Abstract
In yeast, microtubules are organized by the spindle pole body (SPB). The SPB is a disk-like multilayered structure that is embedded in the nuclear envelope via its central plaque, whereas the outer and inner plaques are exposed to the cytoplasm and nucleoplasm, respectively. How the SPB assembles is poorly understood. We show that the inner/central plaque is composed of a stable SPB subcomplex, containing the γ-tubulin complex-binding protein Spc110p, calmodulin, Spc42p, and Spc29p. Spc29p acts as a linker between the central plaque component Spc42p and the inner plaque protein Spc110p. Evidence is provided that the calmodulin-binding site of Spc110p influences the binding of Spc29p to Spc110p. Spc42p also was identified as a component of a cytoplasmic SPB subcomplex containing Spc94p/Nud1p, Cnm67p, and Spc42p. Spc29p and Spc42p may be part of a critical interface of nucleoplasmic and cytoplasmic assembled SPB subcomplexes that form during SPB duplication. In agreement with this, overexpressed Spc29p was found to be a nuclear protein, whereas Spc42p is cytoplasmic. In addition, an essential function of SPC29 during SPB assembly is indicated by the SPB duplication defect of conditional lethal spc29(ts) cells and by the genetic interaction of SPC29 with CDC31 and KAR1, two genes that are involved in SPB duplication.
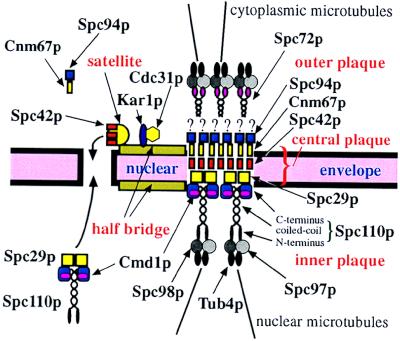
In the yeast Saccharomyces cerevisiae, microtubule-organizing functions are provided by the spindle pole body (SPB) (for review, see ref. 1). The SPB is a multilayered structure that duplicates once per cell cycle and is embedded in the nuclear envelope through the central plaque structure (ref. 2; see Fig. 6). The outer plaque is located on the cytoplasmic side of the SPB, whereas the inner plaque extends into the nucleoplasm. An additional SPB substructure is the half bridge, a one-sided extension of the central plaque.
Figure 6.
Model for the functions of Spc29p. Kar1p, and Cdc31p are components of half bridge (5, 7) that function in SPB duplication (4, 6). Spc72p anchors the yeast γ-tubulin complex containing Tub4p, Spc98p, and Spc97p to the outer plaque (11). The yeast γ-tubulin complex is bound to the inner plaque via Spc110p (20). Spc42p forms a layer within the SPB (2). Cnm67p and Spc94p/Nud1p are components of the cytoplasmic outer plaque (26). Based on our results, we propose that Cnm67p binds to Spc42p and Spc94p/Nud1p interacts with Cnm67p. Spc29p is bridging C–Spc110p and N–Spc42p. During SPB duplication, Spc29p, N–Spc42p, and C–Spc110p form a critical border of cytoplasmic and nuclear assembled SPB subcomplexes. Spc42p may also be part of the satellite that forms in G1 phase of the cell cycle (3).
Some insight into the SPB duplication process came from electron microscopic studies and from the phenotype of conditional-lethal mutants. In early G1 phase of the cell cycle, the half bridge elongates and develops an appendix toward the cytoplasmic side, which was named the satellite. The satellite is probably a precursor of the newly formed SPB. After start of the cell cycle, the SPB fully duplicates, resulting in two SPBs side by side (3). CDC31 (4, 5) and KAR1 (6, 7) code for components of the SPB half bridge required early in SPB duplication. In mps2-1 and ndc1-1 cells, a fully functional SPB is associated with a partially duplicated SPB containing an outer and a central plaque, sitting on top of the cytoplasmic side of the nuclear envelope (8, 9). The mps2/ndc1 phenotype suggests that the outer plaque and some part of the central plaque are built from cytoplasmic components, whereas the inner plaque is probably assembled in the nucleus. This view is further supported by the fact that SPB components of the inner plaque such as Spc110p (10) are imported into the nucleus, whereas the outer plaque component Spc72p is not (11). Finally, the cytoplasmic and nuclear assembled SPB subcomplexes have to be brought together by opening the nuclear envelope.
We recently identified an SPB subcomplex containing the SPB components calmodulin (Cmd1p), Spc110p, Spc42p, and an uncharacterized protein of 35 kDa (12). Here we describe that the 35-kDa protein is encoded by SPC29. Spc29p is an essential SPB component that fulfills a crucial role during the SPB duplication process.
MATERIALS AND METHODS
SPC29 Mutants, Yeast Strains, and Plasmid Constructions.
Conditional lethal alleles of SPC29 were selected as described (13). spc29-2 and spc29-3 carry single missense mutations resulting in R161S and L158Q, respectively. spc29-9 carries multiple mutations. Yeast strains are derivatives of YPH499/500 (14). Temperature-sensitive (ts) mutants of CMD1 (15), SPC42 (16), SPC110 (10, 17), SPC98 (18), SPC97 (12), and TUB4 (19) and strain ESM172 (ProA–SPC110) (20) have been described. CNM67–GFP, SPC29–3HA, SPC29–GFP, SPC42–GFP, SPC94–ProA, and SPC110–GFP gene fusions were constructed by using PCR-based methods (21). For overexpression studies SPC42, SPC29, and SPC110 were under control of the Gal1 promoter. For multicopy suppression, the indicated genes were cloned into the 2-μm vectors. Two-hybrid interactions were tested as described (18).
Electron Microscopy, Immunofluorescence, and Flow Cytometry.
Electron microscopy (22), immunofluorescence of formaldehyde-fixed cells (20), and flow cytometry (23) were performed as described.
Antibodies.
Anti-Cnm67p antibodies were produced against glutathione S-transferase–N–Cnm67p (amino acids 1–214) in rabbits. Other antibodies against SPB components have been described (5, 7, 11, 19, 20). The mouse monoclonal anti-hemagglutinin antibody (12CA5) was from Hiss Diagnostics. The mouse mAb WA3 was used to stain microtubules (19). Secondary antibodies were from Jackson ImmunoResearch.
RESULTS
Spc29p Is a Subunit of a Complex Containing Cmd1p, Spc110p, and Spc42p.
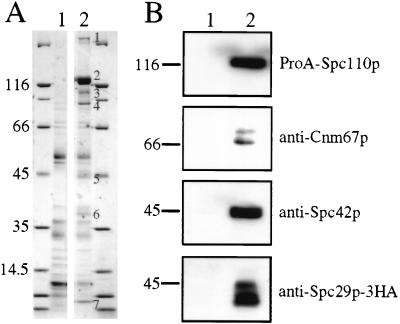
Spc110p is a key SPB component because its N-terminal domain interacts with the yeast γ-tubulin complex (ref. 20; see Fig. 6), and in addition, it is important for SPB integrity (24). To understand how Spc110p fulfills these essential functions, we purified an Spc110p-containing subcomplex after salt fragmentation of SPBs (Fig. 1A) (12). Seven protein bands were identified on a Coomassie blue-stained gel. Bands 1–4 were derived from Spc110p-ProA, whereas band 5 was identified as Spc42p by immunoblotting (Fig. 1B). Spc42p is an essential SPB component that forms a central crystal within the SPB (2). Analysis of the additional subunits of the Spc110p-containing complex by high mass-accuracy matrix-assisted laser desorption ionization peptide mapping (25) identified band 7 as Cmd1p, which has been shown to bind to the C terminus of Spc110p (15, 17). The 35-kDa protein (band 6) was identified as Spc29p, an essential SPB component (26). Spc29p’s association with Spc110p was confirmed by performing a coimmunoprecipitation experiment (Fig. 1B). Furthermore, antibodies against the SPB components Cdc31p, Kar1p, Tub4p, Cnm67p, Spc72p, Spc97p, and Spc98p were used to test whether any of these proteins are associated with the Spc110p complex in substoichiometric amounts. Only small amounts of Cnm67p were detectable (Fig. 1B), raising the possibility that Cnm67p physically interacts with one of the components of the Spc110p complex. Thus, the Spc110p complex contains Spc110p, Spc42p, Spc29p, Cmd1p, and low amounts of Cnm67p.
Figure 1.
An Spc110p-containing SPB subcomplex. (A) An Spc110p-containing SPB subcomplex was purified from cells with a protein A-tagged SPC110 (ProA–SPC110) (lane 2) by using IgG Sepharose. The protein bands were subjected to matrix-assisted laser desorption ionization (MALDI) MS analysis. Wild-type cells, containing Spc110p without a ProA tag (lane 1), were treated identically to identify proteins that bound nonspecifically to the IgG resin. The major proteins are encoded by SPC110 (band 1, ProA–Spc110p homodimer; band 2, ProA-Spc110p; bands 3 and 4, ProA–Spc110p degradation products), SPC42 (band 5), SPC29 (band 6), and CMD1 (band 7). (B) Coimmunoprecipitation of Spc29p and Spc110p. High-salt extracts from cells expressing SPC110 SPC29–3HA (lane 1) or ProA–SPC110 SPC29–3HA (lane 2) were incubated with IgG Sepharose. The immunoblots were probed with the indicated antibodies.
Genetic Interactions Between CMD1, SPC29, SPC42, and SPC110.
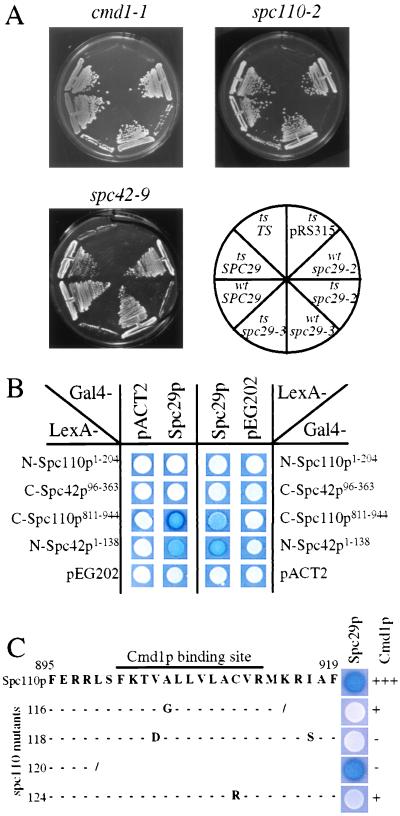
The functional relationship between SPC29 and genes coding for SPB components was verified by performing genetic studies. Synthetic lethality was observed when spc29-2 or spc29-3 were combined with cmd1-1, spc110-2 or spc42-9 (Fig. 2A) but not when combined with tub4-1, spc97-14, spc97-21, spc98-2, or spc98-7 (data not shown), coding for proteins of the Tub4p complex. A genetic interaction between SPC29 and SPC42 also is indicated by the allele-specific, high-dosage suppression of the temperature-dependent growth defect of spc42-10 but not of spc42-9 by SPC29 (Table 1). In contrast, high gene dosage of SPC42 or SPC110 increased the growth defect of some spc29(ts) alleles (Table 1). Thus, SPC29 shows genetic interaction with SPC110, SPC42, and CMD1 but not with genes coding for subunits of the yeast γ-tubulin complex.
Figure 2.
Genetic interactions between SPC29 and CMD1, SPC110, and SPC42. (A) The chromosomal SPC29 of cmd1-1, spc110-2, and spc42-9 (sectors “ts”) mutant or wild-type cells (sector “wt”) was deleted in the presence of a plasmid containing SPC29 (URA3-based). Growth of mutant and wild-type cells with the control plasmid pRS315 (sector “pRS315”), or spc29-2 (sectors “spc29-2”), spc29-3 (sectors “spc29-3”), or SPC29 (sectors “SPC29”) on pRS315 were tested at 23°C on plates containing 5-fluoroorotic acid (5-FOA), which selects against the URA3-based pRS316-SPC29 plasmid. Lack of growth on 5-FOA in the presence of pRS315 (sector “ts/pRS315”) shows that SPC29 is an essential gene. All cells grew with SPC29 on pRS315 (sectors “ts/SPC29 ” or “wt/SPC29”). The failure of cmd1-1, spc110-2, or spc42-9 cells to grow with pRS315–spc29-2 (ts/spc29-2) or pRS315–spc29-3 (ts/spc29-3) on 5-FOA indicates synthetic lethality. In contrast, wild-type cells with spc29-2 (“wt/spc29-2”) or spc29-3 (“wt/spc29-3”) on pRS315 grew on 5-FOA. As a control, the strains were complemented with CMD1, SPC110, or SPC42 (sector named “ts/TS”) on pRS315, which allowed growth on synthetic complete medium at 37°C (data not shown). However, these cells did not grow on 5-FOA, because SPC29 is essential for growth. (B) Two-hybrid interactions of Spc29p with C–Spc110p and N–Spc42p. The indicated pACT2 and pEG202 derivatives were transformed into the indicator strain SGY37. β-galactosidase activity was determined by using a plate assay (18). Blue colony color indicates interaction. (C) SPC110811–944 in plasmid pEG202 of wild type or of SPC110 mutants carrying the indicated mutations in the Cmd1p-binding site were tested in the two-hybrid system for interaction with SPC29 in pACT2. Cmd1p binding to the Spc110p mutants has been determined before by using an in vitro binding assay (18).
Table 1.
Multicopy suppression analysis
| Mutant | Multicopy gene | Temperature, °C
|
||||
|---|---|---|---|---|---|---|
| 23 | 30 | 33 | 35 | 37 | ||
| spc29-2 | SPC29 | +++ | +++ | +++ | +++ | +++ |
| SPC42 | +++ | +++ | +++ | − | − | |
| SPC110 | +++ | +++ | +++ | − | − | |
| CDC31 | +++ | +++ | +++ | +++ | +++ | |
| KAR1 | +++ | +++ | − | − | ||
| control | +++ | +++ | +++ | − | − | |
| spc29-3 | SPC29 | +++ | +++ | +++ | +++ | +++ |
| SPC42 | +++ | +++ | − | − | − | |
| SPC110 | +++ | + | − | − | − | |
| CDC31 | +++ | +++ | − | − | − | |
| KAR1 | +++ | − | − | − | − | |
| control | +++ | +++ | − | − | − | |
| spc29-9 | SPC29 | +++ | +++ | +++ | +++ | +++ |
| SPC42 | +++ | + | − | − | − | |
| SPC110 | +++ | + | − | − | − | |
| CDC31 | +++ | +++ | − | − | − | |
| KAR1 | +++ | +++ | − | − | − | |
| control | +++ | +++ | − | − | − | |
| spc42-9 | SPC42 | +++ | +++ | +++ | +++ | +++ |
| SPC29 | +++ | +++ | +++ | +++ | − | |
| SPC110 | +++ | +++ | +++ | +++ | − | |
| control | +++ | +++ | +++ | +++ | − | |
| spc42-10 | SPC42 | +++ | +++ | +++ | +++ | +++ |
| SPC29 | +++ | +++ | +++ | +++ | +++ | |
| SPC110 | +++ | +++ | +++ | − | − | |
| control | +++ | +++ | +++ | − | − | |
| spc110-118 | SPC110 | +++ | +++ | +++ | +++ | +++ |
| SPC29 | +++ | +++ | + | − | − | |
| CMDI | +++ | +++ | +++ | +++ | +++ | |
| control | +++ | +++ | +++ | + | − | |
| spc110-124 | SPC110 | +++ | +++ | +++ | +++ | +++ |
| SPC29 | +++ | +++ | +++ | − | − | |
| CMDI | +++ | +++ | +++ | +++ | +++ | |
| control | +++ | +++ | +++ | +++ | − | |
The conditional-lethal mutants spc29-2, spc29-3, spc29-9, spc42-9, spc42-10, spc110-118, and spc110-124 were transformed with either an empty 2-μm plasmid (control) or with a 2-μm plasmid carrying one of the indicated genes. Only KAR1 was on the centromere-based plasmid pRS315. Transformants were tested for growth at various temperatures. +++, good growth, +, reduced growth, −, no growth.
We used the yeast two-hybrid system to investigate which proteins of the Spc110p complex interact with each other. Binding of Cmd1p to the C terminus of Spc110p has previously been demonstrated by using a two-hybrid approach (15). In addition, two-hybrid interactions between Spc29p and C-Spc110p and Spc29p with N-Spc42p were observed (Fig. 2B). In contrast, no binding of Spc29p with N-Spc110p, C-Spc42p (Fig. 2B), the central domain of Spc110p, components of the Tub4p complex, or the outer plaque protein Cnm67p (26) was detected (data not shown).
The Cmd1p-binding site of Spc110p has been mapped to amino acids 900–914 of Spc110p (15, 17), whereas Spc29p showed interaction with amino acids 811–944 of Spc110p (Fig. 2C), raising the possibility that the Spc29p and Cmd1p-binding sites overlap. To test this possibility, a C-terminal Spc110p truncation (spc110–120) lacking the Cmd1p-binding site was tested for its interaction with Spc29p (Fig. 2C). This Spc110p811–898 still showed binding to Spc29p, indicating that Spc29p binds adjacent to the Cmd1p-binding site of Spc110p.
Previous studies suggested that the Cmd1p-binding site of Spc110p has an inhibitory role, which is either relieved by Cmd1p binding or by its deletion (15, 17), raising the possibility that the Cmd1p-binding site of Spc110p regulates the binding of Spc29p. This notion was tested by studying the interaction of Spc29p with Spc110p proteins carrying mutations in the Cmd1p-binding site (Fig. 2C) that reduced Cmd1p binding (17). We observed that the mutated Spc110 proteins were impaired in their interaction with Spc29p, suggesting that Cmd1p binding to Spc110p plays a critical role in regulating the binding between Spc110p and Spc29p. We reasoned that overexpression of SPC29 in cells in which Spc110p has a defective Cmd1p-binding site may be toxic, because the Cmd1p-regulated binding of Spc29p to Spc110p is impaired. Indeed, the growth defect of such SPC110 mutants was increased by high gene dosage of SPC29, whereas it was suppressed by CMD1 (Table 1; ref. 17). Taken together, our genetic data suggest that Spc29p binds to Spc110p and Spc42p and that Spc29p binding to Spc110p may be regulated by the adjacent Cmd1p-binding site.
Spc29p Is a Nuclear Protein, Whereas Spc42p Is Cytoplasmic.
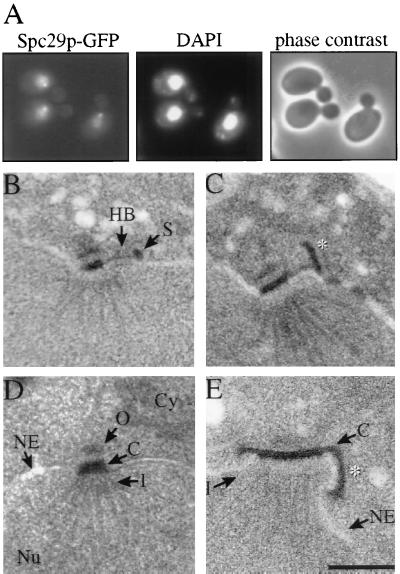
To understand whether Spc29p is assembled into the Spc110p complex from the cytoplasmic or nuclear side, the localization of overexpressed Spc29p was determined. Overexpressed Spc29p–GFP accumulated inside the nucleus and to some extent at the SPB, most likely the nuclear side, indicating that Spc29p is a nuclear protein (Fig. 3A). However, this analysis does not exclude the possibility that some Spc29p functions on the cytoplasmic side of the SPB.
Figure 3.
Cellular localization of Spc29p and Spc42p. (A) Localization of Spc29p-GFP after its overexpression from the Gal1 promoter for 3 hours at 30°C was determined by using fluorescence microscopy. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). (B and C) Electron microscopic picture of an SPB of mps2-1 cells with (C) or without (B) overexpression of SPC42. Cells of mps2-1 with or without Gal1–SPC42 were shifted to 37°C for 30 min in 2% raffinose medium followed by the induction of the Gal1 promoter by the addition of 2% galactose. ∗ in C indicates the Spc42p polymer. (D and E) Electron microscopic picture of an SPB from cells with (E) and without (D) cooverexpression of SPC29, SPC42, and SPC110. Cmd1p is probably present in excess and was therefore not overexpressed. ∗ in E indicates the Spc42p polymer. C, central plaque; Cy, cytoplasm; HB, half bridge; I, inner plaque; NE, nuclear envelope; Nu, nucleus; O, outer plaque; S, satellite. (Bar = 0.25 μm.). B–E are all of same magnification.
Overexpression of SPC42 results in the formation of a large Spc42p polymer that extends the already existing Spc42p layer within the SPB (2). This self-assembly property of Spc42p was used to determine whether the central layer of Spc42p is cytoplasmically assembled. We overexpressed SPC42 in mps2-1 cells in which the outer and central plaques of the newly formed SPB sit on top of the nuclear envelope, next to the mother SPB (8). An extension of the Spc42p layer of the partially duplicated SPB would indicate a cytoplasmic localization of Spc42p. We noticed that in our strain background, the mps2-1 phenotype was slightly different. An elongated half bridge was associated with what looked like a satellite (Fig. 3B). Overexpression of SPC42 in mps2-1 cells resulted in the formation of an Spc42p polymer that extended from the satellite into the cytoplasm (Fig. 3C, ∗), indicating the cytoplasmic assembly of Spc42p. This result also suggests that the satellite contains a layer of Spc42p that is enlarged on SPC42 overexpression. In wild-type cells, the Spc42p crystal within the SPB is extended parallel to the nuclear envelope on SPC42 overexpression (2). This difference compared with mps2-1 cells is most easily explained by the assumption that the Spc42p layer within the satellite is ordered perpendicular to the Spc42p layer in the central plaque.
Cooverexpression of SPC29, SPC42, and SPC110 Increases the Size of the Inner Plaque.
The size of the SPB correlates with the DNA content of yeast cells (3), raising the question of how the dimensions of SPB substructures are determined. We investigated whether the expression level of the components of the Spc110p complex contributes to the size regulation of the inner plaque. Single overexpression of any of the Spc110p complex components did not alter the overall dimensions of the inner plaque (refs. 10 and 16, and data not shown). However, cooverexpression of SPC110, SPC42, and SPC29 resulted in an inner plaque with a diameter that was double to triple that of wild-type cells (compare Fig. 3D, wild-type, with Fig. 3E, overexpression). In addition, the Spc42p layer was located on top of the cytoplasmic side of the nuclear envelope (Fig. 3E, ∗). Thus, expression levels of the Spc110p complex components contribute to the size regulation of the inner plaque.
The Outer Plaque Components Cnm67p and Spc94p/Nud1p Form a Complex with Spc42p.
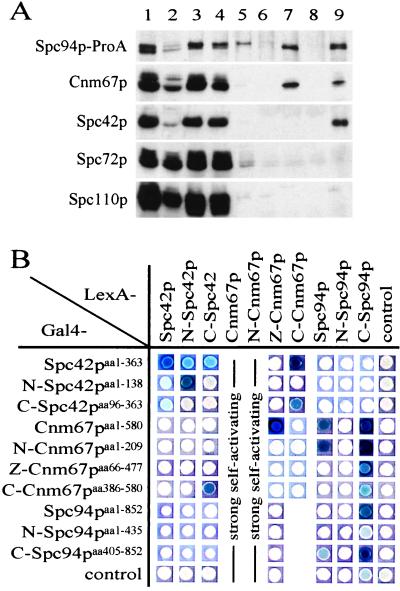
Cnm67p, a component of the cytoplasmic outer plaque (26), was identified as a minor component of the Spc110p complex (Fig. 1B). We investigated how Cnm67p becomes assembled into the Spc110p complex. Because Cnm67p has been localized close to Spc42p and Spc94p/Nud1p within the SPB (26), coimmunoprecipitation of these three proteins was tested. An extract of yeast cells with a functional SPC94–ProA gene fusion was separated into a 40,000 × g supernatant and an SPB-containing pellet. Spc94p–ProA in the supernatant was precipitated with IgG Sepharose, resulting in the coimmunoprecipitation of Cnm67p but not of Spc42p (Fig. 4A, lane 7). Thus, the Spc94p–Cnm67p complexes may form in the cytoplasm before integration into the SPB.
Figure 4.
Spc94p/Nud1p, Cnm67p, and Spc42p are present in common complexes. (A) A lysate of cells expressing SPC94–ProA (lane 1) was separated by centrifuging (40,000 × g, 30 min, 4°C) into a cytoplasmic extract (lane 2) and a SPB-containing pellet (lane 3). This pellet was extracted with 1 M NaCl/1% Triton X-100/50 mM Tris⋅HCl (pH 7.5) followed by centrifuging, resulting in a supernatant (lane 4) and a pellet (lane 5). Both extracts (lanes 2 and 4) were incubated with IgG Sepharose. The bound proteins were eluted (lanes 7 and 9). Lanes 6 and 8 are controls of wild-type cells that were treated identically to lanes 7 and 9, respectively. Fractions were tested by immunoblotting using antibodies directed against the indicated proteins. (B) Two-hybrid interactions between Spc42p, Cnm67p, and Spc94p/Nud1p. Experiment was performed as described in Fig. 2B legend.
The SPB-containing pellet was extracted with a high-salt buffer to fragment the SPBs, followed by the immunoprecipitation of Spc94p–ProA. Cnm67p and Spc42p, but not Spc110p or Spc72p, were coimmunoprecipitated (Fig. 4A, lane 9). Taken together, these results suggest that a complex containing Spc94p/Nud1p and Cnm67p binds to Spc42p within the SPB. In addition, Spc94p/Nud1p may interact with Spc42p via Cnm67p. Further support for the latter conclusion came from two-hybrid interactions between C–Spc42p and C–Cnm67p and N–Cnm67p with C–Spc94p (Fig. 4B). These proteins may interact similarly within the SPB.
Conditional Lethal Mutants of SPC29 Are Defective in SPB Duplication.
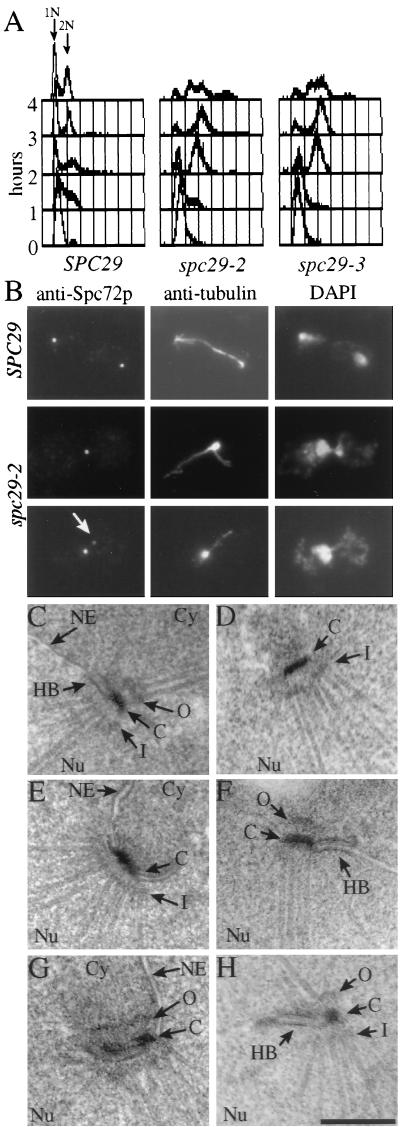
Our results suggest that the interface between the cytoplasmically assembled outer/central plaque and the nuclear inner plaque is formed among Spc29p, N–Spc42p, and C–Spc110p. This interface must form during SPB duplication, raising the possibility that spc29(ts) mutants have a defect in SPB duplication. This possibility was investigated by shifting α-factor-synchronized wild-type and spc29-2 and spc29-3 mutant cells to 37°C, the restrictive temperature of the mutants. Whereas the SPC29 wild-type cells progressed normally, spc29(ts) mutants arrested in the first cell cycle after the temperature shift with a 2n DNA content and a large bud (Fig. 5A). This arrest was only transient, because DNA contents larger than 2n were observed at later time points.
Figure 5.
spc29(ts) cells are defective in SPB duplication. Wild-type cells and cells of spc29-2 and spc29-3 were incubated for 2.5 hours at 23°C with 0.01 mg/ml α-factor. α-Factor was removed by washing with prewarmed (37°C) medium to release the cell-cycle block. Cells were incubated for 3 hours (B–H) or for up to 4 hours (A) at 37°C. (A) The DNA content of cells of SPC29, spc29-2, and spc29-3 was determined by fluorescence-activated cell sorting analysis (FACS) before release or 1, 2, 3 and 4 hours after release of the cell-cycle block. (B) Microtubules and the outer plaque protein Spc72p of spc29-2 and SPC29 cells were visualized by using indirect immunofluorescence. DNA was stained with DAPI. Wild-type cells (C) and cells of spc29-2 (D and E) and of spc29-3 (F–H) were prepared for electron microscopy followed by serial sectioning. Abbreviations are as in Fig. 3. (Bar = 0.25 μm.). C–H are all of same magnification.
In large-budded wild-type cells, two separated SPBs interconnected by one parallel bundle of microtubules that had segregated the chromosomes were observed (Fig. 5B, SPC29). In contrast, the microtubules of large-budded cells of spc29-2 originated from a single focus (about 95%, n = 150; Fig. 5B, Center Middle) and the duplicated chromosomes were not separated (Fig. 5B, Right). SPB staining by using antibodies against the outer plaque component Spc72p (11) identified only one SPB signal in 70 ± 10% of the large-budded spc29-2 cells, indicating that most spc29-2 cells have an SPB duplication or separation defect. In most of the cells with two Spc72p dots (30 ± 10%), one signal was clearly less intense, and this weaker signal was not associated with microtubules (Fig. 5B, arrow). A similar result regarding SPB number and signal intensity was obtained when the SPB of spc29-2 cells was marked with the cytoplasmic Spc42p–GFP or Cnm67p–GFP, whereas with the nuclear Spc110p–GFP, only one SPB signal was observed in ≈95% of the cells (data not shown). spc29-3 mutant cells behaved similarly (data not shown). Taken together, the immunofluorescence data suggests that spc29(ts) mutant cells are defective in either SPB duplication or separation of side-by-side SPBs.
To understand the SPB defect of cells of spc29-2 and spc29-3, serial sections through 15 spc29(ts) nuclei were investigated. These revealed only one SPB structure (Fig. 5 D–H), despite the occurrence of two outer plaque signals in 30% of the cells. Either the outer plaque components do not form a defined structure recognizable by electron microscopy or the outer plaque detaches from the nuclear envelope and is then located in the densely packed cytoplasm. A closer inspection of SPBs from spc29(ts) cells revealed various SPB duplication defects. SPBs of spc29-2 (Fig. 5 D and E) or spc29-3 cells (Fig. 5 F–H) were occasionally associated with an elongated half bridge (compare Fig. 5 F and H with Fig. 5C, wild-type) that extended in one case (Fig. 5H) into a central plaque-like structure. In addition, two apparently fused SPBs were recognizable in some spc29(ts) cells containing either two inner plaque like structures (Fig. 5D) or two outer, central, and inner plaques (Fig. 5G). Taken together, cells of spc29(ts) are defective in SPB duplication.
The SPB duplication defect of spc29(ts) cells led us to test whether SPC29 genetically interacts with genes involved in SPB duplication. High dosage of CDC31 suppressed the growth defect of spc29-2 cells, but not of spc29-3 or spc29-9 cells. In contrast, a single additional copy of KAR1 increased the temperature sensitivity of spc29-2 and spc29-3 but not of spc29-9 cells (Table 1).
DISCUSSION
We have identified a subcomplex from salt-extracted SPBs containing mainly Spc110p, Cmd1p, Spc29p, and Spc42p (Fig. 6). The simple fact that these four proteins are present in one complex suggests that at least some of the components interact directly. The C terminus of Spc110p has been shown to be located near the layer of Spc42p (2, 27, 28), suggesting that either C–Spc110p interacts directly with Spc42p or that it interacts via an additional protein. Because C–Spc110p899–944 containing the Cmd1p-binding site can be deleted without consequence (15), it is unlikely that this domain of Spc110p is directly involved in anchoring Spc110p to the Spc42p layer. By using the two-hybrid system, Spc29p was shown to bind to C–Spc110p811–898 next to the Cmd1p-binding site of Spc110p and to N–Spc42p. These data, together with multiple genetic interactions, suggest that Spc29p interconnects Spc42p with Spc110p (Fig. 6). In support of our conclusion, the observation that the region of Spc110p next to the Cmd1p binding-site, most likely the domain that binds Spc29p, has been implicated to function in the stable attachment of Spc110p to the central plaque (24).
The observations that mutations in Spc110p that reduce Cmd1p binding are lethal or conditional lethal for yeast cells and the deletion of the entire Cmd1p-binding site is not lethal and suppressed the cmd1-1 mutation led to the suggestion that the Cmd1p-binding site of Spc110p has an inhibitory function that is either relieved by its deletion or by Cmd1p binding (15, 17). Our results are consistent with this model and raise the possibility that Spc29p binding to Spc110p is regulated by Cmd1p. It is likely that Cmd1p binds first to Spc110p to relieve the inhibitory role of the C terminus of Spc110p. Whether Cmd1p binding to Spc110p is regulated by Ca2+ is unclear, mainly because Cmd1p’s essential functions in yeast have been reported to be Ca2+-independent (29), whereas in vitro binding of Cmd1p to Spc110p was found to be Ca2+-dependent (7). In any case, the ordered assembly of the Spc110p/Cmd1p/Spc29p-containing complex may be important before or during SPB duplication.
Diploid cells have SPBs that are twice the size of SPBs in haploid cells (3), and the size of the SPB correlates with the number of nuclear microtubules organized. Cooverexpression of components of the Spc110p complex increased the diameter of the inner plaque 2- to 3-fold, suggesting that the expression levels of some of the components of the Spc110p complex contribute to the size regulation of the inner plaque.
We have provided evidence that the cytoplasmic Cnm67p and Spc94p/Nud1 (26) form a complex in the cytoplasm that then binds to the cytoplasmically assembled Spc42p layer via the interaction of Cnm67p with Spc42p (Fig. 6). In contrast, Spc110p (10) and overexpressed Spc29p were identified as nuclear proteins. The distinct subcellular localization of Spc29, Spc42p, and Spc110p, together with their interaction, suggests that Spc29p is part of a critical border of cytoplasmic and nuclear assembled SPB subcomplexes that forms during SPB duplication. This model is further supported by the SPB duplication defect of spc29(ts) cells and by the genetic interactions of SPC29 with CDC31 and KAR1, two genes involved in SPB duplication (4, 6).
Acknowledgments
We thank M. Winey for strain Wx193-7b, J. Kilmartin for plasmids carrying spc42-9, spc42-10, and spc110-2, and M. Stark for spc110 mutants. We are very grateful to the laboratory of P. Silver for plasmids containing NIP29/SPC29. This work was carried out with support from the Cancer Research Campaign.
ABBREVIATIONS
- Cmd1p
yeast calmodulin
- SPB
spindle pole body
- GFP
green fluorescent protein
- ts
temperature sensitive
References
- 1.Winey M, Byers B. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- 2.Bullitt E, Rout M P, Kilmartin J V, Akey C W. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- 3.Byers B, Goetsch L. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum P, Furlong C, Byers B. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose M D, Fink G R. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- 7.Spang A, Courtney I, Grein K, Matzner M, Schiebel E. J Cell Biol. 1995;128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winey M, Goetsch L, Baum P, Byers B. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winey M, Hoyt M A, Chan C, Goetsch L, Botstein D, Byers B. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilmartin J V, Goh P Y. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- 11.Knop M, Schiebel E. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knop M, Pereira G, Geissler S, Grein K, Schiebel E. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhlrad D, Hunter R, Parker R. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 14.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiser J R, Sundberg H A, Chang B H, Muller E G, Davis T N. Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson D, Kilmartin V. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stirling D A, Welch K A, Stark M J. EMBO J. 1994;13:4329–4342. doi: 10.1002/j.1460-2075.1994.tb06753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissler S, Pereira G, Spang A, Knop M, Souès S, Kilmartin J, Schiebel E. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- 19.Spang A, Geissler S, Grein K, Schiebel E. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knop M, Schiebel E. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Byers B, Goetsch L. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- 23.Hutter K J, Eipel H E. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- 24.Sundberg H A, Davis T A. Mol Biol Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A, Jensen O N, Podtelejnikov A V, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigge P A, Jensen O N, Holmes S, Souès S, Mann M, Kilmartin J V. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spang A, Grein K, Schiebel E. J Cell Sci. 1996;109:2229–2237. doi: 10.1242/jcs.109.9.2229. [DOI] [PubMed] [Google Scholar]
- 28.Sundberg H A, Goetsch L, Byers B, Davis T N. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiser J R, Tuinen v D, Brockerhoff S E, Neff M M, Davis T N. Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]