Abstract
Xylem vessel elements are hollow cellular units that assemble end-to-end to form a continuous vessel throughout the plant body; the xylem vessel is strengthened by the xylem elements' reinforced secondary cell walls (SCWs). This work aims to unravel the contribution of unknown actors in xylem vessel differentiation using the model in vitro cell culture system of Zinnia elegans differentiating cell cultures and the model in vivo system of Arabidopsis thaliana plants. Tracheary Element Differentiation-Related6 (TED6) and TED7 were selected based on an RNA interference (RNAi) screen in the Zinnia system. RNAi reduction of TED6 and 7 delayed tracheary element (TE) differentiation and co-overexpression of TED6 and 7 increased TE differentiation in cultured Zinnia cells. Arabidopsis TED6 and 7 were expressed preferentially in differentiating vessel elements in seedlings. Aberrant SCW formation of root vessel elements was induced by transient RNAi of At TED7 alone and enhanced by inhibition of both TED6 and 7. Protein–protein interactions were demonstrated between TED6 and a subunit of the SCW-related cellulose synthase complex. Our strategy has succeeded in finding two novel components in SCW formation and has opened the door for in-depth analysis of their molecular functions.
INTRODUCTION
Recent genome sequencing has defined the number of genes responsible for plant life: ∼27,000 genes in Arabidopsis thaliana, ∼45,000 genes in Populus species, and ∼41,000 genes in Oryza sativa (Sterck et al., 2007). Associated microarray expression analyses have allowed us to identify subsets of genes specifically associated with developmental or stress-related processes. Nevertheless, a large proportion (∼25% in the Arabidopsis genome) of genes present in plant genomes still have no attributed function (Berardini et al., 2004). The goal is now to actively find the roles of such unknown genes for a better understanding of the overall processes.
To define the gene functions required for a distinct developmental process, many studies have been conducted on the differentiation of typical cell types. An example is the Zinnia elegans in vitro differentiation system in which freshly isolated mesophyll cells can be specifically induced to transdifferentiate into tracheary elements (TEs) by the application of auxin and cytokinin (Fukuda and Komamine, 1980). TEs form xylem vessel strands through the tissues of vascular plants to transport water, nutrients, and signaling molecules (Fukuda, 1996). To be fully functional, mature vessel elements are first strengthened with thick cell walls, called secondary cell walls (SCWs), which are laid down beneath thin primary cell walls in some specialized cells and are composed mostly of cellulose, hemicellulose, and lignin; they then lose their cellular content and are finally perforated, thereby forming a reinforced hollow cylinder with an accessible lumen suitable for xylem sap conduction (Aloni, 1987; Turner et al., 2007). To unravel the specific gene expression associated with TE formation, EST sequencing and microarray expression profiling have been performed (Demura et al., 2002; Milioni et al., 2002; Pesquet et al., 2005). Although the Zinnia TE differentiation system represents an excellent source of gene candidates, functional analysis of these genes during the differentiation process remained inaccessible until recently because an effective method of direct transformation of the in vitro system has only just been developed (Endo et al., 2008). Using this method, nucleic acids, including plasmid DNAs and double-stranded RNAs (dsRNAs), can be introduced and subsequently either activate gene expression or specifically silence the expression of target genes, respectively, in differentiating Zinnia cells (Endo et al., 2008).
Combining this recently developed transient transformation method and the global transcriptome analyses of in vitro Zinnia TE differentiation, we performed an RNA interference (RNAi) screen with selected genes of unknown function expressed at specific stages of TE differentiation to identify gene candidates that directly affect TE differentiation. The candidates were further analyzed using both the Zinnia in vitro system and Arabidopsis whole plants to define the role of these unknown genes in TE differentiation. In this study, we focused on two closely related unknown genes that are expressed when TEs are forming their SCWs: TE Differentiation-Related6 (TED6) and TED7, designated according to Demura and Fukuda (1994). Through the over- and underexpression of these genes in both differentiating Zinnia cell cultures and transgenic Arabidopsis seedlings, we unraveled their function in SCW deposition and their potential interaction with the SCW synthesis machinery.
RESULTS
Screening Genes with Unknown Function by RNAi in the Zinnia TE Differentiation System
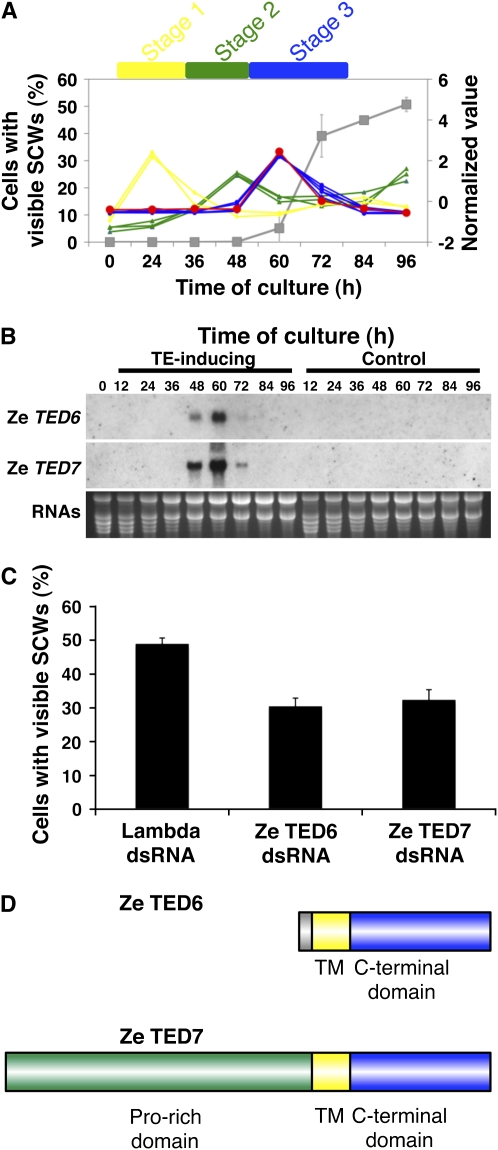
Using a recently developed dsRNA-mediated RNAi method (Endo et al., 2008), a preliminary genetic screen to identify gene candidates directly affecting the rate of TE differentiation was performed using genes with specifically upregulated expression at different time points along the TE differentiation time course. These genes were identified by microarray analysis and can be classified into stage 1, 2, and 3 genes based on their expression patterns during TE differentiation (Figures 1A and 1B; Demura et al., 2002). Z1943 is a Zinnia EST clone selected in the preliminary screening as one candidate from the stage 3 genes and whose corresponding unique gene was named TED6 (Figure 1A). dsRNA-mediated RNAi for TED6 resulted in a significant decrease in the number of cells with visible SCWs, which represented TEs, in the in vitro TE differentiation culture, as compared with a control using dsRNA corresponding to a partial sequence of λDNA (Figure 1C). Since TED6 encodes a previously uncharacterized protein with no attributed function, we decided to analyze the function of TED6 further. Another EST clone (Z16653) was identified with significant sequence similarity to TED6 (Figure 1D; see Supplemental Figure 1A online), and its corresponding gene was thereafter named TED7. TED7 also has stage 3–specific expression (Figure 1B), and dsRNA-mediated RNAi for TED7 also significantly reduced the rate of SCW formation (Figure 1C). Although two alleles of TED7, TED7-1 and TED7-2, were found in the genome of Z. elegans (cv Canary Bird) (see Supplemental Figure 1A online), genotyping revealed that most individual plants have TED7-1 (see Supplemental Figure 1B online). The predominant TED7-1 sequence was therefore used as the Zinnia TED7 gene in all of the following experiments.
Figure 1.
Selection of Ze TED6 and Ze TED7 Genes Using a Zinnia TE Differentiating Culture System.
(A) Time line of marker gene expression and TE differentiation in the Zinnia system. The graph was drawn from previously published microarray data (Demura et al., 2002). The expression data were modified to show changes in expression level as average = 0 and sd = 1 for each gene on the right-side y axis. Stage 1 markers (yellow diamonds: PI1 and PI2), stage 2 markers (green triangles: TED2, TED3, and TED4), stage 3 markers (blue circles: ZEN1, ZRNaseI, ZCP4, and CesA genes), and TED6 (Z1943; large red circles). Gray squares on the left-side y axis show TE differentiation in native Zinnia culture conditions. TE differentiation is represented by the numbers of cells with visible SCWs, which are seen only in differentiating and mature TEs.
(B) RNA gel blot analysis of TED6 and TED7 transcripts. RNAs were prepared from cells cultured for 0, 12, 24, 36, 48, 60, 72, 84, and 96 h in the TE-inducing medium and a control medium (in which no TE differentiation occurs). The blotted RNAs were probed with labeled cRNA of Z1943 and Z16653 for TED6 and TED7, respectively. The bottom panel shows ethidium bromide staining of RNAs on gel as a loading control.
(C) Effects of TED6 and TED7 dsRNAs on SCW formation. Zinnia cells that had been electroporated with indicated dsRNAs were induced to transdifferentiate into TEs, and the numbers of cells with visible SCWs were counted at 96 h of culture. Each data point presents the mean of three independent cultures ± sd.
(D) Structures of TED6 and TED7 proteins. Amino acid sequences deduced from cDNAs show that they have single transmembrane (TM) domains and relatively similar C-terminal domains (see Supplemental Figure 1A online). TED7 has a Pro-rich sequence at the N terminus.
Functional Analysis of TED6 and TED7 Genes in Zinnia TEs
Bioinformatic analysis (with SOSUI, TMHMM, and SignalP) of TED6 and TED7 proteins predicts that both are type I membrane proteins (single-pass transmembrane proteins with extracellular or luminal N termini and cytoplasmic C termini). They both contain a single transmembrane domain-like hydrophobic region in common (representing 23 of 95 and 23 of 300 amino acids of TED6 and TED7 proteins, respectively), and their C termini are predicted to be at the cytoplasm side (Figure 1D). Nevertheless, no functional domains were predicted by ProDom, PROSITE, or Pfam. Only a Pro-rich region was identified in the N terminal region of the TED7 protein. Among extracellular proteins in plants, Pro-rich sequences are a well-known characteristic of hydroxyproline-rich glycoproteins (HRGPs). Although TED7 could be an HRGP, it does not have any of the repetitive motifs typically found in HRGP families.
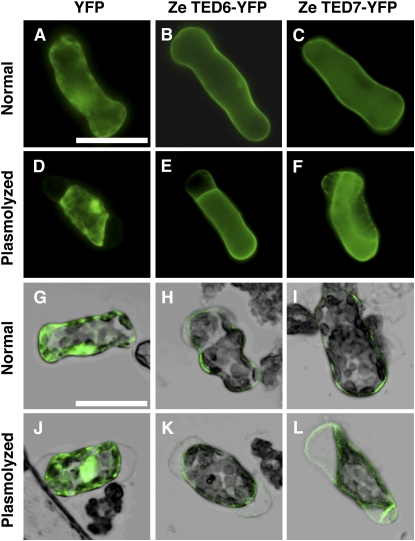
The subcellular localization of TED6 and TED7 was experimentally examined by introducing plasmids for the cauliflower mosaic virus (CaMV) 35S promoter-driven fusion proteins with a yellow fluorescent protein (YFP) at their C termini into Zinnia mesophyll cells by electroporation (Endo et al., 2008). The fluorescent signal for both fusion proteins was only detected at the peripheral region of the cells, suggesting a plasma membrane and/or cell wall localization (Figure 2). Plasmolyzing cells revealed that they were mainly localized to the plasma membrane and also to the cell walls to some extent. It should be noted that more TED7-YFP fusion protein was retained by the cell wall than TED6-YFP fusion protein (cf. Figures 2E and 2F, and 2K and 2L), which could suggest the possible interaction of the Pro-rich N-terminal domain of TED7 protein with cell walls.
Figure 2.
Localization of TED6- and TED7-YFP Fusion Proteins in Zinnia Mesophyll Cells.
Normal fluorescent images ([A] to [F]) and laser scanning confocal images merged with bright-field images ([G] to [L]) of cells cultured for 48 h after electroporation. Cells were observed before ([A] to [C] and [G] to [I]) or after plasmolysis ([D] to [F] and [J] to [L]) in medium supplemented with twofold mannitol content of the normal medium. Bars in (A) for (A) to (F) and in (G) for (G) to (L) = 50 μm.
(A), (D), (G), and (J) Cells electroporated with 35SPro:YFP.
(B), (E), (H), and (K) Cells electroporated with 35SPro:Ze TED6-YFP.
(C), (F), (I), and (L) Cells electroporated with 35SPro:Ze TED7-YFP.
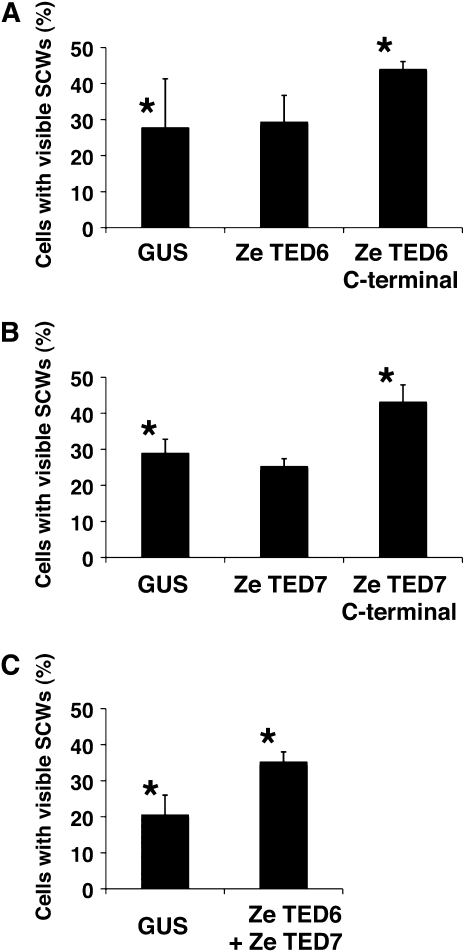
The effect of overexpression of full-length TED6 and TED7 proteins on TE SCW formation was examined using the same conditions as used for the dsRNA-mediated RNAi method. In addition, the C-terminal domains of these proteins were also overexpressed to estimate the function of the predicted cytoplasmic domains in SCW formation. As a result, we found only a slight but significant increase (a few percent increase from controls overexpressing β-glucuronidase [GUS]) in the rate of SCW formation with the C-terminal domains but no increase with the full-length proteins. Therefore, to further observe this positive effect on SCW formation, we used a lower cell density culture (0.5 × 105 cells/mL), which usually exhibits a low rate of SCW formation in the control (<30%). Results in the lower-density cultures confirmed a statistically significant increase in the rate of SCW formation with overexpression of the TED6 and 7 C-terminal domains (Figures 3A and 3B). Similarly, simultaneous overexpression of both TED6 and TED7 full-length proteins increased the rate of SCW formation (Figure 3C), suggesting a functional interaction of TED6 and TED7 proteins during SCW formation of TEs.
Figure 3.
Effects of Overexpression of TED6 and TED7 on SCW Formation.
Zinnia cells were induced to transdifferentiate into TEs after electroporation with the indicated constructs for overexpression under control of the CaMV 35S promoter. GUS was used as an overexpression control. Cell numbers with visible SCWs, which were differentiating and mature TEs, were counted at 96 h of culture. Each data point presents the mean of three independent cultures ± sd. Asterisks show pairs of samples that are significantly different by the two-sample t test (P < 0.05).
(A) Overexpression of the full-length and the C-terminal domain (Leu-27 to Ala-95) of TED6.
(B) Overexpression of the full-length and the C-terminal domain (Trp-109 to Gly-300) of TED7.
(C) Co-overexpression of TED6 and TED7. Cells had been electroporated with a mixture of both full-length constructs.
Functional Analysis of Arabidopsis TED6 and TED7 in Whole Plants
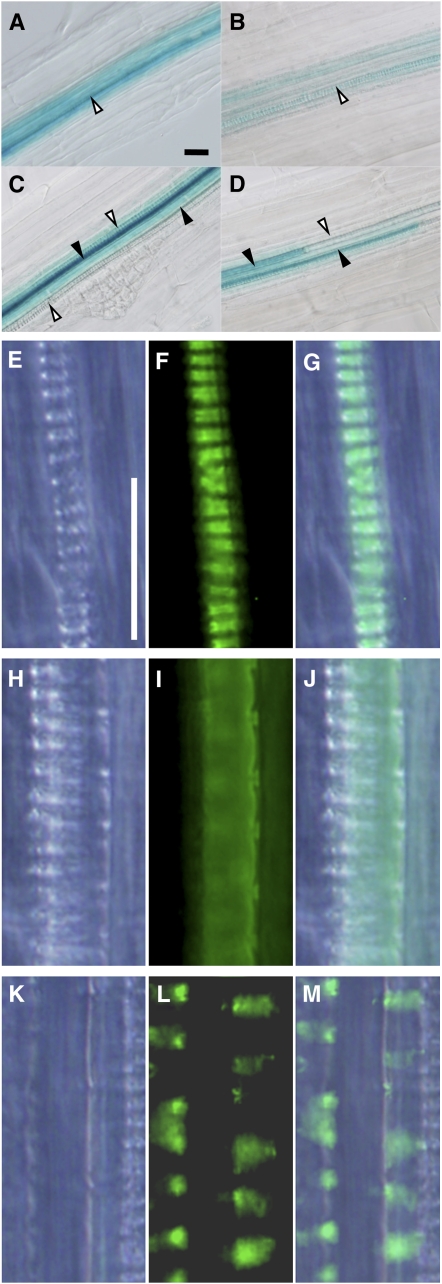
The homologs of Ze TED6 and Ze TED7 genes in Arabidopsis were found to be At1g43790 (At TED6) and At5g48920 (At TED7), respectively (see Supplemental Figure 2 online). Promoter activities of At TED6 and At TED7 were examined with 1- and 0.5-kb upstream sequences of At TED6 and At TED7, respectively, fused to the GUS reporter gene. Reporter transgene expression in Arabidopsis seedlings was restricted to differentiating vessel elements for both genes, thus further confirming the functional homology with Ze TED6 and Ze TED7 (Figures 4A to 4D; see Supplemental Figures 3A and 3B online). The coding sequences of At TED6 and At TED7 were N-terminally fused to the YFP reporter gene, respectively, and expressed under the control of their own promoters (Figures 4E to 4M; see Supplemental Figures 3C to 3H online). The At TED6- and TED7-YFP signals were both specifically observed only beneath developing, patterned SCWs in the differentiating vessel elements of protoxylem (Figures 4E to 4G) and metaxylem (Figures 4H to 4M). In some cases, patchy localization of these signals was recognized in metaxylem vessel elements (Figures 4K to 4M), the biological meaning of which remains unclear.
Figure 4.
Expression Pattern of TED6 and TED7 Genes in Arabidopsis Roots.
(A) to (D) Differentiating vessel element-specific expression of At TED6Pro:GUS ([A] and [C]) and At TED7Pro:GUS ([B] and [D]). Open arrowheads, protoxylem vessels at the outermost position of the vascular system; closed arrowheads, metaxylem vessels inside.
(E) to (M) Differentiating vessel element-specific expression of At TED6Pro:At TED6-YFP and At TED7Pro:At TED7-YFP in the protoxylem ([E] to [G], At TED7Pro:At TED7-YFP) and metaxylem ([H] to [M], At TED6Pro:At TED6-YFP). Both constructs gave similar YFP signals (see Supplemental Figures 3C and 3H online).
Bright-field ([E], [H], and [K]), fluorescent field ([F], [I], and [L]), and merged images ([G], [J], and [M]) are aligned from the left to the right. Protoxylem vessel elements have annular and spiral SCW thickenings, whereas metaxylem vessel elements usually have reticulate and pitted SCW thickenings. Bars in (A) for (A) to (D) and in (E) for (E) to (M) = 25 μm.
To investigate the function of At TED6 and At TED7, transgenic Arabidopsis plants expressing the full-length proteins, C-terminal domains only, or producing transcripts with inverted repeats (for RNAi) of At TED6 and At TED7 driven by the CaMV 35S promoter were prepared. No drastic morphological change was observed in the transgenic lines, except for those harboring the inverted repeat of At TED7. These transgenic plants were not able to grow on germination medium but could be retained alive on a callus-inducing culture, suggesting that constitutive loss of function of the At TED7 gene results in seedling lethality.
To examine the loss-of-function phenotypes of TED6 and TED7 further in Arabidopsis, we searched for insertional mutants in the SALK T-DNA database (http://signal.salk.edu/cgi-bin/tdnaexpress). While no lines were available for TED6, several T-DNA/transposon insertion lines (SALK_084115, SALK_089549, and SM_2_30444) for TED7 were available. However, none of these insertion lines were knockout mutants for TED7 because the insertions were not located in the TED7 coding region. Rather, they were either in the 5′ or 3′ flanking regions or in another gene (data not shown).
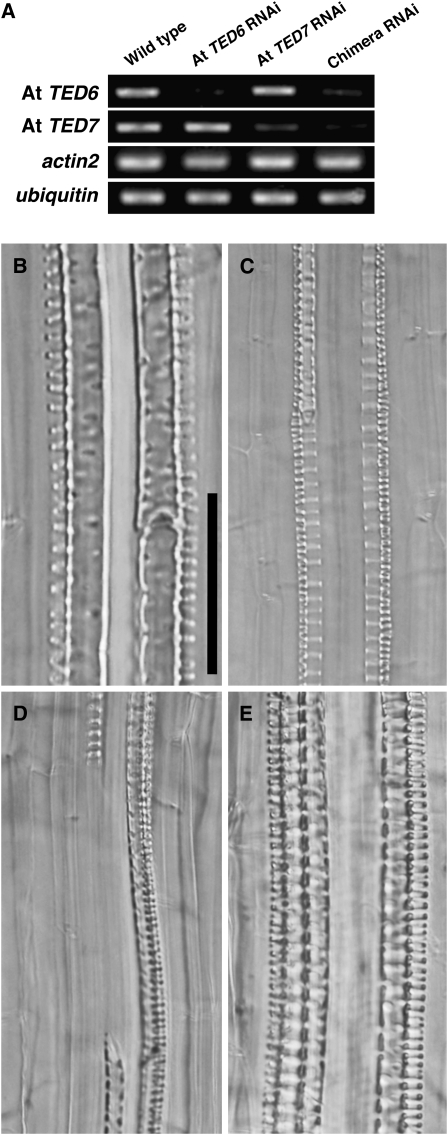
Because of the lethal phenotype produced using constitutive RNAi for At TED7, we next adopted an inducible RNAi technique using the dexamethasone (DEX)-inducible promoter (Aoyama and Chua, 1997). Constructs producing inverted repeats of At TED6, At TED7, and a chimera of At TED6 and At TED7 as well as of YFP (as a control) driven by the inducible promoter were introduced into Arabidopsis plants. The selection of transformants was established by choosing healthy plants carrying the inducible RNAi constructs under noninducing conditions. Only a small number of transformants for the At TED7 RNAi and At TED6–TED7 chimera RNAi constructs could be obtained, probably due to residual leaky expression of the inverted repeat of At TED7 resulting in seedling lethality. The efficiency of TED6 and TED7 suppression by the inducible RNAi constructs was confirmed by RT-PCR on each of the target transcripts in the young seedlings of selected lines after 5 h exposure with 10 μM DEX (Figure 5A). The RNAi was induced on 3-week-old Arabidopsis plants by incubating the plants on growth medium supplemented with 10 μM DEX for 5 d (Figures 5B to 5E; see Supplemental Figure 4 online). Unexpectedly, we found that the DEX-inducible RNAi of control YFP itself has intrinsic effects, including the formation of root metaxylem vessels with aberrant large pits on SCWs (Figure 5E; see Supplemental Figure 4C online) and occasional inhibition of root protoxylem vessel formation. However, beyond the intrinsic effects, apparent defects in SCW formation of root vessel elements were caused in the inducible RNAi lines for At TED7 and the At TED6–TED7 chimera (Figures 5B and 5C; see Supplemental Figures 4A and 4B online). In both lines, aberrant vessel elements with unusual, scalariform (ladder-shaped) SCWs (Figure 5C) were formed in place of metaxylem vessel elements normally exhibiting reticulated or pitted SCWs (Figure 5B). Furthermore, At TED6–TED7 chimera RNAi lines showed discontinuous or gapped vessels in the metaxylem (Figures 5D). Transmission electron microscopy performed on roots of At TED6–TED7 chimera RNAi lines showed that, in contrast with the YFP RNAi control, vessels with thin or incomplete SCWs, which might represent the scalariform SCWs, existed at the metaxylem position (see Supplemental Figure 5 online). These results strongly suggest the involvement of TED6 and TED7 in the SCW formation of vessel elements.
Figure 5.
Transient RNAi Suppression of TED6 and TED7 Genes in Arabidopsis Roots.
(A) Expression of At TED6 and At TED7 genes in transiently induced RNAi conditions. Inverted repeat sequences corresponding to At TED6, At TED7, and both were transiently expressed in transgenic Arabidopsis plants under control of the glucocorticoid-mediated induction system. Total RNAs were extracted from 1-week-old Arabidopsis seedlings that had been incubated on growth medium supplemented with 10 μM DEX for 5 h. All RT-PCR samples were prepared under the same conditions, except for PCR cycles: 25 cycles for At TED6 and actin2, and 30 cycles for At TED7 and ubiquitin.
(B) to (E) Phenotype of RNAi lines for At TED6 and At TED7 genes. Three-week-old transgenic Arabidopsis plants were incubated on growth medium supplemented with 10 μM DEX for 5 d to express inverted repeat sequences under control of the glucocorticoid-mediated induction system. Effects of RNAi were examined on vessel formation in later developed roots during induction. Bar in (B) for (B) to (G) = 50 μm.
(B) Wild type (Columbia-0). Metaxylem vessel elements with pitted SCW thickenings, distinguished from spiral SCW thickenings of protoxylem vessel elements aligned in the outsides.
(C) At TED7 RNAi line. Scalariform (ladder-shaped) metaxylem vessel elements.
(D) At TED6–TED7 chimera RNAi line. Gap in metaxylem vessel strand.
(E) YFP RNAi line. Large-pitted metaxylem vessel elements.
Interaction of At TED6 with a Member of the SCW–Cellulose Synthase (CesA) Complex
Several genes have been implicated in the SCW formation of vessel elements in Arabidopsis, such as FRAGILE FIBER8 (putative glucuronyltransferase; Zhong et al., 2005), IRREGULAR XYLEM2 (IRX2) (KORRIGAN; Szyjanowicz et al., 2004), IRX1 (At CesA8; Taylor et al., 2000), IRX3 (At CesA7; Taylor et al., 1999), and IRX5 (At CesA4; Taylor et al., 2003), whose mutant lines all show a reduction/alteration in SCW thickenings. Among them, IRX1, IRX3, and IRX5 encode CesA subunit proteins with transmembrane domains that have been suggested to be delivered to the site of SCW synthesis (Gardiner et al., 2003; Wightman and Turner, 2008). The characteristics of TED6 and TED7 described above could be correlated with a direct or indirect interaction of these proteins with the CesA complex participating in SCW formation.
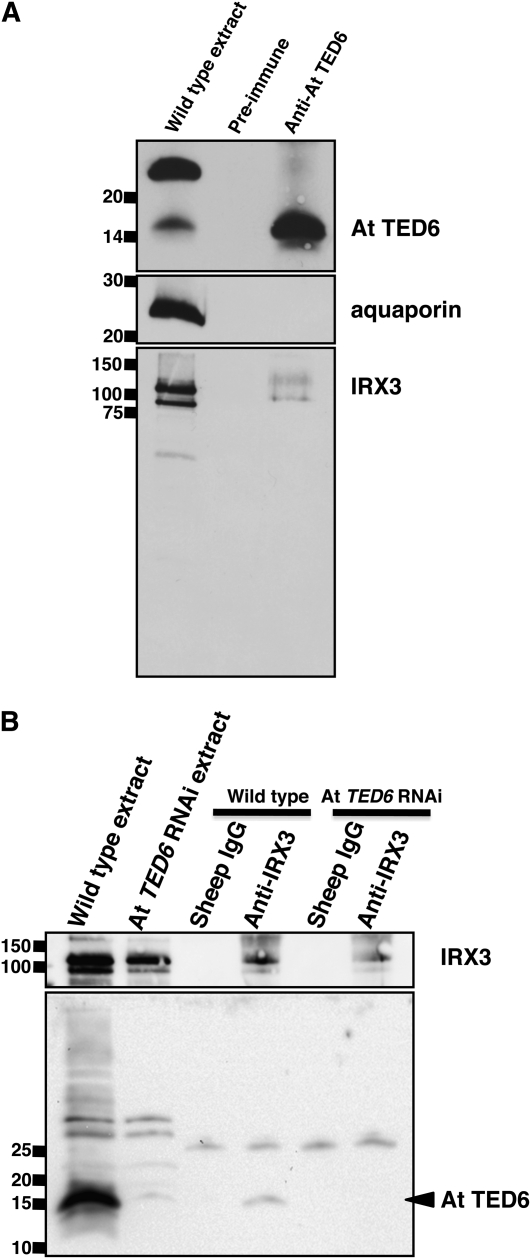
To investigate such possible interactions, copurification and coimmunoprecipitation assays were conducted (Figure 6; see Supplemental Figure 6 online). A rabbit polyclonal antibody was raised against the C-terminal domain of At TED6 protein fused to glutathione S-transferase (GST), and the antiserum was partially purified with a Protein A column as described in Methods. Crude Arabidopsis protein extracts were prepared according to the experimental conditions established by Taylor et al. (2003) in which IRX1, IRX3, and IRX5 proteins form a complex. In the crude protein extracts prepared from wild-type plants, the Protein A–purified anti-TED6 antibody recognized a band with a molecular mass consistent with At TED6 protein (∼15 kD) as well as a band with a molecular mass of ∼25 kD (see Supplemental Figure 6 online). The 15-kD signal was diminished in seedlings of At TED6 RNAi lines and was significantly increased in seedlings overexpressing VND7 in which ectopic TE SCW formation was induced under the control of an estrogen receptor-based chemical-inducible system (Zuo et al., 2000), indicating that the 15-kD signal represents the endogenous TED6 protein. By contrast, the 25-kD signal was equally detected in all samples tested (see Supplemental Figure 6 online), suggesting that the 25-kD signal corresponds to background band, not TED6. An affinity column of Protein A–purified anti-At TED6 antibody was prepared to purify At TED6 protein. This column successfully concentrated At TED6 protein, along with a small amount of IRX3 protein. However, the column did not enrich for the 25-kD protein nor the abundant plasma membrane protein, aquaporin (Figure 6A), suggesting the existence of a specific interaction between TED6 and IRX3.
Figure 6.
Interaction between At TED6 and IRX3 Proteins on Copurification and Coimmunoprecipitation Assays.
(A) Copurification of IRX3 protein with At TED6 protein by Protein A–purified anti-At TED6 antibody-based affinity column purification. IgGs from preimmune and anti-At TED6 sera were cross-linked to Protein A-sepharose. The fractions bound to IgG-Protein A-sepharose were subjected to immunoblot analysis using Protein A–purified anti-At TED6, anti-aquaporin (anti-all PIP), and anti-IRX3 antibodies.
(B) Coimmunoprecipitation of At TED6 protein by the anti-IRX3 antibody. Normal sheep IgG was used as a control precipitant. Total crude extract and precipitated fractions from wild-type plants and At TED6 RNAi lines were subjected to immunoblot analysis.
Antibodies used are shown on the right. Numbers on the left indicate size markers (kD).
To exclude possibility that the 25-kD protein affects the At TED6–IRX3 interaction, the anti-TED6 antibody was further purified by a GST-At TED6 column in combination with a GST column as described in Methods; this purification resulted in the extinction of the 25-kD signal (see Supplemental Figure 6 online), confirming that this signal was not related to TED6. The affinity-purified antibody was next used for immunoblot analysis of the output of coimmunoprecipitation assays. Using anti-IRX3 antibody as a precipitant, a small but significant amount of At TED6 protein was coprecipitated with IRX3 (Figure 6B), confirming the possible interaction of At TED6 with the SCW CesA complex.
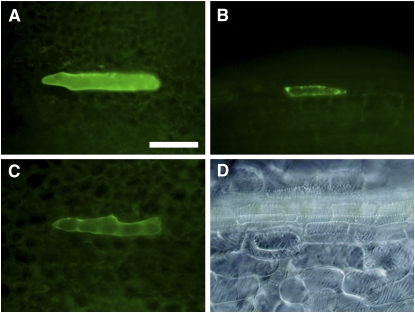
The possible interaction was further examined by bimolecular fluorescence complementation (BiFC) assay (Walter et al., 2004). Considering the interaction with the other CesA subunits, IRX1, IRX3, and IRX5 were all included (Figure 7; see Supplemental Table 1 online). Constructs for At TED6 N-terminally fused to YFP (At TED6-YFP), YFP N-terminally fused to IRX1, IRX3, or IRX5 (YFP-IRX1, IRX3, or IRX5) under the control of the CaMV 35S promoter were introduced into Arabidopsis leaves by particle bombardment to first examine the expression level of the fusion proteins. No YFP-IRX signal was detected in wild-type leaves, whereas At TED6-YFP was detected as expected (see Supplemental Table 1 online). Therefore, to test if YFP-IRX expression was maintained in SCW-forming cells, seedlings overexpressing VND7 under the estrogen-inducible system (Zuo et al., 2000) were used. Ectopic expression of VND7 drives TE differentiation in leaf tissue. In the VND7-overexpressing seedlings, the YFP-IRX signals were detectable, especially a clear YFP-IRX3 signal (Figure 7B; see Supplemental Table 1 online), which could agree with a recent suggestion that CesA proteins that are not participating in the CesA complex could be targeted for degradation (Taylor, 2007). Consequently, a significant BiFC signal appeared at some frequency in the combination of At TED6 N-terminally fused to the C-terminal half of YFP (At TED6-cYFP) and the N-terminal half of YFP N-terminally fused to IRX3 (nYFP-IRX3) constructs (Figure 7C; see Supplemental Table 1 online). The signal, however, was dramatically decreased within 48 h after the induction of VND7 expression probably due to the completion of ectopic TE differentiation. Taken together with the copurification and coimmunoprecipitation assays, our results strongly suggest a biological interaction of TED6 with the SCW CesA complex.
Figure 7.
Interaction between At TED6 and IRX3 Proteins on BiFC Assay in SCW-Forming Cells.
Full-length YFP fusion constructs and the mixture of nYFP and cYFP fusion constructs were biolistically introduced into leaves of transgenic Arabidopsis in which ectopic TE differentiation was induced by VND7 expression. Bar in (A) for (A) to (D) = 100 μm.
(A) Expression of 35SPro:At TED6-YFP (18 h after the induction).
(B) Expression of 35SPro:YFP-IRX3 (18 h after the induction).
(C) BiFC signal in the combination of 35SPro:At TED6-cYFP and 35SPro:nYFP-IRX3 (18 h after the induction).
(D) Ectopic TEs induced by VND7 expression in leaves (3 d after the induction).
DISCUSSION
Our challenge to define the function of unknown genes expressed during in vitro TE differentiation has successfully provided new components implicated in the mechanisms of SCW formation in vessel elements. These new actors, named TED6 and TED7, were characterized using a dual approach on both in vitro Zinnia TE differentiating cell cultures and Arabidopsis plants. This allowed us not only to confirm the conserved function of these genes in different plant species, but also to pinpoint their role in the vessel differentiation process.
Role of TED6 and TED7 Genes during SCW Formation
As previously mentioned, Zinnia differentiating cells undergo three consecutive stages of differentiation (Fukuda, 1996): stage 1, where mesophyll cells lose their photosynthetic capacity; stage 2, where xylogenic potency is acquired; and stage 3, where the TE formation processes occur sensu stricto. Time scaling of these events in the Zinnia system is enabled by the expression of stage-specific markers (Figure 1A). Consequently, temporal expression of specific markers pinpoints distinct cellular processes.
The TED6 gene was selected as a stage 3 gene (Figure 1), and the inhibitory effect of Ze TED6 RNAi on TE differentiation can be explained only by a defect in TE SCW formation and not by the loss of initial TE differentiation potential. Although no visible phenotype was observed for At TED6 RNAi lines, the involvement of the At TED6 gene in the SCW formation processes was indicated by the much stronger phenotype of At TED6–TED7 chimera RNAi lines compared with At TED7 single RNAi lines (Figure 5). In the TED7 single RNA lines, the vessel elements showed defects in SCW morphology, but in the TED6–TED7 chimera RNAi lines, SCWs of metaxylem vessels disappeared, causing gaps in the vessel strands.
Based on the TED6 sequence, Ze TED7 was then identified and determined to be similarly involved in SCW formation, being a stage 3 gene and altering the SCW formation rate by RNAi (Figure 1). Knocking down the expression of the At TED7 gene in Arabidopsis plants also resulted in the defect in SCW formation (Figure 5) and, hence, probably resulted in the defect in overall plant viability.
In addition, our results of the co-overexpression and the chimera RNAi analyses in Zinnia cells and the Arabidopsis plants, respectively (Figures 3 and 5), clearly indicate a functional interaction between TED6 and TED7 in SCW formation. Moreover, both At TED6- and At TED7-YFP fusion proteins expressed under the control of their native promoters localize to the site of SCW formation in differentiating vessel elements (Figures 4E to 4M).
Role of the N-Terminal and C-Terminal Domains
The main difference between the TED6 and 7 proteins is the N-terminal Pro-rich sequence present in TED7 (Figure 1C). Because a part of Ze TED7-YFP fusion protein remained localized in the cell wall in plasmolyzed Zinnia cells compared with Ze TED6-YFP, the N-terminal domain must play an essential role in allowing TED7 to physically anchor to the cell wall (Figure 2).
With the exception of the TED7 Pro-rich N-terminal domain, TED6 and TED7 proteins in both Zinnia and Arabidopsis share very similar overall amino acid sequence structure with single transmembrane domains and highly similar C-terminal domains that are predicted to be cytoplasmic (Figure 1D; see Supplemental Figure 2 online). The increase in the number of cells with visible SCWs when co-overexpressing Ze TED6 and Ze TED7 could be explained as an enhanced efficiency of SCW formation in Zinnia cells (Figure 3C). Overexpressing the C-terminal domain of either Ze TED protein produced similar results, suggesting that TED6 and TED7 promote SCW formation through their cytoplasmic domains. However, such effects promoting SCW formation were not visible in transgenic Arabidopsis plants overexpressing the C-terminal domain of either At TED6 or At TED7 protein. This clearly shows that these C-terminal protein domains cannot shift cell fate in the entire organism toward TE differentiation and confirms that both TED6 and TED7 are implicated in SCW formation and not in the acquisition of xylogenic potency.
Homologs in Other Species
Cell wall–related enzymes appear to be conserved between Arabidopsis, poplar, and rice even though some variation in their gene structures exists. No other homologs of TED6 and TED7 were found in the entire Arabidopsis genome, but homologs in both poplar and rice were identified as probable counterparts of TED6 and TED7 (see Supplemental Figure 2 online). Poplar presents at least one TED6 and two TED7 homolog genes. Nevertheless, the poplar TED6 homolog exhibits a short Pro-rich arm in addition to the previously described TED6 structure. Comparatively, rice presents one homolog gene encoding an N-terminal Pro-rich arm, a single transmembrane domain, and duplicated C-terminal domains. Although they show differences with Zinnia and Arabidopsis TED6 and TED7, they share domains closely related to those characterized in this study. This opens new prospects as to how these differences in protein structure contribute in the SCW synthesis machinery.
Interaction with SCW CesA Complex Subunits
Manipulating TED6 and TED7 expression both in the Zinnia system and in Arabidopsis plants suggests that TED6 and TED7 are involved in SCW formation of TEs. Because both At TED6- and At TED7-YFP fusion proteins localize to the site of SCW formation in vessel elements similar to the localization previously shown for an IRX3–green fluorescent protein fusion (Gardiner et al., 2003), an interaction between these proteins could occur in vivo to determine the location and/or progression of the SCW deposition. This interaction is confirmed by the copurification, coimmunoprecipitation, and BiFC assays, which showed an association between At TED6 and IRX3 proteins. These experiments support the active role of TED6 and TED7 proteins in SCW formation and suggest that they could be part of the SCW CesA complex (Figures 6 and 7).
METHODS
Culture Conditions and Transformation Procedure of Zinnia elegans Cells
Normal culture of Zinnia (Z. elegans cv Canary Bird) cells was performed as described (Fukuda and Komamine, 1980; Demura et al., 2002). Isolated mesophyll cells were cultured in a TE-inducing medium containing 0.1 mg/L naphthylacetic acid and 0.2 mg/L benzyladenine or in a control medium containing 0.1 mg/L naphthylacetic acid. RNA samples were prepared from cells cultured for indicated periods and subjected to RNA gel blot analysis according to a previous report (Demura et al., 2002). For electroporation, Zinnia plants were grown to harvest the first true leaves (2 to 3 cm) at 14 d after imbibition. Zinnia mesophyll cells were isolated from the leaves by vigorous maceration using an ACE Homogenizer (Nissei) at 10,000 rpm for 60 s. Electroporation of the isolated Zinnia cells was performed as previously described (Endo et al., 2008). Total amounts of dsRNAs and plasmid DNAs were adjusted to 40 and 80 μg, respectively, in 0.8-mL reactions when electropulsated. The parameters were set at 325 v/cm and 900 μF, yielding time constants around 50 ms. Cells were cultured with initial densities of 1.2 to 1.5 × 105 cells/mL in dsRNA-mediated RNAi and 0.5 × 105 cells/mL in overexpression experiments.
Cells with visible SCWs (percentage) were measured as the percentage of cells with SCWs per total living cells and TEs. The cells with visible SCWs were differentiating and mature TEs with thick SCWs that were visible under normal bright microscopy. Zinnia cells were stained with 50 μM Evan's blue just before observation to determine cell viability.
Bioinformatic Analysis
Protein structure predictions were performed using online SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and SignalP (http://www.cbs.dtu.dk/services/SignalP/). Identification of protein domains was performed using online ProDom (http://prodom.prabi.fr/prodom/current/html/home.php), PROSITE (http://www.expasy.ch/prosite/), and Pfam (http://www.sanger.ac.uk/Software/Pfam/).
Primers, dsRNAs and Plasmid DNAs
All primer sequences are shown in Supplemental Table 2 online. Zinnia EST clones (Demura et al., 2002) were subjected to PCR to generate fragments containing a T7 or SP6 promoter at each end. The PCR products were purified and used directly as templates for the in vitro synthesis of dsRNAs as described by Endo et al. (2008). Full-length cDNAs of Ze TED6 and Ze TED7 genes were obtained using a SMART RACE cDNA amplification kit (Clontech). Coding sequences were inserted into pH35GY (Kubo et al., 2005) and pY35GS (Endo et al., 2008) to examine protein localization and overexpression, respectively, in Zinnia cells. pY35GUS constructed by inserting GUS into pY35GS was used as a control. Sequences encoding from Leu-27 to Ala-95 of Ze TED6 and from Trp-109 to Gly-300 of Ze TED7 with start and stop codons were inserted into pY35GS. At 5′ upstream from the start, codons of At TED6 and At TED7 genes, 1 and 0.5 kb, respectively, were used as promoter regions. The fragments were inserted into pBGGUS (Kubo et al., 2005) to examine their promoter activities in Arabidopsis thaliana plants. Genomic fragments containing promoter regions and open reading frames (ORFs) were inserted into pHGY, which was derived from pH35GY by deleting its CaMV 35S promoter sequence. The first intron of the FAD2 (At3g12120) gene was used as a linker of inverted repeat sequences of full-length At TED6, At TED7 ORFs, and At TED6–TED7 chimera sequence. The chimera was constructed by inserting the At TED7 ORF into the PshAI site of the At TED6 ORF. Inverted repeats were placed into pH35GS (Kubo et al., 2005) and/or pTA7002 (Aoyama and Chua, 1997) for constitutive and transient RNAi lines, respectively. VND7 ORF (Yamaguchi et al., 2008) was inserted into pER8 (Zuo et al., 2000) for ectopic TE-inducible lines. The pH35GY-derived 35SPro:YFP construct (Yamaguchi et al., 2008) was used as a YFP expression control. The ORF of At TED6 or At CesA genes was inserted into pH35GY, pH35YG (a binary vector containing the CaMV 35S promoter with YFP followed by the sense GATEWAY cassette), pEMT-C2 (for the sense GATEWAY cassette followed by the 155 to 238 amino acid part of YFP), or pEMT-NO (for the 1 to 154 amino acid part of YFP followed by the sense GATEWAY cassette) (T. Ueda, personal communication).
Histology
Fluorescent and/or differential interference contrast images of Zinnia cells and Arabidopsis seedlings and leaves were taken under a BX51 microscope with a DP70 or DP71 camera (Olympus). An FV500 (Olympus) was used for confocal laser scanning microscopy of Zinnia cells.
Procedures for ultrastructural analysis were conducted according to Toyooka et al. (2000). Ultrathin sections were stained with 4% uranyl acetate and lead citrate solution and then examined with a transmission electron microscope (JEM-1011; JEOL). Images were acquired using a Gatan DualView CCD camera and Gatan Digital Micrograph software or films.
RT-PCR
RNAs were prepared from mixtures of two independent lines for each and used for cDNA synthesis as described previously (Endo et al., 2008). PCR was performed with primers located at 5′- and 3′-untranslated region of At TED6 and At TED7, respectively. The number of PCR cycles was determined for each gene: 25 cycles for At TED6 and actin2 (At3g18780) and 30 cycles for At TED7 and ubiquitin (At5g57860). All primer sequences are shown in Supplemental Table 2 online.
Protein Extraction and Immunoblot Analysis
Pulverized inflorescence stems of Arabidopsis plants were briefly macerated in a solution containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 2% (w/v) glycerol, and a protease inhibitor cocktail (Sigma-Aldrich). After washing away the solution following centrifuging at 10,000g, detergent-soluble proteins were extracted from the pellets by incubating them in a solution supplemented with Triton X-100 to a final concentration of 2% (w/v) for 30 min at 4°C.
Anti-At TED6 serum (BioGate) was raised against GST-At TED6 C-terminal domain fusion protein that had been prepared using pGEX6P-1 for a GST fusion system (GE Healthcare). For affinity purification of the specific antibody, IgGs were collected by Protein G-sepharose (GE Healthcare) from 8 mL of the serum and then further purified with 2.2 mg of GST-At TED6 covalently linked to NHS-activated sepharose (GE Healthcare). After passing the purified serum through 6.4 mg of GST-linked sepharose to absorb anti-GST antibodies, the obtained fraction was concentrated to ∼400 μL. Immunoblot analysis was performed as described (Endo et al., 2001). For detection of the monomers of aquaporins, protein samples were incubated at 65°C for 5 min in SDS-PAGE loading buffer.
Both Protein A–purified and affinity-purified anti-At TED6 antibody were used at 1 in 2000, and anti-IRX3 antibody (Taylor et al., 2000) was used at 1 in 5000 as the first antibody. Anti-all PIP (aquaporin) antibody (Ohshima et al., 2001) was used at 1 in 2000. Anti-rabbit horseradish peroxidase–linked antibody (Santa Cruz Biotechnology) for anti-At TED6 antibody and anti-sheep IgG horseradish peroxidase–linked antibody (Sigma-Aldrich) for the anti-IRX3 antibody were used at 1 in 10,000 as the second antibody. Signals were detected with SuperSignal (Thermo Scientific) on a ChemiDoc system (Bio-Rad).
Purification of At TED6 Protein
Each 2-mL sample of antiserum against At TED6 protein and preimmune serum was diluted in PBST (PBS and 0.05% Tween 20) and then mixed with 1 mL of nProteinA-sepharose (GE Healthcare) at 4°C overnight. The sepharose was washed with 0.2 M borate-NaOH, pH 8.6, and adjusted to a total of 3 mL. Dimethyl pimelimidate dihydrochloride (47 mg) was dissolved in 6 mL of 0.2 M triethanolamine-HCl, pH 8.3, and immediately added to the 3-mL sepharose solution. After incubation at room temperature for 30 min, the sepharose was washed with 0.2 M monoethanolamine-HCl, pH 8.2, and incubated at room temperature for 1 h. The sepharose was packed into a column and washed with a washing buffer (10 mM Tris-HCl, pH 7.5, and 0.5 M NaCl). The column was also washed with an elution buffer (100 mM glycine-HCl, pH 2.5), immediately followed by 0.1 M Tris-HCl, pH 8.0, at 4°C.
Crude proteins extracted from inflorescence stems were added to the antibody-linked column that had been equilibrated with a solution containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2% (w/v) Triton X-100 at 4°C. After extensively washing the column with the washing buffer supplemented with a final 2% of Triton X-100, proteins bound to the column were eluted with the elution buffer supplemented with a final 2% of Triton X-100 and neutralized with a one-tenth volume of 1.5 M Tris-HCl, pH 8.0.
Immunoprecipitation of IRX3 Proteins
Crude proteins (1 mg) extracted from inflorescence stems were incubated with 20 μL of Protein G sepharose (GE Healthcare) and 10 μL of anti-IRX3 antibody or 3 μg of normal sheep IgG (Sigma-Aldrich) in a solution containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 2% (w/v) glycerol, and a protease inhibitor cocktail (Sigma-Aldrich) at 4°C for 1 h. After the sepharose was washed extensively with the same solution, proteins precipitated with the sepharose were solubilized in SDS-PAGE loading buffer.
Particle Bombardment
Transgenic Arabidopsis plants harboring pER8-VND7 (Yamaguchi et al., 2008) were grown for 2 to 3 weeks. BiFC constructs were introduced into the Arabidopsis leaves by IDERA GIE-III (Tanaka) with the following parameters: vacuum, 665 mmHg; distance, 10.5 cm; helium press, 4.0 kgf/cm2; releasing time, 0.075 s. After incubating the bombarded plants on normal culture medium for 3 h, VND7 expression was triggered by the addition of 10 μM β-estradiol to induce ectopic SCW formation.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL/DDBJ data libraries, the Arabidopsis Genome Initiative database, or the Department of Energy Joint Genome Institute Populus trichocarpa v1.1 database under the following accession numbers: AB377514 (Ze TED6), AB377515 (Ze TED7-1), AB377516 (Ze TED7-2), At1g43790 (At TED6), At5g48920 (At TED7), Os08g0108300, eugene3.00020671, eugene3.00070382, and fgenesh1_pg.C_LG_V000008.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Ze TED6, Ze TED7-1, and Ze TED7-2.
Supplemental Figure 2. Comparison of Ze TED6 and Ze TED7 and Counterparts in Arabidopsis, Populus, and Oryza.
Supplemental Figure 3. Expression Pattern of At TED6 and At TED7.
Supplemental Figure 4. Transient RNAi Analysis on At TED6 and At TED7 in Arabidopsis Roots.
Supplemental Figure 5. Transmission Electron Micrographs of the At TED6–TED7 chimera RNAi Line.
Supplemental Figure 6. Affinity Purification of Anti-At TED6 Antibody.
Supplemental Table 1. Summary of Single YFP and BiFC Signals in Wild Type and SCW-Forming Arabidopsis Cells.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to Supplemental Figure 1.
Supplemental Data Set 2. Text File of Alignment Corresponding to Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank Takashi Aoyama and Nam-Hai Chua for providing pTA7002 and pER8, Simon R. Turner for providing the anti-IRX3 antibody, Masayoshi Maeshima for providing the anti-all PIP antibody, Takashi Ueda for providing BiFC constructs, Takako Kawai and Mayumi Wakazaki for technical assistance in electron microscopy, and Ayumi Ihara, Mitsutaka Araki, and Sachiko Oyama for excellent technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (Grant 20061029 to T.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Taku Demura (demura@riken.jp).
Online version contains Web-only data.
References
- Aloni, R. (1987). Differentiation of vascular tissues. Annu. Rev. Plant Physiol. 38 179–204. [Google Scholar]
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Berardini, T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura, T., and Fukuda, H. (1994). Novel vascular cell-specific genes whose expression is regulated temporally and spatially during vascular system development. Plant Cell 6 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura, T., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. USA 99 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, S., Demura, T., and Fukuda, H. (2001). Inhibition of proteasome activity by the TED4 protein in extracellular space: a novel mechanism for protection of living cells from injury caused by dying cells. Plant Cell Physiol. 42 9–19. [DOI] [PubMed] [Google Scholar]
- Endo, S., Pesquet, E., Tashiro, G., Kuriyama, H., Goffner, D., Fukuda, H., and Demura, T. (2008). Transient transformation and RNA silencing in Zinnia tracheary element differentiating cell cultures. Plant J. 53 864–875. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (1996). Xylogenesis: Initiation, progression, and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 299–325. [DOI] [PubMed] [Google Scholar]
- Fukuda, H., and Komamine, A. (1980). Establishment of an experimental system for the tracheary element differentiation from single cells isolated from the mesophyll cells of Zinnia elegans. Plant Physiol. 65 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Taylor, N.G., and Turner, S.R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioni, D., Sado, P.E., Stacey, N.J., Roberts, K., and McCann, M.C. (2002). Early gene expression associated with the commitment and differentiation of a plant tracheary element is revealed by cDNA-amplified fragment length polymorphism analysis. Plant Cell 14 2813–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima, Y., Iwasaki, I., Suga, S., Murakami, M., Inoue, K., and Maeshima, M. (2001). Low aquaporin content and low osmotic water permeability of the plasma and vacuolar membranes of a CAM plant Graptopetalum paraguayense: Comparison with radish. Plant Cell Physiol. 42 1119–1129. [DOI] [PubMed] [Google Scholar]
- Pesquet, E., Ranocha, P., Legay, S., Digonnet, C., Barbier, O., Pichon, M., and Goffner, D. (2005). Novel markers of xylogenesis in zinnia are differentially regulated by auxin and cytokinin. Plant Physiol. 139 1821–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck, L., Rombauts, S., Vandepoele, K., Rouzé, P., and Van de Peer, Y. (2007). How many genes are there in plants (…and why are they there)? Curr. Opin. Plant Biol. 10 199–203. [DOI] [PubMed] [Google Scholar]
- Szyjanowicz, P.M.J., McKinnon, I., Taylor, N.G., Gardiner, J., Jarvis, M.C., and Turner, S.R. (2004). The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 37 730–740. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G. (2007). Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites – A potential role in regulating protein degradation. Plant Mol. Biol. 64 161–171. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.-R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka, K., Okamoto, T., and Minamikawa, T. (2000). Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J. Cell Biol. 148 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S., Gallois, P., and Brown, D. (2007). Tracheary element differentiation. Annu. Rev. Plant Biol. 58 407–433. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55 652–664. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Scütze, K., Batistic, O., Weckermann, K., Näke, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wightman, R., and Turner, S.R. (2008). The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54 794–805. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Peña, M.J., Zhou, G.-K., Nairn, J., Wood-Jones, A., Richardson, E.A., Morrison III, W.H., Darvill, A.G., York, W.S., and Ye, Z.-H. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., and Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.