Abstract
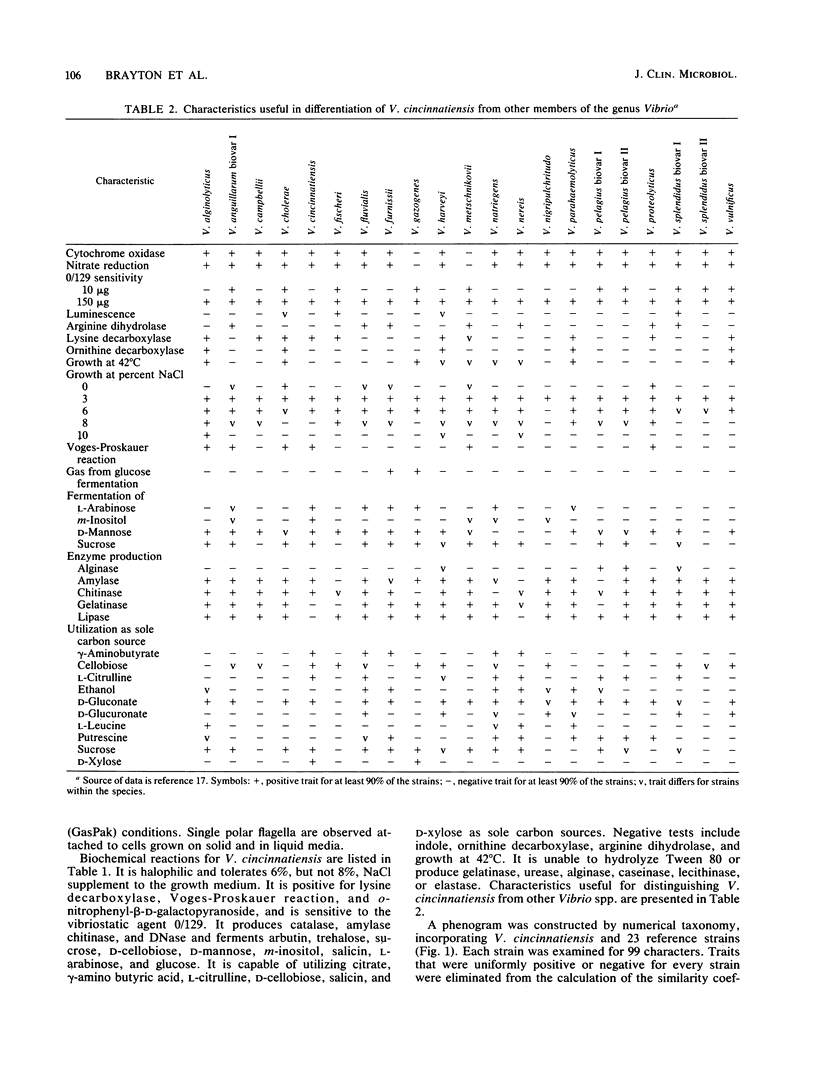
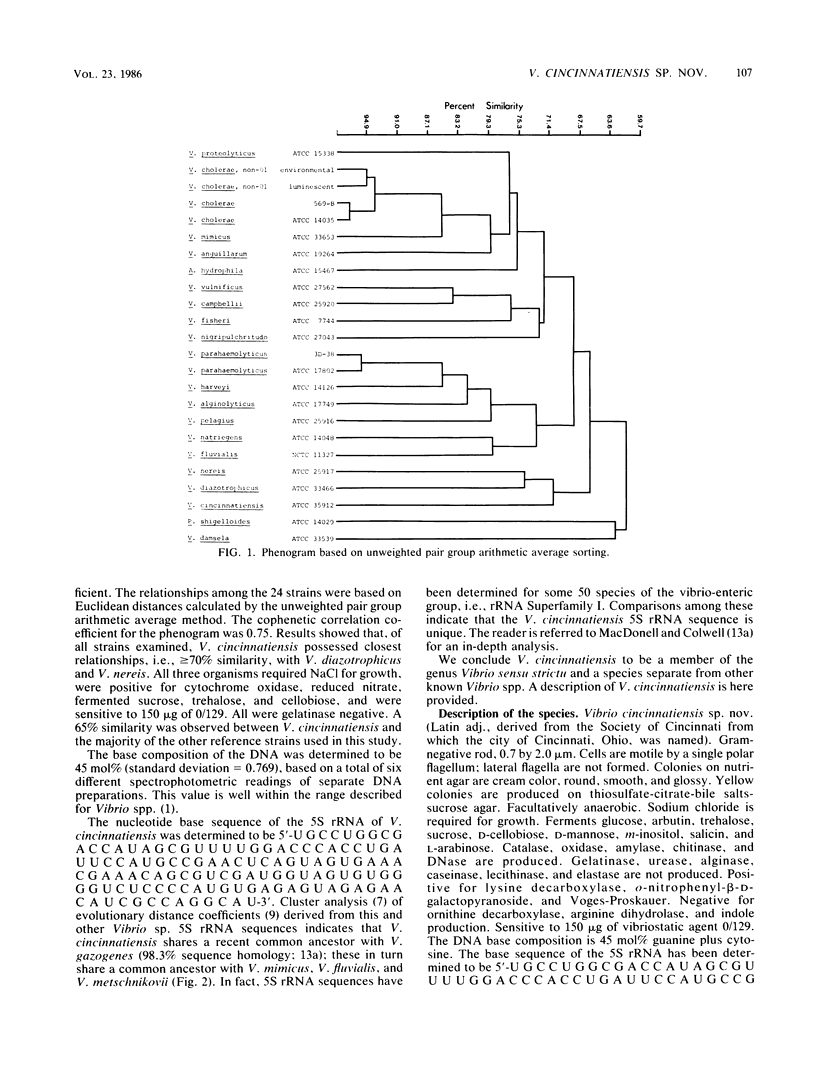
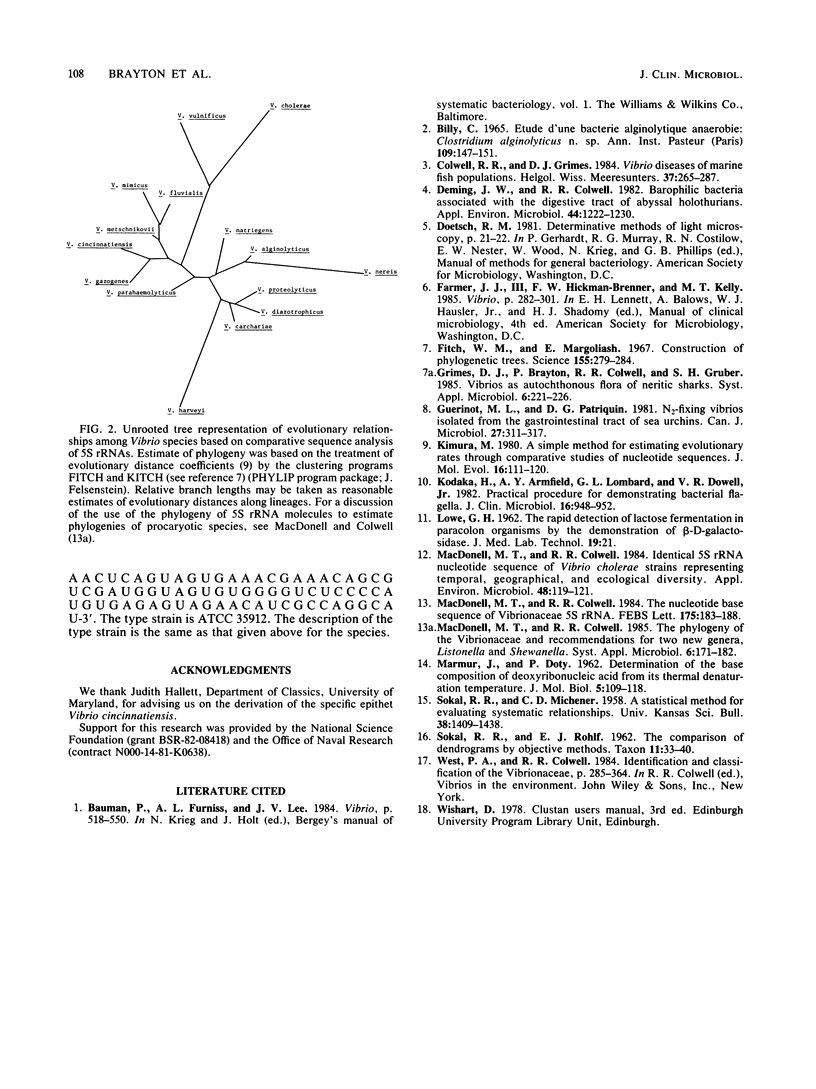
A halophilic gram-negative rod was isolated from blood and cerebrospinal fluid collected from a 70-year-old male having no known contact with seafood or salt water. Positive biochemical tests included oxidase, sensitivity to 0/129, O-nitrophenyl-beta-D-galactopyranoside, lysine decarboxylase and fermentation of glucose, salicin, n-inositol, sucrose, L-mannose, L-arabinose, and arbutin. Negative tests included indole, ornithine decarboxylase, arginine dihydrolase fermentation of lactose, and production of gelatinase and urease. The DNA base composition was 45.0 mol% guanine plus cytosine. Numerical taxonomy indicated 70% similarity with known reference Vibrio sp. strains. The 5S rRNA sequence for this strain has been determined: 5'-U G C C U G G C G A C C A U A G C G U U U U G G A C C C A C C U G A U U C C A U G C C G A A C U C A G U A G U G A A A C G A A A C A G C G U C G A U G G U A G U G U G G G G U C U C C C C A U G U G A G A G U A G A A C A U C G C C A G G C A U-3'. Based on the phenetic, molecular genetic, and nucleic acid sequencing data, it is concluded that Vibrio cincinnatiensis represents a new species of the genus Vibrio sensu strictu (as defined by 5S rRNA sequencing results). On a basis of 5S rRNA comparative sequence analysis, the organism appears to share a recent common ancestor with V. gazogenes (98% homology) and close ancestry with V. mimicus, V. fluvialis, and V. metschnikovii.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billy C. Etude d'une bactérie alginolytique anaérobie: Clostridium alginolyticum n. sp. Ann Inst Pasteur (Paris) 1965 Jul;109(1):147–151. [PubMed] [Google Scholar]
- Deming J. W., Colwell R. R. Barophilic bacteria associated with digestive tracts of abyssal holothurians. Appl Environ Microbiol. 1982 Nov;44(5):1222–1230. doi: 10.1128/aem.44.5.1222-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Guerinot M. L., Patriquin D. G. N2-fixing vibrios isolated from the gastrointestinal tract of sea urchins. Can J Microbiol. 1981 Mar;27(3):311–317. doi: 10.1139/m81-048. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kodaka H., Armfield A. Y., Lombard G. L., Dowell V. R., Jr Practical procedure for demonstrating bacterial flagella. J Clin Microbiol. 1982 Nov;16(5):948–952. doi: 10.1128/jcm.16.5.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWE G. H. The rapid detection of lactose fermentation in paracolon organisms by the demonstration of beta-D-galactosidase. J Med Lab Technol. 1962 Jan;19:21–25. [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- MacDonell M. T., Colwell R. R. Identical 5S rRNA nucleotide sequence of Vibrio cholerae strains representing temporal, geographical, and ecological diversity. Appl Environ Microbiol. 1984 Jul;48(1):119–121. doi: 10.1128/aem.48.1.119-121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonell M. T., Colwell R. R. Nucleotide base sequence of vibrionaceae 5 S rRNA. FEBS Lett. 1984 Sep 17;175(1):183–188. doi: 10.1016/0014-5793(84)80595-5. [DOI] [PubMed] [Google Scholar]


