Abstract
Epigenetic silencing involving the aberrant DNA methylation of promoter-associated CpG islands is one mechanism leading to the inactivation of tumor suppressor genes in human cancers. However, the molecular mechanisms underlying this event remains poorly understood. TMS1/ASC is a novel proapoptotic signaling factor that is subject to epigenetic silencing in human breast and other cancers. The TMS1 promoter is embedded within a CpG island that is unmethylated in normal cells and is spanned by three DNase I-hypersensitive sites (HS). Silencing of TMS1 in cancer cells is accompanied by local alterations in histone modification, remodeling of the HS, and hypermethylation of DNA. In this study, we probed the functional significance of the CpG island-specific HS. We identified a methylation-sensitive complex that bound a 55-bp intronic element corresponding to HS2. Affinity chromatography and mass spectrometry identified a component of this complex to be the GA-binding protein (GABP) α. Supershift analysis indicated that the GABPα binding partner, GABPβ1, was also present in the complex. The HS2 element conferred a 3-fold enhancement in TMS1 promoter activity, which was dependent on both intact tandem ets binding sites and the presence of GABPα/β1 in trans. GABPα was selectively enriched at HS2 in human cells, and its occupancy was inversely correlated with CpG island methylation. Down-regulation of GABPα led to a concomitant decrease in TMS1 expression. These data indicate that the intronic HS2 element acts in cis to maintain transcriptional competency at the TMS1 locus and that this activity is mediated by the ets transcription factor, GABPα.
Methylation of DNA in the human genome is tightly controlled during development by the action of DNA methyltransferases. DNA methyltransferases catalyze the transfer of a methyl group from S-adenosylmethionine to the carbon-5 position of cytosine in the dinucleotide 5′-CpG-3′. This epigenetic mark is copied after DNA synthesis, providing a heritable memory of transcriptional status. Overall, CpG dinucleotides are depleted in the human genome except in CpG islands (1). CpG islands are ∼200 bp to several kb in length and are found mainly in the 5′-regions of 70% of human genes (2). Most CpG dinucleotides in the genome are heavily methylated whereas, in contrast, the CpG sites in the CpG islands, especially those associated with gene promoters, are usually unmethylated. Aberrant methylation of such CpG islands is associated with inappropriate gene silencing and has been implicated in the inactivation of tumor suppressor genes in human cancers.
At present, the mechanisms underlying cancer-associated CpG island methylation are unknown. There are at least two mechanisms in which aberrant DNA methylation is thought to contribute to stable gene repression. DNA methylation can affect gene expression through its affects on local chromatin structure. Methylated DNA is recognized by methyl-CpG-binding domain proteins, which are components of repressor complexes that contain histone deacetylases and other chromatin modifying activities (1). Histone deactylase-mediated deacetylation of lysine residues (e.g. H3K9) allows for their subsequent methylation by histone methyltransferases and recruitment of methyl lysine-binding proteins such as HP1 leading to a compact chromatin configuration. Indeed, cancer-associated methylation of CpG islands is associated with a shift to a more compact chromatin structure characterized by hypoacetylated histones H3 and H4 and a shift in the histone methylation pattern from H3 methylated at K4 to H3 methylated at K9 (3) and/or K27 (4). DNA methylation can also play a more direct role in the transcriptional response by interfering with the binding of some transcription factors (e.g. AP-2, E2F, c-myc, NF-κB) (5) or by influencing long-range chromatin interactions, for example, by blocking the binding of the chromatin insulator and enhancer blocker CTCF (6).
In previous work, we identified a novel CpG-island associated gene called TMS14 (target of methylation-induced silencing-1) that is subject to aberrant methylation and epigenetic silencing in breast and other cancers (7–12). Also known as ASC, TMS1 encodes an intracellular signaling adapter with roles in apoptosis and inflammation. TMS1/ASC is highly expressed in monocytes/macrophages where it plays a critical role in the innate immune response (13, 14). Oligomerization of TMS1/ASC induced by interaction with intracellular pathogen sensors mediates the assembly of a multiprotein complex that ultimately leads to the activation of caspase-1, maturation and release of the cytokines interleukin-1β and interleukin-18, and the induction of “pyroptosis,” a proinflammatory form of programmed cell death (15, 16). TMS1/ASC is also widely expressed in epithelial cells, where it is up-regulated in response to cell stress stimuli, such as the death ligands TNFα and TRAIL (17). Overexpression or forced oligomerization of TMS1 in epithelial cells promotes a caspase-8-dependent cell death (17–19). Recently, we showed that TMS1 plays a critical role in anoikis, or apoptosis induced in response to the loss of integrin-mediated contacts with the substratum, in breast epithelial cells (20). Aberrant methylation of the TMS1/ASC CpG island is frequently observed in primary breast cancers, and loss of TMS1 expression accompanies the transition from ductal carcinoma in situ to invasive carcinoma in primary breast lesions (20). Together these data suggest that epigenetic silencing of TMS1 may contribute to the pathogenesis of human cancers by allowing cells to bypass normal cell death cues in the early stages of cancer progression.
Little is known about the transcriptional regulation of TMS1, and the mechanisms underlying its epigenetic silencing during tumorigenesis remain unclear. In normal cells and breast cancer cells that retain TMS1 expression, the TMS1 CpG island is unmethylated, and exhibits an “active” chromatin signature characterized by hyperacetylated histones H3 and H4, enrichment of H3K4me2, and positioned nucleosomes (21, 22). Three DNase I-hypersensitive sites (HS) span the CpG island in the active state; HS1 and HS3 mark the boundaries between unmethylated CpG island DNA and densely methylated flanking DNA, whereas HS2 forms at the center of the CpG island (21). Epigenetic silencing of TMS1 in breast cancer is accompanied by the remodeling of the CpG island-associated HS sites, hypoacetylation of histones, a shift in histone methylation status, and hypermethylation of DNA (21, 22). These findings have led us to propose that there may be an activity and/or a structural barrier occurring at the level of chromatin, and marked by the HS sites, that protects the TMS1 CpG island from methylation in normal cells. Loss of function at these sites might allow for aberrant methylation, changes in chromatin structure and gene silencing.
In this study, we sought to characterize the functional significance of the TMS1 CpG island-associated HS sites. We identified a novel protein complex that binds to HS2, located within intron 1 of the TMS1 gene. Using DNA affinity chromatography and mass spectrometry we identified a component of this complex to be the ets family transcription factor GA-binding protein α (GABPα). We find that GABPα binds TMS1 at HS2 in a methylation-sensitive manner in vitro and in vivo, and acts as a positive mediator of TMS1 expression. These data indicate that GABPα acts in trans to maintain transcriptional competency at the TMS1 locus, and suggest that methylation-mediated inhibition of GABP binding may be one factor contributing to the stable repression of TMS1 in cancer cells.
EXPERIMENTAL PROCEDURES
Preparation of Nuclear Extracts—Cells were washed in 1× phosphate-buffered saline and resuspended in 20 packed cell volumes of ice-cold 0.67× phosphate-buffered saline, swollen on ice 10 min, pelleted, and resuspended in 5 packed cell volumes of ice-cold hypotonic buffer A (10 mm HEPES, pH 7.9, 1 mm MgCl2, 10 mm NaCl, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 mm DTT, 0.7 μg/ml pepstatin, 0.5 μg/ml leupeptin). Cells were Dounce homogenized with a Type B pestle and nuclei were harvested by centrifugation at 3300 × g. Nuclei were resuspended in ½ packed cell volume of Buffer A, followed by the addition of Buffer C (20 mm HEPES, pH 7.9, 25% glycerol, 1.5 mm MgCl2, 1.0–1.5 m NaCl, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 mm DTT, 0.7 μg/ml pepstatin, 0.5 μg/ml leupeptin) to a final concentration of 750 mm NaCl. Nuclei were extracted for 30 min at 4 °C. Clarified nuclear extracts were dialyzed (6,000–8,000 Mr) against Buffer D (20 mm HEPES, pH 7.9, 20% glycerol, 100 mm NaCl, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 mm DTT, 0.7 μg/ml pepstatin, 0.5 μg/ml leupeptin) for 5 h, clarified by centrifugation, and stored at –80 °C. Protein concentrations were determined by Bradford assay (Bio-Rad).
Mobility Shift Assays—The HS2–236bp probe was amplified by PCR using the “HS2” primers from Stimson-Crider et al. (21). All other probes were either amplified by PCR followed by purification on Promega Wizard® Minicolumns (catalog number A7170) or were synthesized as single-stranded oligonucleotides and annealed. Probes were end-labeled with [γ-32P]ATP (Amersham Biosciences, 3000 Ci/mmol, 10 mCi/ml) and T4 polynucleotide kinase. Binding reactions (20 μl) were performed at room temperature for 30 min and contained nuclear extract (5–10 μg) plus 1.5 μg of poly(dI-dC) and ∼20 fmol of end-labeled double-stranded DNA (∼50,000 cpm) in 1× Binding Buffer (50 mm NaCl, 20 mm HEPES, pH 7.9, 12% glycerol, 0.2 mm EDTA, 1 mm DTT). Competitor DNA was included at 100-fold molar excess unless otherwise indicated. For supershift analysis, antibodies (GABPβ1, sc-13444, Santa Cruz) were added to binding reaction after an initial 30 min and followed by an additional 20-min incubation. Samples were electrophoresed at 25 mA on 4% polyacrylamide 0.5× TBE gels, dried onto Whatman paper, and exposed to x-ray film (Biomax-MS, Kodak). Probe and competitor sequences are listed in supplemental Table S1.
DNase I Footprinting—A 107-bp fragment of HS2 (supplemental Table 1) was amplified by PCR, gel purified, and end-labeled with [γ-32P]ATP. Unincorporated radioactivity was removed and the labeled product (4 ng) was digested with either HhaI or NlaIV followed by gel purification to generate fragments labeled on either the sense or antisense strand. Binding conditions were as described above and contained ∼20 fmol (20,000 cpm) of labeled probe and 25 μg of nuclear extract per 20 μl of reaction. After 30 min, MgCl2 (5 mm) and DNase I (0 to 30 ng) were added, and the reaction was carried out for another 2 min. Reactions were stopped by the addition of 1% SDS, 30 mm EDTA, 200 mm NaCl, and DNA was recovered by phenol: chloroform extraction and ethanol precipitation. Pellets were resuspended in 2× loading buffer (98% formamide, 10 mm EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue), denatured at 95 °C for 10 min, and electrophoresed on an 8 m urea, 8% polyacrylamide (19:1), 1× TBE sequencing gel at 80 watts. The gels were dried onto Whatman paper and exposed to x-ray film (Biomax MR, Kodak).
Cation Exchange Chromatography—HeLa S3 cells (24 liters) were grown as spinner cultures to a density of 106 cells/ml. Nuclear extract (207 mg) was prepared as described above and applied to a 50-ml SP-Sepharose Fast Flow (17-0729-01, Amersham Biosciences) cation exchange column in 1× binding buffer (20 mm HEPES, pH 7.9, 20% glycerol, 50 mm NaCl, 0.2 mm EDTA, 1 mm DTT, 0.5 μg/ml leupeptin, 0.5 μg/ml pepstatin). The column was eluted stepwise with 3 column volumes each of 50 mm, 100 mm, 250 mm, 500 mm, and 1 m NaCl in 1× Binding Buffer. Fractions (8.5 ml) were collected, and binding activity monitored by mobility shift assays using the HS2–55bp fragment as a probe in the presence of 100-fold molar excess of HS2–55bp methylated in vitro with M.SssI. Binding activity corresponding to the methylation-sensitive Complex A eluted in 250 mm NaCl between fractions 46 and 48.
DNA Affinity Purification—To prepare DNA affinity matrices, oligonucleotides (5′-phospho) corresponding to each strand of the HS2–55bp binding site plus a 5′-GATC overhang were synthesized and annealed. The resulting double-stranded DNA was oligomerized by ligation at room temperature for 4 h to an average length of 200–500 bp, and coupled to CNBr-activated Sepharose-4B (∼200 μg of DNA per 1.5-ml bed volume) according to the manufacturer's instructions (Amersham Biosciences). After successive washes in 1 m ethanolamine, pH 8.0, 10 mm potassium phosphate, pH 8.0, 1 m potassium phosphate, pH 8.0, 1 m KCl, and H2O, the affinity columns were equilibrated with 10 column volumes of Binding Buffer (20 mm HEPES, pH 7.9, 20% glycerol, 50 mm NaCl, 0.2 mm EDTA, 1 mm DTT, 0.5 μg/ml leupeptin, 0.5 μg/ml pepstatin). Unmethylated and methylated affinity columns were prepared by incorporating oligonucleotides that were either unmethylated or methylated on both strands at the two CpG sites.
Fractions containing peak HS2 binding activity from the cation exchange column were pooled and dialyzed against 1× Binding Buffer. Poly(dI-dC) (180 μg/15 ml of sample) was added and insoluble material removed by centrifugation. The cleared sample was allowed to bind in bulk to a 1-ml affinity matrix consisting of the fully methylated HS2–55bp oligomer for 2 h at 4 °C. The flow-through fractions were then applied to a second 1-ml affinity column consisting of the unmethylated HS2–55bp oligomer. The column was washed with 10 column volumes of 1× Binding Buffer and eluted with 3 column volumes of 500 mm NaCl. Fractions (250 μl) were collected and methylation-sensitive binding activity monitored as described above. Two additional rounds of binding and elution (500 mm NaCl) from the unmethylated HS2–55bp column were performed, with an additional 250 mm NaCl step added to the third elution. HS2 binding activity (Complex A) eluted in the 250 mm step. Fractions containing peak binding activity were precipitated in trichloroacetic acid, washed with acetone, dried at 37 °C, and resuspended in 2× Laemmli buffer. The pH was adjusted to ∼8.0 with 1 m Tris base. Samples were separated on an SDS-10% polyacrylamide gel and visualized with Gel Code Blue (Pierce 24590). Bands co-eluting with HS2 binding activity were excised and submitted to the Emory University Microchemical Facility for MALDI tandem MS/MS mass spectroscopy analysis.
Chromatin Immunoprecipitation (ChIP)—Cells were cross-linked for 10 min in 1% formaldehyde at room temperature. ChIP was carried out essentially as described in the Acetyl-histone H3 Immunoprecipitation Assay Kit (Millipore) except that MCF7 and MDA-MB231 cells were subject to a two-step lysis procedure in which nuclei were first harvested in Cell Lysis Buffer (5 mm PIPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40, 1× protease inhibitor mixture) followed by lysis of nuclei in Nuclei Lysis Buffer (50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1% SDS, 1× protease inhibitor mixture). Immunoprecipitated DNA was analyzed by quantitative real-time PCR using SYBR Green detection. Reactions (25 μl) contained 1 μl of DNA, 0.2 μm primers, and 12.5 μl of IQ SYBR Green Supermix (Bio-Rad). The reaction was subjected to a hot start for 3 min at 95 °C and cycles of 95 °C, 10 s; 55–60 °C, 60 s. Melt curve analysis was performed to verify a single product species. Starting quantities were determined relative to a common standard curve generated using MCF7 genomic DNA. Fold enrichment is calculated as the percent of input DNA immunoprecipitated by the GABPα antibody relative to that of a non-specific control antibody at each primer pair. Primers are listed in supplemental Table S1. Antibodies used were GABPα (sc-22810, Santa Cruz), rabbit IgG (sc-2027, Santa Cruz), or ApafI (AB16941, Chemicon).
Plasmid Constructs and Luciferase Reporter Assays—A genomic SmaI-NcoI fragment containing 263 bp upstream of the TMS1 translation start codon was cloned in-frame into the pGL3-Basic luciferase reporter plasmid (Promega) to generate p-min-TMS1. A 236-bp fragment containing HS2 was amplified by PCR and cloned into the SmaI site of p-min-TMS1 upstream of the TMS1 promoter to create pTMS1-HS2sense. Constructs in which one or the other GABP binding site (pTMS1-HS2-m1, pTMS1-HS2-m2), or both GABP binding sites (pTMS1-HS2-dm) were created by deleting the 4-bp central core (GGAA/T) of each site using the QuikChange® Site-directed Mutagenesis Kit (Stratagene). All clones were verified by sequencing. Primers used to generate the mutation are listed in supplemental Table S1.
MCF7 cells (2 × 105) were seeded in 6-well plates and transfected the next day with 1 μg of the reporter plasmids using the Lipofectamine (Invitrogen) reagent. A Renilla luciferase reporter (200 ng, pRL-TK, Promega) was included as a control for transfection efficiency. After 48 h, cells were lysed and firefly and Renilla luciferase activities were determined using the Dual Luciferase Reporter Assay system (Promega). For experiments incorporating GABPα/β siRNA, MCF7 cells (5 × 104) were seeded in 24-well plates and transfected the following day with 200 ng of reporter plasmids and 40 ng of pRL-TK control plus 10 pmol each of GABPα and GABPβ siRNA (Invitrogen Stealth™ Select, supplemental Table S1) or 20 pmol of scrambled siRNA using 1.5 μl of Lipofectamine 2000. Luciferase activities were determined after 48 h as described above.
Immunoblotting and Antibodies—Whole cell lysates were prepared in RIPA buffer supplemented with protease inhibitors (Complete Mini Protease Inhibitor Mixture, Roche), 1 mm sodium orthovanadate, and 10 mm sodium fluoride. Total protein (25 μg) was separated on a 12% SDS-polyacrylamide gel, transferred to polyvinylidene difluoride (Bio-Rad), and probed with the indicated primary antibody. Immunocomplexes were detected by incubation with horseradish peroxidase-conjugated secondary antibody and chemiluminescence detection (Pierce). For the analysis of luciferase lysates, cell lysates were prepared by 15 min incubation with 1× Passive Lysis Buffer (Dual Luciferase Reporter Assay System, Promega). The antibodies used were: anti-ASC (ProteinTech), GABPα (Santa-Cruz), GABPβ1/2 (Santa-Cruz), and GAPDH (Abcam).
Lentiviral shRNA Production and Infection—293T cells were transfected with the viral packaging plasmid, psPAX2 (900 ng); viral envelope plasmid, pMD2G-VSV-G (100 ng); and the pLKO.1 vector (1 μg) or pLKO.1 containing a short hairpin sequence targeting GABPα (number 18288–18292, Open Bio-systems) using the FuGENE 6 reagent. After 18 h, the transfection medium was replaced with high serum growth media (Dulbecco's modified Eagle's medium + 30% fetal bovine serum). Supernatant containing lentiviral particles was harvested at 24 and 48 h, combined, and stored at –80 °C. MCF7 cells (2 × 105) were seeded in 6-well plates and infected with 0.5 ml of viral supernatant in the presence of 8 μg/ml Polybrene. At 24 h post-infection, cells were placed under puromycin selection (0.5 μg/ml) and harvested 5 days later for immunoblot analysis.
RESULTS
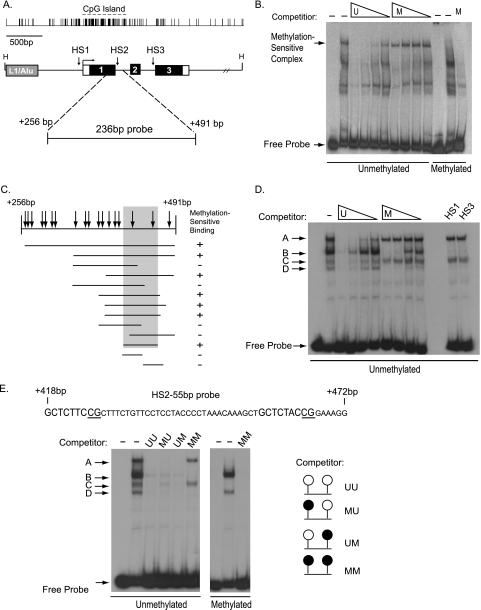
Identification of a Methylation-sensitive HS2 DNA Binding Activity—The human TMS1 gene spans about 3.5 kb on chromosome 16. A 5′ CpG island spans the promoter, first exon, and part of intron 1 (Fig. 1A). In normal diploid fibroblasts and breast epithelial cells where TMS1 is expressed, the CpG island is encompassed by a 1.2-kb unmethylated domain flanked by heavily methylated DNA (21). In previous work (21, 22), we have shown that this domain is spanned by positioned nucleosomes and marked by hyperacetylated histones H3 and H4 (H3Ac, H4Ac), and dimethylated at histone H3 lysine 4 (H3K4me2). Four DNase I-hypersensitive sites were identified, three of which are within the CpG island domain and form only when the CpG island is unmethylated. HS1 and HS3 demarcate the boundaries between the unmethylated CpG island and heavily methylated flanking DNA, whereas HS2 resides in the center of the CpG island, near the exon 1/intron 1 interface (Fig. 1A). Silencing of TMS1 in DNA methyltransferase 1 overexpressing cells or certain breast cancer cell lines (e.g. MDA-MB231 cells) is associated with dense methylation of the CpG island, hypoacetylation of H3 and H4, a shift in histone methylation from H3K4me2 to H3K9me2/3, and remodeling of the CpG island-associated DNase I-hypersensitive sites (HS1–HS3) (21–23). A fourth HS maps to a region 1 kb downstream of the CpG island and forms independently of the methylation status of the CpG island or gene expression. We hypothesized that there may be DNA-binding proteins defined by the HS sites that protect the CpG island from methylation in normal cells, and that loss of function at these sites might contribute to aberrant methylation, changes in chromatin structure, and gene silencing.
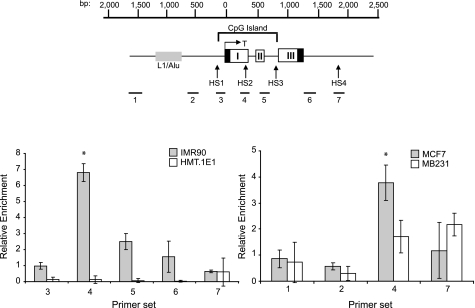
FIGURE 1.
Identification of a methylation-sensitive binding activity against HS2. A, schematic of the TMS1 locus. The TMS1 gene is encompassed within a 5.5-kb HindIII (H) fragment and consists of three exons (I, II, and III). CpG density is shown above the gene. A ∼1.1-kb CpG island spans the promoter and 5′ end of the gene. CpG island-specific DNase I-hypersensitive sites (HS1, HS2, and HS3) and an upstream repeat element (L1/Alu) are shown. Open boxes, noncoding regions. The relative position of the HS2–236bp probe used for electrophoretic mobility shift assay (EMSA) studies is shown. The nucleotide positions are numbered with respect to the transcription start site. B, identification of a methylation-sensitive binding activity. EMSA experiments were performed as described under “Experimental Procedures” using HMT.1E1 nuclear extract and the HS2–236bp probe. Lanes 1–10, unmethylated HS2–236bp probe; lanes 11–13, HS2–236bp probe methylated in vitro with M.SssI. Binding reactions were performed in the absence (–) or presence of a 100-fold molar excess of cold competitor (HS2–236bp) that was either methylated (M) in vitro with M.SssI or left unmethylated (U). The concentration of cold competitor was titrated by serial 3-fold dilutions. From left to right, lane 1, unmethylated probe alone; lane 2, no competition; lanes 3–6, competition with unmethylated HS2–236bp; lanes 7–10, competition with HS2–236bp in vitro methylated; lane 11, methylated probe alone; lane 12, no competition; lane 13, competition with 100-fold molar excess of in vitro methylated HS2–236bp. C, localization of the methylation-sensitive element. Thirteen sequences spanning the HS2–236bp were amplified by PCR and used as probes in EMSA experiments. Parallel binding reactions were performed in the presence of 100-fold molar excess of unmethylated or in vitro methylated cold competitor of the same sequence as the labeled probe. Methyl-sensitive DNA binding was assessed as described in panel A. Probes for which binding activity was blocked by the unmethylated competitor but not by the methylated competitor were considered positive (+) for methylation-sensitive binding. Arrows indicate the position of CpG dinucleotides. D, methylation-sensitive binding to HS2–55bp. EMSA experiments were performed using MCF7 nuclear extract and the HS2–55bp probe containing 2 CpG sites. Binding reactions were performed in the absence (–) or presence of a 100-fold molar excess of cold competitor (HS2–55bp) that was either in vitro methylated (M) or left unmethylated (U). The concentration of cold competitor was titrated by serial 3-fold dilutions. From left to right, lane 1, unmethylated probe alone; lane 2, no competition; lanes 3–6, competition with unmethylated HS2–55bp; lanes 7–10, competition with methylated HS2–55bp; lanes 11 and 12, empty; lanes 13 and 14, competition with 50-bp fragments of CpG-rich DNA from hypersensitive sites 1(HS1) and 3 (HS3) of the TMS1 locus. E, binding to HS2–55bp is sensitive to the methylation status of either CpG site. Top, sequence of the HS2–55 probe. The 9-bp consensus ets sequence (GCTCTNCCG) is shown, and the CpG residues tested are underlined. EMSA were performed using MCF7 nuclear extract and the labeled HS2–55bp probe that was either unmethylated (lanes 1–6) or methylated at both CpG sites (lanes 7–9). Competition analysis was performed using a 100-fold molar excess of cold competitor oligonucleotide (HS255bp) that was unmethylated (UU) or methylated at the 5′ CpG (UM), the 3′ CpG (MU), or both (MM) CpG sites. From left to right, lane 1, unmethylated probe alone; lane 2, no competition; lane 3, competed with UU; lane 4, competed with MU; lane 5, competed with UM; lane 6, competed with MM; lane 7, methylated probe alone; lane 8, no competition; lane 9, MM competition.
As an initial screen, we utilized mobility shift assays to examine the pattern of nuclear factor binding to probes encompassing the regions surrounding HS1, HS2, or HS3, and compared nuclear extracts prepared from cells that are unmethylated at the CpG island and express TMS1 (IMR90) and cells in which TMS1 is methylated and silent (HMT.1E1) (Fig. 1A). We also examined complex binding to HS probes either unmethylated or methylated in vitro with M.SssI and S-adenosylmethionine. There was little difference in the pattern of nuclear factor binding to the HS1 and HS3 probes between nuclear extracts from IMR90 and HMT.1E1 cells, and methylation of the probes had no impact on nuclear factor binding (data not shown). In contrast, we observed a reduced mobility species that bound to a 236-bp probe encompassing HS2 (HS2–236bp) that was sensitive to the methylation status of the probe (Fig. 1B). This complex could be competed by a 100-fold excess of unmethylated, unlabeled (cold) competitor but not by a similar level of cold competitor methylated in vitro with M.SssI (Fig. 1B). There were several additional reduced mobility species that bound to the HS2–236bp probe, but these were not sensitive to DNA methylation and could be competed by similar concentrations of unlabeled methylated and unmethylated competitor DNA (Fig. 1B). We examined nuclear extracts from fibroblast-derived (IMR90, SV40-immortalized IMR90 (90SV)) and breast cancer cell lines (MCF7, MDA-MB231, MDA-MB468, and HS578T) for the presence of this methylation-sensitive binding activity. All cell lines tested contained the methylation-sensitive binding activity (data not shown). Similar results were observed in cell lines in which the endogenous TMS1 was unmethylated and expressed (IMR90, 90SV, MCF7) or methylated and silent (HMT.1E1, MDA-MB231).
Characterization of a HS2 Binding Activity—Our initial experiments used a 236-bp probe surrounding HS2. To narrow the region of interest, we fragmented the HS2–236bp sequence and used mobility shift assays to localize the methylation-sensitive DNA binding activity. The HS2–236bp probe covers the end of the first exon and the begining of the first intron of TMS1, and contains a total of 18 CpG sites (Fig. 1, A and C). Fragments of this region were amplified by PCR and incubated with MCF7 cell nuclear extracts. Methylation-sensitive binding was determined in competition experiments in which binding reactions were performed in the presence of 100-fold molar excess of cold, unmethylated probe or cold probe methylated in vitro at all CpG sites with M.SssI methyltransferase. Thirteen different probes were tested (Fig. 1C). A 55-bp minimal fragment (HS2–55bp), containing two CpGs separated by 33 bp, was sufficient for complex formation (Fig. 1C). Fragments lacking either CpG showed no methylation-sensitive binding activity, indicating that the nucleotides surrounding two CpG residues were necessary for complex formation (complex A, see below).
Four distinct species formed on the unmethylated HS2–55bp probe (Fig. 1D). The formation of complex A and complex C were sensitive to the methylation status of the DNA in that a cold, unmethylated HS2–55bp competitor efficiently competed for binding to these complexes, whereas a cold, methylated competitor did not (Fig. 1D). In contrast, the binding of complexes B and D to HS2–55bp was competed by both the unmethylated and methylated competitor (Fig. 1D), indicating that their formation is unaffected by CpG methylation. We also tested the specificity of binding to HS2–55bp by using two unrelated regions of the TMS1 locus that contain multiple CpG sites as competitors in mobility shift assays (Fig. 1D, HS1 and HS3). We found that binding of complexes A and C were not competed by 100-fold molar excess of either region, but binding of complexes B and D were efficiently competed. This suggested that the methylation-sensitive complexes A and C are specific for CpGs in the sequence context of the HS2–55bp probe, whereas complexes B and D are not sequence-specific, and may represent more general DNA binding activities.
Next we determined the impact of methylation on complex formation. Oligonucleotides corresponding to HS2–55bp were synthesized such that either one or both CpG sites was methylated (UM, MU, MM), and these were then used as competitors in mobility shift assays using the unmethylated HS2–55bp as a probe (Fig. 1E). As expected, a 100-fold molar excess of the fully unmethylated HS2–55bp was sufficient to compete for binding of all complexes A–D. In contrast, HS2–55bp methylated at both CpG sites only competed for binding with complexes B and D, but not with complexes A and C. Interestingly, when either the 5′ or 3′ CpG site was methylated, all four complexes were competed, similar to the unmethylated competitor. These data suggest that complexes B and D represent nonspecific binding activities that are unaffected by the methylation status of the CpG sites, whereas complexes A and C represent sequence-specific binding activities that are sensitive to the methylation status of both CpG sites. This conclusion is further supported by the finding that only the nonspecific complexes B and D formed on a fully methylated HS2–55bp probe fragment (Fig. 1E).
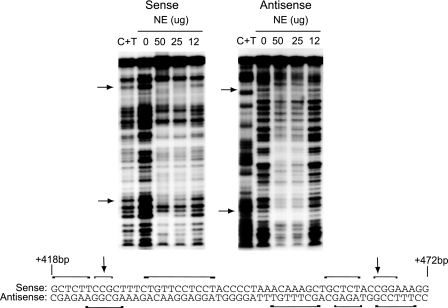
To determine a more exact binding site for the methylation-sensitive complex, we performed in vitro DNase I footprinting. A 107-bp probe containing the HS2–55bp sequence was end-labeled with 32P and then digested with different restriction enzymes to generate probes labeled on either the sense or antisense strand. Footprint analysis showed that the sequences surrounding the CpG dinucleotides are protected (Fig. 2). The footprint suggests that a complex in the nuclear extract binds to the sequence surrounding each of the CpGs. Strikingly, each CpG is located within a 9-base pair sequence with the consensus GCTCTNCCG (Fig. 1E). Taken together these data suggest a model in which the components of the same complex bind to sequences surrounding either the 5′ or 3′ CpG sites, causing the formation of methylation-sensitive complex C, and that when both ends are bound, the methylation-sensitive complex A is formed.
FIGURE 2.
DNase I footprint analysis. A 107-bp probe encompassing the region containing HS2–55bp was end-labeled and then digested with either HpaII or NALV4 to generate a double-stranded probe labeled on either the sense (left) or antisense (right) strand. Labeled probes (∼20 fmol) were incubated with 0, 50, 25, or 12μg of MCF7 cell nuclear extract in the presence of DNase I. Arrows indicate the position of the CpG sites. Maxim and Gilbert sequencing was used to generate a C-T ladder (C+T). A portion of the probe sequence (+418 to +472 bp) is shown below and the relative regions protected on the sense and antisense strand are indicated by the brackets.
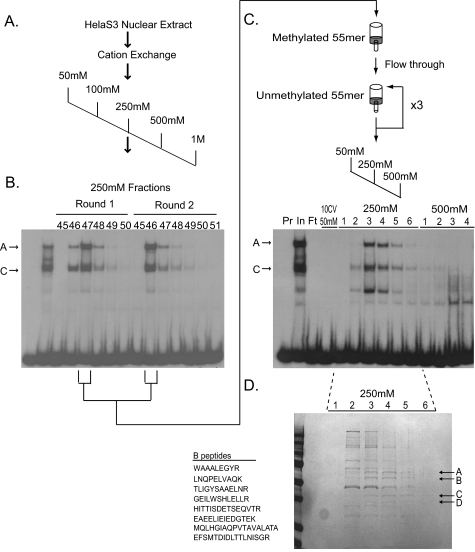
Isolation of the Methylation-sensitive Complex—Next we employed DNA affinity chromatography to identify the factor(s) involved in the methylation-sensitive HS2 binding activity. Nuclear extracts prepared from HeLa S3 cells were subject to cation exchange chromatography followed by a two-step DNA affinity protocol (Fig. 3). Methylation-sensitive binding activity was monitored throughout the purification procedure by mobility shift assays using HS2–55bp as the probe. To monitor specifically the formation of the methylation-sensitive complex A and C, binding reactions were carried out in the presence of 100-fold molar excess of cold, methylated, HS2–55bp to compete away the nonspecific DNA binding. Peak fractions from the cation exchange column were subjected to a negative selection on a DNA affinity column consisting of a multimerized, in vitro methylated version of the HS2–55bp fragment (Fig. 3C). The flow-through from this column was then bound to a second DNA affinity column of an oligomerized, unmethylated version of HS2–55bp. Two additional rounds of binding and elution were performed and binding activity was eluted with the 250 mm NaCl fractions (Fig. 3C). Four polypeptide species that co-eluted with methylation-sensitive binding activity were selected for analysis (Fig. 3D). MALDI MS/MS analysis revealed that bands A, C, and D best matched to hnRNP L (60 kDa), hnRNP D (37 kDa), and the small isoform of the elongation factor EEF1D (31 kDa), respectively. Band “B” showed an 11 (8 unique) peptide match with the ets family transcription factor, GABPα (51 kDa). As known RNA binding factors and regulators of mRNA translation and processing, the functional significance of the co-purification of the hnRNPs/EEF1D is unclear. However, we decided to focus our attention on GABPα.
FIGURE 3.
Purification of the HS2–55bp binding activity. A, the HS2–55bp binding activity was isolated from HeLa S3 cell nuclear extract by cation exchange chromatography and a two-step DNA affinity chromatography scheme. HeLa S3 nuclear extract (415 mg) was divided in half, and each half was independently bound to a SP-Sepharose cation exchange column in 50 mm NaCl binding buffer and step eluted in 50 mm, 100 mm, 250 mm, 500 mm, and 1 m NaCl. Methylation-sensitive binding activity in the fractions was monitored by electrophoretic mobility shift assay (EMSA) using the HS-55bp probe in the presence of 100-fold molar excess of unlabeled HS2–55bp methylated at both CpG sites as a competitor. B, methylation-sensitive binding activity from each round of purification eluted with the 250 mm fractions. Representative EMSA analysis is shown. Methylation-sensitive complexes A and C are indicated. C, fractions containing peak activity from the cation exchange column were combined and negatively selected on a DNA affinity column containing the oligomerized, methylated HS2–55bp oligonucleotide. Flow-through from the methylated column was then bound to a DNA affinity column containing the oligomerized, unmethylated, HS2–55bp oligonucleotide, and eluted in 500 mm NaCl. Binding and elution from the unmethylated column were repeated twice, with a final step elution in NaCl. Fractions were analyzed by EMSA using the HS-55bp probe in the presence of 100-fold molar excess of unlabeled HS2–55bp methylated at both CpG sites as competitor. A representative analysis of the third round of purification is shown. Lanes 1, probe alone (Pr); lane 2, input (In); lane 3, flow-through (Ft); lane 4, 10 column volumes of 50 mm NaCl wash, 250 mm NaCl, and 500 mm NaCl eluted fractions. D, fractions 1–6 (250 mm NaCl) from the third round of affinity purification (see C) were trichloroacetic acid precipitated, separated on an SDS-10% polyacrylamide gel, and stained with Gel Code Blue (Pierce). Bands A–D (arrows) co-eluting with HS2 binding activity were excised and subject to MALDI MS/MS.
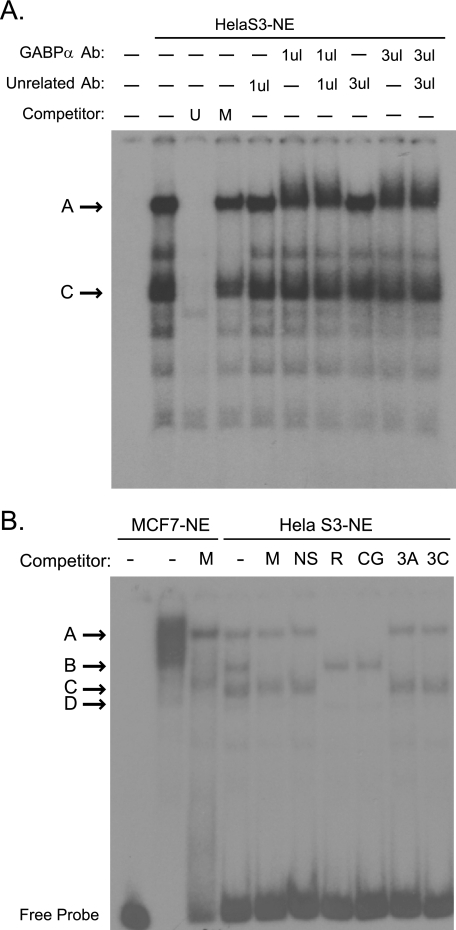
GABPα is a member of the ets transcription factor family. Previous studies have shown that the GABP complex interacts with tandem binding sites as a heterotetrameric (α2β2) complex with the DNA binding activity and transactivation activities contributed by the separate α and β subunits, respectively (24). Supershift analysis confirmed the presence of GABPβ1 in the methylation-sensitive complex that binds HS2–55bp (Fig. 4A). The two CpG sites of interest in the HS2–55bp fragment lay within a 9-bp consensus GCTCTNCCG, a known consensus for ets transcription factors. Interestingly, the 5′ binding site in the HS2–55bp (GCTCTTCCG) is identical to a known binding site for GABPα in the rat cytochrome c oxidase IV gene (25). Consistent with a role for GABPα in TMS1 HS2 binding activity, an oligonucleotide containing known binding sites for GABPα in the rat cytochrome c oxidase IV locus was capable of competing with the HS2–55bp probe for binding to methylation-sensitive complexes A and C (Fig. 4B).
FIGURE 4.
GABP complex binds HS2–55bp. A, electrophoretic mobility shift assays (EMSA) were performed using HeLa S3 nuclear extract and the HS2–55bp probe. Binding reactions were performed in the absence (–) or presence of 100-fold molar excess of unmethylated (U) or methylated (M) cold HS-55bp competitor. Where indicated, 1 or 3 μl of an unrelated antibody (NRF2) or antibody against GABPβ1 was added to the binding reaction after 30 min, after which the reaction was allowed to proceed for an additional 20 min. Note the decreased mobility in the presence of the GABPβ1 antibody. B, EMSA were performed using MCF7 cell or HeLa S3 nuclear extract and the HS2–55bp probe. Binding reactions were performed in the absence (–) or presence of 100-fold molar excess of the indicated competitor. M, methylated HS-55bp; NS, 55-bp region of HS2 that does not contain the 9-bp ets consensus sequences; R, fragment of the RCO4 locus containing a known GABP binding site; CG, fragment of the CGBP locus containing a known GABP binding site; 3A, CG-rich 5′ fragment of TMS1-HS3; 3C, CG-rich 3′ fragment of TMS1-HS3. Competitor sequences are listed in supplemental Table S1.
GABPα Binds TMS1 HS2 in Vivo in a Methylation-sensitive Manner—We next sought to determine whether GABP plays a functional role at the TMS1 locus in vivo. Previous reports from our laboratory have shown that the two CpG dinucleotides within consensus GABPα binding sites are unmethylated in the MCF7 breast cancer cells and IMR90 diploid fibroblasts in which TMS1 is expressed but are fully methylated in MDA-MB231 and HMT.1E1 cells in which TMS1 is silent (21, 23). ChIP was performed with chromatin from all four cell lines using an antibody specific to GABPα and analyzed by quantitative real-time PCR using primers spanning the TMS1 locus (Fig. 5). We found that GABPα was selectively enriched at the HS2 region of the TMS1 locus in cells in which TMS1 is unmethylated and expressed (IMR90 and MCF7) but not in cell lines in which TMS1 is methylated and silent (MDA-MB231 and HMT.1E1). These data indicate that, GABP associates with the TMS1 locus in vivo, and that this association is inversely correlated with the methylation state of the CpG island.
FIGURE 5.
GABP occupancy at the TMS1 locus is dependent on the methylation status of the DNA. Top, schematic of the TMS1 locus. The nucleotide positions are numbered with respect to the transcription start site (T) and are shown above the gene. The location of the CpG island is denoted and spans from approximately –100 to +900 bp. Primer sets used (1–7) for chromatin immunoprecipitation assays are shown. Bottom, occupancy of GABP at the TMS1 locus was analyzed by chromatin immunoprecipitation in human fibroblasts (IMR90,HMT.1E1) and human breast cancer cell lines (MCF7, MDA-MB231) differing in their expression and methylation status at the TMS1 locus. ChIP was performed using an antibody against GABPα or a nonspecific control. Immunoprecipitated DNA was quantified by real-time PCR using the indicated primer sets. Data are presented as the fold enrichment in the GABPα immunoprecipitation relative to that of the control. Error bars represent the ± S.D. of three independent experiments. *, significant difference compared with all other points analyzed (p < 0.01, Student's t test). Primer set 4 flanks the HS2–55bp probe DNA.
GABP and HS2 Regulate TMS1 Promoter Activity—HS2 is located within the center of the TMS1 CpG island and forms only when the CpG island is unmethylated (21). The above data indicate that GABP is recruited to the locus when it is unmethylated and the gene is expressed. These findings suggest that the GABP complex is responsible for the formation of HS2, and may serve as a regulator of TMS1 transcription. To test this hypothesis, we performed transient transfection assays to determine the impact of the HS2 DNA sequence on TMS1 promoter activity. Luciferase reporter constructs were developed that contained the minimal TMS1 promoter driving expression of luciferase in the presence and absence of a 236-bp fragment encompassing HS2. The resulting constructs, as well as a control pGL3 construct, were transfected into the MCF7 breast cancer cell line and luciferase activity measured as an indication of TMS1 promoter activity. We found that the HS2 sequences conferred a ∼3-fold increase in TMS1 promoter activity relative to the promoter alone (Fig. 6A).
FIGURE 6.
Impact of GABP on TMS1 promoter activity. A, MCF7 cells were transfected with the indicated plasmids (5 μg) and a Renilla luciferase reporter plasmid (pRL-TK) (50 ng) as an internal transfection control. pGL3, control vector; min-TMS1, minimal TMS1 promoter (–263 bp to +77 relative to transcription start); HS2-sense, HS2–236bp fused to min-TMS1 in the sense orientation with respect to the promoter. Luciferase activity was measured after 48 h using the Dual Luciferase reporter assay. Data are represented as fold over pGL3 control after normalization to Renilla luciferase activity. Error bars represent ± S.D. of six independent experiments performed in triplicate. *, significant compared with min-TMS1 (p < 0.05, Student's t test). B, MCF7 cells were transfected with pGL3 or pGL3 constructs containing the TMS1 promoter alone (min-TMS1), in the presence of a 236-bp HS2 fragment (HS2-sense) or HS2 fragment in which either the 5′ (HS2-sense m1), 3′ (HS2-sense m2), or both (HS2-sense dm) GABP binding sites have been deleted. A Renilla luciferase reporter plasmid (pRL-TK) was included as an internal transfection control. Luciferase activity was determined after 48 h. Data are represented as fold over pGL3 control after normalization to Renilla luciferase activity. Error bars represent ± S.D. of three independent experiments performed in triplicate. *, significant compared with HS2-sense (p < 0.05, Student's t test). C, MCF7 cells were transfected with the indicated plasmid (5 μg), and siRNAs targeting GABPα and GABPβ (100 nmol/liter each) or a scrambled siRNA control (200 nmol/liter). Renilla luciferase reporter (pRL-TK, 50 ng) was included as an internal control. Left, luciferase activity was measured after 48 h. Data are represented as fold over pGL3 after normalization to the Renilla luciferase control. Error bars represent ± S.D. of three independent experiments performed in triplicate. Right, representative immunoblot showing GABPα, GABPβ1, and GAPDH levels in cells transfected with scrambled or both GABPα-1 and GABPβ siRNA in the same cell lysates used for luciferase assays.
To determine whether this effect was mediated by GABP binding sites, constructs were also created in which one or both of the tandem GABPα binding sites in HS2 were deleted. Disruption of either GABPα binding site inhibited HS2-mediated enhancement of TMS1 promoter activity (Fig. 6B). Interestingly, whereas deletion of the 5′ ets site (HS2-sense m1) completely blocked HS2 enhancer activity, reporter constructs in which the 3′ site (HS2-sense m2) retain some enhancer activity. This latter site deviates from the ets consensus in a key core position (from “GGAA” to “GGTA”) shown to be critical for optimal DNA binding (24). These results indicate that the HS2 region acts in cis to positively regulate the TMS1 locus and that this activity is dependent on the GABP binding sites.
To further test that the effect of HS2 on TMS1 promoter activity is mediated by GABP, luciferase assays were performed in cells treated with siRNA targeting GABPα and GABPβ1. Depletion of endogenous GABPα/β1 abrogated the stimulation of promoter activity conferred by HS2 (Fig. 6C). These results indicate that the HS2 region acts in cis to positively regulate the TMS1 promoter and this activity is dependent on both the GABP binding sites, and the activity of GABPα/β1 in trans.
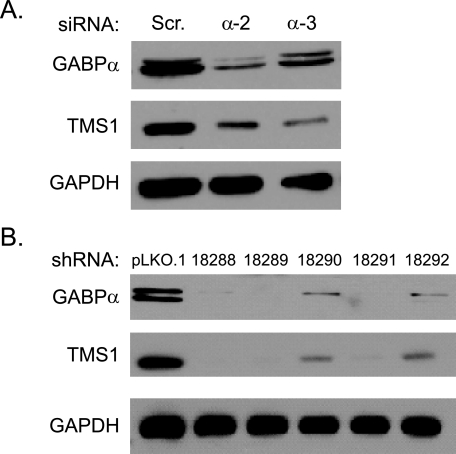
Knockdown of GABPα Results in a Concomitant Decrease in TMS1 Expression—We next examined the impact of GABPα on endogenous TMS1 expression in MCF7 cells. Transient transfection of MCF7 cells with siRNA targeting GABPα resulted in a >75% reduction in GABPα protein levels and a corresponding decrease in TMS1 expression levels (Fig. 7). Similar results were obtained using an infection approach in which MCF7 breast cancer cells were infected with lentiviral shRNA constructs targeting different sites on the GABPα mRNA. There was a direct correlation between the degree of GABPα knockdown and levels of TMS1 expression (Fig. 7). Taken together, these data indicate that the intronic HS2 element acts in cis to positively regulate the TMS1 promoter and that this is mediated by the ets transcription factor GABP.
FIGURE 7.
GABPα regulates TMS1 expression in vivo. A, MCF7 cells were transfected with 200 nmol/liter of two independent siRNAs targeting GABPα or a scrambled siRNA control. Cells were harvested 48 h later and analyzed for GABPα, TMS1, or GAPDH (loading control) protein expression by Western blot analysis. A similar level of depletion was observed with a third independent GABPα siRNA. B, MCF7 cells were infected with lentivirus carrying the empty pLKO.1 vector (pLKO.1) or pLKO.1 expressing GABPα shRNA (five independent shRNA constructs, 88–92) and selected with 0.5 μg/ml puromycin for 5 days. Cells were harvested and analyzed for GABPα, TMS1, or GAPDH (loading control) protein expression by Western blot analysis. The doublet observed in the GABPα blot most likely arises from post-translational modification as GABPα is known to be phosphorylated.
DISCUSSION
In this study, we demonstrate a role for GABP in the regulation of TMS1 gene expression. We used a biochemical approach to purify and identify GABP as a factor that bound a 55-bp fragment coincident with a DNase I-hypersensitive site (HS2) located in intron 1 of the TMS1 gene. The intronic HS2 region containing GABP binding sites acts as an enhancer of TMS1 promoter activity and promotes a 3–5-fold increase in transcription when placed in the sense orientation relative to the promoter. This enhancement is dependent on both the GABPα binding sites and the presence of the GABPα/GABPβ complex. We further showed that CpG methylation blocks the binding of GABP to the TMS1–55bp fragment in vitro, and correlates with a loss of GABP occupancy and down-regulation of TMS1 expression in vivo. Taken together these data indicate that GABP positively regulates TMS1 in a manner that is dependent on the methylation state of the DNA.
The ets transcription factor family consists of more than 30 members that are related by a conserved “winged helix-loop-helix” DNA binding domain that recognizes a core binding site of GGA(A/T). GABPα is unique among the ets factors in that its transcriptional activity requires complexation with an unrelated factor, GABPβ, which contributes the transcriptional activation domain as well as a nuclear localization signal and influences the efficiency of GABPα nuclear transport as well as its DNA binding affinity (26). GABPα is the only ets factor that can recruit GABPβ to DNA (24). The GABPα/β complex exists in solution as a heterodimer but often binds tandem sites separated by 10–30 bp. Homotypic interactions between the leucine zipper domains of GABPβ allow for the formation of a heterotetramer with 10–20-fold greater affinity for DNA (24, 26). In fact, recent genome-wide studies of GABPα occupancy indicate that most sites of GABP enrichment contain two ets consensus motifs (27). Indeed, we found that the methylation-sensitive complex bound to HS2–55bp existed in two forms, a faster migrating form (“C”) and a slower migrating form (“A”) (see Fig. 1E), both of which were competed by oligonucleotides containing known GABPα binding sites, as well as an excess of HS2–55bp methylated at one or the other ets binding site, but not both. We interpret these data to mean that the same complex, likely the GABPα/β heterodimer, can bind either available ets consensus site, but occupancy of both sites is necessary for the formation of the heterotetramer represented by complex A. Consistent with this interpretation, we found that an antibody to GABPβ1 supershifted complex A. These data strongly suggest that the GABPα/β1 heterotetramer binds the TMS1 locus at HS2.
GABPα was originally identified in studies of viral gene transcription (28–30). However, it has been discovered to play a critical role in the regulation of a wide array of genes involved in core cellular processes (31), and more specialized functions, including several genes with known roles in breast cell biology and breast carcinogenesis (e.g. prolactin, oxytocin receptor, BRCA1, and Her2/neu) (32–34). The importance of GABP is underscored by the phenotype of Gabpα knock-out mice that show an early preimplantation lethal phenotype (35).
Recent studies have examined the genome-wide occupancy of ets transcription factors, including GABPα, using ChIP-chip and ChIP-seq approaches (27, 36). These studies indicate considerable redundancy in the binding of different ets factors to the same site. These redundant sites correlated with the strongest match to the ets consensus and tended to lie in close proximity to promoters of widely expressed genes (36). In contrast, sites more selectively bound by one ets factor or another were more divergent from consensus and tended to lie further from the transcription start sites of genes with more specialized roles. Our data suggest that TMS1 may be representative of this latter class. HS2 lies within intron 1 of the TMS1 gene and ChIP experiments confirm enrichment of GABP over this region in cells in which TMS1 is expressed. One of the two tandem ets consensus sites in HS2 deviates from the GABPα consensus GGAA in a key core position (to GGTA) shown to be critical for optimal DNA binding (24). Interestingly, our reporter assays showed that deletion of the 5′ perfect ets consensus site completely abolished enhancement of TMS1 promoter activity, whereas deletion of this latter site had less of an impact.
TMS1 is normally highly expressed in cells of the monocyte/macrophage lineage and most epithelial cell types (13, 14, 37), however, little is known about the regulation of TMS1 transcription. Previous studies have shown that TMS1 is up-regulated in response to inflammatory stimuli, such as interleukin 1β, interferon-γ, and lipopolysaccharide in macrophage (15, 16), although the mechanism of that regulation has not been determined. In breast epithelial cells, TMS1 is up-regulated in response to stress stimuli, including proinflammatory cytokines, such as TNFα and TRAIL (17), and in response to detachment from the substratum (20). Whereas the former is dependent on the NF-κB and c-Jun NH2-terminal kinase (JNK) pathways; up-regulation in response to detachment was independent of these pathways (20). One study indicated that TMS1 may be directly regulated by p53, and identified a putative p53 binding site in the TMS1 promoter region (38). However, subsequent studies suggest that the regulation of TMS1 by p53 may be more complex. For example, although treatment of breast epithelial cells or breast cancer cells wild-type for p53 with DNA damaging agents leads to a modest increase in TMS1 protein expression (17), this up-regulation appeared to be independent of p53 in siRNA experiments, and similar treatments had little impact on the activity of TMS1 promoter fragments containing the putative p53 element in reporter assays.5
The data presented here indicate that the GABPα/β1 complex plays a role in the regulation of TMS1 and is necessary to maintain basal expression at the TMS1 locus. GABPα/β1 may interact directly with the basal transcriptional machinery and/or other factors bound at the promoter to enhance transcription. For example, binding of the GABP complex at a distal enhancer mediates loop formation and interaction with the RAR/RXR complex at the proximal promoter during retinoic acid-induced activation of CD18 in myeloid cells (39). Consistent with this model, the HS2 element enhanced TMS1 promoter activity in a manner that was dependent on the presence of the tandem ets sites and GABP expression. Alternatively, GABP binding at HS2 could promote the recruitment of chromatin modifying complexes to establish a permissive mark on the CpG island chromatin that either prevents the recruitment of repressive factors or provides a signal that opposes the spread of heterochromatin and DNA methylation. GABP is known to interact with the histone acetyltransferase CBP/p300 (39, 40), and may serve to establish a zone of H3 acetylation across the TMS1 CpG island. We have previously shown that the TMS1 CpG island is marked by hyperacetylated histones H3 and H4, and H3K4me3 in cells expressing TMS1, including the IMR90 fibroblasts and MCF7 cells utilized here (21, 22). Epigenetic silencing in cancer cells is associated with the loss of these marks and the gain of H3K9me2/3 and DNA methylation.
Recently, we showed that the histone acetyltransferase hMOF and the MSL complex are recruited to, and play a critical role in, nucleosome positioning and maintenance of gene expression at the TMS1 locus (22). hMOF-mediated acetylation of H4K16 at two strongly positioned nucleosomes flanking the TMS1 CpG island and coincident with HS1 and HS3, is necessary to maintain nucleosome positioning and gene expression (see Fig. 1). Interestingly, knockdown of MSL1 or hMOF in MCF7 cells led to a loss of H4K16 acetylation and reversible silencing of TMS1 expression, but had no impact on H3K9/14 acetylation or H3K4me2 at the CpG island, suggesting that other factors are involved in maintaining a permissive chromatin structure at the TMS1 CpG island (22). The work described here identifies the GABP complex as one potential factor. GABP is enriched at HS2 in the center of the CpG island in cells where the gene is expressed, and down-regulation of GABP leads to a concomitant loss in TMS1 expression. Whether long-term absence of GABP binding leads to a subsequent shift in histone modification patterns and the acquisition of DNA methylation has proved difficult to test, as we have been unable to establish cell lines stably knocked down for GABPα, likely due to its central role in cell cycle progression and cell viability (31, 35, 41).
Previous work has shown that the interaction of GABPα with its recognition sequence is blocked by CpG methylation (42–45). Yokomori et al. (44) suggested that the sensitivity of GABPα to DNA methylation may be a mechanism for regulating sex-specific expression at the mouse Cyp 2d-9 locus, which is differentially methylated in males and females. The same group demonstrated an inverse relationship between DNA methylation, GABP binding, and gene expression at the THSR gene in thyroid cells (45). Methylation of GABPα binding sites within a downstream enhancer has also been implicated in the regulation of the mouse M-lysozyme gene. During macrophage differentiation, the formation of a DNase I-hypersensitive site at a downstream enhancer correlates with DNA demethylation, binding of the GABPα/β1 heterotetramer, and transcriptional activation (43, 46, 47). We similarly find that binding of GABP to the HS2–55bp element is blocked by methylation. Methylation of either ets site blocked complex formation in binding assays, and, whereas GABP was enriched at HS2 in cells where the TMS1 CpG island is unmethylated, it was absent from cells in which the TMS1 CpG island is densely methylated. Interestingly, transgene experiments in mice have shown that the M-lysozyme downstream enhancer containing the GABP binding site, genetically programmed to remain unmethylated during mouse development maintains a zone of open chromatin marked by hyperacetylated histones H3/H4 and H3K4me2 (48). Programmed de novo methylation of the element allows for the hypoacetylation of histones H3/H4, hypermethylation of H3K9, and silencing of a linked transgene.
Epigenetic silencing of TMS1 has been implicated in the pathogenesis of breast and a number of other tumor types, including glioblastomas, prostate carcinomas, non-small cell lung cancers, and melanomas (7–11). It is possible that loss of GABP binding due to spurious methylation of the CpG sites in its recognition sequence results in transcriptional down-regulation and potentially a loss of histone acetylation, a prerequisite to the acquisition of other silencing marks (e.g. H3K9 methylation), putting the locus at risk of further methylation and stable silencing. Such seeding of methylation and transcriptional down-regulation has been shown to promote de novo methylation of the GSTP1 CpG island in prostate cancer cells (49). In this regard, it is noteworthy that in a comprehensive bisulfite sequencing analysis of normal human mammary epithelial cells, one of the CpGs in the GABP binding sites was methylated at twice the frequency of any other CpG site in the CpG island (in 25% of alleles analyzed; no other CpG in the CpG island was methylated in more than 12%) and in the MCF7 cells shown here, it was one of only two CpG sites that showed any methylation (23). Loss of GABP binding also may be a consequence of aberrant methylation of the CpG island domain and serve to ensure the maintenance of a hypoacetylated state. These and our future studies will help to elucidate the role of cis-acting sequences and trans-acting factors in preventing genes from aberrant methylation and silencing, and may be representative of the events at other CpG-island associated genes silenced by methylation in cancer.
Supplementary Material
Acknowledgments
We thank Dr. Paul Wade for advice on the purification of the GABP complex and Jan Pohl and the Microchemical Core Facility of Emory University for the mass spectrometry analysis.
This work was supported, in whole or in part, by National Institutes of Health NCI Grant 2RO1 CA077337.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: TMS1, target of methylation-induced silencing-1; HS, hypersensitive site; hnRNP, heterogeneous nuclear ribonucleoprotein; GABP, GA-binding protein; DTT, dithiothreitol; TNFα, tumor necrosis factor α; TRAIL, tumor necrosis factor–related apoptosis-inducing ligand; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; ChIP, chromatin immunoprecipitation; PIPES, 1,4-piperazinediethanesulfonic acid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interfering RNA.
P. Kapoor-Vazirani, M. J. Parsons, and P. M. Vertino, unpublished data.
References
- 1.Plass, C. (2002) Hum. Mol. Genet. 11 2479–2488 [DOI] [PubMed] [Google Scholar]
- 2.Cross, S. H., and Bird, A. P. (1995) Curr. Opin. Genet. Dev. 5 309–314 [DOI] [PubMed] [Google Scholar]
- 3.Herman, J. G., and Baylin, S. B. (2003) N. Engl. J. Med. 349 2042–2054 [DOI] [PubMed] [Google Scholar]
- 4.Swigut, T., and Wysocka, J. (2007) Cell 131 29–32 [DOI] [PubMed] [Google Scholar]
- 5.Tate, P. H., and Bird, A. P. (1993) Curr. Opin. Genet. Dev. 3 226–231 [DOI] [PubMed] [Google Scholar]
- 6.Zhao, H., Kim, A., Song, S. H., and Dean, A. (2006) J. Biol. Chem. 281 30573–30580 [DOI] [PubMed] [Google Scholar]
- 7.Collard, R. L., Harya, N. S., Monzon, F. A., Maier, C. E., and O'Keefe, D. S. (2006) Prostate 66 687–695 [DOI] [PubMed] [Google Scholar]
- 8.Conway, K. E., McConnell, B. B., Bowring, C. E., Donald, C. D., Warren, S. T., and Vertino, P. M. (2000) Cancer Res. 60 6236–6242 [PubMed] [Google Scholar]
- 9.Guan, X., Sagara, J., Yokoyama, T., Koganehira, Y., Oguchi, M., Saida, T., and Taniguchi, S. (2003) Int. J. Cancer 107 202–208 [DOI] [PubMed] [Google Scholar]
- 10.Stone, A. R., Bobo, W., Brat, D. J., Devi, N. S., Van Meir, E. G., and Vertino, P. M. (2004) Am. J. Pathol. 165 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virmani, A., Rathi, A., Sugio, K., Sathyanarayana, U. G., Toyooka, S., Kischel, F. C., Tonk, V., Padar, A., Takahashi, T., Roth, J. A., Euhus, D. M., Minna, J. D., and Gazdar, A. F. (2003) Int. J. Cancer 106 198–204 [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama, T., Sagara, J., Guan, X., Masumoto, J., Takeoka, M., Komiyama, Y., Miyata, K., Higuchi, K., and Taniguchi, S. (2003) Cancer Lett. 202 101–108 [DOI] [PubMed] [Google Scholar]
- 13.Shiohara, M., Taniguchi, S., Masumoto, J., Yasui, K., Koike, K., Komiyama, A., and Sagara, J. (2002) Biochem. Biophys. Res. Commun. 293 1314–1318 [DOI] [PubMed] [Google Scholar]
- 14.Stehlik, C., Lee, S. H., Dorfleutner, A., Stassinopoulos, A., Sagara, J., and Reed, J. C. (2003) J. Immunol. 171 6154–6163 [DOI] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri, T., Wu, J., Yu, J. W., Datta, P., Miller, B., Jankowski, W., Rosenberg, S., Zhang, J., and Alnemri, E. S. (2007) Cell Death Differ. 14 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J., and Alnemri, E. S. (2009) Nature 458 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons, M. J., and Vertino, P. M. (2006) Oncogene 25 6948–6958 [DOI] [PubMed] [Google Scholar]
- 18.Masumoto, J., Dowds, T. A., Schaner, P., Chen, F. F., Ogura, Y., Li, M., Zhu, L., Katsuyama, T., Sagara, J., Taniguchi, S., Gumucio, D. L., Nunez, G., and Inohara, N. (2003) Biochem. Biophys. Res. Commun. 303 69–73 [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa, M., Kawase, K., Inohara, N., Imamura, R., Yeh, W. C., Kinoshita, T., and Suda, T. (2007) Oncogene 26 1748–1756 [DOI] [PubMed] [Google Scholar]
- 20.Parsons, M. J., Patel, P., Brat, D. J., Colbert, L., and Vertino, P. M. (2009) Cancer Res. 69 1706–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stimson, K. M., and Vertino, P. M. (2002) J. Biol. Chem. 277 4951–4958 [DOI] [PubMed] [Google Scholar]
- 22.Kapoor-Vazirani, P., Kagey, J. D., Powell, D. R., and Vertino, P. M. (2008) Cancer Res. 68 6810–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine, J. J., Stimson-Crider, K. M., and Vertino, P. M. (2003) Oncogene 22 3475–3488 [DOI] [PubMed] [Google Scholar]
- 24.Rosmarin, A. G., Resendes, K. K., Yang, Z., McMillan, J. N., and Fleming, S. L. (2004) Blood Cells Mol. Dis. 32 143–154 [DOI] [PubMed] [Google Scholar]
- 25.Virbasius, J. V., and Scarpulla, R. C. (1991) Mol. Cell Biol. 11 5631–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawa, C., Goto, M., Suzuki, F., Watanabe, H., Sawada, J., and Handa, H. (1996) Nucleic Acids Res. 24 4954–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valouev, A., Johnson, D. S., Sundquist, A., Medina, C., Anton, E., Batzoglou, S., Myers, R. M., and Sidow, A. (2008) Nat. Methods 5 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aurrekoetxea-Hernandez, K., and Buetti, E. (2000) J. Virol. 74 4988–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown, T. A., and McKnight, S. L. (1992) Genes Dev. 6 2502–2512 [DOI] [PubMed] [Google Scholar]
- 30.Flory, E., Hoffmeyer, A., Smola, U., Rapp, U. R., and Bruder, J. T. (1996) J. Virol. 70 2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Z. F., Mott, S., and Rosmarin, A. G. (2007) Nat. Cell Biol. 9 339–346 [DOI] [PubMed] [Google Scholar]
- 32.Atlas, E., Stramwasser, M., and Mueller, C. R. (2001) Oncogene 20 7110–7114 [DOI] [PubMed] [Google Scholar]
- 33.Hoare, S., Copland, J. A., Wood, T. G., Jeng, Y. J., Izban, M. G., and Soloff, M. S. (1999) Endocrinology 140 2268–2279 [DOI] [PubMed] [Google Scholar]
- 34.Schweppe, R. E., Melton, A. A., Brodsky, K. S., Aveline, L. D., Resing, K. A., Ahn, N. G., and Gutierrez-Hartmann, A. (2003) J. Biol. Chem. 278 16863–16872 [DOI] [PubMed] [Google Scholar]
- 35.Ristevski, S., O'Leary, D. A., Thornell, A. P., Owen, M. J., Kola, I., and Hertzog, P. J. (2004) Mol. Cell Biol. 24 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollenhorst, P. C., Shah, A. A., Hopkins, C., and Graves, B. J. (2007) Genes Dev. 21 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McConnell, B. B., and Vertino, P. M. (2004) Apoptosis 9 5–18 [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuka, T., Ryu, H., Minamishima, Y. A., Macip, S., Sagara, J., Nakayama, K. I., Aaronson, S. A., and Lee, S. W. (2004) Nat. Cell Biol. 6 121–128 [DOI] [PubMed] [Google Scholar]
- 39.Resendes, K. K., and Rosmarin, A. G. (2006) Mol. Cell Biol. 26 3060–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang, H. S., Nelson, M. L., Mackereth, C. D., Scharpf, M., Graves, B. J., and McIntosh, L. P. (2008) J. Mol. Biol. 377 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imaki, H., Nakayama, K., Delehouzee, S., Handa, H., Kitagawa, M., Kamura, T., and Nakayama, K. I. (2003) Cancer Res. 63 4607–4613 [PubMed] [Google Scholar]
- 42.Cardot, P., Pastier, D., Lacorte, J. M., Mangeney, M., Zannis, V. I., and Chambaz, J. (1994) Biochemistry 33 12139–12148 [DOI] [PubMed] [Google Scholar]
- 43.Nickel, J., Short, M. L., Schmitz, A., Eggert, M., and Renkawitz, R. (1995) Nucleic Acids Res. 23 4785–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokomori, N., Kobayashi, R., Moore, R., Sueyoshi, T., and Negishi, M. (1995) Mol. Cell Biol. 15 5355–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokomori, N., Tawata, M., Saito, T., Shimura, H., and Onaya, T. (1998) Mol. Endocrinol. 12 1241–1249 [DOI] [PubMed] [Google Scholar]
- 46.Ammerpohl, O., Short, M. L., Asbrand, C., Schmitz, A., and Renkawitz, R. (1997) Gene (Amst.) 200 75–84 [DOI] [PubMed] [Google Scholar]
- 47.Schmitz, A., Short, M., Ammerpohl, O., Asbrand, C., Nickel, J., and Renkawitz, R. (1997) J. Biol. Chem. 272 20850–20856 [DOI] [PubMed] [Google Scholar]
- 48.Hashimshony, T., Zhang, J., Keshet, I., Bustin, M., and Cedar, H. (2003) Nat. Genet. 34 187–192 [DOI] [PubMed] [Google Scholar]
- 49.Song, J. Z., Stirzaker, C., Harrison, J., Melki, J. R., and Clark, S. J. (2002) Oncogene 21 1048–1061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.