and properties of cytosolic and mitochondrial malic enzyme isolated from human brain. Int. J. Biochem. Cell Biol. 1995;27:47–54. doi: 10.1016/1357-2725(94)00057-3. [DOI] [PubMed] [Google Scholar]
- R6.Chang G.G., Tong L. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry. 2003;42:12721–12733. doi: 10.1021/bi035251+. [DOI] [PubMed] [Google Scholar]
- R7.Coleman D.E., Rao G.S., Goldsmith E.J., Cook P.F., Harris B.G. Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 Å resolution. Biochemistry. 2002;41:6928–6938. doi: 10.1021/bi0255120. [DOI] [PubMed] [Google Scholar]
- R8.Ezaki S., Miyaoku K., Nishi K.I., Tanaka T., Fujiwara S., Takagi M., Atomi H., Imanaka T. Gene analysis and enzymatic properties of thermostable β-glycosidase from Pyrococcus kodakaraensis KOD1. J. Biosci. Bioeng. 1999;88:130–135. doi: 10.1016/s1389-1723(99)80190-x. [DOI] [PubMed] [Google Scholar]
- R9.Fukuda W., Fukui T., Atomi H., Imanaka T. First characterization of an archaeal GTP-dependent phosphoenolpyruvate carboxykinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2004;186:4620–4627. doi: 10.1128/JB.186.14.4620-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Goldberg R.N., Tewari Y.B., Bell D., Fazio K., Anderson E. Thermodynamics of enzyme-catalyzed reactions: Part 1. Oxidoreductases. J. Phys. Chem. Ref. Data. 1993;22:515–579. [Google Scholar]
- R11.Gourdon P., Baucher M.F., Lindley N.D., Guyonvarch A. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 2000;66:2981–2987. doi: 10.1128/aem.66.7.2981-2987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Hansen E.J., Juni E. Isolation of mutants of Escherichia coli lacking NAD- and NADP-linked malic enzyme activities. Biochem. Biophys. Res. Commun. 1975;65:559–566. doi: 10.1016/s0006-291x(75)80183-5. [DOI] [PubMed] [Google Scholar]
- R13.Hong S.H., Lee S.Y. Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol. Bioeng. 2001;74:89–95. doi: 10.1002/bit.1098. [DOI] [PubMed] [Google Scholar]
- R14.Imanaka T., Atomi H. Catalyzing “hot” reactions: enzymes from hyperthermophilic archaea. Chem. Rec. 2002;2:149–163. doi: 10.1002/tcr.10023. [DOI] [PubMed] [Google Scholar]
- R15.Kawai S., Suzuki H., Yamamoto K., Inui M., Yukawa H., Kumagai H. Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing of its gene. Appl. Environ. Microbiol. 1996;62:2692–2700. doi: 10.1128/aem.62.8.2692-2700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Kobayashi K., Doi S., Negoro S., Urabe I., Okada H. Structure and properties of malic enzyme from Bacillus stearothermophilus . J. Biol. Chem. 1989;264:3200–3205. [PubMed] [Google Scholar]
- R17.Kuo C.C., Tsai L.C., Chin T.Y., Chang G.G., Chou W.Y. Lysine residues 162 and 340 are involved in the catalysis and coenzyme binding of NADP+-dependent malic enzyme from pigeon. Biochem. Biophys. Res. Commun. 2000;270:821–825. doi: 10.1006/bbrc.2000.2502. [DOI] [PubMed] [Google Scholar]
- R18.Mitsch M.J., Voegele R.T., Cowie A., Osteras M., Finan T.M. Chimeric structure of the NAD(P)+- and NADP+-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti . J. Biol. Chem. 1998;273:9330–9336. doi: 10.1074/jbc.273.15.9330. [DOI] [PubMed] [Google Scholar]
- R19.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Nakatani M., Ezaki S., Atomi H., Imanaka T. A DNA ligase from a hyperthermophilic archaeon with unique cofactor specificity. J. Bacteriol. 2000;182:6424–6433. doi: 10.1128/jb.182.22.6424-6433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- R22.Park S.H., Harris B.G., Cook P.F. pH dependence of kinetic parameters for oxalacetate decarboxylation and pyruvate reduction reactions catalyzed by malic enzyme. Biochemistry. 1986;25:3752–3759. doi: 10.1021/bi00361a004. [DOI] [PubMed] [Google Scholar]
- R23.Rahman R.N., Fujiwara S., Takagi M., Imanaka T. Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and comparison of the enzymatic characteristics of native and recombinant GDHs. Mol. Gen. Genet. 1998;257:338–347. doi: 10.1007/s004380050655. [DOI] [PubMed] [Google Scholar]
- R24.Sambrook J., Russell D.W. Cold Spring Harbor: 3rd Edn. Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: a laboratory manual.2344 [Google Scholar]
- R25.Sato T., Fukui T., Atomi H., Imanaka T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2003;185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Stols L., Donnelly M.I. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Thauer R.K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Verhees C.H., Kengen S.W., Tuininga J.E., Schut G.J., Adams M.W., De Vos W.M., Van Der Oost J. The unique features of glycolytic pathways in archaea. Biochem. J. 2003;375:231–246. doi: 10.1042/BJ20021472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Wierenga R.K., Terpstra P., Hol W.G. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- R30.Yang Z., Floyd D.L., Loeber G., Tong L. Structure of a closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 2000;7:251–257. doi: 10.1038/73378. [DOI] [PubMed] [Google Scholar]
- R31.Yang Z., Zhang H., Hung H.C., Kuo C.C., Tsai L.C., Yuan H.S., Chou W.Y., Chang G.G., Tong L. Structural studies of the pigeon cytosolic NADP+-dependent malic enzyme. Protein Sci. 2002;11:332–341. doi: 10.1110/ps.38002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Ziegler I. Malate dehydrogenase in Zea mays: properties and inhibition by sulfite. Biochim. Biophys. Acta. 1974;364:28–37. doi: 10.1016/0005-2744(74)90129-6. [DOI] [PubMed] [Google Scholar]