Abstract
Among the 78 eukaryotic ribosomal proteins, eleven are specific to Eukarya, 33 are common only to Archaea and Eukarya and 34 are homologous (at least in part) to those of both Bacteria and Archaea. Several other translational proteins are common only to Eukarya and Archaea (e.g., IF2a, SRP19, etc.), whereas others are shared by the three phyla (e.g., EFTu/EF1A and SRP54).
Although this and other analyses strongly support an archaeal origin for a substantial fraction of the eukaryotic translational machinery, especially the ribosomal proteins, there have been numerous unique and ubiquitous additions to the eukaryotic translational system besides the 11 unique eukaryotic ribosomal proteins. These include peptide additions to most of the 67 archaeal homolog proteins, rRNA insertions, the 5.8S RNA and the Alu extension to the SRP RNA. Our comparative analysis of these and other eukaryotic features among the three different cellular phylodomains supports the idea that an archaeal translational system was most likely incorporated by means of endosymbiosis into a host cell that was neither bacterial nor archaeal in any modern sense. Phylogenetic analyses provide support for the timing of this acquisition coinciding with an ancient bottleneck in prokaryotic diversity.
Keywords: Crenarchaea, eukaryotic origin, evolution, ribosome
Introduction
There have been a number of studies since the original work of Woese et al. (1990) on ribosomal RNA showing that many components of the eukaryotic translational system are closely related to those of the archaeal system. Given recent work showing that the ribosomal proteins common to both Archaea and Bacteria have an unusual phylodomain-specific block or segmental structure (Vishwanath et al. 2004), we have undertaken a detailed study of the eukaryotic ribosomal proteins. In particular, we have investigated their relationship to ribosomal proteins of the two current major divisions of Archaea. Such an investigation has the potential to provide some insight into fundamental questions on the origin of the complex structures of the eukaryotic cell. Although the origin of mitochondria via an ancient endosymbiotic event (Margulis and Bermudes 1985) is generally accepted, there are many competing theories on the origin and timing of the other eukaryotic components (Hartman 1984, Sogin 1991, Lake and Rivera 1994, Gupta and Golding 1996, Brown and Doolittle 1997, Moreira and Lopez-Garcia 1998, Horiike et al. 2001, Hartman and Fedorov 2002). Our results not only support the closeness of the eukaryotic to the archaeal ribosome but, in combination with other data (Hartman and Fedorov 2002, Vishwanath et al. 2004), support the idea that the eukaryotic ribosome was brought into a non-bacterial, non-archaeal host cell from a crenarchaeal ancestor very early in the history of Eukarya.
Materials and methods
Detailed multiple comparative sequence alignments across a wide taxonomic range of Eukarya (excluding reduced genome parasites), Archaea and Bacteria were carried out for the ribosomal proteins and several other translation-associated proteins. Only species for which there existed a draft of the entire genome were included in the study (for a list of species, see the caption to Figure 4). Each of the sequences annotated as ribosomal protein was searched through the set of genomes for both potential homologs and paralogs using PsiBlast (Altschul et al. 1997) and all suggestive matches were followed up. Next, all of the identified probable ribosomal proteins were clustered using CLUSTAL (Thompson et al. 1994). The primary results of these initial analyses was two-fold. First, no additional ribosomal proteins common to all three major phylodomains were identified; and second, when all three phylodomains were included, most of the alignments were only partial. However, within each phylodomain the alignments were nearly full-length, except for a few of the N- and C-terminal regions. To generate alignments that were as complete as possible, initial statistical profiles (Das and Smith 2000) for each of the identified segments common within and between the three phylodomains were constructed and used to generate initial full-species multiple alignments. These were then extended and refined by hand by exploiting the available structural and functional information (Ban et al. 2000, Brodersen et al. 2001, Klein et al. 2004) in constraining and adjusting the computer-generated alignments. In particular, the information was used to restrict alignment gaps to surface loop regions, maintain the alignment of probable rRNA-contacting, functionally equivalent residues and to maintain likely structurally equivalent patterns across all sequences whenever possible. The latter included maintaining hydrophobicity profiles and regions containing amino acids of high turn propensity. These procedures are described in Vishwanath et al. (2004). Finally, new statistical profiles were constructed for all segments or blocks (see Results) that were clearly alignable, and thus were assumed truly homologous among each phylodomain, for Bacteria and Archaea, for Eukarya and Archaea and for all three phylodomains. This was followed by full-sequence genome searches to ensure that apparent conserved patterns were truly characteristic of the various phylodomains over the widest possible set of sequenced genomes.
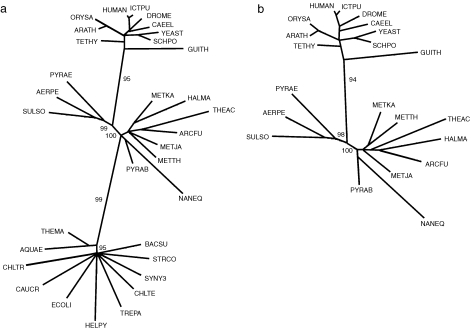
Figure 4.
Maximum-likelihood unrooted trees constructed from the positional variation among aligned sets of ribosomal proteins. (a) Tree constructed from the concatenated set of alignment blocks common in the 34 universal ribosomal proteins from the three phylodomains: Bacteria, Archaea and Eukarya, labeled by orange bars in Figure 3. (b) Tree constructed from the concatenated set of alignment blocks common to both the eukaryotic and archaeal representatives and labeled by purple bars in Figures 1 and 2. The trees were constructed by the maximum likelihood approach with the TREE-PUZZLE procedure, Quartet Puzzling, 1000 steps and the JTT model of substitution. Abbreviations are Swiss-Prot codes: Eukarya (top set): ARATH = Arabidopsis thaliana, CAEEL = Caenorhabditis elegans, DROME = Drosophila melanogaster, GUITH = Guillardia theta, HUMAN = Homo sapiens, ICTPU = Ictalurus punctatus, ORYSA = Oryza sativa, SCHPO = Schizosaccharomyces pombe, TETHY = Tetrahymena thermophila and YEAST = Saccharomyces cerevisiae; Archaea (middle set): AERPE = Aeropyrum pernix, ARCFU = Archaeoglobus fulgidus, HALMA = Haloarcula marismortui, METJA = Methanococcus jannischii, METKA = Methanopyrus kandleri, METTH = Methanobacterium thermoautotrophicum, NANEQ = Nanoarchaeum equitans, PYRAB = Pyrococcus abyssi, PYRAE = Pyrobaculum aerophilum, SULSO = Sulfolobolus solfataricus and THEAC = Thermoplasma acidophilum; Bacteria (bottom set): AQUAE = Aquifex aeolicus, BACSU = Bacillus subtilis, CAUCR = Caulobacter crescentus, CHLTR = Chlamydia trachomatis, CHLTE = Chlorobium tepidum, ECOLI = Escherichia coli, HELPY = Helicobacter pylori, STRCO = Streptomyces coelicolor, SYNY3 = Synechocystis sp. (strain PCC 6803), THEMA = Thermotoga maritima and TREPA = Treponema pallidum.
Full phylogenetic reconstruction was carried out based on the sequence variation within the ribosomal protein alignment sets. Considerable effort was expended to ensure that the implied results are robust. This involved comparing the results of multiple methodologies (Strimmer and von Haeseler 1997, Katoh et al. 2001, Kolaczkowski and Thornton 2004) and various combinations of subsets of the alignments. In particular, trees were constructed from the sequence variation for each of the three classes of ribosomal taxon-specific segments or blocks concatenated into various subsets, ranging from those of individual proteins, to the two ribosomal subunits separately, to the full set of all common blocks among the 34 universal ribosomal proteins.
Implied trees were assumed to be compatible if all the nested sets of one were either identical to or contained within those of the other. In other words, many of the implied trees constructed from single proteins showed little resolution within one or more of the phylodomains, yet did not cluster a mix of phylodomain members. For each ribosomal protein or its universal concatenated blocks, or both, statistics were obtained on the length and the number of conserved and informative positions (see Table 1). Here, informative position was defined as any position with two or more aligned distinct amino acids, each present in at least two species, implying a division of the species into at least one cluster and all of the rest.
Table 1.
Phylogenetic information statistics for individual ribosomal proteins and translation factors. The statistics were calculated from the multialignments generated as described in Materials and methods for the concatenation of the blocks within each protein common to all phylodomains considered. Five subsets of representative species were considered: AEB (Archaea, Eukarya, Bacteria), AE (Archaea, Eukarya), AB (Archaea, Bacteria), EC (Eukarya, Crenarchaea) and EY (Eukarya, Euryarchaea). For each subset, the table reports the percentage of informative (Inform.) and absolutely conserved (Abs. cons.) positions in the concatenated universal blocks (ccblk) of each protein separately. An informative position is defined as a position in the alignment where at least two different amino acids are observed, each in at least two different species. Phylogenetic tree reconstructions performed on each individual protein show resolution among the three phylodomains (B-A-E), resolution between the two archaeal divisions (C-Y) and clustering of the Eukarya with the Crenarachaea (E + C).
| Length | AEB (%) | AE (%) | AB (%) | EC (%) | EY (%) | Tree resolution | ||||||
| ccblk | Inform. | Abs. cons. | Inform. | Abs. cons. | Inform. | Abs. cons. | Abs. cons. | Abs. cons. | B-A-E | C-Y | E + C | |
| S2 | 197 | 93.9 | 3.0 | 85.3 | 7.1 | 87.3 | 5.1 | 10.7 | 8.6 | yes | yes | no |
| S3 | 158 | 91.8 | 2.5 | 83.5 | 5.7 | 89.9 | 3.2 | 12.0 | 6.3 | yes | yes | no |
| S4 | 104 | 95.2 | 1.9 | 78.8 | 11.5 | 91.3 | 5.8 | 19.2 | 17.3 | yes | no | no |
| S5 | 149 | 90.6 | 6.0 | 79.9 | 12.8 | 85.9 | 6.7 | 22.1 | 13.4 | yes | yes | yes |
| S7 | 133 | 91.0 | 4.5 | 71.4 | 16.5 | 88.7 | 5.3 | 30.8 | 17.3 | yes | partial | no |
| S8 | 97 | 91.8 | 6.2 | 79.4 | 7.2 | 88.7 | 7.2 | 15.5 | 9.3 | yes | partial | no |
| S9 | 138 | 84.8 | 7.2 | 77.5 | 13.0 | 84.1 | 8.0 | 18.1 | 13.8 | yes | yes | no |
| S10 | 94 | 92.6 | 5.3 | 88.3 | 5.3 | 87.2 | 7.4 | 9.6 | 5.3 | yes | partial | no |
| S11 | 125 | 81.6 | 7.2 | 60.0 | 20.0 | 79.2 | 8.8 | 39.2 | 21.6 | yes | yes | no |
| S12 | 121 | 86.0 | 7.4 | 57.9 | 22.3 | 81.8 | 11.6 | 32.2 | 26.4 | yes | yes | no |
| S13 | 123 | 89.4 | 7.3 | 79.7 | 11.4 | 87.8 | 8.9 | 17.9 | 12.2 | yes | yes | no |
| S14 | 49 | 89.8 | 8.2 | 67.3 | 12.2 | 87.8 | 12.2 | 16.3 | 12.2 | B-A-E | partial | no |
| S15 | 59 | 94.9 | 3.4 | 84.7 | 8.5 | 93.2 | 3.4 | 15.3 | 8.5 | yes | no | no |
| S17 | 85 | 89.4 | 1.2 | 80.0 | 7.1 | 85.9 | 2.4 | 15.3 | 14.1 | yes | yes | no |
| S19 | 29 | 93.1 | 0.0 | 82.8 | 10.3 | 89.7 | 0.0 | 13.8 | 10.3 | yes | no | no |
| TOTAL | 1661 | 90.1 | 4.9 | 77.1 | 11.6 | 86.8 | 6.6 | 19.7 | 13.3 | |||
| L1 | 198 | 95.5 | 2.0 | 92.9 | 4.5 | 91.9 | 4.0 | 6.6 | 5.6 | yes | no | no |
| L2 | 168 | 88.1 | 8.3 | 78.0 | 13.1 | 85.1 | 10.1 | 22.6 | 14.9 | yes | yes | yes |
| L3 | 157 | 90.4 | 5.1 | 80.9 | 11.5 | 87.9 | 6.4 | 16.6 | 13.4 | yes | partial | no |
| L4 | 202 | 99.0 | 0.5 | 87.6 | 6.4 | 99.0 | 0.5 | 7.9 | 7.4 | yes | no | no |
| L5 | 147 | 93.9 | 4.8 | 81.0 | 9.5 | 90.5 | 6.1 | 19.7 | 9.5 | yes | no | no |
| L6 | 191 | 95.8 | 1.6 | 90.1 | 5.2 | 91.6 | 1.6 | 10.5 | 6.3 | yes | yes | no |
| L10 | 102 | 98.0 | 1.0 | 95.1 | 1.0 | 97.1 | 2.9 | 2.9 | 1.0 | no | no | no |
| L11 | 143 | 97.2 | 1.4 | 93.7 | 2.1 | 90.9 | 1.4 | 4.2 | 8.4 | yes | partial | no |
| L12 | 82 | 98.8 | 0.0 | 96.3 | 1.2 | 91.5 | 1.2 | 1.2 | 3.7 | yes | partial | no |
| L13 | 109 | 92.7 | 3.7 | 89.0 | 4.6 | 90.8 | 6.4 | 7.3 | 8.3 | yes | partial | no |
| L14 | 124 | 83.9 | 8.1 | 71.8 | 14.5 | 79.8 | 9.7 | 23.4 | 15.3 | yes | yes | no |
| L15 | 137 | 96.4 | 1.5 | 84.7 | 7.3 | 92.7 | 2.2 | 15.3 | 8.0 | yes | yes | no |
| L18 | 115 | 93.0 | 3.5 | 80.9 | 13.9 | 92.2 | 3.5 | 19.1 | 13.9 | yes | partial | no |
| L22 | 112 | 96.4 | 0.9 | 85.7 | 6.3 | 96.4 | 1.8 | 8.0 | 7.1 | yes | partial | no |
| L23 | 79 | 97.5 | 0.0 | 83.5 | 5.1 | 92.4 | 0.0 | 10.1 | 6.3 | yes | no | no |
| L24 | 84 | 94.0 | 2.4 | 85.7 | 3.6 | 90.5 | 3.6 | 6.0 | 3.6 | yes | no | no |
| L29 | 63 | 96.8 | 1.6 | 92.1 | 3.2 | 95.2 | 1.6 | 9.5 | 3.2 | no | no | no |
| L30 | 57 | 98.2 | 0.0 | 94.7 | 3.5 | 91.2 | 0.0 | 8.8 | 3.5 | yes | yes | yes |
| TOTAL | 2270 | 94.5 | 2.8 | 86.4 | 7.0 | 91.4 | 3.8 | 11.7 | 8.3 | |||
| EF1 | 334 | 79.3 | 16.5 | 64.4 | 27.5 | 77.25 | 17.66 | 34.13 | 31.44 | yes | yes | yes |
| EFG | 548 | 87.4 | 9.3 | 78.6 | 15.7 | 78.83 | 12.77 | 20.44 | 17.52 | yes | yes | yes |
| IF2 | 260 | 87.7 | 7.7 | 79.6 | 10.0 | 80.00 | 12.69 | 12.69 | 11.54 | yes | yes | yes |
Results
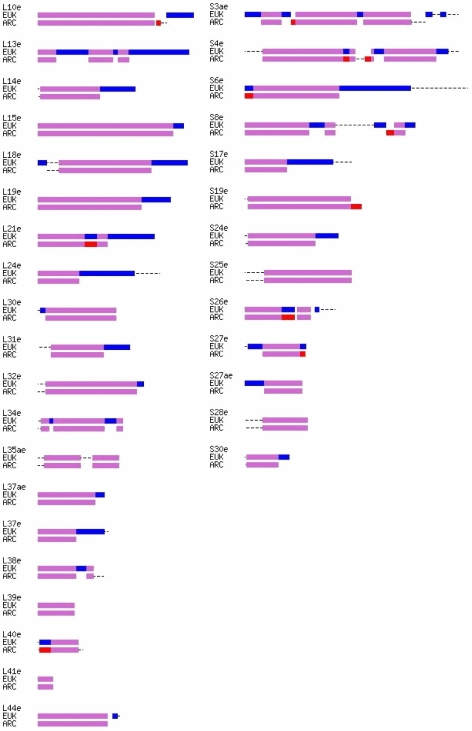
Several key points were made clear by the resulting multi-sequence alignments and database searches. As previously reported (Lecompte et al. 2002), there are 11 unique and ubiquitous eukaryotic ribosomal proteins with no archaeal or bacterial homologs; there is a clear homology of 67 eukaryotic ribosomal proteins with those of Archaea; and 33 of these 67 eukaryotic-archaeal homologs have no bacterial homologs (Figure 1). Finally, the remaining 34 of those 67 are universal ribosomal proteins found in all three phylodomains (Figure 2). What is new and supported by this work is that the eukaryotic members among these 34 clearly show closer similarity to the crenarchaeal division of Archaea (Table 1). These results were obtained through analysis of the positional conservation from the alignments of the 34 universal proteins across all three phylodomains. The alignments show a well-defined block or segmental structure associated uniquely to one, two or all three of the phylodomains (Vishwanath et al. 2004) (Figure 3). Critically, the current analyses show that eukaryotic homologous proteins contain only the archaeal-specific blocks and no bacterial-specific blocks, with the possible but unclear exception of S3p (Figure 3). In addition, the vast majority of the 67 eukaryotic-archaeal ribosomal protein homologs contain eukaryotic-specific sequence additions, many of which are ubiquitous to all known Eukarya, as indicated in Figure 4. The full detailed amino-acid-level alignments are available on the web at http://bmerc.bu.edu.
Figure 1.
A schematic representation of the multiple alignments of the 33 ribosomal proteins common to Archaea and Eukarya. Purple marks segments alignable, and thus homologous, across both phylogenetic domains; blue marks segments unique and alignable only within Eukarya; red marks segments alignable only within Archaea. Dotted lines represent sequence regions of varying length that are unalignable across the full sets of either Eukarya or Archaea. The representative species are listed in the legend of Figure 4.
Figure 2.
A schematic representation of the multiple alignments of the archaeal and eukaryotic species for the 34 universal ribosomal proteins. Purple marks segments alignable, and thus homologous, across both the eukaryal and archaeal phylogenetic domains; blue marks segments unique and alignable only among Eukarya; red marks segments alignable only among Archaea. Dotted lines represent sequence regions of varying length that are unalignable across the full sets of either Eukarya or Archaea. The representative species are listed in the legend of Figure 4. Ribosomal protein L7ae has been identified in some, but not all, major representative Bacteria, and is therefore neither universal nor eukaryotic-archaeal specific.
Figure 3.
A schematic representation of the multiple alignments of the 34 universal ribosomal proteins common to Eukarya, Archaea and Bacteria. Orange marks segments alignable, and thus homologous, across all three phylogenetic domains; purple marks blocks alignable across Eukarya and Archaea; blue marks those blocks alignable only with in the Eukarya; red marks those blocks alignable only within the Archaea; dark green marks blocks alignable only across Archaea and Bacteria, while light green marks those alignable only within the Bacteria. The dotted lines represent sequence regions of varying length unalignable across the full sets of any of the three groups. The representative species are listed in the legend of Figure 4. Ribosomal protein L7ae has been identified in some, but not all, major representative Bacteria.
Using the alignable regions of all 67 ribosomal proteins common to Eukarya and Archaea, as well as various subsets, probable phylogenetic relationships were constructed, as described in Materials and methods. First, as noted by others (Lecompte et al. 2002), there is a clear separation of the three major phylodomains for all subsets of the data, excluding the individual ribosomal proteins for which there were insufficient numbers of aligned informative positions (Table 1). In addition, the concatenation of the alignable regions among both the full 67- and the 33-protein subset of only those common to Archaea and Eukarya provided a clear three-way separation between Eukarya, Crenarchaea and Euryarchaea (Figure 4b). The same relationship was obtained using only the blocks common to both Eukarya and Archaea found in the 34 universal ribosomal proteins. The implied close association between Eukarya and Crenarchaea is more clearly seen in the numbers of conserved positions between the two archaeal divisions on the one hand and Eukarya on the other hand, as displayed in Table 1. This is also supported by the presence of five proteins (S25e, S26e, S30e, L13e, L38e) that are apparently shared universally only by Crenarchaea and Eukarya (Lecompte et al. 2002). The concatenated alignable common sequence segments in the 34 universal ribosomal proteins provided a highly statistically significant and consistent separation among Eukarya, Crenarchaea, Euryarchaea and Bacteria for both the large and small subunits independently. For phylogenetic analyses based on many individual proteins, the limited number of informative variable positions either failed to fully resolve these divisions or produced low bootstrap values. The positional variation statistics and clustering resolutions are listed in Table 1. A number of other translation-related proteins, including the elongation and initiation factors EF2/EFG and IF2P/IF2, not only showed a similar taxon-specific block structure, but displayed significantly higher eukaryotic similarity to Crenarchaea than to Euryarchaea, as observed earlier (Rivera and Lake 1992). Phylogenetic reconstruction with these proteins, combined with other translational proteins (e.g., EFTu/EF1a, SRP54 and SRP19), again generated the same four-division clustering shown in Figure 4a as a concatenated set as well as individual proteins.
The two archaeal divisions coalesce for the concatenated set of ribosomal proteins, L23p, L29p, L5p and L18p, which interact respectively with the SRP complex and the 5S rRNA (Speek and Lind 1982), as happens with SRP54 and SRP19. These were the only small ribosomal protein sets with significant statistical resolving power that did not resolve the two major archaeal divisions relative to Eukarya. There are other proteins that clearly support the closeness of Eukarya and Archaea (Puhler et al. 1989), such as the DNA clamp protein with its common six-domain trimer symmetry, that also fail to resolve these two archaeal divisions. These results may provide information on the relative age of these components within Eukarya.
Only 23 of the total 67 common archaeal-eukaryotic ribosomal proteins contain neither N- nor C-terminal eukaryotic ubiquitous extensions, nor insertions relative to their archaeal homologs (Figure 3). Eleven proteins in the large subunit, L4p, L29p, L13p, L13e, L14e, L18e, L19e, L21e, L24e, L31e and L37e, show significant eukaryotic-specific C-terminal extensions, whereas only S6e and S17e have such C-terminal extensions within the small subunit. Large eukaryotic-specific N-terminal extensions are rarer and are found only in L23p, L30p, S27ae and S27e. As in many other protein families, there are also substantial additional length variations at both eukaryotic protein termini that have very limited eukaryotic phylogenetic correlation. With the exception of L13e, the eukaryotic-specific internal sequence insertions are relatively small. Finally, there are three eukaryotic-archaeal homologs, L21ae, S3p and S26e, that appear to have commensurate insertions in both Eukarya and Archaea, these being regions that show no sequence similarity yet are found between clearly alignable segments. Whether these blocks are actually homologous remains unclear without full comparative structural information.
To complete the study, the mitochondrial ribosomal proteins homologous to the 34 universal ribosomal proteins were aligned and subjected to phylogenetic analysis. The 34 mitochondrial ribosomal proteins can be aligned with the bacterial proteins, although they display considerable modification and variation. In general, they are not alignable with their archaeal and eukaryotic homologs over any significant contiguous regions. This is not only the result of the inclusion of archaeal-specific blocks, but of the increased variation within the blocks common to all. Thus, no consistent, statistically significant trees could be obtained when the mitochondrial ribosomal proteins were added to those of the three major phylodomains, due largely to the limited variation in reliably alignable regions.
Discussion
That 67 out of 68 archaeal ribosomal proteins have clear eukaryotic homologs and that 34 of these eukaryotic ribosomal proteins within the universal set contain the archaeal-specific block structure, implies that these eukaryotic ribosomal proteins are predominantly of archaeal origin. The inclusion of the archaeal-specific blocks at the exclusion of bacterial-specific blocks also implies that the incorporation of the 34 universal proteins into the eukaryotic cell must have occurred long after the last common ancestor of modern Archaea and Bacteria. The latter is in part represented by the common blocks within these universal ribosomal proteins. A comparison of the sequence divergence within the three different phylodomain-specific block types suggests that, within the large uncertainties common to such analyses, the time from the last common ancestor of Archaea and Bacteria to the origin of the modern eukaryotic cell is nearly the same as the time from the origin of the eukaryotic cell to the present. The set of ribosomal proteins with no recognizable bacterial or archaeal homologs, unique and ubiquitous to modern eukaryotes, suggests an additional source.
It has long been noted that many of the eukaryotic metabolic proteins are closely related to those found in modern Bacteria. This has been explained as the result of a genetic transfer from the endosymbiotic precursors of the mitochondria and chloroplasts. If this is correct, the subtraction of those components from the eukaryotic proteome provides insight into an original host cell’s makeup, perhaps largely living by the phagocytosis of Bacteria or Archaea or both. The same subtractive logic should be applicable to the closest archaeal related components. The resulting remainders from the translational system are: (1) the 11 eukaryotic-specific ribosomal proteins; (2) the eukaryotic ubiquitous ribosomal protein extensions; (3) the insertions and additions to the eukaryotic rRNA; and (4) a number of other translational proteins. All suggest a non-archaeal, non-bacterial origin for these translational components.
This same subtractive logic was used in the identification of eukaryotic signature proteins (ESPs) found ubiquitously in eukaryotic cells having no obvious bacterial or archaeal homologs (Hartman and Fedorov 2002). In addition to the eukaryotic-specific ribosomal proteins, these ESPs include a considerable number of RNA processing proteins, many signaling proteins, cytoskeleton proteins such as actin and tubulin and motor proteins such as kinesin and myosin. Eukarya use membrane proteins and a calcium ion gradient to control their unique ability to phagocytize prokaryotes and to control motility (Berridge et al. 2003). This control by the calcium ion is mediated through Eukarya-unique cytoskeleton (microtubules and actin filaments) and sliding filament motility by a motor protein (i.e., myosin). Bacteria and Archaea use membrane proteins and a proton gradient for motility and ATP synthesis (Harold 1977). This suggests that the direct ancestors of modern Archaea or modern Bacteria were not the source of these proteins and processes. One must assume that, like many differences between Bacteria and Archaea, the far more distinctive features of the eukaryotic cell accumulated over a long time period. Yet the mitochondria, and now the apparent crenarchaean hybrid ribosome, support the view that much of the complexity of Eukarya arose through symbiosis on a short time scale.
There are several current hypotheses concerning the origin of the eukaryotic cell and its various components, in particular its nucleus: (i) the nucleus formed autogenously, perhaps in a bacterial ancestral cell through a membrane surrounding the bacterial DNA, as currently observed in Planctomycetes (Lindsay et al. 2001); (ii) the nucleus formed through fusion of a bacterium and an archaeon (Lake and Rivera 1994, Gupta and Golding 1996, Martin and Müller 1998, Moreira and Lopez-Garcia 1998); and (iii) the nucleus formed through endosymbiosis, rather than fusion, of an archaeon or a bacterium into a distinct host cell. At least three candidates have been proposed for a potential host cell: a bacterium (Lake and Rivera 1994, Brown and Doolittle 1997, Horiike et al. 2001); an archaeon (Brown and Doolittle 1997, Martin and Müller 1998, as a strict hydrogen-dependent, autotrophic); and a chronocyte, an RNA-based cell (Hartman 1984, Sogin 1991, Hartman and Fedorov 2002). In all such hypotheses, it has been assumed that the appearance of mitochondria and chloroplasts was the result of later or possibly concurrent endosymbioses (Margulis and Bermudes 1985).
The simple fusion hypotheses or mutual endosymbiosis of a bacterium and an archaeon do not provide a direct explanation for the presence of ESPs. In particular, the hypothesis that a bacterial host or a bacterial membrane surrounding its DNA to form the nucleus would not explain why the eukaryotic ribosomal protein set has no bacterial-like blocks independent of those shared with Archaea. Thus, even though the majority of the mitochondrial ribosomal proteins are encoded in the modern eukaryotic nuclear genome, there is no detectable recombination between these mitochondrial riboproteins or their bacterial precursor and their nuclear-encoded archaeal or eukaryotic homologs. The latter appears to set limits on the interchangeability or compatibility of the two prokaryotic ribosomal protein components, and thus on the likely success of any horizontal transfers. This is in contrast with the evidence of such transfers of tRNAs, their synthetases and some enzymes. It has been assumed that successful horizontal gene transfers not only require significant selective forces, but the transferred proteins have few required complex interactions. Whereas the tRNAs interact with the full ribosomal complex, they are notable in being nearly identical in structure and function in all three domains. It is possible, but unlikely, that all modern Bacteria and Archaea acquired their unique and ubiquitous ribosomal components via massive horizontal gene transfer across their wide ranges of environments.
We therefore suggest that the eukaryotic ribosome is a modified ancient archaeal ribosome that was brought into a distinct host and was complemented by the addition of extra proteins, additional RNAs and peptide and RNA insertions. It is now clear that archaeal endosymbiosis can happen, as observed in the endosymbiosis of a methanogenic archaeon by an anaerobic ciliate (Van Hoek et al. 2000). The reason for suggesting a possibly more ancient original RNA-based host, a chronocyte, is the large role played by RNA in eukaryotic cells, such as in retroposons, spliceosomes and the large number of small and large non-protein coding RNAs, many of which are thought to be essential in gene regulation. However, such an RNA-based host would require a protein synthesis system compatible (e.g., having the same genetic code and tRNA) with that of the acquired ancestral crenarchaeal ribosome at the time of such an endosymbiotic takeover. This in turn points to an early common ancestry for this common protein encoding system, among the earliest cellular life that later led to Bacteria, Archaea and the pre-eukaryotic host.
As shown in this work, the eukaryotic ribosomal proteins are more similar to those of Crenarchaea than of Euryarchaea. On the other hand it is Euryarchaea, containing an archaeal histone associated with its DNA, that is a probable precursor to the eukaryotic histones (Bailey et al. 2000). This implies that there were at least two endosymbioses of Archaea into a host cell, one leading to the formation of a nucleus with histone-packaged DNA (from a euryarchaeon as endosymbiont) and the other leading to the takeover of the cytoplasmic translational apparatus (from a crenarchaeon as a endosymbiont). Although the majority of eukaryotic riboproteins clearly show closer relatedness to those of Crenarchaea than of Euryarchaea, there are a few informational proteins that show no clear separation between Crenarchaea and Euryarchaea. These include SRP54 and those ribosomal proteins contacting the 5S RNA and the SRP. It is thus possible that SRP54 and its RNA-bound loop (Nagai et al. 2003), the 5S RNA and even the 5.8S and their ribosomal-contacting proteins, are more ancient and represent parts of an older protein synthesis system.
The block structure observed in the majority of proteins related to polypeptide synthesis suggests that the aforementioned proposed endosymbiotic events occurred after a catastrophe that enveloped the cellular biosphere. In our recent paper on the block structure of the 34 universal ribosomal proteins among the Prokaryota (Vishwanath et al. 2004), we noted that phylogenetic analyses implied the occurrence of some series of catastrophic events or the equivalence of an evolutionary bottleneck that led to a major reduction in prokaryotic cellular diversity after the divergence of the lines of descent leading to the common ancestors of modern Bacteria and Archaea. Steitz and co-workers (Klein et al. 2004) recently identified a set of ribosomal proteins in Bacteria and Archaea that show no sequence or structural similarity, but bind to the ribosome in identical positions and have loops or terminal extensions making nearly identical contact with their respective rRNAs (L44e/L33, L21e/L27, L31e/L17, L37e/ L34, L15e/L31, L24e/L19). We observed a similar situation in two universal ribosomal proteins, S8 and L4, where sequence blocks showed no similarity but the contacts of these blocks with the rRNA were almost identical (Vishwanath et al. 2004). This implies that multiple alternative solutions to the roles played by individual ribosomal proteins, such as stabilizing the structure and restricting the RNA fold space, were possible (Favaretto et al. 2005). Given the significant differences in these proteins and protein blocks there appears little reason to assume they were the only successful solutions. Yet only two types have survived to the present: one solution adopted by the last common ancestor of all modern Archaea, and the other by the ancestor of all modern Bacteria. Neither cross-species coalescence nor massive horizontal gene transfers seem as likely as one or more diversity-reducing events.
The present analysis suggests that the eukaryotic cell incorporated its modern translational machinery from a crenarchaeal ancestor at about the same time as the probable prokaryotic bottleneck noted above. It is possible that besides the proposed radical cooling of the earth, i.e., the snowball hypothesis (Kirschvink et al. 2000), the introduction of oxygen represented a major cause of this evolutionary bottleneck. Thus, because extreme selective pressures would have been acting at that time, only those pre-eukaryotic host organisms (i.e., Chronocyte) that took up Bacteria (Martin et al. 2001, Esser et al. 2004) for oxygen protection and utilization and that possessed the more selectively advantageous crenarchaeal translational system, may have survived. That pre-eukaryotic host would be one of the last representatives of the very ancient RNA-based cellular organism with an older protein coding system.
Acknowledgments
This paper was supported in part by NSF Grant No. DBI-0205512. The authors would like to thank Prashanth Vishwanath for helpful discussions.
References
- R1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Bailey K., Pereira S.L., Widom J., Reeve J.N. Archaeal histone selection of nucleosome positioning sequences and the prokaryotic origin of histone-dependent genome evolution. J. Mol. Biol. 2000;303:25–34. doi: 10.1006/jmbi.2000.4128. [DOI] [PubMed] [Google Scholar]
- R3.Ban N., Nissen P., Hansen J., Moore P., Steitz T. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- R4.Berridge M., Bootman M., Roderick H. Calcium signalling: dynamics, homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- R5.Brodersen D.E., Clemons W.E., Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16S RNA. J. Mol. Biol. 2001;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- R6.Brown J.R., Doolittle W.F. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Das S., Smith T. Identifying nature’s protein lego set. In: Kim P., editor. Advances In Protein Chemistry. San Diego: Academic Press; 2000. pp. 159–183. [DOI] [PubMed] [Google Scholar]
- R8.Esser C., Ahmadinejad N., Wiegand C., et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- R9.Favaretto P., Bhutkar A.J., Smith T. Constraining ribosomal RNA conformational space. Nucleic Acids Res. 2005;33:5106–5111. doi: 10.1093/nar/gki805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Gupta R., Golding G. The origin of the eukaryotic cell. Trends Biochem. Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- R11.Harold F. Ion currents and physiological functions in microorganisms. Annu. Rev. Microbiol. 1977;31:181–203. doi: 10.1146/annurev.mi.31.100177.001145. [DOI] [PubMed] [Google Scholar]
- R12.Hartman H. The origin of the eukaryotic cell. Specul. Sci. Technol. 1984;7:77. [PubMed] [Google Scholar]
- R13.Hartman H., Fedorov A. The origin of the eukaryotic cell: a genomic investigation. Proc. Natl. Acad. Sci. USA. 2002;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Horiike T., Hamada K., Kanaya S., Shinozawa T. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria is revealed by homology-hit analysis. Nat. Cell Biol. 2001;3:210–214. doi: 10.1038/35055129. [DOI] [PubMed] [Google Scholar]
- R15.Katoh K., Kuma K., Miyata T. Genetic algorithm-based maximum-likelihood analysis for molecular phylogeny. J. Mol. Evol. 2001;53:477–484. doi: 10.1007/s002390010238. [DOI] [PubMed] [Google Scholar]
- R16.Kirschvink J.L., Gaidos E.J., Bertani L.E., Beukes N.J., Gutzmer J., Maepa L.N., Steinberger R.E. Paleoproterozoic snowball earth: extreme climatic and geochemical global change and its biological consequences. Proc. Natl. Acad. Sci. USA. 2000;97:1400–1405. doi: 10.1073/pnas.97.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Klein D., Moore P., Steitz T. The roles of ribosomal proteins in the structure, assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- R18.Kolaczkowski B., Thornton J. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- R19.Lake J., Rivera M. Was the nucleus the first endosymbiont? Proc. Natl. Acad. Sci. USA. 1994;91:2880–2881. doi: 10.1073/pnas.91.8.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Lecompte O., Ripp R., Thierry J.-C., Moras D., Poch P. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Lindsay M., Webb R., Strous M., Jetten M., Butler M., Forde R., Fuerst J. Cell compartmentalization in planctomycetes: novel types of structural organization for the bacterial cell. Arch. Microbiol. 2001;175:413–429. doi: 10.1007/s002030100280. [DOI] [PubMed] [Google Scholar]
- R22.Margulis L., Bermudes D. Symbiosis as a mechanism of evolution: status of cell symbiosis theory. Symbiosis. 1985;1:101–124. [PubMed] [Google Scholar]
- R23.Martin W., Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- R24.Martin W., Hoffmeister M., Rotte C., Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol. Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- R25.Moreira D., Lopez-Garcia P. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J. Mol. Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- R26.Nagai K., Oubridge C., Kuglstatter A., Menichelli E., Isel C., Jovine L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Puhler G., Leffers H., Gropp F., Palm P., Klenk H.P., Lottspeich F., Garrett R.A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl. Acad. Sci. USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Rivera M.C., Lake J.A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992;257:74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- R29.Sogin M. Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- R30.Speek M., Lind A. Structural analyses of E.coli 5S RNA fragments, their associates and complexes with proteins L18 and L25. Nucleic Acids Res. 1982;10:947–965. doi: 10.1093/nar/10.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R31.Strimmer K., von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Van Hoek A., van Alen T., Sprakel V., Leunissen J., Brigge T., Vogels G., Hackstein J. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 2000;17:215–258. doi: 10.1093/oxfordjournals.molbev.a026304. [DOI] [PubMed] [Google Scholar]
- R34.Vishwanath V., Favaretto P., Hartman H., Mohr S., Smith T. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol. Phylogenet. Evol. 2004;33:615–625. doi: 10.1016/j.ympev.2004.07.003. [DOI] [PubMed] [Google Scholar]
- R35.Woese C., Kandler O., Wheelis M. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]