Summary
The acquisition and maintenance of final neuronal identity depends in part upon the implementation of fate-specification programs in postmitotic neurons; however, the mechanisms involved remain unclear. In the developing spinal cord, retinoic acid (RA) signaling pathways specify the columnar and divisional identities of postmitotic motoneurons (MNs). Here we show that RA signals induce expression of the NET transcriptional regulator Nolz1 in differentiated chick MNs, where it regulates the progressive specification of prospective Lim3-negative motor columns. Nolz1 controls the initial formation of forelimb and thoracic Lim3-negative motor columns by downregulating Lim3 expression and maintaining the expression of key homeodomain proteins necessary for MN identity and survival. At forelimb levels, Nolz1 specifies lateral motor column (LMC) identity by inducing the expression of the postmitotic LMC determinant Hoxc6, and implements the partial specification of lateral LMC identity through Lim1 induction. The specificity of Nolz1 function depends upon distinct repressor activities that require, in part, the modulatory activity of Grg5, an atypical member of the Gro-TLE family of co-repressors. Thus, RA signals regulate diverse events in MN subtype specification by inducing the expression of a key transcriptional regulator that controls multiple developmental pathways via functionally distinct repressor complexes.
Keywords: Retinoids, Nolz1, Grg5, Motoneuron, Identity, Repressor
INTRODUCTION
The establishment of a functional nervous system depends upon the coordinated generation and maintenance of diverse neuronal subtypes during embryonic and postnatal development. In progenitor cells, cross-repressive transcriptional mechanisms elicit the expression of distinct transcription factors that confer subsequent neuronal fate (Jessell, 2000; Vallstedt et al., 2001). However, neurons retain a degree of plasticity in their fates after cell cycle exit, implying that the induction or maintenance of cell fate-specification programs in postmitotic neurons is crucial for the acquisition and preservation of final neuronal identity (Gross et al., 2002; Moran-Rivard et al., 2001; Muller et al., 2002; Pearson and Doe, 2004). While the mechanisms that regulate progenitor patterning and differentiation are emerging, less is known about the molecular pathways that shape and maintain identity in postmitotic neurons.
The study of spinal motoneuron (MN) development has helped elucidate the transcriptional mechanisms that generate and consolidate postmitotic neuronal fate (Jessell, 2000). Spinal MNs derive from a discrete ventral progenitor domain termed pMN. In the chick, MNR2 is expressed in the S phase of the terminal cell cycle of pMN progenitors and triggers the expression of a cascade of factors essential for somatic MN development (Tanabe et al., 1998). All newly differentiating MNs coexpress Islet1 and Lim3, two transcription factors crucial for maintaining somatic MN properties. Islet1 is required for MN survival and, together with Lim3, regulates the expression of HB9, a homeodomain (HD) protein necessary for suppressing intrinsic interneuron specification programs in developing MNs (Pfaff et al., 1996; Arber et al., 1999; Thaler et al., 1999; Thaler et al., 2002). These `generic' MNs subsequently diversify into distinct neuronal subtypes that are manifest by the organization of their cell bodies into specific motor columns, their characteristic axonal projection patterns, and their distinct LIM-HD protein expression profiles (Landmesser, 1978; Tosney et al., 1995; Tsuchida et al., 1994; Jessell, 2000). MNs forming the medial division of the median motor column (MMCm) span all rostral-caudal levels and innervate axial muscles, whereas the preganglionic MNs of the column of Terni (CT) and MNs of the lateral MMC (MMCl) are found at thoracic regions (Prasad and Hollyday, 1991; Jessell, 2000). At limb levels, lateral motor column (LMC) MNs that innervate the limb form medial and lateral divisions, which innervate ventrally and dorsally derived limb muscles, respectively. MN diversification is first apparent by the downregulation of Lim3 in prospective CT, MMCl and LMC MNs (Sharma et al., 2000). Within Lim3- MNs, cross-repressive interactions between different Hox HD proteins consolidate rostralcaudal Hox protein distributions, which activate the expression of distinct LIM-HD proteins that dictate MN subtype identity and connectivity (Liu et al., 2001; Dasen et al., 2003; Sharma et al., 2000; Kania et al., 2000). For example, cross-repressive interactions between Hoxc9 and Hoxc6 define their expression domains at thoracic and forelimb levels, where they respectively regulate the formation of CT and MMCl, and of forelimb LMC columnar identities (Dasen et al., 2003).
Retinoic acid (RA) signaling pathways play central roles in regulating generic MN differentiation and in the specification and maintenance of forelimb LMC identity and lateral LMC divisional character (Solomin et al., 1998; Sockanathan and Jessell, 1998; Diez del Corrall et al., 2003; Novitch et al., 2003; Sockanathan et al., 2003; Vermot et al., 2005; Ji et al., 2006). RA signals directly regulate gene transcription through the activity of nuclear receptors; however, the factors that mediate RA responses to regulate postmitotic MN identity remain unknown (Maden, 2002). One candidate is Nlz2/Nolz1, a member of the Noc, Elbow and Tlp-1 (NET) family of atypical zinc-finger-containing transcriptional regulators (Nakamura et al., 2004; Runko and Sagerstrom, 2004; Hoyle et al., 2004). Both zebrafish Nlz2 (also known as Znf503 - ZFIN) and its mouse homolog Nolz1 (also known as Zfp503 - Mouse Genome Informatics) are expressed at known sites of RA synthesis and function in the brain (Runko and Sagerstrom, 2004; Hoyle et al., 2004; Chang et al., 2004). Nlz2 is found in the hindbrain, where it is required for the specification of rhombomeric identity, a process known to be dependent upon RA signaling, whereas Nolz1 is coincidently expressed with Raldh3 in the lateral ganglionic eminence (LGE), a region of the telencephalon that gives rise to the striatum. Nolz1 expression in cell culture is RA-inducible, a finding supported by the identification of a functional direct repeat 5 retinoic acid response element (DR5-RARE) upstream of the Nolz1 translational start site (Chang et al., 2004). Based on these collective observations, we examined if and how Nlz2/Nolz1 mediates RA-dependent events regulating postmitotic MN development.
Here, we show that Nolz1 regulates the diversification of Lim3- motor columns from Lim3+ MNs, consolidates their formation and controls the specification and diversification of LMC MNs by regulating Hox and LIM-HD protein expression. These divergent functions of Nolz1 require distinct repressor activities that depend in part on modulatory functions of the Gro-TLE protein, Grg5 (Fisher and Caudy, 1998). This study thus reveals that RA signals regulate the progressive specification of postmitotic MN columnar and divisional identity by inducing a single pivotal molecule that executes multiple fate-specification transcriptional programs through the assembly of functionally distinct repressor complexes.
MATERIALS AND METHODS
In ovo electroporation and luciferase transcriptional assays
All cDNAs were cloned into pCAGGS or a 3 kb Hb9 promoter-based vector (Lee et al., 2004) for in ovo electroporation (William et al., 2003). For siRNA experiments, Nolz1 and Grg5 siRNA duplexes were electroporated as previously described (Rao et al., 2004). Target sequences are as follows: Nolz1, 5′-AAACTTGCTCGCAGATAGGAA-3′ and 5′-AATTGCTATGATTGTATCGTA-3′; Grg5, 5′-GCGAGAAATCGGAGATGCA-3′ and 5′-TGGCCAAGGAGGACAAGAA-3′. DsRed siRNA sequences are as published (Rao et al., 2004). Luciferase assays were carried out as described (Novitch et al., 2001; Nishihara et al., 2003).
Immunohistochemistry and in situ hybridization
Chicken embryos were prepared for immunohistochemistry and in situ hybridization as described (Sockanathan and Jessell, 1998). Primary antibodies used were as follows. Polyclonal rabbit antiserum against chick Nolz1 was generated by immunizing rabbits with a GST fusion of the first 373 amino acids of Nolz1 (Covance). Primary antibodies were used at the following dilutions: rabbit anti-Nolz1, 1:1000; K5 (rabbit anti-Isl1/2), 1:2500; T4 (rabbit anti-Lim1/2), 1:3000; guinea pig anti-Isl1/2, 1:10,000 (provided by T. M. Jessell, HHMI, Colombia University, New York, NY); monoclonal antibodies 4F2 (anti-Lim1/2), 1:2; 4H9 (anti-Isl2), 1:100; 674E12 (anti-Lim3), 1:100; 81.5C10 (anti-HB9/MNR2), 1:100 (Developmental Studies Hybridoma Bank); rabbit anti-MNR2, 1:8000 (provided by B. Novitch, UCLA, Los Angeles, CA); goat anti-Hoxc6 (provided by J. Dasen, Smilow Institute, New York, NY); rabbit anti-Lhx3 (Abcam), 1:500; goat anti-β-galactosidase (Arnel), 1:3000; GFP, mouse anti-SC1, 1:40; rabbit anti-Chx10, 1:2000; mouse anti-En1 (4G11), 1:100; mouse anti-Evx1/2 (99.1-32A), 1:50; and mouse anti-HA (12CA5), 1:1000.
Images were captured using a Zeiss LSM 5 Pascal confocal microscope. In situ hybridization was performed as described (Schaeren-Wiemers and Gerfin-Moser, 1993). Quantitation of neuronal number was carried out using ten sections per embryo from four to ten embryos.
Yeast two-hybrid screen
Yeast two-hybrid screens were carried out using the Matchmaker Two-Hybrid System (Clontech). The library was prepared from RNA generated from St 23 chick spinal cords and cotransformed into yeast strain AH109 with the GAL4 activation domain plasmid pGADT7-Rec and bait construct pGBKT7-Nolz1. Only colonies growing on SD/-Ade/-His/-Leu/-Trp/X-α-Gal selective medium were picked for further analysis.
Co-immunoprecipitation
Flag or HA epitope tags were fused to the N-terminus of Nolz1 or Grg5 and cloned into pCAGGS or pCS2 vectors. Transiently transfected HEK293T cells were harvested and homogenized in lysis buffer using standard procedures. Lysates were precleaned by incubation with GammaBind G Sepharose beads (GE Healthcare) followed by centrifugation, and were mixed with anti-Flag M2 (Sigma)-bound beads overnight at 4°C under constant rotation. After extensive washing, the precipitated proteins were analyzed by SDS-PAGE and western blot using anti-HA antibodies (Santa Cruz).
Reverse northern blot analysis
Two different chick Nolz1 EST clones (University of Delaware Chick EST Database) were electrophoresed in agarose gels and blotted according to standard procedures. Filters were probed with cDNAs derived from Hamburger Hamilton St 19 brachial chick neural explants grown for 18 hours in the absence or presence of retinol. Retinol was used to take advantage of the endogenous expression of the retinoid synthetic enzyme Raldh2, which is expressed in brachial MNs. Retinol is metabolized within the explants to substrates for Raldh2, which subsequently catalyzes the formation of endogenous retinoid metabolites (Sockanathan and Jessell, 1998). RNA from 100 explants was isolated using Trizol (Gibco BRL). cDNAs were generated and amplified using the Marathon cDNA PCR Amplification Kit (Stratagene).
RESULTS
Nolz1 expression is induced by RA in the developing spinal cord
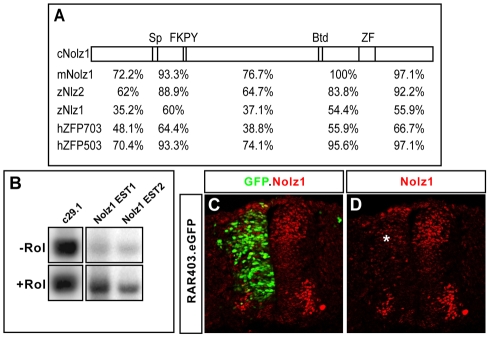
We cloned the chick homolog of Nolz1 by RT-PCR using RNA prepared from dissected Hamburger Hamilton stage (St) 21-23 chick spinal cords. The predicted Nolz1 protein shares similar domains with zebrafish, mouse and human Nolz1 homologs, and is 74% and 78% identical to zebrafish and mouse Nolz1, respectively (Fig. 1A) (Nakamura et al., 2004). To test whether RA signals can regulate Nolz1 expression in the chick spinal cord, we performed reverse northerns. Densitometry analysis showed a 5-fold increase of Nolz1 expression in explants exposed to retinol (Fig. 1B). Consistent with a role for RA signals in regulating Nolz1 expression, electroporation of constructs expressing RAR403, a potent dominant-negative retinoid receptor that disrupts RA signaling, caused a decrease in Nolz1 expression in the dorsal spinal cord (Fig. 1C,D) (Novitch et al., 2003; Sockanathan et al., 2003).
Fig. 1.
Nolz1 expression is induced by RA signals in spinal cord explants. (A) Conserved domains between chick (c), mouse (m), zebrafish (z) and human (h) predicted Nolz1 ORFs. Sp, Sp1-related domain; FKPY, putative Groucho consensus binding site; Btd, Drosophila Buttonhead domain; ZF, zinc-finger domain. Numbers indicate percentage identity in the regions between these domains. (B) Reverse northern of two different chick Nolz1 EST clones (EST1, EST2) probed with pools of cDNAs generated from St 19 ventral chick embryonic spinal cord explants grown in the presence (+) or absence (-) of retinol (Rol). c29.1 is a cDNA fragment from an unrelated gene known to be transcriptionally unresponsive to RA signals (Rao and Sockanathan, 2005). (C,D) Confocal images of chick spinal cords electroporated with RAR403 showing decreased dorsal Nolz1 expression (asterisk). Motoneurons are reduced as their generation is dependent on RA signaling.
Nolz1 is expressed in subsets of developing spinal MNs
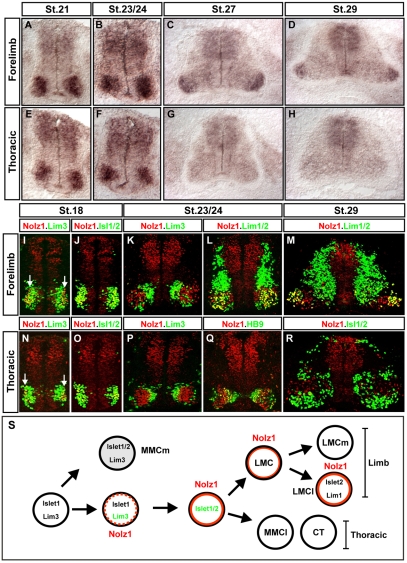
To examine the spatial and temporal distribution of Nolz1 in the chick spinal cord, we carried out a developmental time course of Nolz1 mRNA expression. Prior to St 24, MNs at all rostral-caudal levels express Nolz1; however, from St 25 onwards, Nolz1 is restricted to a subset of laterally located forelimb and lumbar MNs (Fig. 2A-H; data not shown). To determine whether Nolz1 expression correlates with the development of specific motor columns, we examined the expression of Nolz1 protein in relation to molecular markers that define the major spinal motor columns and divisions (Tsuchida et al., 1994). All newly differentiated spinal MNs coexpress Islet1 and Lim3. Lim3 expression is maintained in MMCm neurons spanning all axial levels, but it is rapidly downregulated in LMC MNs at limb levels, and in CT and MMCl neurons at thoracic regions of the spinal cord (Tsuchida et al., 1994). Nolz1 expression was initiated in a narrow band of laterally located Islet1+ Lim3+ neurons at St 18, but rapidly segregated with forelimb and thoracic Islet1/2+ MNs that had downregulated Lim3 expression (Fig. 2I,J,N,O). By St 24, no MNs coexpressed Nolz1 and Lim3, and there was a clear demarcation between Nolz1- Lim3+ and Nolz1+ Lim3- MNs at all axial levels (Fig. 2K,P). MMCm MNs marked by Islet1/2 and Lim3 coexpression do not express Nolz1 (Fig. 2K,P; data not shown). By contrast, Nolz1 expression at limb levels was confined to medial (Islet1/2+, Lim3-, Lim1-) and lateral (Lim3-, Islet2+, Lim1+) divisions of the LMC, while at thoracic regions, Nolz1 was detected in MMCl neurons (Islet1/2+, HB9+, Lim3-) and prospective CT MNs (Islet1/2+, HB9-, Lim3-) (Fig. 2K,L,P,Q; data not shown). From St 25 onwards, Nolz1 expression was downregulated in thoracic MNs; however, at limb levels, Nolz1 expression was maintained in lateral LMC (LMCl) neurons and in a subset of medial LMC (LMCm) neurons (Fig. 2M,R; data not shown). Thus, Nolz1 expression initiates in a narrow band of newly differentiated Islet1+ Lim3+ MNs, rapidly segregates with Lim3- motor columns at all axial levels, and subsequently localizes to subsets of limb-level LMC neurons (Fig. 2S).
Fig. 2.
Nolz1 is expressed in subsets of postmitotic spinal MNs. (A-H) In situ hybridization of Nolz1 mRNA expression on transverse sections of embryonic chick spinal cords. (I-R) Confocal micrographs showing analysis of Nolz1 protein expression as compared with molecular markers of specific spinal motor columns. Arrows mark Nolz1 and Lim3 coexpressing MNs. (S) Summary of Nolz1 expression in relation to motor column development. The dashed orange line indicates transient Nolz1 expression. MMC, median motor column; MMCm, medial division of the MMC; MMCl, lateral division of the MMC; LMC, lateral motor column; LMCm, medial division of the LMC; LMCl, lateral division of the LMC; CT, column of Terni.
Nolz1 loss-of-function results in reduced numbers of Lim3- spinal MNs
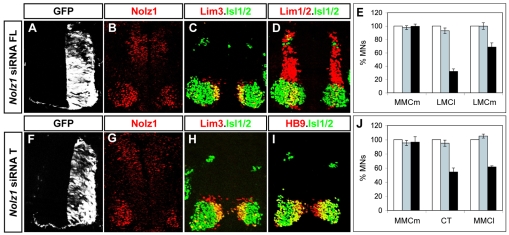
To define the requirement for Nolz1 in MN development, we ablated Nolz1 expression using short interfering RNAs (siRNAs). Oligonucleotides (21 bp) designed against the Nolz1 ORF and 3′UTR were electroporated into chick spinal cords at St 12, prior to the onset of Nolz1 expression and MN generation (Rao et al., 2004). Plasmids expressing GFP were coelectroporated to mark the transfected side (Fig. 3A,F). Electroporated embryos showed a ∼50% reduction of Nolz1 mRNA and protein when compared with the contralateral non-electroporated side, or when electroporated with unrelated siRNAs (Fig. 3A,B,F,G; data not shown; see Fig. S1 in the supplementary material). However, no changes in the number of Lim3+ MMCm MNs at either forelimb or thoracic levels of the spinal cord were observed, consistent with the lack of Nolz1 expression in MMCm MNs (Fig. 3C,E,H,J). Lim3- MNs give rise to medial and lateral divisions of the LMC at forelimb levels, while at thoracic regions they generate MMCl and CT motor columns (Tsuchida et al., 1994; Jessell, 2000). At forelimb levels, knockdown of Nolz1 resulted in a 30% loss of LMCm neurons and a 70% loss of LMCl cells, as compared with the contralateral non-electroporated side or when control unrelated siRNAs were electroporated, while at thoracic levels there was a reduction of CT and MMCl MNs by 50% and 40%, respectively (Fig. 3D,E,I,J). The percentage decrease in CT and MMCl MNs correlates well with the extent of Nolz1 knockdown; at forelimb levels, however, LMCm generation appeared less affected by the knockdown of Nolz1 than neurons of the LMCl subtype. This difference might reflect the relative distribution of Nolz1 in LMC neurons, in that all LMCl neurons, but only a subset of LMCm neurons, normally express Nolz1 (Fig. 2M). These results suggest that Nolz1 is specifically required for the formation of Lim3- motor columns, consistent with its early expression in Lim3- MN populations in the spinal cord.
Fig. 3.
Nolz1 ablation causes a decrease of Lim3- motor columns. (A-D,F-I) Confocal micrographs of sectioned St 23/24 chick spinal cords electroporated with Nolz1 siRNAs and examined for Nolz1 expression and motor column markers by immunohistochemistry at forelimb (FL) and thoracic (T) levels of the spinal cord. (E,J) Bar charts showing the percentage of MNs corresponding to different motor columns in electroporated Nolz1 siRNAs (black bars), control siRNAs (gray bars) and non-electroporated contralateral (white bars, set as 100%) sides of the spinal cord at forelimb (E) and thoracic (J) regions. n=4-5 embryos, mean ± s.e.m.
Grg5 interacts with Nolz1
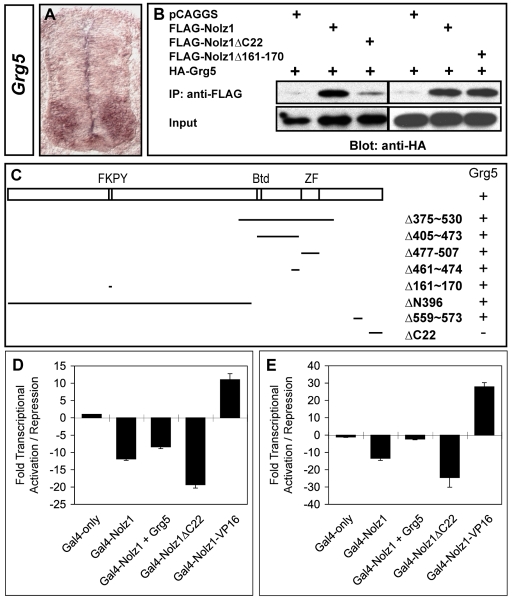
Nolz1 contains a single, atypical zinc finger and is thus unlikely to bind DNA directly; however, NET proteins can activate or repress transcription through the formation of multimeric complexes (Nakamura et al., 2004; Runko and Sagerstrom, 2004). We reasoned that it would be more informative to evaluate the role of Nolz1 in Lim3- motor column development by investigating its function in the absence and presence of its associated proteins. To identify proteins that interact with Nolz1, we performed a yeast two-hybrid screen using full-length Nolz1 fused to the GAL4 DNA-binding domain as bait, together with a cDNA library generated from St 23 ventral chick spinal cords fused to the GAL4 activation domain. We identified one clone that overlapped with Nolz1 expression in chick spinal MNs (Fig. 4A). This clone corresponded to Grg5, a member of the Gro-TLE family of co-repressors that functions as a `derepressor' of other Groucho (Grg) proteins owing to its inability to complex with histone deacetylases (HDACs) (Fisher and Caudy, 1998; Brantjes et al., 2001). The overlap of Grg5 and Nolz1 expression in the ventral spinal cord suggests that Grg5 is a viable candidate for mediating Nolz1 function in spinal MNs. We confirmed that Grg5 interacts with Nolz1 by co-immunoprecipitation (co-IP) assays using extracts prepared from HEK293T cells transfected with tagged versions of Nolz1 and Grg5 (Fig. 4B,C). To identify the Grg5 interaction sites on Nolz1, we generated a series of deletions within Nolz1 and examined which of these deletions lacked the ability to interact with Grg5 by co-IP (Fig. 4C). Deletion of a putative Grg consensus binding site (FKPY, Δ161-170) within Nolz1 did not abolish the interaction between Nolz1 and Grg5 (Fig. 4B,C). Instead, deletion of the C-terminal 22 amino acids significantly reduced Nolz1-Grg5 complex formation (Nolz1ΔC22) (Fig. 4B,C). Together, these results show that Grg5 can specifically interact with the C-terminus of Nolz1.
Fig. 4.
Nolz1 and Grg5 interact and function as transcriptional repressors. (A) In situ hybridization of Grg5 mRNA expression in transverse sections of chick embryonic spinal cords. (B) Co-immunoprecipitation (IP) analyses of Grg5 protein interactions with Nolz1. (C) Schematic of the Nolz1 coding region. FKPY, putative Grg consensus binding site; Btd, Buttonhead domain; ZF, atypical zinc-finger domain. Lines beneath indicate the regions deleted within the Nolz1 coding sequence; the numbers indicate the corresponding amino acids that are deleted. Whether Nolz1 retains (+) or loses (-) its ability to interact with Grg5 in co-IP is indicated alongside. (D) Luciferase-based transcriptional assays using extracts from transfected COS-7 cells. Data are from three replicate experiments, mean ± s.e.m. Student's t-test compared with Nolz1: Nolz1+Grg5, P=0.0055; Nolz1ΔC22, P=0.0016. (E) Luciferase-based transcriptional assays using extracts from electroporated chick spinal cords. n=6-8 embryos, mean ± s.e.m. Student's t-test compared with Nolz1: Nolz1+Grg5, P<0.000001; Nolz1ΔC22, P=0.0029. Compared with GAL4-only: Nolz1+Grg5, P=0.0003.
Nolz1 repressor function is modulated by Grg5 activity
To investigate the transcriptional properties of Nolz1, we assayed its function using an in vitro reporter-based assay. Full-length Nolz1 was fused to the GAL4 DNA-binding domain (GalNolz1) and cotransfected into COS-7 cells with a reporter plasmid containing GAL4 DNA-binding sequences cloned upstream of a basal E1B promoter and the luciferase reporter gene (GAL4x5-E1b-luciferase) (Novitch et al., 2001). Transfection of GAL4x5-E1b-luciferase into COS-7 cells generated basal transcriptional activity that was repressed ∼11-fold by GalNolz1 (Fig. 4D). Cotransfection of Grg5 expression plasmids caused a reproducible decrease in Nolz1-dependent repressor activity (Fig. 4D). Consistent with this function, transfection of Nolz1ΔC22 fused to the GAL4 DNA-binding domain resulted in a marked increase of Nolz1 repressor activity (GalNolz1ΔC22; Fig. 4D). To determine the transcriptional properties of Nolz1 and Grg5 in vivo, we repeated these experiments using extracts prepared from dissected chick spinal cords electroporated with the same series of expression plasmids (Nishihara et al., 2003). However, in this case we utilized MH100, which contains GAL4 DNA-binding sequences upstream of a minimal thymidine kinase promoter and the luciferase reporter gene (Muhr et al., 2001). Similar to the assays carried out in COS-7 cells, GalNolz1 repressed basal MH100 expression (Fig. 4E). But, coelectroporation of Grg5 caused a marked `derepression' of GalNolz1-dependent transcriptional activity (Fig. 4E). GalNolz1ΔC22 showed increased repressor activity compared with GalNolz1, consistent with the ability of Grg5 to modulate Nolz1 repressor function.
Taken together, these data suggest that Nolz1 functions as a repressor molecule, and that Grg5 interactions with Nolz1 serve to modulate Nolz1 repressor function. Mechanistically, Grg5 is known to modulate Grg co-repressor activity, raising the possibility that classical Grg proteins mediate Nolz1-dependent transcriptional repression (Fisher and Caudy, 1998; Brantjes et al., 2001; Muhr et al., 2001).
Nolz1 overexpression leads to the downregulation of Lim3 and to MN loss
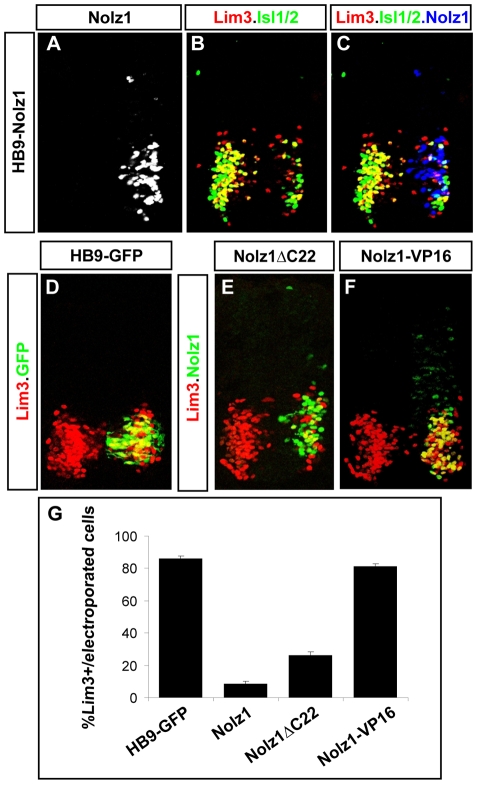
To investigate how Nolz1 regulates the formation of Lim3- MNs, we overexpressed Nolz1 under the control of a 3 kb promoter derived from the mouse Hb9 (Mnx1 - Mouse Genome Informatics) gene, which initiates expression in newly differentiating MNs (HB9-Nolz1) (Lee et al., 2004). Embryos electroporated with plasmids expressing GFP under the Hb9 promoter (HB9-GFP) expressed Lim3 in ∼80% of electroporated MNs (Fig. 5D,G). By contrast, less than 5% of electroporated MNs expressing Nolz1 were Lim3+, suggesting that Nolz1 is sufficient to cause the downregulation of Lim3 expression (Fig. 5A-C,G). Although the Nolz1-expressing cells had initiated MN differentiation, which was evident by the induction of Nolz1 expression from the Hb9 promoter, they lacked Islet1/2 and HB9 expression (Fig. 5B,C; data not shown). Furthermore, a number of the cells were labeled by TUNEL, indicating that they had initiated apoptosis (data not shown).
Fig. 5.
Nolz1 represses Lim3 expression in postmitotic MNs. (A-F) Confocal micrographs of St 21 chick spinal cord sections electroporated on the right. The high levels of Nolz1 elicited by overexpression preclude the detection of endogenous Nolz1 in the spinal cord under these imaging conditions. (G) Bar chart quantifying the percentage of electroporated cells expressing Lim3. n=10 embryos, mean ± s.e.m.
To examine whether the ability of Nolz1 to downregulate Lim3 expression utilizes endogenous Grg5 activity, we electroporated Hb9 promoter constructs driving Nolz1ΔC22 expression into chick spinal cords. Quantification of the percentage of Lim3+ cells expressing Nolz1 and Nolz1ΔC22 showed that removal of the Grg5-interaction domain resulted in a minor increase in the number of Nolz1 and Lim3 coexpressing cells, but did not restore Lim3 coexpression to control levels (Fig. 5E,G). This observation suggests that the Nolz1-dependent repression of Lim3 expression does not require its interaction with Grg5. In vivo transcriptional assays indicated that Nolz1 functions as a transcriptional repressor (Fig. 5E). To confirm that Nolz1 downregulates Lim3 through repressor activity and not through activator functions derived from interactions with unidentified binding partners in vivo, we fused the VP16 activator domain to the C-terminus of full-length Nolz1 (Nolz1-VP16). We verified that Nolz1-VP16 functions as an activator using in vitro and in vivo transcriptional reporter assays (Fig. 4D,E). HB9-Nolz1-VP16 constructs failed to downregulate Lim3 expression in electroporated chick spinal cords (Fig. 5F,G). These collective observations indicate that Nolz1 repressor activity downregulates Lim3 expression in spinal MNs and that this repressor function is not dependent on its association with Grg5. These findings suggest that Nolz1 repressor function plays crucial roles in the early segregation of Lim3- and Lim3+ MN populations by its ability to suppress Lim3 expression in postmitotic MNs.
Nolz1 and Grg5 coexpression induces ectopic MNs
The loss of Lim3- motor columns in Nolz1 knockdown embryos is unlikely to result from Lim3 derepression, as forced expression of Lim3 in MNs is not detrimental to MN survival (Sharma et al., 2000; William et al., 2003). These observations suggest additional roles for Nolz1 in regulating Lim3- motor column development that are distinct from its function in downregulating Lim3 expression. MN generation and maintenance are orchestrated by key HD transcription factors that include MNR2, Lim3, Islet1 and HB9 (Pfaff et al., 1996; Tanabe et al., 1998; Sharma et al., 1998; Arber et al., 1999; Thaler et al., 1999; William et al., 2003; Thaler et al., 2002). We reasoned that Nolz1 might function to maintain the expression of these key transcription factors in prospective Lim3- MNs. If so, then Nolz1 should be capable of regulating their transcription, and thus might induce their expression in gain-of-function assays. We tested this possibility by expressing Nolz1 in the dorsal spinal cord by in ovo electroporation of Nolz1 expression constructs driven by the chick β-actin promoter (pCAGGS-Nolz1).
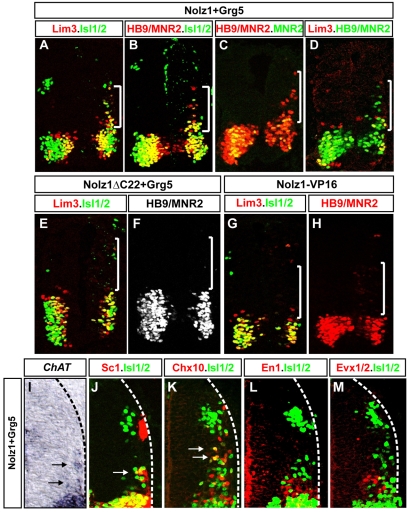
Electroporation of Nolz1 alone did not lead to the ectopic expression of MN factors dorsally (see Fig. S2 in the supplementary material). By contrast, coexpression of Nolz1 and Grg5 induced the cell-autonomous expression of Lim3, Islet1 and HB9 (Fig. 6A-D). A small number of cells expressed Islet2 2 days after electroporation, reflecting the temporal profile of Islet2 expression in mature MNs (data not shown) (Tsuchida et al., 1994). Nolz1 and Grg5 coexpression failed to induce MNR2, consistent with the restriction of endogenous Nolz1 and Grg5 expression to postmitotic MNs (Fig. 4A, Fig. 6C). Consistent with previous reports, Grg5 overexpression alone did not lead to ectopic MNR2, HB9, Islet1, Islet2 or Lim3 expression (see Fig. S2 in the supplementary material) (Muhr et al., 2001). Coelectroporation of Nolz1ΔC22 and Grg5 did not elicit the ectopic expression of MN HD proteins, suggesting that Nolz1-Grg5 complex formation is necessary for these events (Fig. 6E,F; data not shown). Furthermore, Nolz1-VP16 expression did not induce Lim3, HB9, Islet1 or Islet2, consistent with the ability of Nolz1-Grg5 complexes to implement MN determinant expression through repressor activities (Fig. 6G,H).
Fig. 6.
Nolz1-Grg5 complexes induce postmitotic MNs. Transverse sections of St 21 embryonic chick spinal cords electroporated on the right. White brackets denote ectopic MNs. (A-H,J-M) Immunohistochemical analyses using antibodies against MN and ventral interneuron markers. Arrows indicate ectopic MNs expressing the MN surface marker SC1 (J) or Chx10 (K). (I) Arrows mark ectopic MNs expressing transcripts for choline acetyltransferase (ChAT). Dashed lines mark the lateral extent of the spinal cord.
HB9 expression is directly induced by Islet1-Lim3 multimers, and HB9 can upregulate the expression of Islet1 and Lim3 (Arber et al., 1999; Thaler et al., 1999; Thaler et al., 2002; William et al., 2003). To examine the hierarchy of MN determinant expression induced by Nolz1-Grg5, we compared the expression profiles of these markers relative to each other. Nolz1 and Grg5 coexpression induced many HB9+ cells in the absence of Lim3 or Islet1, suggesting that Nolz1-Grg5 provides an alternative pathway to upregulate HB9 expression that does not require Islet1 and Lim3 (Fig. 6B-D). Furthermore, 35% of ectopic Islet1+ cells and 62% of ectopic Lim3+ cells lacked detectable HB9 expression (Fig. 6B,D; n=126 Islet1+ cells, n=42 Lim3+ cells analyzed). Taken together, these results suggest that Nolz1-Grg5 complexes can independently upregulate the expression of HB9, Islet1 and Lim3.
Dorsal cells that express the MN HD transcription factors when Nolz1 and Grg5 are coexpressed consistently occupied lateral positions, suggesting that the induction of these HD proteins occurred in the context of normal neurogenesis (Fig. 6A-D). Ectopic cells expressed choline acetyltransferase (ChAT), the enzyme responsible for acetylcholine synthesis, and the MN surface marker SC1, consistent with their adopting MN fates (Fig. 6I,J) (Tanabe et al., 1998). In support of their MN identity, none of the ectopic Islet1+ cells expressed the V0 or V1 interneuron markers Evx1/2 or En1 (Fig. 6L,M) (Pierani et al., 1999). However, 1-2 ectopic cells expressed the V2 marker Chx10, probably owing to the absence of HB9, which is required to repress V2-specific fates in MNs (Fig. 6K) (Arber et al., 1999; Thaler et al., 1999). To determine whether Nolz1-Grg5-induced MNs adopt a Lim3- columnar identity, we examined electroporated embryos 48 hours after coelectroporation of Nolz1 and Grg5. We found that 98% of ectopic Islet1+ MNs lacked Lim3 expression (n=120 Islet1+ cells analyzed).
Thus, Nolz1/Grg5 repressor activity can initiate MN differentiation programs downstream of MNR2, resulting in MNs with a Lim3- expression profile. The transient expression of Lim3 mediated by Nolz1 and Grg5 coexpression reflects the profile of Nolz1 expression in developing spinal MNs, where Nolz1 is coexpressed with Lim3 in a lateral band of newly generated MNs, prior to its rapid segregation with Lim3- spinal MNs (Fig. 2S).
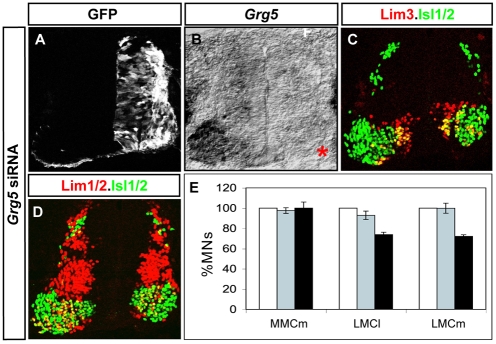
Grg5 is required for Lim3- motor column development
Our gain-of-function studies suggest that Grg5 plays central roles in regulating the development and formation of Lim3- motor columns. To examine whether Grg5 is necessary for Lim3- motor column development, we electroporated 21 bp siRNAs designed against the Grg5 ORF into chick embryonic spinal cords at St 14, and analyzed electroporated embryos at St 23/24 (Rao et al., 2004). Grg5 mRNA levels were efficiently decreased in siRNA-treated embryos; however, owing to the lack of available antibodies against chick Grg5, we were unable to assess the efficiency of reducing Grg5 protein (Fig. 7A,B). Nevertheless, we evaluated embryos electroporated with Grg5 siRNAs for the formation of Lim3+ MMCm MNs and Lim3- LMCl and LMCm MNs at forelimb levels of the spinal cord. No changes in the number of Lim3+ MMCm MNs were detected in embryos that showed efficient knockdown of Grg5 mRNA. By contrast, we consistently detected a 30% reduction in the number of Lim3- LMCl and LMCm MNs, as compared with the non-electroporated contralateral side, or when control siRNAs were electroporated (Fig. 7C-E). These results indicate that Grg5 is required for the formation of Lim3- MNs, and further supports our model that Nolz1-Grg5 complexes regulate the generation of Lim3- motor columns in the spinal cord.
Fig. 7.
Grg5 knockdown decreases the number of Lim3- motor columns. (A-D) St 24 chick embryos electroporated with Grg5 siRNAs. (A) The right-hand half of the spinal cord is electroporated, as indicated by GFP expression. (B) In situ hybridization of Grg5 mRNA showing efficient ablation of Grg5 mRNA (asterisk) on the Grg5 siRNA-electroporated side. (C,D) Confocal micrographs of sectioned chick spinal cords showing immunohistochemical analysis of motor column markers. (E) Bar chart showing the percentage of motor columns in electroporated Grg5 siRNAs (black bars), control siRNAs (gray bars) and contralateral non-electroporated (white bars, set as 100%) sides of the spinal cord at forelimb regions. n=4-5 embryos, mean ± s.e.m.
Nolz1 interacts with Grg5 to induce Hoxc6 expression in the spinal cord
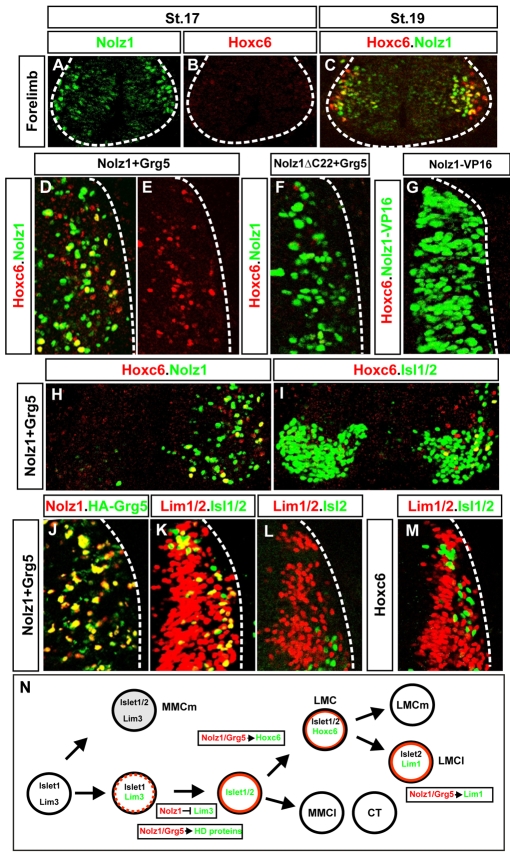
The retention of Nolz1 expression in LMC neurons suggests that Nolz1 might be required for the development of LMC motor columns. Since RA signaling pathways are necessary for specifying forelimb LMC identity, we examined whether Nolz1 mediates the transcriptional programs required for triggering forelimb LMC specification (Sockanathan et al., 2003). We first examined the onset of Nolz1 expression in prospective forelimb LMC neurons in relation to that of Hoxc6, a key postmitotic determinant of forelimb LMC identity (Dasen et al., 2003). At St 17, we detected many Nolz1+ MNs at forelimb levels, but very few, weakly expressing Hoxc6+ cells (Fig. 8A,B). By St 19, many Nolz1+ MNs were distributed along the mediolateral axis, a lateral subset of which coexpressed Hoxc6 (Fig. 8C). These data indicate that Nolz1 expression initiates prior to the induction of Hoxc6 in prospective forelimb LMC neurons.
Fig. 8.
Coexpression of Nolz1 and Grg5 induces Hoxc6 expression and partial lateral LMC specification. (A-C) Confocal micrographs of ventral chick embryonic spinal cords showing that the onset of Nolz1 expression occurs prior to that of Hoxc6. The dashed line outlines the spinal cord. (D-G) Confocal micrographs of electroporated dorsal right halves of St 23 chick spinal cords. Grg5 expression is not shown but is ∼95-98% coincident with exogenous Nolz1. The dashed line marks the lateral boundary of the spinal cord. (H,I) Confocal micrographs of the ventral MN domain of St 23/24 chick embryonic spinal cords electroporated on the right. Yellow cells in panel I mark thoracic MNs that express Hoxc6. (J-M) Confocal micrographs of electroporated dorsal right halves of St 23 chick spinal cords. The dashed line marks the lateral boundary of the spinal cord. The right-hand side is electroporated in all cases. The high levels of Nolz1 elicited by overexpression preclude the detection of endogenous Nolz1 under these imaging conditions. (N) Model for Nolz1-dependent specification of MN subtype identity. Orange lines mark cells expressing Nolz1. Broken orange line refers to transient Nolz1 expression. Nolz1 downregulates Lim3 expression in prospective Lim3-negative MNs and, in a complex with Grg5, maintains the expression of key MN determinants. At limb levels, Nolz1-Grg5 complexes confer forelimb LMC identity through induction of Hoxc6, and induce Lim1 expression in prospective LMCl MNs.
To test whether Nolz1 or Grg5 is capable of inducing Hoxc6 expression, we electroporated pCAGGS-Nolz1 into chick spinal cords and analyzed dorsal thoracic spinal cords at St 24, when Hoxc6 is normally absent. Expression of Nolz1 or Grg5 alone failed to elicit Hoxc6 expression (see Fig. S2 in the supplementary material). By contrast, coelectroporation of Nolz1 and Grg5 induced many dorsal Hoxc6+ cells, and upregulated Hoxc6 expression in a small number of thoracic MNs (Fig. 8D,E,H,I). Notably, dorsal Hoxc6 expression rarely coincided with cells coexpressing Grg5 and Nolz1, implying that Nolz1/Grg5-dependent induction of Hoxc6 occurs non-cell-autonomously. Grg5 and Nolz1ΔC22 coexpression did not induce Hoxc6, suggesting that Nolz1 and Grg5 interact to upregulate Hoxc6 expression (Fig. 8F; see Fig. S2 in the supplementary material). Electroporation of Nolz1-VP16 failed to elicit Hoxc6 expression (Fig. 8G). Taken together, these findings suggest that Nolz1-Grg5 complexes induce Hoxc6 expression through transcriptional repressor functions, and provide a mechanism that links Hoxc6-dependent specification of forelimb LMC identity with RA signaling pathways.
Grg5 and Nolz1 cooperate to induce Lim1 expression in ectopic MNs
At later stages of development, Nolz1 is localized to LMCl neurons, suggesting potential roles for Nolz1 in regulating their development (Fig. 2L,M). RA signaling pathways induce LMCl identity in particular by upregulating Lim1, which directs the dorsal trajectories of LMCl axons in the limb (Sockanathan and Jessell, 1998; Kania et al., 2000; Vermot et al., 2005; Ji et al., 2006). Nolz1 and Grg5 coexpression induces Lim3- MNs in the dorsal spinal cord (Fig. 6). To examine whether a subset of these neurons acquires lateral LMC fates, we examined embryos 2 days after coelectroporation with Nolz1 and Grg5. LMCl neurons are distinguished by their coexpression of Islet2 and Lim1, and by their downregulation of Islet1 (Tsuchida et al., 1994). Sixty-three percent of ectopic Islet1/2+ MNs coexpressed Lim1 when embryos were coelectroporated with Nolz1 and Grg5 (Fig. 8J,K; n=374 Islet1/2+ cells). However, staining with an Islet2-specific antibody showed that the majority of these cells did not express Islet2, demonstrating that they had induced Lim1 expression but failed to downregulate Islet1 (Fig. 8K,L). Ectopic Islet1/2+ cells did not express interneuron markers such as Evx1/2, En1 and Chx10, but were ChAT+ and SC1+ (Fig. 6I-M). These observations suggest that Nolz1 and Grg5 coexpression results in Islet1+ Islet2- MNs that had initiated a partial LMCl specification program. Hoxc6 triggers a temporal sequence of LMC formation, whereby Raldh2-derived RA induces LMCl subtype identity (Sockanathan and Jessell, 1998; Dasen et al., 2003; Vermot et al., 2005). Nolz1 and Grg5 coexpression does not induce dorsal Raldh2 expression, suggesting that Lim1 induction in ectopic MNs arises independently of Raldh2 function (data not shown). Moreover, many dorsal Lim1+ MNs did not coexpress Hoxc6 and overexpression of Hoxc6 in the dorsal spinal cord did not induce Islet1/2+ Lim1+ cells (Fig. 8M; see Fig. S3 in the supplementary material). Taken together, these results argue that Nolz1 can induce MNs that are capable of initiating partial programs of LMCl specification downstream of Hoxc6 and Raldh2 activity.
DISCUSSION
Here, we show that RA signals induce Nolz1 expression in the chick ventral spinal cord, and that Nolz1 mediates novel and known RA-dependent events that specify columnar and divisional identities in postmitotic MNs (Fig. 8N). Our results suggest a model in which paraxial mesoderm-derived RA signals induce Nolz1 in postmitotic MNs, where it regulates the diversification and maintenance of Lim3- MNs, prior to triggering forelimb LMC specification through Hoxc6 induction. At limb levels, RA synthesized in paraxial mesoderm and in MNs maintains Nolz1 expression, which acts to upregulate Lim1 expression in late-born prospective LMCl neurons. We discuss below how Nolz1 regulates the specification, function and survival of postmitotic Lim3- MNs in the context of known transcriptional hierarchies that orchestrate these events.
Nolz1 regulates the segregation and development of Lim3- MNs
We show here that Nolz1 is sufficient to downregulate Lim3 expression in postmitotic MNs by Grg5-independent repressor functions. Lim3 downregulation is an essential prerequisite for the acquisition of LMC, CT and MMCl columnar fates, as sustained Lim3 expression suppresses CT and LMC motor column formation, and causes MNs to adopt cell body settling positions, gene expression profiles and axonal projections characteristic of MMCm MNs (Sharma et al., 2000; William et al., 2003). Previous studies have shown that HB9 also functions to suppress Lim3 expression (Arber et al., 1999; Thaler et al., 1999). We find that Nolz1 can upregulate HB9 expression through a separate repressor activity involving association with Grg5. Thus, our studies support a model in which Nolz1 implements at least two pathways that converge to control the timely repression of Lim3 expression in prospective LMC, CT and MMCl MNs. These findings invoke crucial roles for Nolz1 in regulating the early segregation of Lim3- motor columns from Lim3+ MMCm MNs.
Ablating Nolz1 causes the loss of Lim3- MNs, suggesting that Nolz1 has additional functions in the development and survival of CT, MMCl and LMC motor columns besides downregulating Lim3 expression. We find that Nolz1-Grg5 repressor complexes can induce terminally differentiated MNs with a Lim3- molecular identity, through upregulating a series of key HD transcription factors. Since endogenous Nolz1 expression initiates in postmitotic MNs and not in progenitor cells, this finding is consistent with the model that Nolz1-Grg5 complexes implement HD-dependent transcriptional programs that are necessary for consolidating and/or maintaining the properties and survival of postmitotic Lim3- MNs. Accordingly, Nolz1-Grg5 complexes are not sufficient to activate MNR2 expression, consistent with their role in MN consolidation or maintenance rather than induction (Tanabe et al., 1998). The involvement of Nolz1-Grg5 repressor complexes in this regard parallels the requirement for Nkx-Grg complexes in derepressing MN differentiation programs during MN generation (Muhr et al., 2001). These observations suggest that continuous derepression strategies are required in progenitors and postmitotic MNs to generate and actively preserve the properties and survival of postmitotic MNs. The function of Nolz1-Grg5 complexes might differ between limb-level and thoracic motor columns. The prolonged expression of Nolz1 at limb levels suggests that Nolz1-Grg5 complexes play more sustained roles in the specification and maintenance of LMC neurons as compared with CT and MMCl neurons, in which Nolz1 is downregulated after St 24. Indeed, prolonged retinoid receptor activation is detrimental to thoracic MN survival, suggesting that retinoid-dependent pathways operate transiently during thoracic motor column development (Sockanathan et al., 2003).
Nolz1 regulates forelimb LMC specification
Disrupting RA signals in prospective forelimb MNs causes putative LMC MNs to adopt thoracic identities, suggesting that RA signals have dual roles in specifying LMC fates and suppressing thoracic-specific columnar differentiation programs (Sockanathan et al., 2003). We show that Nolz1-Grg5 repressors induce Hoxc6, which can drive LMC-specific programs of differentiation and suppress thoracic columnar identity in limb-level MNs (Liu et al., 2001; Dasen et al., 2003). These findings provide a mechanism that links RA signaling with the specification of forelimb LMC identity through the Nolz1-dependent induction of Hoxc6 expression. How do Nolz1-Grg5 repressor complexes upregulate Hoxc6? Hoxc9 can repress Hoxc6 expression cell-autonomously in spinal MNs (Dasen et al., 2003). However, our results argue against the possibility that Nolz1 derepresses Hoxc6 expression through inhibition of Hoxc9, as Nolz1 functions non-cell-autonomously to upregulate Hoxc6. Instead, Nolz1 might induce Hoxc6 through the derepression of additional signaling pathways that remain to be identified.
A second question is how Nolz1-Grg5 complexes might regulate the restricted expression of Hoxc6 at forelimb levels, when Nolz1 and Grg5 are expressed at all axial levels at the time of Hoxc6 induction. One option is that the intermediary components that relay Nolz1-Grg5 effects are restricted primarily to spinal MNs at forelimb levels. In support of this idea, very few thoracic MNs induce Hoxc6 when Nolz1 is overexpressed, as compared with those in the dorsal spinal cord. Alternatively, the restricted expression of Hoxc6 by Nolz1-Grg5 activity might depend on the function of other Hox proteins. Hoxc9 is induced by FGFs and is expressed in thoracic progenitors and spinal MNs (Dasen et al., 2003). Thus, early Hoxc9 expression in progenitors and MNs could override the later inductive capabilities of Nolz1-Grg5 complexes, thereby repressing Hoxc6 at thoracic levels. This Hox-dependent mechanism could potentially operate at lumbar regions via Hox10 proteins, thus confining the ability of Nolz1-Grg5 complexes to induce Hoxc6 expression to forelimb levels of the spinal cord (Dasen et al., 2003; Wu et al., 2008).
Nolz1 induces Lim1 expression in prospective lateral LMC neurons
Our expression analyses and functional experiments collectively support a role for Nolz1 in lateral LMC neuronal development. Ectopic MNs induced by Nolz1-Grg5 complexes coexpress Lim1, a LIM-HD protein that partly defines LMCl molecular identity and directs their dorsal axonal trajectory (Tsuchida et al., 1994; Kania et al., 2000). However, the Lim1+ MNs induced by Nolz1-Grg5 complexes coexpress Islet1, which is normally downregulated in LMCl neurons (Tsuchida et al., 1994). These observations indicate that Nolz1-Grg5 complexes implement a subset of LMCl differentiation pathways, specifically those regulating LMCl axonal projections. They also imply that Lim1 induction can be uncoupled from the downregulation of Islet1, suggesting that independent pathways function to execute these previously linked events. We show that the Lim1+ MNs induced by Nolz1-Grg5 complexes are generated independently of prior Raldh2 and Hoxc6 function. These results support a mechanism whereby Nolz1-Grg5 repressors function downstream of forelimb LMC specification and Raldh2 activity, to induce Lim1 expression in prospective LMCl MNs. Nolz1 is expressed in LMCl neurons until at least St 29, suggesting that Nolz1-Grg5 complexes might function to maintain Lim1 expression in LMCl cells.
In conclusion, our study suggests that RA signals specify multiple events governing MN subtype identity through the induction of Nolz1. Nolz1 exhibits strong transcriptional repressor activity, and this repressor function is modulated by interactions with Grg5. Importantly, Nolz1 repressor complexes are functionally distinct, suggesting that their individual components differ. Notably, our observation that Grg5 modulates Nolz1 repressor activity invokes the possibility that graded Nolz1 repressor activities are a crucial component in implementing selective transcriptional pathways that direct different aspects of MN development. The induction of a single key factor with functional flexibility, such as Nolz1, circumvents the need to generate large numbers of proteins that individually execute separate developmental pathways, and provides a useful and effective strategy to regulate diverse events in neuronal development.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/2/231/DC1
Supplementary Material
We thank Shaneka Lawson for technical assistance; Jeremy Dasen, Tom Jessell, Ben Novitch and Sam Pfaff for antibodies and reagents; Alex Kolodkin, Ben Novitch and Ye Yan for comments on the manuscript; and members of the Sockanathan laboratory for scientific discussions. Funding for this work was provided by grants from NINDS (NIH), the Robert Packard Center for ALS Research and the Wings over Wall Street Campaign by the Muscular Dystrophy Association. Deposited in PMC for release after 12 months.
References
- Arber, S., Han, B., Mendelsohn, M., Smith, M., Jessell, T. M. and Sockanathan, S. (1999). Requirement for the homeobox gene Hb9 in the consolidation of MN identity. Neuron 23, 659-674. [DOI] [PubMed] [Google Scholar]
- Brantjes, H., Roose, J., van de Wetering, M. and Clevers, C. (2001). All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29, 1410-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. W., Tsai, C. W., Wang, H. F., Tsai, H. C., Chen, H. Y., Tsai, T. F., Takahashi, H., Li, H. Y., Fann, M. J., Yang, C. W. et al. (2004). Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc. Natl. Acad. Sci. USA 101, 2613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen, J. S., Liu, J. P. and Jessell, T. M. (2003). MN columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926-933. [DOI] [PubMed] [Google Scholar]
- Diez del Corral, R., Olivera-Martinez, I., Goriely, A., Gale, E., Maden, M. and Storey, K. (2003). Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 65-79. [DOI] [PubMed] [Google Scholar]
- Fisher, A. L. and Caudy, M. (1998). Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 12, 1931-1940. [DOI] [PubMed] [Google Scholar]
- Gross, M. K., Dottori, M. and Goulding, M. (2002). Lbx1 specifies somatosensory association interneurons in the spinal cord. Neuron 34, 535-549. [DOI] [PubMed] [Google Scholar]
- Hoyle, J., Tang, Y. P., Wielllette, E. L., Wardle, F. C. and Sive, H. (2004). nlz gene family is required for hindbrain patterning in the zebrafish. Dev. Dyn. 229, 835-846. [DOI] [PubMed] [Google Scholar]
- Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20-29. [DOI] [PubMed] [Google Scholar]
- Ji, S. J., Zhuang, B., Falco, C., Schneider, A., Schuster-Gossler, K., Gossler, A. and Sockanathan, S. (2006). Mesodermal and neuronal retinoids regulate the induction and maintenance of limb innervating spinal MNs. Dev. Biol. 297, 249-261. [DOI] [PubMed] [Google Scholar]
- Kania, A., Johnson, R. L. and Jessell, T. M. (2000). Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell 102, 161-173. [DOI] [PubMed] [Google Scholar]
- Landmesser, L. (1978). The distribution of motoneurones supplying chick hindlimb muscles. J. Physiol. 284, 371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. K., Jurata, L. W., Funahashi, J., Ruiz, E. C. and Pfaff. S. L. (2004). Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development 131, 3295-3306. [DOI] [PubMed] [Google Scholar]
- Liu, J. P., Laufer, E. and Jessell, T. M. (2001). Assigning the positional identity of spinal MNs: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 32, 997-1012. [DOI] [PubMed] [Google Scholar]
- Maden, M. (2002). Retinoid signalling in the development of the central nervous system. Nat. Rev. Neurosci. 3, 843-853. [DOI] [PubMed] [Google Scholar]
- Moran-Rivard, L., Kagawa, T., Saueressig, H., Gross, M. K., Burrill, J. and Goulding, M. (2001). Evx1 is a postmitotic determinant of v0 interneuron identity in the spinal cord. Neuron 29, 385-399. [DOI] [PubMed] [Google Scholar]
- Muhr, J., Andersson, E., Persson, M., Jessell, T. M. and Ericson, J. (2001). Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861-873. [DOI] [PubMed] [Google Scholar]
- Muller, T., Brohmann, H., Pierani, A., Heppenstall, P. A., Lewin, G. R., Jessell, T. M. and Birchmeier, C. (2002). The homeodomain protein lbx1 distinguished two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34, 551-562. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., Runko, A. P. and Sagerstrom, C. G. (2004). A novel family of zinc finger genes involved in embryonic development. J. Cell. Biochem. 93, 887-895. [DOI] [PubMed] [Google Scholar]
- Nishihara, S., Tsuda, L. and Ogura, T. (2003). The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem. Biophys. Res. Commun. 7, 55-63. [DOI] [PubMed] [Google Scholar]
- Novitch, B. G., Chen, A. I. and Jessell, T. M. (2001). Coordinate regulation of MN subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773-789. [DOI] [PubMed] [Google Scholar]
- Novitch, B. G., Wichterle, H., Jessell, T. M. and Sockanathan, S. (2003). A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and MN specification. Neuron 40, 81-95. [DOI] [PubMed] [Google Scholar]
- Pearson, B. J. and Doe, C. Q. (2004). Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20, 619-647. [DOI] [PubMed] [Google Scholar]
- Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. and Jessell, T. M. (1996). Requirement for LIM homeobox gene Isl1 in MN generation reveals a MN-dependent step in interneuron differentiation. Cell 84, 309-320. [DOI] [PubMed] [Google Scholar]
- Pierani, A., Brenner-Morton, S., Chiang, C. and Jessell, T. M. (1999). A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell 97, 903-915. [DOI] [PubMed] [Google Scholar]
- Prasad, A. and Hollyday, M. (1991). Development and migration of avian sympathetic preganglionic neurons. J. Comp. Neurol. 307, 237-258. [DOI] [PubMed] [Google Scholar]
- Rao, M. and Sockanathan, S. (2005). Transmembrane protein GDE2 induces MN differentiation in vivo. Science 309, 2212-2215. [DOI] [PubMed] [Google Scholar]
- Rao, M., Baraban, J., Rajaii, F. and Sockanathan, S. (2004). In vivo comparative study of RNAi methodologies by in ovo electroporation in the chick embryo. Dev. Dyn. 231, 592-600. [DOI] [PubMed] [Google Scholar]
- Runko, A. P. and Sagerstrom, P. (2004). Isolation of nlz2 and characterization of essential domains in Nz family proteins. J. Biol. Chem. 279, 11917-11925. [DOI] [PubMed] [Google Scholar]
- Shaeren-Wiemers, N. and Gerfin-Moser, A. (1993). A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431-440. [DOI] [PubMed] [Google Scholar]
- Sharma, K., Sheng, H. Z., Lettieri, K., Li, H., Karavanov, A., Potter, S., Westphal, H. and Pfaff, S. L. (1998). LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for MNs. Cell 95, 817-828. [DOI] [PubMed] [Google Scholar]
- Sharma, K., Leonard, A. E., Lettieri, K. and Pfaff, S. L. (2000). Genetic and epigenetic mechanisms contribute to MN pathfinding. Nature 406, 515-519. [DOI] [PubMed] [Google Scholar]
- Sockanathan, S. and Jessell, T. M. (1998). MN-derived retinoid signaling specifies the subtype identity of spinal MNs. Cell 94, 503-514. [DOI] [PubMed] [Google Scholar]
- Sockanathan, S., Perlmann, T. and Jessell, T. M. (2003). Retinoid receptor signaling in postmitotic MNs regulates rostrocaudal positional identity and axonal projection pattern. Neuron 40, 97-111. [DOI] [PubMed] [Google Scholar]
- Solomin, L., Johansson, C. B., Zetterstrom, R. H., Bissonette, R. P., Heyman, R. A., Olson, L., Lendahl, U., Frisen, J. and Perlmann, T. (1998). Retinoid X receptor signaling in the developing spinal cord. Nature 395, 398-402. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y., William, C. and Jessell, T. M. (1998). Specification of MN identity by the MNR2 homeodomain protein. Cell 95, 67-80. [DOI] [PubMed] [Google Scholar]
- Thaler, J. P., Harrison, K., Sharma, K., Lettieri, K., Kehrl, J. and Pfaff, S. L. (1999). Active suppression of interneuron programs within developing MNs revealed by analysis of homeodomain factor HB9. Neuron 23, 675-687. [DOI] [PubMed] [Google Scholar]
- Thaler, J. P., Lee, S. K., Jurata, L. W., Gill, G. N. and Pfaff, S. L. (2002). LIM factor Lhx3 contributes to the specification of MN and interneuron identity through cell-type-specific protein-protein interactions. Cell 110, 237-249. [DOI] [PubMed] [Google Scholar]
- Tosney, K. W., Hotary, K. B. and Lance-Jones, C. (1995). Specifying the target identity of motoneurones. BioEssays 17, 379-382. [DOI] [PubMed] [Google Scholar]
- Tsuchida, T. N., Ensini, M., Morton, S. B., Baldessare, M., Edlund, T., Jessell, T. M. and Pfaff, S. L. (1994). Topographic organization of embryonic MNs defined by expression of LIM homeobox genes. Cell 79, 957-970. [DOI] [PubMed] [Google Scholar]
- Vallstedt, A., Muhr, J., Pattyn, A., Pierani, A., Mendelsohn, M., Sander, M., Jessell, T. M. and Ericson, J. (2001). Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in MN and interneuron specification. Neuron 31, 743-755. [DOI] [PubMed] [Google Scholar]
- Vermot, J., Schuhbaur, B., Le Mouellic, H., McCaffery, P., Garnier, J. M., Hentsch, D., Brulet, P., Niederreither, K., Chambon, P., Dolle, P. and Le Roux, I. (2005). Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development 132, 1611-1621. [DOI] [PubMed] [Google Scholar]
- William, C., Tanabe, Y. and Jessell, T. M. (2003). Regulation of MN subtype identity by repressor activity of Mnx class Homeodomain proteins. Development 130, 1523-1536. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Wang, G., Scott, S. A. and Capecchi, M. R. (2008). Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development 135, 171-182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.