Abstract
Cellular membrane lipids, of which phospholipids are the major constituents, form one of the characteristic features that distinguish Archaea from other organisms. In this study, we focused on the steps in archaeal phospholipid synthetic pathways that generate polar lipids such as archaetidylserine, archaetidylglycerol, and archaetidylinositol. Only archaetidylserine synthase (ASS), from Methanothermobacter thermautotrophicus,has been experimentally identified. Other enzymes have not been fully examined. Through database searching, we detected many archaeal hypothetical proteins that show sequence similarity to members of the CDP alcohol phosphatidyltransferase family, such as phosphatidylserine synthase (PSS), phosphatidylglycerol synthase (PGS) and phosphatidylinositol synthase (PIS) derived from Bacteria and Eukarya. The archaeal hypothetical proteins were classified into two groups, based on the sequence similarity. Members of the first group, including ASS from M. thermautotrophicus, were closely related to PSS. The rough agreement between PSS homologue distribution within Archaea and the experimentally identified distribution of archaetidylserine suggested that the hypothetical proteins are ASSs. We found that an open reading frame (ORF) tends to be adjacent to that of ASS in the genome, and that the order of the two ORFs is conserved. The sequence similarity of phosphatidylserine decarboxylase to the product of the ORF next to the ASS gene, together with the genomic context conservation, suggests that the ORF encodes archaetidylserine decarboxylase, which may transform archaetidylserine to archaetidylethanolamine. The second group of archaeal hypothetical proteins was related to PGS and PIS. The members of this group were subjected to molecular phylogenetic analysis, together with PGSs and PISs and it was found that they formed two distinct clusters in the molecular phylogenetic tree. The distribution of members of each cluster within Archaea roughly corresponded to the experimentally identified distribution of archaetidylglycerol or archaetidylinositol. The molecular phylogenetic tree patterns and the correspondence to the membrane compositions suggest that the two clusters in this group correspond to archaetidylglycerol synthases and archaetidylinositol synthases. No archaeal hypothetical protein with sequence similarity to known phosphatidylcholine synthases was detected in this study.
Keywords: archaetidylcholine, archaetidylglycerol, archaetidylinositol, archaetidylserine, genomic context, phylogenetic tree
Introduction
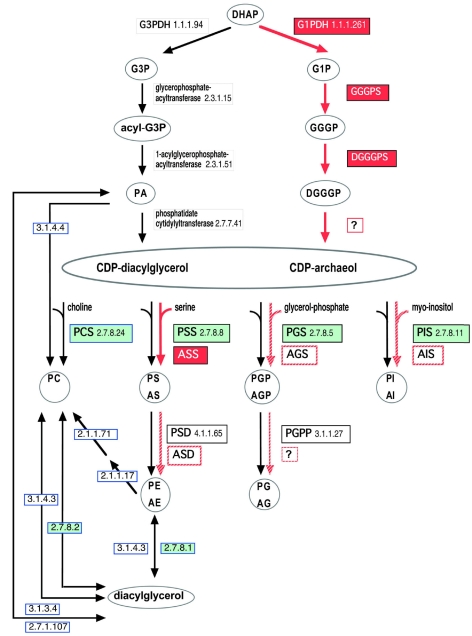
Many studies have revealed the distinctions among the three domains of life. One of the most striking characteristics that distinguishes Archaea from other organisms is the structure of the cellular membrane lipids. Phospholipids are the major membrane constituent in all living organisms. Knowledge of the archaeal phospholipid synthesis pathway has been obtained mainly from experimental studies with methanogens. The synthesis pathways in other Archaea have not yet been fully examined. Metabolic pathways for phospholipid synthesis in Bacteria, Eukarya and Archaea are shown in Figure 1. Glycerophosphate forms the phospholipid backbone and its stereochemistry in Archaea is enantiomeric when compared with that in Eukarya and Bacteria. In Archaea, the molecule exists as sn-glycerol-1-phosphate (G1P), whereas it exists as sn-glycerol-3-phosphate (G3P) in Bacteria and Eukarya (Kates 1978). Both G1P and G3P are generated from a common substrate, dihydroxyacetonephosphate (DHAP), in the first step of the phospholipid synthesis pathway and the chirality of each product is determined by organism-specific enzyme functions. In Archaea, formation of G1P is catalyzed by sn-glycerol-1-phosphate dehydrogenase and in Bacteria and Eukarya, formation of G3P is catalyzed by sn-glycerol-3-phosphate dehydrogenase (Nishihara and Koga 1995, Nishihara et al. 1999). As shown in Figure 1, the three steps in the archaeal pathway following DHAP are distinct, in terms of enzymes and reactions, from the corresponding steps in eukaryotic and bacterial pathways. These reactions are dependent on the substrate stereochemistry. During the second and third steps in Eukarya and Bacteria, the acyl groups form ester bonds at the sn-1 and -2 positions of the glycerol moiety, whereas in Archaea, ether bonds with geranylgeranyl groups are generated at the sn-2 and -3 positions.
Figure 1.
Synthetic pathways for polar lipids. The black arrows indicate the bacterial and eukaryotic pathway, and the red arrows denote the archaeal pathway. The red striped arrows show hypothetical steps in the archaeal pathway. The names or EC numbers in rectangles indicate the enzymes responsible for the corresponding steps. Experimentally identified archaeal enzymes are indicated by characters reverse-printed on red. Bacterial and eukaryotic enzymes belonging to the CDP alcohol phosphatidyltransferase family are shown in green rectangles. Enzymes in rectangles with blue frames are involved in the synthesis of lipids with choline. Abbreviations: DHAP = dihydroxyacetonephosphate; G3PDH = sn-glycerol-3-phosphate dehydrogenase; G1PDH = sn-glycerol-1-phosphate dehydrogenase; G3P = sn-glycerol-3-phosphate; G1P = sn-glycerol-1-phosphate; GGGPS, GGGP synthase; GGGP = 3-O-geranylgeranyl-sn-glycerol phosphate; DGGGPS, DGGGP synthase; PA, phosphatidic acid; DGGGP = 2,3-di-O-geranylgeranyl-sn-glycerol phosphate; PCS = phosphatidylcholine synthase; PSS = phosphatidylserine synthase; ASS = archaetidylserine synthase; PGS = phosphatidylglycerol synthase; AGS = archaetidylglycerol synthase; PIS = phosphatidylinositol synthase; AIS = archaetidylinositol synthase; PC = phosphatidylcholine; PS = phosphatidylserine; AS = archaetidylserine; PGP = phosphatidylglycerolphosphate; AGP = archaetidylglycerolphosphate; PI =phosphatidylinositol; AI = archaetidylinositol; PSD = phosphatidylserine decarboxylase; ASD = archaetidylserine decarboxylase; PGPP = PGP phosphatase; PE = phosphatidylethanolamine; AE = archaetidylethanolamine; PG = phosphatidylglycerol; and AG = archaetidylglycerol.
Following the fourth step, polar lipid synthetic reactions in Archaea and other organisms become independent of stereochemistry and ligand ether/ester bonds. In Eukarya and Bacteria, the fourth synthetic step involves CDP-diacylglycerol synthase, which transfers cytidine phosphate from cytidine triphosphate to generate CDP-diacylglycerol (Dowhan 1997). An experiment with the membrane fraction of a methanoarchaeon suggests that the corresponding step in Archaea is catalyzed by a similar mechanism (Morii et al. 2000). However, the archaeal enzyme involved in the step has not yet been isolated.
The phospholipid synthetic pathway diverges in Eukarya and Bacteria after the fourth step. The cytidine phosphate is substituted with several kinds of polar head groups, such as choline, serine, glycerol and inositol. These reactions produce the membrane components phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylglycerol (PG) and phosphatidylinositol (PI) (Cronan 2003). It should be noted that all the different types of polar lipids are not always present in a living organism, because each organism displays a distinctive phospholipid distribution. Consistent with the variation in the fifth step of phospholipid synthesis, several different transferases, such as phosphatidylcholine synthase (PCS), phosphatidylserine synthase (PSS), phosphatidylglycerol synthase (PGS) and phosphatidylinositol synthase (PIS), have been identified in Eukarya and Bacteria. The formation of phosphatidylglycerolphosphate (PGP) is catalyzed by PGS, which is sequentially dephosphorylated by PGP phosphatase (PGPP) to become PG. Experimental studies have revealed the presence of PGPP in Bacteria, but the enzyme has not been identified in Eukarya. The enzymes PCS, PSS, PGS and PIS all belong to the same protein family (Sohlenkamp et al. 2003). In the Pfam database (Bateman et al. 2004), the family code is ‘CDP-OH_P_transf’ and the Accession Number is PF01066. In accordance with the Pfam descriptions, this paper refers to the protein family as the CDP alcohol phosphatidyltransferase family. The members are diverse in both sequence and length, but they share a common sequence motif, D-G-x(2)-A-R-x(8)- G-x(3)-D-x(3)-D, classified as PS00379 in the PROSITE motif database (Hulo et al. 2004). A recent study suggested that enzymes in this family contain several membrane spanning helices (Katayama et al. 2004). Ethanolaminephosphotransferase and diacylglycerol cholinephosphotransferase, which are known to catalyze similar reactions, are also members of this family.
Archaetidylcholine (AC), archaetidylserine (AS), archaetidylglycerol (AG) and archaetidylinositol (AI) have been identified in archaeal membranes as the counterparts of the phosphatidyl derivatives, PC, PS, PG and PI. As in Eukarya and Bacteria, an archaebacterium does not always utilize all of the possible types of polar lipids (Figure 2). For instance, AC is only detected in Methanopyrus kandleri (Nishihara et al. 2002). At present, the archaeal pathway for polar lipid synthesis is obscure. The enzymes involved in the reactions have not been identified, with the exception of archaetidylserine synthase (ASS), derived from Methanothermobacter thermautotrophicus (Morii and Koga 2003). This enzyme also belongs to the CDP alcohol phosphatidyltransferase family, and shares significant sequence similarity to PSS. The complete genome sequences of several Archaea have revealed the presence of some hypothetical proteins, which share high sequence similarity to the PSSs or PGSs of Eukarya and Bacteria. These observations suggest that Archaea use PSS homologues to introduce polar head groups to phospholipids.
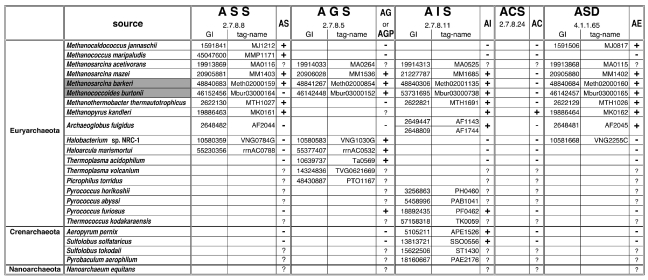
Figure 2.
List of candidate enzymes for polar lipid synthesis. The classifications and the sources are shown in the first and second columns, respectively. The complete genome sequences of the sources, except for those of M. barkeri and M. burtonii, are available. A candidate enzyme obtained by the database search is indicated by the GI number and the gene tag-name for each archaeal species. A blank indicates that the corresponding enzyme was not detected from the source. The presence or absence of a polar lipid in the membrane is shown by ‘+’, or ‘–’. The symbol, ‘?’, indicates that the lipid content of the species has not been examined. Information about lipid distributions was obtained from Kates 1978, De Rosa et al. 1986, Ferrante et al. 1990, Koga et al. 1993, 1998, Nishihara and Koga 1995, Ihara et al. 1997, Sprott et al. 1997, 2003, Morii et al. 1999, Nishihara et al. 2002, Shimada et al. 2002 and Nichols et al. 2004. The lipid composition of A. fulgidus is our unpublished data. Abbreviations: ASS = archaetidylserine synthase; AGS = archaetidylglycerol synthase; AIS = archaetidylinositol synthase; ACS = archaetidylcholine synthase; ASD = archaetidylserine decarboxylase; AS = archaetidylserine; AG = archaetidylglycerol; AGP = archaetidylglycerolphosphate; AI = archaetidylinositol; AC = archaetidylcholine; and AE = archaetidylethanolamine.
This study focused on the fifth step of archaeal phospholipid synthesis. We investigated the molecular evolution of PSS, PGS and PIS, together with their archaeal counterparts. The tree topologies obtained through the analyses were compared with polar lipid distribution over archaeal membranes. We also compared the genomic arrangements of the putative genes encoding the enzymes. The molecular phylogenetic tree patterns, together with membrane composition and the conservation of genomic contexts, provide insight into the archaeal phospholipid synthesis pathways.
Materials and methods
Sequence data collection
To obtain the sequences of the bacterial and eukaryotic enzymes involved in polar lipid synthesis and their archaeal counterparts, we searched the nucleotide and genome databases of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) with FASTA version 3.4 (Pearson and Lipman 1988). The queries used for the similarity-based search are described in the Results and discussion section. When completely or nearly completely identical sequences were detected, we selected only a single representative sequence and discarded the others.
Comparison of gene arrangement
To compare the gene arrangement of phospholipid biosynthesis enzymes from each organism, the gene annotation table for each genome was obtained from the NCBI database (ftp://ftp.ncbi.nih.gov/genbank/genomes) along with the genome sequence and the gene products.
Sequence alignment
The collected amino acid sequences were aligned by MAFFT (Katoh et al. 2002, 2005) with the FFT-NS-i option. The alignment thus obtained was slightly modified by visual inspection. Unambiguously aligned sites of a multiple alignment were subjected to phylogenetic tree analyses.
Phylogenetic analysis
An unrooted molecular phylogenetic tree was constructed using a program with the maximum likelihood (ML) method (Katoh et al. 2001), which uses a genetic algorithm to search for the ML topology. The default parameter values were used to estimate the ML topology. The number of generations for the genetic algorithm was 30,000. The evolutionary rate heterogeneity among the sites was expressed as a gamma distribution, with the optimal shape parameter alpha (Yang 1994). The statistical significance of a cluster or a subtree of the ML tree was evaluated by the reliability index (Katoh et al. 2001).
Results and discussion
Database searching of archaetidylserine synthase, archaetidylglycerol synthase, archaetidylinositol synthase and archaetidylcholine synthase
The sequences of the archaeal enzymes, which are presumably involved in polar head group addition through phospholipid synthesis pathways, were collected by database searching. For the first search, ASS from M. thermautotrophicus, whose function has been experimentally determined, was used as a query to collect ASS candidates. In the case of Bacteria, two PSS types have been reported (Matsumoto 1997), which are hereafter referred to as PSS-I and -II. They have evolved from different evolutionary origins and belong to different protein families. The members of PSS-I, which include two phospholipase D domains in their primary structures, were mainly identified from gammaproteobacteria. On the other hand, the members of PSS-II, which belong to the CDP alcohol phosphatidyltransferase family, have been detected in a wide variety of Bacteria. The distribution of the two types of PSS in Bacteria is discussed below. The ASS from M. thermautotrophicus belongs to PSS-II. We obtained 11 ASS homologous sequences derived from different archaeal species. They were annotated as PSS or ASS, based on the sequence similarity. The bacterial PSS-IIs were also detected after the archaeal sequences. In addition to these proteins, PGS and PIS from Bacteria and Eukarya and other hypothetical proteins from Archaea were detected, although the sequence similarities were very low. To confirm the close relationship between the ASS candidates and PSS-II, database searches with the amino acid sequences of the PSS-IIs from Mesorhizobium loti and Xanthomonas campestris were performed. The results were consistent with those from the previous search. Furthermore, we examined whether the PSS-I homologues existed in the archaeal genome. However, no archaeal protein showed significant sequence similarity to Escherichia coli PSS-I. This result suggests that Archaea use only PSS-II orthologues for AS production.
Next, we collected the amino acid sequences of archaetidylglycerol synthase (AGS) candidates. Under the assumption that AGS is similar to PGS from Bacteria, a preliminary database search with the amino acid sequence of PGS from Bacillus subtilis was performed. Among the detected archaeal hypothetical proteins, the amino acid sequence derived from Pyrococcus abyssi was selected as a query to collect AGS candidates. The top matches of the detected proteins were occupied by a mixture of PGSs and PISs from Bacteria and Eukarya, with 32 hypothetical proteins from Archaea. Most of the archaeal proteins thus obtained were annotated as PGS, PIS, or CDP-alcohol phosphatidyltransferase. This result suggests that these archaeal proteins are close to both the PGSs and PISs. In addition to the 32 proteins, the 11 ASS homologues described above were also detected with low sequence similarity. To confirm this observation, database searches with the PISs from Saccharomyces cerevisiae and Mycobacterium smegmatis were performed. As expected, the top matches were occupied by the mixture of PISs, PGSs, and 32 archaeal proteins. Therefore, it was possible that the detected archaeal proteins included both AGS and archaetidylinositol synthase (AIS). At this stage, it was difficult to distinguish between the two types of enzymes with the database search results. This problem was further examined by the molecular phylogenetic analyses with the archaeal proteins, PGSs and PISs, as discussed below.
The choline derivative, AC, has been identified only from M. kandleri, as described above (Nishihara et al. 2002). Several different pathways involved in PC biosynthesis for Bacteria and Eukarya have been identified thus far (Lopez-Lara and Geiger 2001). The enzyme PCS, which belongs to the CDP-alcohol phosphatidyltransferase family, is involved in one of the pathways in some bacteria, including Sinorhizobium meliloti (Sohlenkamp et al. 2000). We first performed database searches to find archaeal counterparts, with the amino acid sequence of the PCS from S. meliloti as a query. However, no sequence from Archaea, including M. kandleri, showed significant sequence similarity to the bacterial PCS. We subsequently searched for an archaeal protein with sequence similarity to any enzyme involved in the different pathways, such as the CDP-choline pathway and the methylation pathway. Eight bacterial and eukaryotic amino acid sequences were used as queries, according to the description in the section of glycerophospholipid metabolism in the KEGG pathway database (http://www. genome.ad.jp/kegg/pathway.html) (Figure 1). Among them, only the homologues for phosphatidylethanolamine N-methyltransferase (PmtA, EC 2.1.1.17) were detected from 13 Archaea, including M. kandleri. However, no homologue of phosphatidyl-N-methylethanolamine N-methyltransferase (EC 2.1.1.71), which sequentially works after PmtA in the methylation pathway, was detected in any Archaea. Therefore, the 13 archaeal homologues of PmtA are not believed to participate in PC synthesis. The results of the database search suggest that M. kandleri has a pathway for PC synthesis different from those known.
Using the collected sequences, two multiple alignments were constructed. One of them consisted of the ASS homologues and the PSSs from Bacteria and Eukarya, whereas the other alignment included the PGSs, PISs and the close relatives from Archaea. Each alignment was subjected to a molecular phylogenetic analysis. The alignments are available at the web site, http://timpani.genome.ad.jp/~lipid/, as supplemental materials.
Molecular phylogeny of archaetidylserine synthases and phosphatidylserine synthase-IIs
The ASS homologues were detected within a part of Euryarchaeota. As described above, these proteins, with the exception of one from M. thermautotrophicus, have not yet been experimentally characterized. If the homologues work as ASSs, then the Archaea that encode these proteins in their genomes would have AS in their membranes. Until recently, the polar lipid compositions of only a few archaeal membranes have been reported. We compared the ASS homologue distribution with that of AS in the membranes (Figure 2). Out of 11 Archaea with ASS homologues, the membrane compositions of 10 species have been investigated. Six species have AS in their membranes, whereas ASs have not been identified in the remaining four species, Methanococcoides burtonii, Archaeoglobus fulgidus, Halobacterium sp. NRC-1, and Haloarcula marismortui (Figure 2). It is possible that the ASs of M. burtonii and A. fulgidus are immediately transformed into archaetidylethanolamine (AE), as observed in E. coli (Saha et al. 1996). As discussed below, AE synthesis from AS is considered to be catalyzed by archaetidylserine decarboxylase (ASD), by analogy to the bacterial pathway. The ASD homologues are detected in various Archaea, including M. burtonii and A. fulgidus. There is no known reasonable explanation for the inconsistency in Halobacterium sp. and H. marismortui. The two species will be discussed again in the next section. Four other Archaea, Thermoplasma acidophilum, Pyrococcus furiosus, Aeropyrum pernix, and Sulfolobus solfataricus, lacked ASS homologues and AS components. The consistency in the distribution of the ASS homologues and AS, together with the high sequence similarity to ASS from M. thermautotrophicus, suggested that these homologues work as ASSs. Hereafter, we will refer to the 11 proteins as ASSs.
An unrooted phylogenetic tree of the ASSs and PSS-IIs, generated by the ML method, is shown in Figure 3. The ASSs formed a single cluster in the ML tree, although the reliability index was too low to suggest the significance of the clustering. We also constructed an unrooted tree with the neighbor joining (NJ) method (Saitou and Nei 1987) (data not shown). As in the case of the ML tree, the ASSs formed a single cluster in the NJ tree, although the bootstrap probability was small. The phylogeny of the archaeal sequences is similar to the evolutionary relationship among Archaea inferred from 16S rRNA comparison. In other words, the hypothetical proteins are considered to be orthologous to each other, which suggests that the 11 proteins are the ASSs for each species.
Figure 3.
An unrooted maximum likelihood (ML) tree of phosphatidylserine synthase/archaetidylserine synthase (PSS/ASS). Each sequence is indicated by the source name and the GI number from the National Center for Biotechnology Information. The archaeal, bacterial and eukaryotic proteins are colored red, blue and green, respectively. The reliability index of a cluster is only shown at the root node of the cluster when the value is equal to or greater than 0.25. The log likelihood (lnL) of the ML tree was –12209.94 and the optimal shape parameter alpha for gamma distribution was 0.84 (129 sites were compared). The bar under the tree corresponds to 0.1 amino acid substitutions/site. The presence or absence of phosphatidylserine decarboxylase-A or -B (PSD-A or PSD-B) in each species is indicated by a symbol, ‘+’ or ‘–’, after the source name. The symbol, ‘+’, colored red, shows that the genes or open reading frames for PSS/ASS and PSD/archaetidylserine decarboxylase (ASD) are tandemly encoded in the genome.
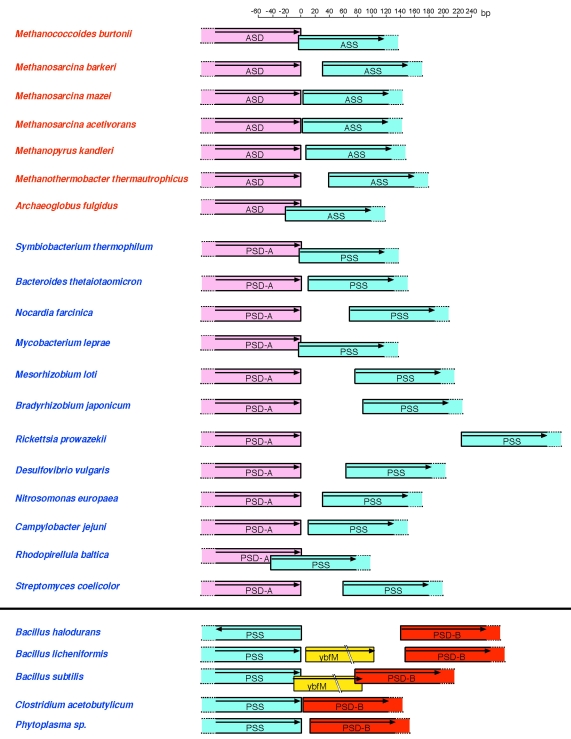
Two bacterial hypothetical proteins, annotated as PSS, were included in the cluster of ASSs in the ML tree. One of them was derived from Streptomyces coelicolor. This protein was also found in the ASS cluster in the NJ tree (data not shown). In both trees, the bacterial protein showed a close relationship to the ASSs from halobacteria, although the relationship was not statistically significant for either tree. As discussed below, the gene arrangement around the PSS-II gene of S. coelicolor was similar to those found in archaeal genomes. The reproduction of the relationship by the two different tree construction methods, together with the similarity in their operon structures, suggests that PSS-II of S. coelicolor may have been introduced from an archaeon by horizontal gene transfer. The other PSS-II sequence in the ASS cluster of the ML tree was from Rhodopirellula baltica. Its operon structure for the PSS-II gene is also conserved, although the bacterial protein did not cluster with the ASSs in the NJ tree. These observations suggest that R. baltica may also have acquired PSS-II by horizontal gene transfer from an archaeon.
Conservation of genomic contexts of phosphatidylserine synthase/archaetidylserine synthase and phosphatidylserine decarboxylase/archaetidylserine decarboxylase
Phosphatidylethanolamine (PE) is a constituent of the biological membranes in some Bacteria and Eukarya. The corresponding polar lipid in the archaeal membranes is AE. Phos
phatidylserine decarboxylase (PSD), which exerts its activity downstream of PSS, generates PE from PS (Figure 1). Based on pulse-chase experiments, Nishihara et al. (1989) suggested that AE is generated from AS in vivo. These observations suggest that AE is generated through a pathway similar to that found in other organisms, whereas ASD has yet not been experimentally identified. Some Archaea have open reading frames (ORFs) with products sharing amino acid sequence similarity to that of PSD. The hypothetical proteins are considered to be ASDs.
Bacterial genes often form transcriptional units called operons. Recent genome comparisons of Bacteria revealed that the order or neighborhood of the genes present in the same operon is conserved among different bacterial genomes when the gene products are involved in the same pathway or process (Overbeek et al. 1999, Snel et al. 2002). Matsumoto et al. (1998) reported that PSS and PSD in B. subtilis are present in the same operon. However, the conservation of the operon structure has not yet been fully examined. Therefore, we investigated the ORF locations for the PSS/ASS and PSD/ASD homologues in archaeal and bacterial genomes.
We collected the amino acid sequences of putative ASDs and PSDs by database searching with the amino acid sequences of the PSDs from M. loti, Campylobacter jejuni, and Fusobacterium nucleatum as queries. Nine archaeal proteins were obtained as PSD homologues. The existence of AE in the membrane has been reported for about seven Archaea, which encode both ORFs for ASS and ASD in the same order (Figure 2). The separate locations of ASS and ASD in the genome of Methanocaldococcus jannaschii are the only exception. The sequence similarity to PSD, the conservation of the gene order, and the consistent distribution of the enzyme homologue and the product suggest that the ORF next to the ASS gene functions as an ASD. Hereafter, we refer to these nine archaeal proteins as ASDs.
Halobacterium sp. and H. marismortui have neither AS nor AE. The former had both ORFs for ASS and ASD, in separate locations, whereas the latter had only the ORF for ASS. According to these results, ASS and ASD may not function in halobacteria.
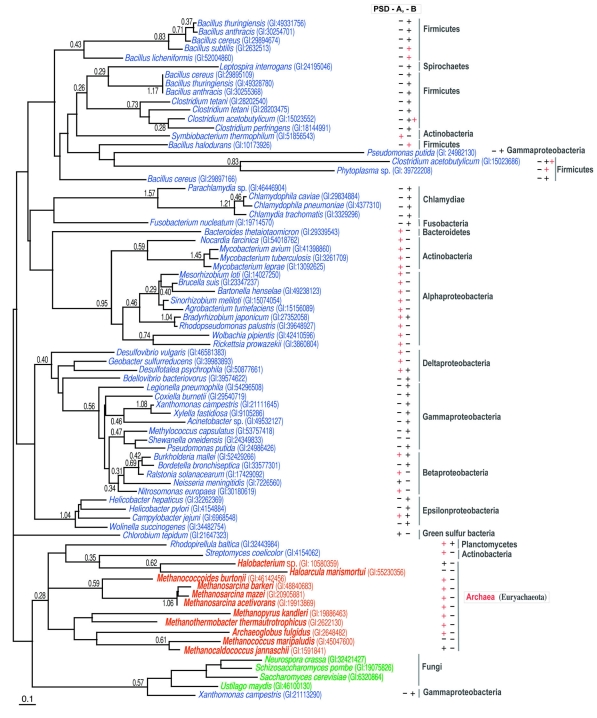
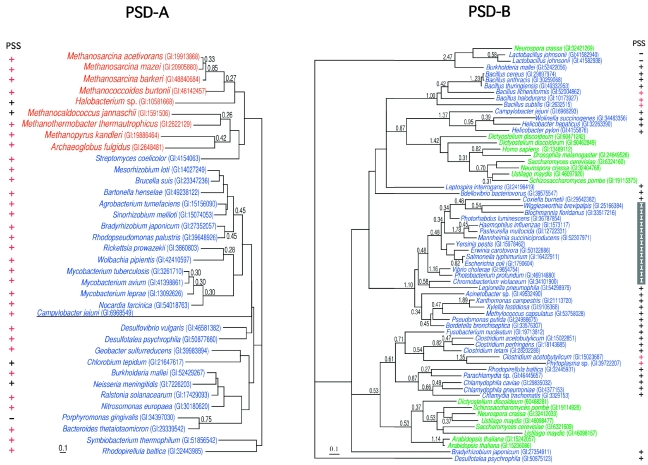
A preliminary analysis suggested that the PSD/ASD family was divided into two groups, with a bootstrap probability of 99% (data not shown). The sequence similarities between the members of the two groups were very weak. Therefore, an unrooted ML tree of each group was drawn, instead of a tree with all of the members (Figure 4). The two groups are referred to here as PSD-A and -B. All of the ASDs and PSDs from the various bacteria belonged to PSD-A. Most of the PSD-A genes were associated in the genomes with the genes for PSS-II with the same order in Archaea (Figure 5). The PSD-B included a large number of bacterial enzymes, as well as enzymes derived from Eukarya, such as vertebrates, arthropods, fungi, slime mold, and plants. In contrast to the locations of the PSD-A members, the locations of the genes for the bacterial PSD-Bs were not close to those of PSSs in the genomes, except in five species. Notably, the genes for PSD-B and PSS-II were in the reverse order compared with those for PSD-A/ASD and PSS-II/ASS (Figure 5).
Figure 4.
Two unrooted maximum likelihood trees of phosphatidylserine synthase/archaetidylserine synthase (PSS/ASS). Each sequence is indicated by the source name and the GI number from the National Center for Biotechnology Information. The colors for each species are the same as in Figure 3. The reliability index of a cluster is only shown when the value is equal to or greater than 0.25. The log likelihoods were –7157.86 and –18485.60 for phosphatidylserine decarboxylase-A and -B (PSD-A or PSD-B), respectively. The optimal shape parameter alpha was 0.85 (145 sites) for PSD-A and 1.13 (197 sites) for PSD-B. The presence or absence of ASS or PSS-II in each species is indicated by a symbol, ‘+’ or ‘–’, near the source name. The symbol, ‘+’, colored red, shows that the genes or open reading frames for PSS/ASS and PSD/ASD are tandemly encoded. When the species uses PSS-I instead of PSS-II, the symbol ‘+’ is replaced with ‘I’.
Figure 5.
Arrangement of the archaetidylserine synthase/phosphatidylserine synthase (ASS/PSS) and archaetidylserine decarboxylase/phosphatidylserine decarboxylase (ASD/PSD) genes. A light blue box indicates a gene or open reading frame (ORF) for ASS or PSS. The ORFs for ASD/PSD-A, PSD-B and ybfM are in pink, red and yellow boxes, respectively. The arrow in the box indicates the coding direction. The scale bar is shown at the top to express the sizes of the spacer regions between the genes. The central region of ybfM, an alkaline phosphatase homolog, is omitted. The sources are shown on the left side, and are colored red for Archaea and blue for Bacteria.
Non-orthologous gene displacement in polar lipid synthesis
Generally, the presence of the PSD-A gene in organisms was negatively correlated with the presence of PSD-B. In other words, when PSD-A is encoded in a genome, the organism lacks a PSD-B gene and vice versa, although a few exceptions exist (Figure 3). The exclusive coding pattern of the PSD-A and -B genes provides a new example of non-orthologous gene displacement (Koonin et al. 1996). That is, the two types of PSDs are involved in the same step of a metabolic pathway, although they are not orthologous.
Another negative correlation was observed for the bacterial PSS genes. As shown in Figure 4, the PSS-I and PSS-II genes are not encoded in the same genome. The exclusive coding pattern suggests that PSS-I and -II have been used for the same reaction in different species by non-orthologous gene displacement.
Molecular phylogeny of phosphatidylglycerol synthases, phosphatidylinositol synthases and their relatives
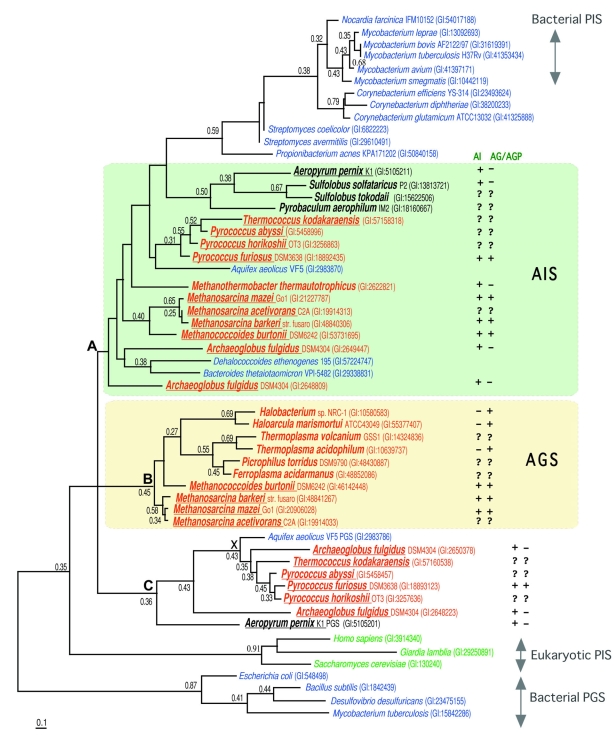
As described above, the annotations for the PGS or PIS homologues from Archaea seemed questionable. To examine the functions of the 32 archaeal proteins based on their evolutionary relationships, we constructed an unrooted ML tree of the PGSs, PISs and their relatives (Figure 6). The archaeal proteins were classified into three groups. Cluster A consisted of 15 proteins from Euryarchaeota and Crenarchaeota, the PISs from mycobacteria, and the bacterial hypothetical proteins. Cluster B was composed of 10 proteins from Euryarchaeota. Cluster C included seven archaeal proteins and a bacterial hypothetical protein. Five archaeal proteins and a bacterial protein, which clustered at node X, have one NTP-transferase domain of about 100 amino acid residues at the N-terminus. Similar clusters were reproduced in the NJ tree with a low bootstrap probability (data not shown).
Figure 6.
An unrooted maximum likelihood tree of phosphatidylglycerol synthase/archaetidylglycerol synthase (PGS/AGS) and phosphatidylinositol synthase/archaetidylinositol synthase (PIS/AIS). Each sequence is indicated by the source name and the GI number from the National Center for Biotechnology Information. The proteins from Euryarchaeota, Crenarchaeota, Bacteria and Eukarya are colored red, black, blue and green, respectively. The underlined species have paralogues in the tree. The reliability index of a cluster is only shown when the value is equal to or greater than 0.25. The log likelihood was –9599.24 and the optimal shape parameter alpha was 1.03 (119 sites). The presence or absence of archaetidylinositol or archaetidylglycerol/archaetidylglycerolphosphoate in each species is indicated by a symbol, ‘+’ or ‘–’, after the source name and GI number.The symbol, ‘?’, indicates that the lipid composition has not been reported for the species. The characters, ‘A’, ‘B’, ‘C’, and ‘X’, are explained in the text. The groups consisting of AIS and AGS candidates are enclosed within light green and light yellow areas, respectively.
To examine the significance of the clusters from a functional viewpoint, we compared the distributions of AGP/AG and AI in the archaeal membranes with the clustering pattern of the phylogeny (Figure 2 and Figure 6). It is believed that AG is generated through AGP. Therefore, we used the presence of AG, in addition to AGP, as circumstantial evidence for the presence of AGS. Four archaeal species, P. furiosus, Methanosarcina mazei, Methanosarcina barkeri, and M. burtonii, contain both AI and AG. The eight Archaea included in Cluster A have lipid data, and all of them contain AI. On the other hand, half of them, S. solfataricus, A. pernix, M. thermautotrophicus and Archaeoglobus fulgidus,do not contain AG. For the members of Cluster B, the membrane compositions of six sources have been reported. Halobacterium sp., H. marismortui and T. acidophilum do not possess AI, whereas AG has been detected in all of the Archaea in Cluster B. As for Cluster C, the phospholipid compositions of three archaeal members have been investigated. All of them have AI, and only P. furiosus also has AG. The presence of polar lipids, together with the close relationship to the PIS of mycobacteria, suggests that the archaeal proteins belonging to Cluster A function as AISs. The membrane compositions of the Archaea in Cluster B suggest that these proteins function as AGSs. The functions of the proteins included in Cluster C could not be predicted by this study because some have another domain and the membrane composition data are insufficient. Archaea with AG and AI in their membranes were found in both Clusters A and B. Therefore, the Archaea are considered to have two enzymes, AGS and AIS.
Archaetidylinositol is reportedly present in the M. kandleri membrane (Sprott et al. 1997), although it is not known whether AG is present in the archaeal membrane. Neither AIS nor AGS is encoded within the M. kandleri genome, according to our database analyses. As described above, AC has been found in the archaebacterium, but the enzyme responsible for its synthesis has not been identified. Therefore, M. kandleri is considered to have unique pathways for polar head group synthesis.
We searched for the archaeal homologues of PGPP using the enzyme from E. coli as a query. The detected proteins included many enzymes and hypothetical proteins from Bacteria. A small number of archaeal hypothetical proteins were also detected, but their sequence similarities were very weak. Some of these Archaea are known to lack AG or AGP in their membranes. On the other hand, the corresponding proteins were not detected in halobacteria and thermoplasma, which have AG in their membranes. Therefore, the hypothetical proteins detected by this search are not considered to be responsible for AGP dephosphorylation. It is possible that a new enzyme is involved in AG synthesis in Archaea.
Conclusions
We have performed a comprehensive comparative study of the enzymes involved in the synthesis of archaeal polar lipids, including examinations of molecular phylogeny, conservation of genomic contexts, and membrane composition. Database searches detected ASS, AGS, and AIS candidates. The proteins belonged to the same protein family, the CDP alcohol phosphatidyltransferase family, like PSS-II, PGS and PIS.
The distribution of genes encoding the archaeal proteins was roughly consistent with the existence of corresponding polar lipids in the membranes. A comparison of the archaeal and bacterial genomes revealed the co-occurrence of the genes or ORFs for PSS-II/ASS and PSD/ASD.
The discovery of ASS (Morii and Koga 2003) suggests that the archaeal pathways for polar lipid synthesis are similar to those of Bacteria and Eukarya. Our study suggests that Archaea use close relatives of bacterial and eukaryotic enzymes in some parts of the pathways. As discussed in the Introduction, methanogens have been major targets for studies of phospholipid synthesis in Archaea. However, our study suggests that many Archaea share pathways similar to those shown in Figure 1. The evolutionary relationships of enzymes involved in polar lipid synthesis are complex. Even within a subfamily whose members are supposed to share the same catalytic activity the evolutionary relationship seems complicated, due to weak sequence similarities, gene duplications and horizontal gene transfers between domains or within a domain. We compensated for the deficiencies in the reliability of the phylogenetic analyses through examination of conservation of genomic contexts and with information about membrane composition. Further development of theoretical treatments for phylogenetic analyses, along with the accumulation of experimental observations, will be required for confirmation of our study.
Acknowledgments
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- R1.Bateman A., Coin L., Durbin R., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Cronan J.E. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- R3.De Rosa M., Gambacorta A., Gliozzi A. Structure, biosynthesis, and physicochemical properties of archaebacterial lipids. Microbiol. Rev. 1986;50:70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Dowhan W. CDP-Diacylglycerol synthase of microorganisms. Biochim. Biophys. Acta. 1997;1348:157–165. doi: 10.1016/s0005-2760(97)00111-2. [DOI] [PubMed] [Google Scholar]
- R5.Ferrante G., Richards J.C., Sprott G.D. Structures of polar lipids from the thermophilic, deep-sea archaeobacterium Methanococcus jannaschii . Biochem. Cell Biol. 1990;68:274–283. doi: 10.1139/o90-038. [DOI] [PubMed] [Google Scholar]
- R6.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:134–137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Ihara K., Watanabe S., Tamura T. Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int. J. Syst. Bacteriol. 1997;47:73–77. doi: 10.1099/00207713-47-1-73. [DOI] [PubMed] [Google Scholar]
- R8.Katayama K., Sakurai I., Wada H. Identification of an Arabidopsis thaliana gene for cardiolipin synthase located in mitochondria. FEBS Lett. 2004;577:193–198. doi: 10.1016/j.febslet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- R9.Kates M. The phytanyl ether-linked polar lipids and isoprenoid neutral lipids of extremely halophilic bacteria. Prog. Chem. Fats Other Lipids. 1978;15:301–342. doi: 10.1016/0079-6832(77)90011-8. [DOI] [PubMed] [Google Scholar]
- R10.Katoh K., Kuma K., Miyata T. Genetic algorithm-based maximum-likelihood analysis for molecular phylogeny. J. Mol. Evol. 2001;53:477–484. doi: 10.1007/s002390010238. [DOI] [PubMed] [Google Scholar]
- R11.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Katoh K., Misawa K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Koga Y., Akagawa-Matsushita M., Ohga M., Nishihara M. Taxonomic significance of the distribution of component parts of polar ether lipids in methanogens. Syst. Appl. Microbiol. 1993;16:342–351. [Google Scholar]
- R14.Koga Y., Morii H., Akagawa-Matsushita M., Ohga M. Correlation of polar lipid composition with 16S rRNA phylogeny in methanogens. Further analysis of lipid component parts. Biosci. Biotechnol. Biochem. 1998;62:230–236. doi: 10.1271/bbb.62.230. [DOI] [PubMed] [Google Scholar]
- R15.Koonin E.V., Mushegian A.R., Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- R16.Lopez-Lara I.M., Geiger O. Novel pathway for phosphatidylcholine biosynthesis in bacteria associated with eukaryotes. J. Biotechnol. 2001;91:211–221. doi: 10.1016/s0168-1656(01)00331-5. [DOI] [PubMed] [Google Scholar]
- R17.Matsumoto K. Phosphatidylserine synthase from bacteria. Biochim. Biophys. Acta. 1997;1348:214–227. doi: 10.1016/s0005-2760(97)00110-0. [DOI] [PubMed] [Google Scholar]
- R18.Matsumoto K., Okada M., Horikoshi Y., Matsuzaki H., Kishi T., Itaya M., Shibuya I. Cloning, sequencing, and disruption of the Bacillus subtilis psd gene coding for phosphatidylserine decarboxylase. J. Bacteriol. 1998;180:100–106. doi: 10.1128/jb.180.1.100-106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Morii H., Koga Y. CDP-2,3-Di-O-geranylgeranyl-sn-glycerol:L-serine O-archaetidyltransferase (archaetidylserine synthase) in the methanogenic archaeon Methanothermobacter thermautotrophicus . J. Bacteriol. 2003;185:1181–1189. doi: 10.1128/JB.185.4.1181-1189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Morii H., Yagi H., Akutsu H., Nomura N., Sako Y., Koga Y. A novel phosphoglycolipid archaetidyl(glucosyl)inositol with two sesterterpanyl chains from the aerobic hyperthermophilic archaeon Aeropyrum pernix K1. Biochim. Biophys. Acta. 1999;1436:426–436. doi: 10.1016/s0005-2760(98)00157-x. [DOI] [PubMed] [Google Scholar]
- R21.Morii H., Nishihara M., Koga Y. CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase in the methanogenic archaeon Methanothermobacter thermautotrophicus . J. Biol. Chem. 2000;275:36568–36574. doi: 10.1074/jbc.M005925200. [DOI] [PubMed] [Google Scholar]
- R22.Nichols D.S., Miller M.R., Davies N.W., Goodchild A., Raftery M., Cavicchioli R. Cold adaptation in the Antarctic Archaeon Methanococcoides burtonii involves membrane lipid unsaturation. J. Bacteriol. 2004;186:8508–8515. doi: 10.1128/JB.186.24.8508-8515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.Nishihara M., Koga Y. Two new phospholipids, hydroxyarchaetidylglycerol and hydroxyarchaetidylethanolamine, from the Archaea Methanosarcina barkeri . Biochim. Biophys. Acta. 1995;1254:155–160. doi: 10.1016/0005-2760(94)00178-2. [DOI] [PubMed] [Google Scholar]
- R24.Nishihara M.H., Morii H,, Koga Y. Heptads of polar ether lipids of an Archaebacterium, Methanobacterium thermoautotrophicum: structure and biosynthetic relationship? Biochemistry. 1989;28:95–102. [Google Scholar]
- R25.Nishihara M., Yamazaki T., Oshima T., Koga Y. sn-glycerol-1-phosphate-forming activities in Archaea: separation of archaeal phospholipid biosynthesis and glycerol catabolism by glycerophosphate enantiomers. J. Bacteriol. 1999;181:1330–1333. doi: 10.1128/jb.181.4.1330-1333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Nishihara M., Morii H., Matsuno K., Ohga M., Stetter K.O., Koga Y. Structural analysis by reductive cleavage with LiAlH4 of an allyl ether choline-phospholipid, archaetidylcholine, from the hyperthermophilic methanoarchaeon Methanopyrus kandleri . Archaea. 2002;1:123–131. doi: 10.1155/2002/682753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Overbeek R., Fonstein M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Saha S.K., Nishijima S., Matsuzaki H., Shibuya I., Matsumoto K. A regulatory mechanism for the balanced synthesis of membrane phospholipid species in Escherichia coli . Biosci. Biotechnol. Biochem. 1996;60:111–116. doi: 10.1271/bbb.60.111. [DOI] [PubMed] [Google Scholar]
- R30.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- R31.Shimada H., Nemoto N., Shida Y., Oshima T. Complete polar lipid composition of Thermoplasma acidophilum HO-62 determined by high-performance liquid chromatography with evaporative light-scattering detection. J. Bacteriol. 2002;184:556–563. doi: 10.1128/JB.184.2.556-563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Snel B., Bork P., Huynen M.A. The identification of functional modules from the genomic association of genes. Proc. Natl. Acad. Sci. USA. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Sohlenkamp C., De Rudder K.E., Rohrs V., Lopez-Lara I.M., Geiger O. Cloning and characterization of the gene for phosphatidylcholine synthase. J. Biol. Chem. 2000;275:18919–18925. doi: 10.1074/jbc.M000844200. [DOI] [PubMed] [Google Scholar]
- R34.Sohlenkamp C., Lopez-Lara I.M., Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003;42:115–162. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- R35.Sprott G.D., Agnew B.J., Patel G.B. Structural features of ether lipids in the archaeobacterial thermophiles Pyrococcus furiosus, Methanopyrus kandleri, Methanothermus fervidus, and Sulfolobus acidocaldarius . Can. J. Microbiol. 1997;43:467–476. [Google Scholar]
- R36.Sprott G.D., Sad S., Fleming L.P., Dicaire C.J., Patel G.B., Krishnan L. Archaeosomes varying in lipid composition differ in receptor-mediated endocytosis and differentially adjuvant immune responses to entrapped antigen. Archaea. 2003;1:151–164. doi: 10.1155/2003/569283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R37.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites; approximate method. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]