Summary
Whereas many molecules that promote cell and axonal growth cone migrations have been identified, few are known to inhibit these processes. In genetic screens designed to identify molecules that negatively regulate such migrations, we identified CRML-1, the C. elegans homolog of CARMIL. Although mammalian CARMIL acts to promote the migration of glioblastoma cells, we found that CRML-1 acts as a negative regulator of neuronal cell and axon growth cone migrations. Genetic evidence indicates that CRML-1 regulates these migrations by inhibiting the Rac GEF activity of UNC-73, a homolog of the Rac and Rho GEF Trio. The antagonistic effects of CRML-1 and UNC-73 can control the direction of growth cone migration by regulating the levels of the SAX-3 (a Robo homolog) guidance receptor. Consistent with the hypothesis that CRML-1 negatively regulates UNC-73 activity, these two proteins form a complex in vivo. Based on these observations, we propose a role for CRML-1 as a novel regulator of cell and axon migrations that acts through inhibition of Rac signaling.
Keywords: Cell migration, Axon guidance, Rac GTPase, CARMIL (Lrrc16a), Trio, Robo
INTRODUCTION
Precise wiring of a nervous system requires that migrating neurons and axonal growth cones navigate to their targets by integrating attractive and repulsive signals. Studies of pathways regulated by Slit and Netrin cues provide a glimpse into the mechanisms of guidance and migration (Yu and Bargmann, 2001). Rho family GTPases and the Ena/VASP family of actin-binding proteins appear to mediate the effects of Slit and Netrin receptors (Guan and Rao, 2003). Rho GTPases cycle between active GTP-bound and inactive GDP-bound states. Guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP for GTP to activate the GTPase, whereas GTPase-activating proteins (GAPs) stimulate GTP hydrolysis. In Drosophila, mutations in genes encoding Rac GTPases (members of the Rho superfamily), their GEFs and GAPs, and Enabled (Ena), all result in misguided axons (Gertler et al., 1995; Hakeda-Suzuki et al., 2002; Hu et al., 2005; Lundstrom et al., 2004; Ng et al., 2002; Yang and Bashaw, 2006).
Rac signaling appears to regulate events not only downstream but also upstream of guidance receptor signaling. Two recent studies suggest that Rac signaling regulates the function of both Netrin and Slit receptors in C. elegans (Levy-Strumpf and Culotti, 2007; Watari-Goshima et al., 2007). The kinesin-like molecule VAB-8L and UNC-73, the homolog of the Rac and Rho GEF Trio, increase the levels of SAX-3 (a homolog of Robo) receptors to promote their function.
We sought to identify genes that regulate the Rac pathway. Since UNC-34/Ena and CED-10/Rac are thought to mediate the effects of UNC-40/DCC independently (Gitai et al., 2003), we reasoned that isolating mutations that suppressed the defects of unc-34 mutants might lead to the identification of genes that inhibit Rac signaling. Here we show that loss of functional CRML-1, the C. elegans homolog of the CARMIL (also known as Lrrc16a) actin-uncapping protein (Jung et al., 2001; Remmert et al., 2004; Uruno et al., 2006; Xu et al., 1997; Yang et al., 2005), suppresses the cell and growth cone migration defects of unc-34 mutants but not the defects of unc-73 mutants. We provide evidence that CRML-1 acts through the Rac GEF of UNC-73. We also find that CRML-1 lowers the levels of the SAX-3 guidance receptor, a feature consistent with the role of CRML-1 as a negative regulator of Rac signaling.
MATERIALS AND METHODS
Nematode strains and genetics
Nematodes were cultured as described (Brenner, 1974). N2 Bristol was the wild-type strain, and experiments were performed at 20°C unless stated otherwise. The following mutations and transgenic arrays were used: LG I: unc-73(e936), unc-73(rh40), bli-4(e937), dpy-5(e61), crml-1(gm326), crml-1(gm331), crml-1(n1960), crml-1(n1962); LG II: rrf-3(pk1426), juIs76 [Punc-25:gfp, lin-15(+)]; LG IV: eri-1(mg366), gmIs14 [Pmec-7::vab-8L::gfp, rol-6(su1006)]; LG V: unc-34(gm114), unc-34(gm104), unc-34(e566). Other strains used: CB4856 Hawaiian isolate, gmIs28 [Pmec-7::sax-3::gfp, Pttx-3::gfp] (LG unknown).
Isolation of crml-1 mutations
The crml-1 alleles gm326, gm331, n1960 and n1962 were isolated in two separate screens for mutations that suppressed the uncoordinated (Unc) phenotype of unc-34 mutants. Mutagenesis was performed as described (Brenner, 1974). n1960 and n1962 were isolated by screening the F2 progeny of mutagenized hermaphrodites, as previously described (Bloom, 1993). gm326 and gm331 were isolated in an F1 clonal screen, in which we screened the F2 progeny from individual F1s for a non-Unc phenotype. We screened the progeny of 5476 F1 hermaphrodites.
Cloning of crml-1
crml-1(gm326) mapped to the interval between 1.66 (snp_uCE1-921) and 1.86 (snp_uCE1-924) (Wicks et al., 2001). We fed bacteria expressing dsRNA complementary to each ORF in this interval as described (Kamath et al., 2001; Timmons and Fire, 1998) to the RNAi-sensitized strains eri-1(mg366); unc-34(gm114) and rrf-3(pk1426); unc-34(gm114) and screened the F1 progeny for suppression of both the Unc and CAN migration phenotypes. We used primers PJV84-PJV91 to determine the sequences of crml-1 in gm326, gm331, n1960 and n1962 mutant animals. The sequences of primers used in this study are shown in Table 1. The sequences obtained covered all exons, splice junctions, 5′ and 3′ UTRs and most introns. All mutations identified were confirmed using a new sample of genomic DNA.
Table 1.
Primers used in this study
| Primer | Application | Sequence (5′ to 3′) |
|---|---|---|
| PJV84 | Sequencing | GTTCGCTCTTCGTGTATCG |
| PJV85 | Sequencing | GTGGTTCAGGTGGATGTGG |
| PJV86 | Sequencing | AATCGATGTGAATCCATCTGC |
| PJV87 | Sequencing | GCCTGCCTCGAGTATTTGG |
| PJV88 | Sequencing | GGCCTTCAAATCGAAAAGC |
| PJV89 | Sequencing | CATTGGCAGCAACTACAACG |
| PJV90 | Sequencing | GGAAATGGAGGGAAATGTGC |
| PJV91 | Sequencing | TTCTTCTGACTTCTCACTGTCTCC |
| PJV92 | Promoter 5′ | TCGTCGTCATCACAGAATCC |
| PJV93 | Promoter 3′ | TTTTAGAATTTAGAATTAAAATGGGCG |
| PJV96 | cDNA creation 5′ | ATATCCCGGGATGTCCATGTCGAGGAGC |
| PJV97 | cDNA creation 3′ | ATATCCCGGGTTTTTTGGAAAATTCTAGCC |
| PJV98 | Genotyping gm326 5′ | ACTTCAACCATTCCGTCTGG |
| PJV99 | Genotyping gm326 3′ | TGATAGACGTTGCCAAATCG |
| PJV154 | Genotyping gm326 5′ | GTATTTTAAGTTTAACCTCTTTTTCAG |
| PJV155 | Genotyping gm326 3′ | AATGAGATACATAATCATCACAGTTG |
| PJV172 | Genotyping gm326 5′ | AGAGTCTGATGATGGTTACATGTCCAT |
| PJV173 | Genotyping gm326 3′ | TTTCGAAAGAATTCTGGCCCT |
| PJV174 | Genotyping WT 5′ | AGAGTCTGATGATGGTTACATGTCCAC |
| PJV175 | Genotyping WT 3′ | TTTCGAAAGAATTCTGGCCCC |
ALM and CAN migration
We scored final ALM or CAN positions in young L1 hermaphrodites relative to the positions of hypodermal nuclei (H1, V1, P1/2, V2, P3/4, V3, P5/6 and V4) using Nomarski optics. If a CAN did not appear posterior to the H1 cell, we inferred that it was somewhere in the head region of the animal as it is difficult to distinguish head neurons from a displaced CAN. A proportion two-sample Z-test was conducted to determine statistical significance between two strains (www.statcrunch.com).
Scoring DD processes
We scored the amount of outgrowth of processes DD2-6 in L1 hermaphrodites using juIs76 [Punc-25::gfp] (Jin et al., 1999). Animals were examined by fluorescence microscopy. We could not score DD1 because GFP expression in the processes of head neurons (RMEs, AVL, RIS) overlapped the DD1 processes. We classified the outgrowth defects by determining whether or not the DD process reached the dorsal nerve cord (DNC). We determined the percentage of DD processes that failed to reach the DNC and used this number for comparison between strains. Statistical significance between two strains was determined as described above.
Scoring ALM processes
We scored the process of ALM in L4 or young adult animals using the GFP signal from the gmIs14 [Pmec-7::vab-8L::gfp] transgene. Processes were scored as rerouted if they were bipolar or posterior. We defined a process as posterior if it extended at least five ALM cell body lengths towards the tail. We scored ALM as bipolar if the two processes extended at least five cell body lengths in each direction. Bipolar and posterior processes were pooled into one group for statistical analysis. Statistical significance between two strains was determined as described above.
gmIs28 scoring
Embryos at the 2-fold stage were identified using Nomarski optics. Embryos were scored for the presence or absence of a GFP signal in ALM and BDU. All strains to be compared in a given experiment were scored on the same day to ensure similar fluorescence bulb intensity. The ALM and BDU borders were traced using a Region-of-Interest tool and epifluorescence intensities were quantified by iVision software (BioVision Technologies). A two-tailed Z-test was conducted to determine statistical significance between two strains (www.statcrunch.com).
Plasmid construction and transgenic strains
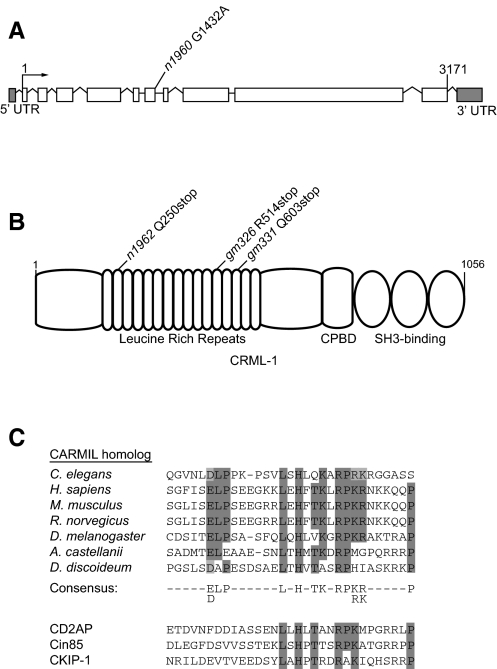
We isolated a full-length cDNA of crml-1 by PCR amplification using an oligo(dT)-primed embryonic cDNA library as template. Determining the sequence of this 3171 bp cDNA revealed ten exons and nine introns (see Fig. 1A).
Fig. 1.
The crml-1 gene and CRML-1 protein. (A) The gene structure of C. elegans crml-1. The n1960 allele is a G-to-A splice donor mutation at the 5′ end of intron 6. White boxes, exons; lines, introns; arrow, transcription start site. (B) CRML-1 contains 16 leucine-rich repeats, a capping protein-binding domain (CPBD), and three PxxP motifs that fit the consensus sequence for SH3-binding domains. The CPBDs of Acanthamoeba and mouse CARMIL can bind capping protein and uncap F-actin (Uruno et al., 2006; Yang et al., 2005). The n1962, gm326 and gm331 alleles are ochre, opal and ochre nonsense mutations in the second, twelfth and fourteenth leucine-rich repeats, respectively. (C) Sequence alignments of the CPBD region were performed using ClustalW. Conserved residues important for binding to capping protein are highlighted in gray, with dark gray indicating identity and light gray indicating similarity. A CARMIL consensus sequence is depicted below. GenBank accession numbers for CARMIL sequences used in phylogenetic analyses are: Caenorhabditis elegans (NP-492024), Homo sapiens (NP-060110), Mus musculus (AAR96060), Rattus norvegicus (XP-225336), Drosophila melanogaster (NP-610316), Acanthamoeba castellanii (AAB57739) and Dictyostelium discoideum (AAK72255). The sequences of three other proteins that contain a CPBD and act as actin-uncappers in vitro are aligned below the CARMIL consensus sequence. GenBank accession numbers for Homo sapiens homologs of CPBD-containing family members used in phylogenetic analyses are: CD2AP (Q9Y5K6), CIN85 (SH3KBP1) (Q96B97), CKIP-1 (PLEKHO1) (CAI14264).
To generate the crml-1 transcriptional fusion (Pcrml-1::gfp), we amplified the intergenic region between crml-1 and the nearest upstream gene and cloned this region into the GFP Fire vector pPD95.75 (A. Fire, personal communication) to generate pPV33. gmEx504, gmEx505 and gmEx506 were generated by injecting pPV33 (50 ng/μL) and the co-injection marker pCL8 [Pmec-7::mcherry] (25 ng/μL) into wild-type hermaphrodites.
To generate the crml-1 translational fusion (Pcrml-1::crml-1cDNA::gfp), we inserted a full-length crml-1 cDNA between the crml-1 promoter and GFP coding sequences of pPV33 to create pPV35. gmEx374 and gmEx375 were generated by injecting pPV35 into wild-type hermaphrodites at 50 ng/μL with 25 ng/μL pRF4 [rol-6(su1006)].
To generate the crml-1 fusion to mCherry (Pmec-7::crml-1cDNA::mCherry), we inserted the full-length crml-1 cDNA between the mec-7 promoter and mCherry coding sequences of pCL8 to create pPV92. gmEx527 and gmEx529 were generated by injecting pPV92 into wild-type hermaphrodites at 50 ng/μL with 6 ng/μL Pmyo-2::gfp.
To integrate gmEx374, we UV-irradiated array-bearing worms using a Stratalinker 2400 (Stratagene) and selected F2 progeny that stably expressed both CRML-1::GFP and the co-injection marker rol-6(su1006). Multiple independent integrants were isolated.
Co-immunoprecipitation and immunoblotting
Co-immunoprecipitation experiments were conducted as described (Chu et al., 2002). We prepared embryo lysates from wild-type and gmIs30 (integrated version of gmEx374) animals. For co-immunoprecipitation experiments, we used 300 μg total protein and added 2 μg of rabbit polyclonal anti-GFP antibody (Abcam, #6556). For immunoblot analysis, 10 mL of immunoprecipitated samples were separated on an SDS-PAGE gel by electrophoresis. Protein was transferred onto a nitrocellulose membrane (Schleicher and Schuell), and the blot was incubated with rabbit anti-UNC-73 antibodies at 1:6000 dilution (Steven et al., 1998). Following hybridization, blots were washed and then incubated with goat anti-rabbit HRP-conjugated secondary antibody (1:10,000, BioRad). The antibody complex was visualized using an ECL Kit (GE Healthcare Amersham) according to the manufacturer's instructions.
Immunohistochemistry
Embryos were fixed and stained as described (Finney and Ruvkun, 1990) using the following antibodies: chicken anti-GFP (Upstate Biotechnology, 1:1000) and rabbit anti-UNC-73 (1:1000). Secondary antibodies were Alexa488-conjugated goat anti-chicken antibody (Molecular Probes) and Cy3-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch Laboratories).
RESULTS
An unc-34 suppressor gene encodes a CARMIL homolog
UNC-34 is a member of the Ena/VASP family of proteins (Yu et al., 2002). These proteins can bind to Robo guidance receptors and to F-actin, suggesting that Ena/VASP family members provide a link between guidance receptors and the actin cytoskeleton (Lebrand et al., 2004). Mutations in unc-34 cause an uncoordinated locomotion phenotype (Unc) (Brenner, 1974), presumably resulting from cell migration and axon guidance defects (Forrester and Garriga, 1997; Hedgecock et al., 1985; McIntire et al., 1992). Gitai et al. (Gitai et al., 2003) provided genetic evidence that UNC-34 and CED-10/Rac signaling mediates the effects of the Netrin receptor UNC-40/DCC. We reasoned that loss of molecules that inhibit Rac signaling would increase Rac activity and might bypass the need for UNC-34. To identify such molecules, we screened for mutations that suppressed the Unc phenotype of unc-34 mutants. Four of the suppressor mutations, gm326, gm331, n1960 and n1962, mapped to chromosome I. Through a combination of genetic mapping, RNAi and DNA sequencing, we identified these mutations as alleles of K07G5.1, a conserved gene that we named crml-1.
Fig. 1B shows the domain structure of CRML-1. Homologs of CRML-1 in Acanthamoeba, Dictyostelium, mouse and human (24, 25, 32 and 33% identical to CRML-1, respectively) have been described (Jung et al., 2001; Xu et al., 1997; Yang et al., 2005). These homologs are known as CARMILs (Capping, Arp2/3, Myosin I Linker), and were first described in Acanthamoeba and Dictyostelium as proteins that bind to the SH3 domain of myosin I (Jung et al., 2001; Xu et al., 1997; Xu et al., 1995). Components of the Arp2/3 complex, which nucleates F-actin branching, also interact with CARMILs (Jung et al., 2001). The loosely defined Arp2/3-binding domain might not be conserved in metazoan CARMILs, as mouse CARMIL failed to activate the Arp2/3 complex (Yang et al., 2005). Acanthamoeba and mammalian CARMILs can uncap actin filaments in vitro (Uruno et al., 2006; Yang et al., 2005). A domain in the C-terminal portion of the protein binds capping protein with high affinity and can remove it from the barbed ends of F-actin. This capping protein-binding domain (CPBD) is conserved in all eukaryotic CARMIL homologs (Fig. 1C).
All four mutations that mapped to chromosome I behave as semi-dominant suppressors, suggesting that they are likely to be different alleles of the same gene. We determined the sequence of the entire crml-1 gene from strains containing the suppressors and identified the mutation(s) in each strain. Two mutations were present in gm326: an opal nonsense mutation in the eighth exon and a missense mutation 80 amino acids further downstream. The crml-1 gene contained an ochre nonsense mutation in the fourth and ninth exons of n1962 and gm331 animals, respectively. The crml-1 gene in n1960 animals was found to contain a splice donor mutation after exon 6 (Fig. 1A,B).
Several observations indicate that these mutations reduce or eliminate crml-1 functions. First, RNAi of crml-1 suppressed the Unc and CAN defects of unc-34 mutants (Fig. 2D; see Fig. S2 in the supplementary material). Second, the mutant proteins should be severely truncated by the three nonsense mutations, and nonsense-mediated decay should degrade the crml-1 transcript produced by these mutants (Pulak and Anderson, 1993). Third, quantitative real-time PCR analysis of the three nonsense mutants indicated that crml-1 mRNA levels are reduced by 50-70% in each mutant (data not shown). Thus, the semi-dominant suppression caused by these alleles could result from haploinsufficiency of crml-1. Alternatively, CRML-1 fragments produced by the mutants could have antimorphic activity. In either case, crml-1 function is reduced in the mutants. At this point, we do not know whether the mutations eliminate all crml-1 functions.
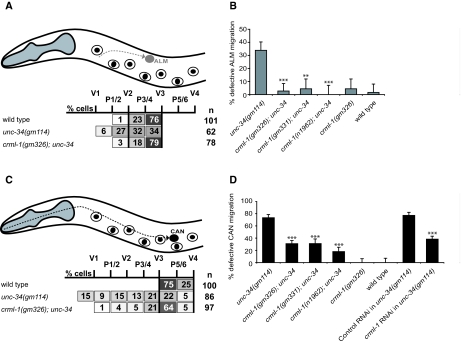
Fig. 2.
Loss of CRML-1 suppresses ALM and CAN migration defects of unc-34 mutants. (A,C) Positions of stationary P and V hypodermal nuclei used to score final ALM (A) or CAN (C) positions in newly hatched L1s. Anterior is to the left, dorsal is up. The gray dashed line in A shows the ALM migration path during embryogenesis, and the black dashed line in C shows the CAN migration path. Each box lists the percentage of cells found in the region indicated. The black vertical lines delineate the region that contains most wild-type ALMs or CANs. (B,D) Graphical representation of the cell positioning data shown in A and C. Any ALM that did not migrate posterior of V2, or any CAN that did not migrate posterior of V3, was scored as defective, and the percentage defective for each genotype examined is shown. Error bars, s.e.m. (B) Statistically significant differences between unc-34 and crml-1; unc-34 strains are marked (***P<0.0001, **P<0.003). For the distribution of ALMs relative to P and V cells for all genotypes and the number of ALMs scored, see Fig. S1 in the supplementary material. At least 45 animals were scored per genotype. (D) Statistically significant differences between unc-34 and crml-1; unc-34 strains and between control RNAi and crml-1 RNAi strains are marked (***P<0.0001). Error bars, s.e.m. For the distribution of CANs relative to P and V cells for all genotypes and the number of CANs scored, see Fig. S2 in the supplementary material. At least 45 animals were scored per genotype.
To our surprise, crml-1 mutants had no obvious abnormal phenotype in the absence of an unc-34 mutation. We considered whether other proteins might function similarly to CRML-1 and compensate for the loss of CRML-1 in wild-type but not sensitized backgrounds, such as in unc-34 mutants. A candidate that might function similarly to CRML-1 is Y44E3A.4, the C. elegans homolog of Cin85 (also known as Sh3kbp1). Two other families that contain the conserved CPBD motif were recently described: the Cin85/CD2AP/CMS family, and the conserved casein kinase-interacting protein CKIP-1 (also known as Plekho1) (Fig. 1C) (Bruck et al., 2006; Canton et al., 2006). The C. elegans genome contains a homolog of Cin85 but not of CKIP-1. In the absence of CRML-1 function, perhaps Y44E3A.4 could serve in the same capacity, partially masking the loss of CRML-1 uncapping function. However, RNAi of Y44E3A.4 in eri-1; crml-1(gm326) animals did not result in locomotion or neuronal migration defects (data not shown).
Suppression of ALM, CAN and DD defects
crml-1 mutants were identified by their ability to suppress the Unc phenotype of unc-34 mutants. If the cell migration and axon guidance defects of unc-34 mutants contribute to the Unc phenotype, then crml-1 mutations should also suppress these defects.
ALMs are a pair of bilaterally symmetric neurons that are generated near the head and migrate during embryogenesis (Sulston et al., 1983). In wild-type larvae, ALMs stop between the V2 and V3 hypodermal cells (Fig. 2A; see Fig. S1 in the supplementary material). In unc-34 mutants, ALMs often stopped prematurely at various positions along their migratory route anterior to V2 (Fig. 2A,B; see Fig. S1 in the supplementary material). We also scored the positions of ALMs in crml-1; unc-34 mutants and found that mutations in crml-1 suppressed the ALM migration defect of unc-34 mutants (Fig. 2A,B; see Fig. S1 in the supplementary material).
We observed a similar suppression of CAN migration. CANs are a pair of bilaterally symmetric neurons that are generated in the head and undergo a posterior migration (Fig. 2C) (Sulston et al., 1983). In unc-34 mutants, CANs were often defective in migration, and crml-1 mutations suppressed this defect (Fig. 2C,D; see Fig. S2 in the supplementary material). In addition, the longer migrations of CANs revealed a semi-dominant effect of crml-1 mutations. Mutating one copy of crml-1 in an unc-34 background weakly suppressed the CAN defect caused by the three alleles (gm326, gm331 and n1962) of crml-1 tested (see Fig. S2 in the supplementary material). Mutating the second copy of crml-1 suppressed the CAN defect further (Fig. 2D; see Fig. S2 in the supplementary material).
We also assessed the morphology of the DD neuronal projections, hereafter referred to as processes. We use the term `process' instead of axon or dendrite because DDs are not polarized with an axon emerging from one part of the cell and dendrites from another. Rather, the branched process of each DD neuron has both presynaptic and postsynaptic domains. During embryogenesis, DDs are generated in the ventral nerve cord (VNC) and extend long anterior processes along the VNC. The anterior processes branch dorsally, reaching the dorsal nerve cord (DNC). In the DNC, the processes branch anteriorly and posteriorly, forming a ladder-like pattern (see Fig. S3A in the supplementary material) (White et al., 1986).
We assessed DD morphology in first larval (L1) stage animals, scoring the percentage of processes that reached the DNC. In unc-34(gm114) mutants, 50% of the processes failed to reach the DNC (see Fig. S3B in the supplementary material) (Forrester and Garriga, 1997). These processes either failed to exit the VNC or extended dorsally but failed to reach the DNC, often branching along the anterior-posterior axis in lateral positions. We quantified the effect of losing one or two copies of crml-1 in unc-34 mutant animals. Lowering crml-1 activity decreased the percentage of processes that failed to reach the DNC in an unc-34 mutant (see Fig. S3B in the supplementary material). Together with the CAN migration data, these findings demonstrate that cell and growth cone migrations are affected in a dose-dependent manner by mutations in crml-1. The suppression affects migrations occurring along both the anterior-posterior and dorsal-ventral axes, indicating that these effects are not specific to a particular direction or cellular environment.
CRML-1 is expressed in the developing nervous system
We examined the expression of crml-1 by constructing two reporters. One was a transcriptional reporter, for which we fused GFP to sequences from the end of the gene upstream to crml-1 to the initiator ATG of crml-1. We also created a translational reporter by fusing a full-length crml-1 cDNA to GFP and driving expression of this fusion from the same crml-1 promoter. Animals bearing the transcriptional and translational reporters had similar GFP expression patterns. As shown in Fig. 3A, L1 animals carrying the translation reporter expressed GFP in many neurons, including CANs, DD-type motoneurons and ALMs. Expression in the nervous system began early in comma-stage embryos and peaked in intensity around the 3-fold stage of embryogenesis (data not shown). Although neuronal expression was much fainter at later larval stages, it persisted in some head and tail neurons through adulthood. Non-neuronal cells that also expressed CRML-1::GFP included the migrating distal tip cells, the pharynx, some vulval epithelial cells, rectal epithelial cells and the excretory canal (data not shown).
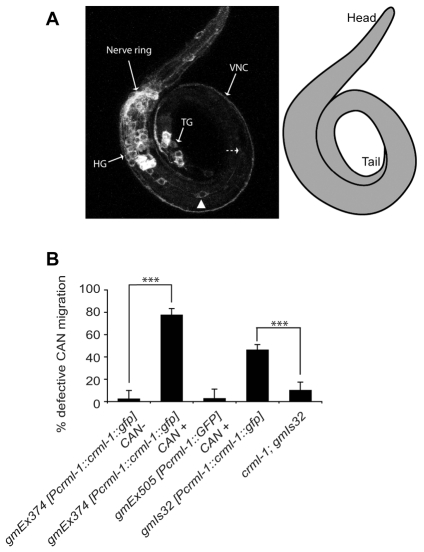
Fig. 3.
Excess CRML-1 disrupts CAN migration. (A) Confocal projection of an L1 C. elegans expressing the extrachromosomal array gmEx374 [Pcrml-1::crml-1::gfp]. CRML-1::GFP is expressed in neurons of the head (HG) and tail (TG) ganglia, and in axons that form the nerve ring, the ventral nerve cord (VNC) and the dorsal nerve cord (DNC) (not shown). The position of a CAN is indicated (arrowhead). This CAN is displaced anterior to its normal position, which is indicated by the dashed arrow. A schematic of the animal is presented on the right. (B) CAN migration defects in CRML-1 transgenic animals. See Fig. 2C legend for details. Extrachromosomal arrays are lost during mitosis, which allowed us to determine differences between animals expressing the array in CAN and those that had lost the array in CAN. For animals carrying gmEx374, we scored CANs in which we detected CRML-1::GFP in the cells (CAN+) and CANs in which we could not (CAN-). Statistically significant differences between CAN+ and CAN-animals and between gmIs32 and crml-1; gmIs32 are marked (***P<0.0001). For the distribution of CANs relative to P and V cells and the number of CANs scored, see Fig. S2 in the supplementary material. Error bars, s.e.m. At least 34 animals were scored per genotype.
Excess CRML-1 causes CAN migration defects
Expression of high levels of the crml-1 cDNA from its native promoter caused a CAN cell migration defect similar to that seen in unc-34 mutants. The transgenic animal shown in Fig. 3A, for example, had a CAN neuron displaced anterior to its normal position. We scored CAN cell body position in animals carrying the extrachromosomal array gmEx374. Extrachromosomal arrays are mitotically unstable, so animals carrying these arrays frequently lose the array in one or more cell lineages. We scored both the position of CAN in transgenic animals and whether or not CAN expressed CRML-1::GFP, which should indicate the presence or absence of the extrachromosomal array in CAN. CANs were usually defective in migration in gmEx374 animals when a GFP signal was present in the CAN (CAN+) (Fig. 3B). If an animal expressed GFP in other cells, but not in the CAN, CAN was in its wild-type position (CAN-) (Fig. 3B). These observations indicate that when expressed in CANs, CRML-1 inhibited their migrations. The migration defect caused by the integrated version of this transgene, gmIs32, became less severe in a crml-1 mutant background (Fig. 3B), consistent with the hypothesis that excess CRML-1 disrupts CAN migration and that the crml-1 mutations reduce gene activity. CANs were in wild-type positions in animals carrying the transcriptional reporter gmEx505, confirming that neither the presence of the crml-1 promoter nor expression of GFP in CANs caused the migration defect seen in gmEx374 or gmIs32 animals (Fig. 3B).
crml-1 and the Rac pathway
The crml-1 alleles identified in our screens bypass the requirement for UNC-34 protein as they suppress not only missense mutations such as gm114, but also nonsense mutations such as gm104 (see Fig. S2 in the supplementary material). One explanation for this type of suppression is that crml-1 inhibits a pathway that acts in parallel to UNC-34. Since Gitai et al. (Gitai et al., 2003) showed that the Rac pathway and the UNC-34 pathway function in parallel to mediate the UNC-40 attractive signaling that guides the AVM process ventrally, we asked whether crml-1 functions in the Rac pathway. All three C. elegans Rac GTPases (ced-10, mig-2 and rac-2/3) function in CAN migration, and loss of each can enhance the cell and growth cone migrations of unc-34 mutants (Lundquist et al., 2001; Shakir et al., 2006).
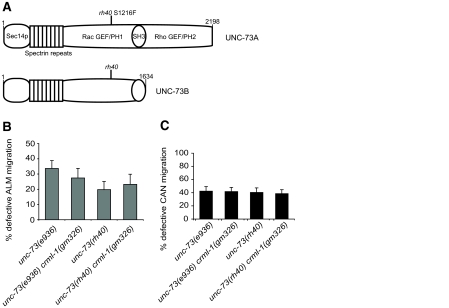
We first asked whether unc-34 and the Racs act in parallel for ALM and CAN migration. The three Racs function redundantly for cell migration, so for simplicity we analyzed the role of the Rac GEF UNC-73, which is homologous to Trio and Kalirin (Steven et al., 1998). UNC-73 acts as a GEF for at least two of the C. elegans Rac proteins (Wu et al., 2002). Although it does not appear to act downstream of unc-40, UNC-73 is required for CAN migration (Forrester and Garriga, 1997; Lundquist et al., 2001; Steven et al., 1998). UNC-73 contains two GEF domains: a Rac GEF required for neuronal cell migrations and axon guidance, and a Rho GEF required for pharyngeal development and synaptic activity (Fig. 4A) (Steven et al., 1998; Steven et al., 2005). At least eight different UNC-73 isoforms are produced. Some isoforms contain the Rac GEF, whereas others contain the Rho GEF, and one isoform contains both. We examined two mutations predicted to disrupt Rac GEF function: e936 is a splice donor mutation that disrupts the two transcripts that encode the Rac GEF-containing isoforms, and rh40 is a missense mutation in the Rac GEF domain. This latter mutation eliminates GDP-GTP exchange activity of the Rac GEF in vitro (Steven et al., 1998). Because rh40 eliminates the Rac GEF function of UNC-73, we assume any effects of this allele reflect a specific defect in Rac pathway function. Both mutations frequently resulted in defective ALM and CAN migration (Fig. 4B,C).
Fig. 4.
crml-1 mutations fail to suppress the CAN and ALM migration defects of unc-73 mutants. (A) Schematic of the two Rac GEF-containing isoforms of C. elegans UNC-73. PH, Pleckstrin homology domain. UNC-73A contains a Rho GEF and a second PH domain that UNC-73B lacks. The rh40 missense mutation eliminates the Rac GEF activity of UNC-73 (Steven et al., 1998). (B) The ALM defects of unc-73 mutants are unaffected by mutation of crml-1. ALM positions were scored as described in Fig. 2A. Error bars indicate s.e.m. For the distribution of ALMs relative to P and V cells and the number of ALMs scored, see Fig. S1 in the supplementary material. At least 41 animals were scored per genotype. (C) The CAN defects of unc-73 mutants are unaffected by mutation of crml-1. CAN positions were scored as described in Fig. 2C. For the distribution of CANs relative to P and V cells and the number of CANs scored, see Fig. S2 in the supplementary material. At least 52 animals were scored per genotype.
To test whether unc-34 and unc-73 act in parallel, we attempted to construct the double mutant. However, double mutants died as embryos, preventing an analysis of their migration defects. RNAi of unc-73 into unc-34(gm114) mutants also resulted in embryonic lethality. If CRML-1 negatively regulates Rac signaling, then loss of crml-1 should not affect the ALM or CAN defects of unc-73 mutants. Unlike the unc-34 interaction, loss of crml-1 did not suppress the ALM or CAN defects of either unc-73 allele (Fig. 4B,C). Similarly, we observed no difference in the DD processes of unc-73 crml-1 and unc-73 mutants (see Fig. S3B in the supplementary material). Although it is possible that crml-1 functions independently of unc-73, our findings are consistent with the hypothesis that CRML-1 negatively regulates the UNC-73 pathway.
CRML-1 and UNC-73 form a complex in vivo
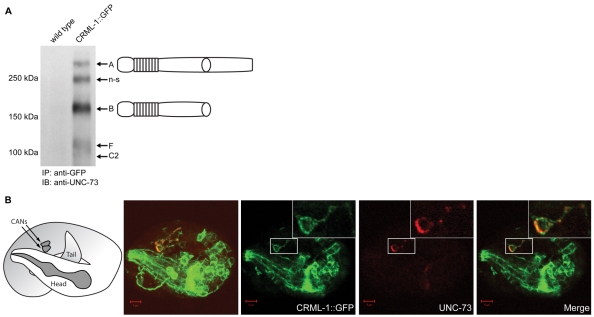
To provide further evidence that CRML-1 acts in the Rac pathway, we tested whether CRML-1 and UNC-73 physically interact in vivo. Using anti-GFP antibodies, we immunoprecipitated CRML-1::GFP from extracts of embryos containing gmIs30, an integrated version of the crml-1 translational reporter, and probed immunoblots of the precipitated proteins with an anti-UNC-73 antiserum. We detected the UNC-73 isoforms predicted to react with this antibody (Fig. 5A) (Steven et al., 2005), showing that the two proteins form a complex in vivo. Consistent with this hypothesis, we found that UNC-73 and CRML-1 were co-expressed in CANs (and their axons) during embryogenesis and partially co-localize (Fig. 5B).
Fig. 5.
UNC-73 forms a complex in vivo with CRML-1. (A) Immunoprecipitation of CRML-1::GFP and UNC-73 from C. elegans embryo extracts. The GFP tag on the CRML-1 fusion protein was used to immunoprecipitate CRML-1 from embryo lysates. Immunoblots were probed with antibodies against UNC-73, which reliably detected four isoforms in the gmIs30 lane, but not in the wild-type lane: the two Rac GEF-containing isoforms A and B, and the C2 and F isoforms; the previously described non-specific (n-s) band (Steven et al., 1998; Steven et al., 2005) is also indicated. (B) Confocal images of a 3-fold stage gmIs30 embryo stained with antibodies against GFP to detect CRML-1::GFP (green) and UNC-73 (red). To the left is a schematic of the embryo orientation in the eggshell, showing the head of the animal folded over the body just posterior to the CANs, which are located near the middle of the animal. Next to the schematic is a projection of the z-stack showing the co-expression of CRML-1 (green) and UNC-73 (red) in CANs (CANs were identified by co-staining with CAN-specific markers, data not shown). The three images to the right are a single 0.9 μm section of the focal plane that includes the lower CAN (boxed, enlarged in insets) and its anteriorly projecting axon. CRML-1 and UNC-73 overlap in the CAN and its axon. Scale bars: 5 μm.
CRML-1 is a negative regulator of SAX-3
After migrating, each ALM extends a single anterior projection that branches in the nerve ring, the main C. elegans nerve bundle (Fig. 2A and data not shown) (Chalfie and Sulston, 1981; White et al., 1986). Because the anterior projections and branches have both dendritic and axonal properties, we refer to them as processes. Recent work demonstrated that in ALMs, UNC-73 acts with VAB-8L, a kinesin-like molecule, as a positive regulator of guidance receptors including SAX-3 (Levy-Strumpf and Culotti, 2007; Watari-Goshima et al., 2007). VAB-8L expression in ALMs produces two phenotypes: ALMs migrate beyond their normal destination and reverse the polarity of their process, causing it to extend toward the tail (Wolf et al., 1998). We call this latter defect ALM rerouting. As with expression of VAB-8L, increased SAX-3 receptor activity leads to ALM rerouting. Furthermore, altering the levels of VAB-8L in ALMs results in corresponding changes in the levels of SAX-3 (Watari-Goshima et al., 2007). These and other observations suggest that VAB-8L signals through UNC-73 to regulate SAX-3 trafficking (Watari-Goshima et al., 2007).
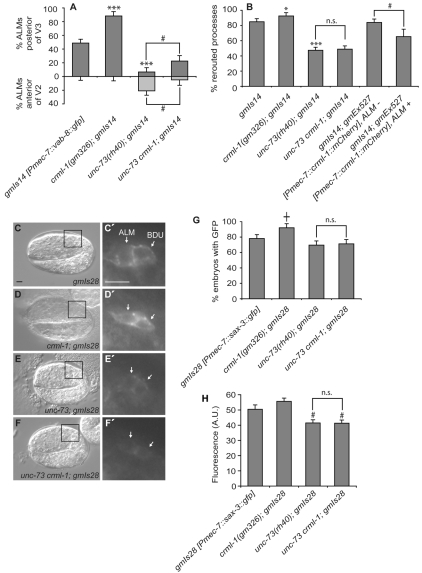
We reasoned that if CRML-1 negatively regulates the UNC-73 pathway, CRML-1 loss should enhance ALM cell migration and rerouting defects caused by VAB-8L misexpression. Using the gmIs14 transgene (Pmec-7::vab-8L::gfp), we found that crml-1(gm326) enhanced both of the defects caused by VAB-8L misexpression (Fig. 6A,B). Using the unc-73(rh40) crml-1(gm326) double-mutant background, we also conducted the rerouting assay to see whether the effect of the crml-1 mutation is dependent on the presence of UNC-73 Rac GEF activity. If crml-1 acts by inhibiting unc-73, then unc-73(rh40) crml-1(gm326); gmIs14 and unc-73(rh40); gmIs14 animals should exhibit similar frequencies of ALM rerouting. We indeed observed such an effect (Fig. 6B), demonstrating that unc-73 functions downstream of crml-1 in this assay. However, when we examined ALM cell migration, we found that ALMs more frequently migrated too far posterior in crml-1 unc-73; gmIs14 animals as compared with unc-73; gmIs14 animals (Fig. 6A). Therefore, the relationship between VAB-8L, UNC-73 and CRML-1 might be more complex in ALM migration than in ALM process rerouting.
Fig. 6.
CRML-1 regulates VAB-8L-induced ALM abnormalities and SAX-3 levels. (A) Excessive ALM migration caused by VAB-8L misexpression is enhanced by loss of CRML-1 and suppressed by loss of UNC-73 Rac GEF activity. The percentage of ALMs found posterior of wild-type positions is depicted in dark gray (above x-axis), and the percentage of ALMs found anterior of wild-type positions is depicted in light gray (beneath x-axis). Statistically significant differences between gmIs14 [Pmec-7::vab-8::gfp] and crml-1; gmIs14 or unc-73; gmIs14 are marked (***P<0.0001). Differences between unc-73 crml-1; gmIs14 and unc-73; gmIs14 are indicated (#, P<0.03). unc-73 crml-1; gmIs14 was compared with unc-73; gmIs14. Error bars, s.e.m. For the distribution of ALMs relative to P and V cells and the number scored, see Fig. S1 in the supplementary material. At least 40 animals were scored per genotype. Animals were raised at 25°C. (B) VAB-8L-dependent ALM process rerouting is enhanced by loss of CRML-1 and suppressed by both the reduction of UNC-73 and overexpression of CRML-1. Statistically significant differences between gmIs14 and other strains are marked (***P<0.0001, *P<0.05); n.s., not significant. The statistically significant difference between gmIs14; gmEx527 strains bearing the array in ALM compared with those bearing the array but not in ALM is marked (#, P<0.03). At least 125 animals were scored per genotype, except gmIs14; gmEx527, ALM+ where n=26. (C-F′) Nomarski (C-F) and GFP (boxed regions enlarged in C′-F′) images of 2-fold C. elegans embryos expressing gmIs28 [Pmec-7::sax-3::gfp] in ALM and BDU neurons (arrows) just prior to cell migration. Anterior is to the left, dorsal is up. Scale bars: 5 μm. (G) Percentage of embryos in which a SAX-3::GFP signal was detected in ALM and BDU neurons during the 2-fold stage of embryogenesis. Strains were scored on the same day to ensure that the GFP signal intensities could be directly compared. The statistically significant difference observed between gmIs28 and crml-1; gmIs28 is marked (†, P<0.009); n.s., not significant. Error bars indicate s.e.m. At least 80 animals were scored per genotype. The increase observed in the crml-1 mutant background was confirmed through an independent experiment that was scored blindly (data not shown). (H) Relative fluorescence intensities of SAX-3::GFP signals in ALM and BDU neurons from G. Only animals with both cells clearly in focus were included; at least 30 animals were scored per genotype. Error bars indicate s.e.m. Statistically significant differences between gmIs28 alone and the two unc-73-containing strains are marked (#, P<0.03).
The model that CRML-1 negatively regulates UNC-73 in ALM rerouting predicts that both proteins function in ALMs. Increasing the levels of CRML-1 in ALMs by driving a crml-1 cDNA tagged with mCherry from the mec-7 promoter decreased the amount of rerouting caused by expression of VAB-8L (Fig. 6B). We also drove an unc-73B cDNA tagged with GFP from the mec-7 promoter in an attempt to test whether UNC-73 expression in ALMs would restore a high level of ALM rerouting in unc-73; gmIs28 animals, but found that the transgene made the animals so sick that we were unable to maintain the transgenic lines. We did find, however, that the transgene caused ALM rerouting in a wild-type background: 20% of ALMs that expressed UNC-73B::GFP had a bipolar process and 2% had a reversed process (n=100). Taken together, these results suggest that CRML-1 and UNC-73 act in ALMs to mediate the effects of VAB-8L.
We also asked whether loss of CRML-1 would change the levels of SAX-3 in ALMs. If CRML-1 negatively regulates UNC-73, loss of crml-1 would be predicted to cause an increase in the expression of SAX-3 in ALMs. To test this prediction, we used the gmIs28 transgene, which results in the expression of SAX-3::GFP in both ALMs and in their lineal sisters, the BDUs. We quantified the number of embryos in which we could detect the SAX-3::GFP signal in ALMs and BDUs at the 2-fold stage and found that crml-1(gm326) enhanced the percentage of cells in which we saw GFP (Fig. 6C-G). Quantification of the amount of SAX-3::GFP in these cells revealed that the levels appeared to be slightly higher in embryos that contained the crml-1 mutation and lower in embryos that contained the unc-73 mutation (Fig. 6H). The unc-73(rh40) crml-1(gm326) double mutant appeared identical to the unc-73 single mutant, suggesting that CRML-1 signals through UNC-73 to alter SAX-3::GFP levels (Fig. 6G,H). Crossing the crml-1 and unc-73 mutations out of the gmIs28 transgenic background and rescoring SAX-3::GFP levels in the original and newly isolated gmIs28 strains showed that the gmIs28 transgene was not altered during construction of the transgenic strains containing the mutations.
DISCUSSION
By conducting a screen for suppressors of the uncoordinated phenotype of unc-34(gm114) mutants, we identified the C. elegans gene crml-1. CRML-1 is a homolog of CARMIL, a protein that binds capping protein and uncaps actin filaments in vitro (Jung et al., 2001; Uruno et al., 2006; Xu et al., 1997; Yang et al., 2005). Our genetic analysis of crml-1; unc-34 mutants demonstrates that CRML-1 acts in parallel to UNC-34 as an inhibitor of cell and axon migrations. This inhibitory function contrasts with the positive role CARMIL homologs are thought to play in promoting glioblastoma cell migrations (Jung et al., 2001; Yang et al., 2005). Our genetic and biochemical analyses indicate a previously unreported role for a CARMIL homolog: inhibiting migration through the Rac pathway.
CRML-1 acts in the same genetic pathway as UNC-73. We detected CRML-1 and UNC-73 in a complex by co-immunoprecipitation, and localization of the proteins overlaps in CAN neurons and their axons during migration. These observations suggest that one function of CRML-1 is to negatively regulate UNC-73 during cell and axon growth cone migrations.
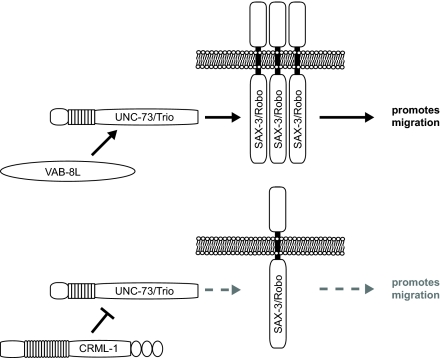
It was recently reported that Rac signaling regulates SAX-3 and Netrin receptors in C. elegans. According to this hypothesis, the kinesin-like molecule VAB-8L acts through UNC-73 to increase guidance receptor levels and promote posterior migrations in C. elegans (Levy-Strumpf and Culotti, 2007; Watari-Goshima et al., 2007). Our results indicate that UNC-73 is not only positively regulated by VAB-8L, but is also negatively regulated by CRML-1, suggesting that UNC-73 might integrate opposing signals to regulate neuronal migration and process outgrowth (Fig. 7).
Fig. 7.
Model of SAX-3/Robo regulation. VAB-8L signals through UNC-73/Trio to positively regulate SAX-3/Robo levels and promote neuronal cell and axon growth cone migration, whereas CRML-1 signals through UNC-73/Trio to negatively regulate SAX-3/Robo levels, reducing its ability to promote migration.
Most models place Rac GTPase signaling downstream of guidance receptors. Our data support a less commonly held view that Rac signaling can also regulate receptor levels, presumably through membrane trafficking. As a negative regulator of the receptor SAX-3 and the Rac GEF UNC-73, CRML-1 could act directly on SAX-3, making it refractory to UNC-73 activity. This type of inhibition might be similar to the role of vertebrate Rig1 (also known as Robo3.1) on Robo1 at the ventral midline of the spinal cord. Rig1 inhibits the ability of Robo1 to respond to Slit, but the precise mechanism of inhibition is unknown (Chen et al., 2008; Sabatier et al., 2004). Alternatively, CRML-1 could act indirectly on SAX-3 by inhibiting UNC-73. Our ability to detect UNC-73 in a complex with CRML-1 supports this latter hypothesis.
Rac signaling can modulate cell surface receptor levels. Rac signaling has been implicated in clathrin-mediated endocytosis of both transferrin and EGF receptors (Jou et al., 2000; Lamaze et al., 1996). These studies demonstrated that activated Rac inhibits receptor endocytosis in cell culture. Our results are consistent with a model in which Rac signaling regulates endocytic events in C. elegans. Controlling the levels of the receptor itself is one way to control the amount of repulsive signaling occurring through the SLT-1/Slit-SAX-3/Robo pathway. Based on our genetic and biochemical data, we favor a model in which CRML-1 and VAB-8L signaling converges on UNC-73, the role of which is to integrate these signals and determine the amount of SAX-3 endocytosis during neuronal development.
It is also possible that these molecules regulate the delivery of SAX-3 to the cell surface, similar to how the transmembrane protein Commissureless (Comm) regulates Drosophila Robo levels at the central nervous system (CNS) midline. Certain neurons ensure that their growth cones cross the midline by expressing Comm, which targets Robo to the lysosome (Keleman et al., 2002; Keleman et al., 2005). By downregulating Robo, Comm blinds these growth cones to the effects of Slit at the CNS midline. Subsequent loss of Comm expression after crossing allows Robo to reach the growth cone and respond to Slit, ensuring that the growth cone does not recross the midline (Keleman et al., 2002; Keleman et al., 2005). Although only insects have comm homologs, similar mechanisms might control Robo levels in other organisms. The inhibition by CRML-1 of Rac signaling could prevent SAX-3 from reaching the cell surface.
The types of receptors and signaling pathways expressed define how migrating cells and growth cones respond to their environment. Our observations are consistent with an emerging view that it is not only signaling downstream of guidance receptors that is regulated, but that receptor function and expression are also regulated. VAB-8L and UNC-73 can positively regulate the Robo and Netrin receptors to control the directional migration of cells and growth cones (Levy-Strumpf and Culotti, 2007; Watari-Goshima et al., 2007). Like CRML-1, Drosophila Comm and vertebrate Rig1 inhibit the function of Robo homologs (Keleman et al., 2005; Sabatier et al., 2004). As new regulators of guidance receptors are identified, the challenge will be to understand how they collaborate to coordinate cell and axon migrations.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/7/1201/DC1
We thank Rob Steven and Terry Kubiseski for the anti-UNC-73 antibody and advice; Lianna Wong for C. elegans cDNAs; Chun-Liang Pan for the pCL8 plasmid; Andy Fire for expression vectors; Steve Ruzin and the UC Berkeley College of Natural Resources Biological Imaging Center for help in quantifying CAN GFP levels; Yishi Jin and the C. elegans Genetics Center for some of the strains used in this study; Matt Vanderzalm for assistance with the figures; Richard Ikegami and Hector Aldaz for constructive comments on the manuscript; and members of the Garriga, Dernburg and Meyer labs for helpful discussions. This work was supported by National Institutes of Health grants NS32057 (G.G.), K08 CA-104890 (M.E.H.) and GM24663 (H.R.H.). H.R.H. is an Investigator of the Howard Hughes Medical Institute. P.J.V. was supported by an NSF predoctoral fellowship. Deposited in PMC for release after 6 months.
References
- Bloom, L. (1993). Genetic and molecular analysis of genes required for axon outgrowth in Caenorhabditis elegans. In Biology, p. 410. Cambridge, MA: Massachusetts Institute of Technology.
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck, S., Huber, T. B., Ingham, R. J., Kim, K., Niederstrasser, H., Allen, P. M., Pawson, T., Cooper, J. A. and Shaw, A. S. (2006). Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J. Biol. Chem. 281, 19196-19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton, D. A., Olsten, M. E., Niederstrasser, H., Cooper, J. A. and Litchfield, D. W. (2006). The role of CKIP-1 in cell morphology depends on its interaction with actin-capping protein. J. Biol. Chem. 281, 36347-36359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M. and Sulston, J. (1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82, 358-370. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Gore, B. B., Long, H., Ma, L. and Tessier-Lavigne, M. (2008). Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58, 325-332. [DOI] [PubMed] [Google Scholar]
- Chu, D. S., Dawes, H. E., Lieb, J. D., Chan, R. C., Kuo, A. F. and Meyer, B. J. (2002). A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 16, 796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney, M. and Ruvkun, G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895-905. [DOI] [PubMed] [Google Scholar]
- Forrester, W. C. and Garriga, G. (1997). Genes necessary for C. elegans cell and growth cone migrations. Development 124, 1831-1843. [DOI] [PubMed] [Google Scholar]
- Gertler, F. B., Comer, A. R., Juang, J. L., Ahern, S. M., Clark, M. J., Liebl, E. C. and Hoffmann, F. M. (1995). Enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 9, 521-533. [DOI] [PubMed] [Google Scholar]
- Gitai, Z., Yu, T. W., Lundquist, E. A., Tessier-Lavigne, M. and Bargmann, C. I. (2003). The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron 37, 53-65. [DOI] [PubMed] [Google Scholar]
- Guan, K. L. and Rao, Y. (2003). Signalling mechanisms mediating neuronal responses to guidance cues. Nat. Rev. Neurosci. 4, 941-956. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki, S., Ng, J., Tzu, J., Dietzl, G., Sun, Y., Harms, M., Nardine, T., Luo, L. and Dickson, B. J. (2002). Rac function and regulation during Drosophila development. Nature 416, 438-442. [DOI] [PubMed] [Google Scholar]
- Hedgecock, E. M., Culotti, J. G., Thomson, J. N. and Perkins, L. A. (1985). Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158-170. [DOI] [PubMed] [Google Scholar]
- Hu, H., Li, M., Labrador, J. P., McEwen, J., Lai, E. C., Goodman, C. S. and Bashaw, G. J. (2005). Cross GTPase-activating protein (CrossGAP)/Vilse links the Roundabout receptor to Rac to regulate midline repulsion. Proc. Natl. Acad. Sci. USA 102, 4613-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y., Jorgensen, E., Hartwieg, E. and Horvitz, H. R. (1999). The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J. Neurosci. 19, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, T. S., Leung, S. M., Fung, L. M., Ruiz, W. G., Nelson, W. J. and Apodaca, G. (2000). Selective alterations in biosynthetic and endocytic protein traffic in Madin-Darby canine kidney epithelial cells expressing mutants of the small GTPase Rac1. Mol. Biol. Cell 11, 287-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., Remmert, K., Wu, X., Volosky, J. M. and Hammer, J. A., 3rd (2001). The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. and Ahringer, J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed]
- Keleman, K., Rajagopalan, S., Cleppien, D., Teis, D., Paiha, K., Huber, L. A., Technau, G. M. and Dickson, B. J. (2002). Comm sorts robo to control axon guidance at the Drosophila midline. Cell 110, 415-427. [DOI] [PubMed] [Google Scholar]
- Keleman, K., Ribeiro, C. and Dickson, B. J. (2005). Comm function in commissural axon guidance: cell-autonomous sorting of Robo in vivo. Nat. Neurosci. 8, 156-163. [DOI] [PubMed] [Google Scholar]
- Lamaze, C., Chuang, T. H., Terlecky, L. J., Bokoch, G. M. and Schmid, S. L. (1996). Regulation of receptor-mediated endocytosis by Rho and Rac. Nature 382, 177-179. [DOI] [PubMed] [Google Scholar]
- Lebrand, C., Dent, E. W., Strasser, G. A., Lanier, L. M., Krause, M., Svitkina, T. M., Borisy, G. G. and Gertler, F. B. (2004). Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron 42, 37-49. [DOI] [PubMed] [Google Scholar]
- Levy-Strumpf, N. and Culotti, J. G. (2007). VAB-8, UNC-73 and MIG-2 regulate axon polarity and cell migration functions of UNC-40 in C. elegans. Nat. Neurosci. 10, 161-168. [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., Reddien, P. W., Hartwieg, E., Horvitz, H. R. and Bargmann, C. I. (2001). Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128, 4475-4488. [DOI] [PubMed] [Google Scholar]
- Lundstrom, A., Gallio, M., Englund, C., Steneberg, P., Hemphala, J., Aspenstrom, P., Keleman, K., Falileeva, L., Dickson, B. J. and Samakovlis, C. (2004). Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons. Genes Dev. 18, 2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire, S. L., Garriga, G., White, J., Jacobson, D. and Horvitz, H. R. (1992). Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron 8, 307-322. [DOI] [PubMed] [Google Scholar]
- Ng, J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., Dietzl, G., Dickson, B. J. and Luo, L. (2002). Rac GTPases control axon growth, guidance and branching. Nature 416, 442-447. [DOI] [PubMed] [Google Scholar]
- Pulak, R. and Anderson, P. (1993). mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7, 1885-1897. [DOI] [PubMed] [Google Scholar]
- Remmert, K., Olszewski, T. E., Bowers, M. B., Dimitrova, M., Ginsburg, A. and Hammer, J. A., 3rd (2004). CARMIL is a bona fide capping protein interactant. J. Biol. Chem. 279, 3068-3077. [DOI] [PubMed] [Google Scholar]
- Sabatier, C., Plump, A. S., Le M., Brose, K., Tamada, A., Murakami, F., Lee, E. Y. and Tessier-Lavigne, M. (2004). The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell 117, 157-169. [DOI] [PubMed] [Google Scholar]
- Shakir, M. A., Gill, J. S. and Lundquist, E. A. (2006). Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172, 893-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven, R., Kubiseski, T. J., Zheng, H., Kulkarni, S., Mancillas, J., Ruiz Morales, A., Hogue, C. W., Pawson, T. and Culotti, J. (1998). UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785-795. [DOI] [PubMed] [Google Scholar]
- Steven, R., Zhang, L., Culotti, J. and Pawson, T. (2005). The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 19, 2016-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., Schierenberg, E., White, J. G. and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. [DOI] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Uruno, T., Remmert, K. and Hammer, J. A., 3rd (2006). CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J. Biol. Chem. 281, 10635-10650. [DOI] [PubMed] [Google Scholar]
- Watari-Goshima, N., Ogura, K., Wolf, F. W., Goshima, Y. and Garriga, G. (2007). C. elegans VAB-8 and UNC-73 regulate the SAX-3 receptor to direct cell and growth-cone migrations. Nat. Neurosci. 10, 169-176. [DOI] [PubMed] [Google Scholar]
- White, J. G., Southgate, E., Thomson, J. N. and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1-340. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., Yeh, R. T., Gish, W. R., Waterston, R. H. and Plasterk, R. H. (2001). Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160-164. [DOI] [PubMed] [Google Scholar]
- Wolf, F. W., Hung, M. S., Wightman, B., Way, J. and Garriga, G. (1998). vab-8 is a key regulator of posteriorly directed migrations in C. elegans and encodes a novel protein with kinesin motor similarity. Neuron 20, 655-666. [DOI] [PubMed] [Google Scholar]
- Wu, Y.-C., Cheng, T.-W., Lee, M.-C. and Weng, N.-Y. (2002). Distinct Rac activation pathways control Caenorhabditis elegans cell migration and axon outgrowth. Dev. Biol. 250, 145-155. [DOI] [PubMed] [Google Scholar]
- Xu, P., Zot, A. S. and Zot, H. G. (1995). Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J. Biol. Chem. 270, 25316-25319. [DOI] [PubMed] [Google Scholar]
- Xu, P., Mitchelhill, K. I., Kobe, B., Kemp, B. E. and Zot, H. G. (1997). The myosin-I-binding protein Acan125 binds the SH3 domain and belongs to the superfamily of leucine-rich repeat proteins. Proc. Natl. Acad. Sci. USA 94, 3685-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Pring, M., Wear, M. A., Huang, M., Cooper, J. A., Svitkina, T. M. and Zigmond, S. H. (2005). Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 9, 209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. and Bashaw, G. J. (2006). Son of sevenless directly links the robo receptor to Rac activation to control axon repulsion at the midline. Neuron 52, 595-607. [DOI] [PubMed] [Google Scholar]
- Yu, T. W. and Bargmann, C. I. (2001). Dynamic regulation of axon guidance. Nat. Neurosci. 4Suppl, 1169-1176. [DOI] [PubMed] [Google Scholar]
- Yu, T. W., Hao, J. C., Lim, W., Tessier-Lavigne, M. and Bargmann, C. I. (2002). Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/Enabled and a Netrin-independent UNC-40/DCC function. Nat. Neurosci. 5, 1147-1154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.