Abstract
Through the reductive divisions of meiosis, sexually reproducing organisms have gained the ability to produce specialized haploid cells called germ cells that fuse to establish the diploid genome of the resulting progeny. The totipotent nature of these germ cells is highlighted by their ability to provide a single fertilized egg cell with all the genetic information necessary to develop the complete repertoire of cell types of the future organism. Thus, the production of these germ cells must be tightly regulated to ensure the continued success of the germ line in future generations. One surprising germ cell development mechanism utilizes variation of the global transcriptional machinery, such as TFIID and TFIIA. Like histone variation, general transcription factor variation serves to produce gonadal-restricted or -enriched expression of selective transcriptional regulatory factors required for establishing and/or maintaining the germ line of diverse organisms. This strategy is observed among invertebrates and vertebrates, and perhaps plants, suggesting that a common theme in germ cell evolution is the diversification of selective promoter initiation factors to regulate critical gonadal-specific programs of gene expression required for sexual reproduction. This review discusses the identification and characterization of a subset of these specialized general transcription factors in diverse organisms that share a common goal of germ line regulation through transcriptional control at its most fundamental level.
Keywords: Transcription, TFIID, TFIIA, Oogenesis, Spermatogenesis, Meiosis
1. Introduction
The precise control of gene expression is a highly coordinated process orchestrated by the integration of numerous cellular and molecular events. Targets of many signaling pathways are found in the nucleus in the form of regulatory elements coded in the genome and their associated transcription factors. Thus, gene regulation is achieved by an abundant and diverse array of protein factors generically referred to here as the chromatin and transcription machinery. The chromatin and transcription machinery is a large conglomeration of individual proteins and multi-protein complexes that collectively determine how active or repressed specific genes are in a given cell [1, 2]. Multiple components of this complex machinery must adeptly maneuver these regulatory DNA sequences in the context of chromatin, where they are often wrapped around nucleosomes (Figure 1). The enzyme responsible for messenger RNA synthesis of the protein encoding genes of eukaryotic organisms is the multi-subunit enzyme RNA polymerase II (Pol II). A large battery of Pol II accessory proteins termed general transcription factors (GTFs) were originally isolated as factors that aid in the recruitment of Pol II to target promoters that are required for basal transcription. A subset of GTFs, includes subunits of transcription factor (TF) IIA, TFIIB and TFIID (referred to as the TFII-DAB complex), that are core promoter recognition factors that associate with DNA sequences encompassing the start site of Pol II transcription (Figure 2A). Rather than directly associating with Pol II, transcriptional activator proteins utilize the GTFs to modulate the rate of Pol II recruitment and transcriptional initiation [3, 4].
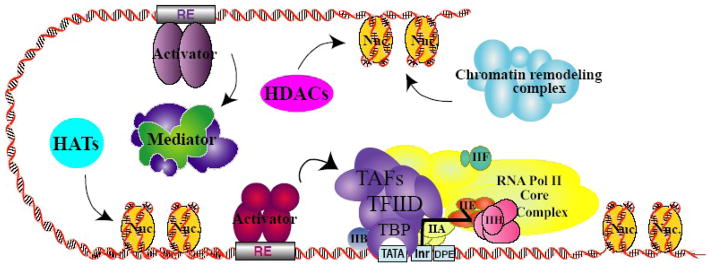
Figure 1.
Integration of multiple regulatory steps in transcription initiation by RNA polymerase II (Pol II). Schematic representation of the proteins required for chromatin-dependent initiation of Pol II transcription. Numerous protein-DNA and protein-protein interactions are depicted by individual factors and multi-protein complexes recruited to regulatory DNA sequences. Transcription initiation by Pol II is dependent on the combinatorial efforts of multiple factors and multi-subunit complexes around the start site of transcription. TBP, TATA-box binding protein; TAFs, TBP-associated factors; Nuc., nucleosomes; HATs, histone acetyltransferases; HDACs, histone deacetylases; RE, recognition element; TATA, TATA-box, Inr, initiator; DPE, downstream promoter element.
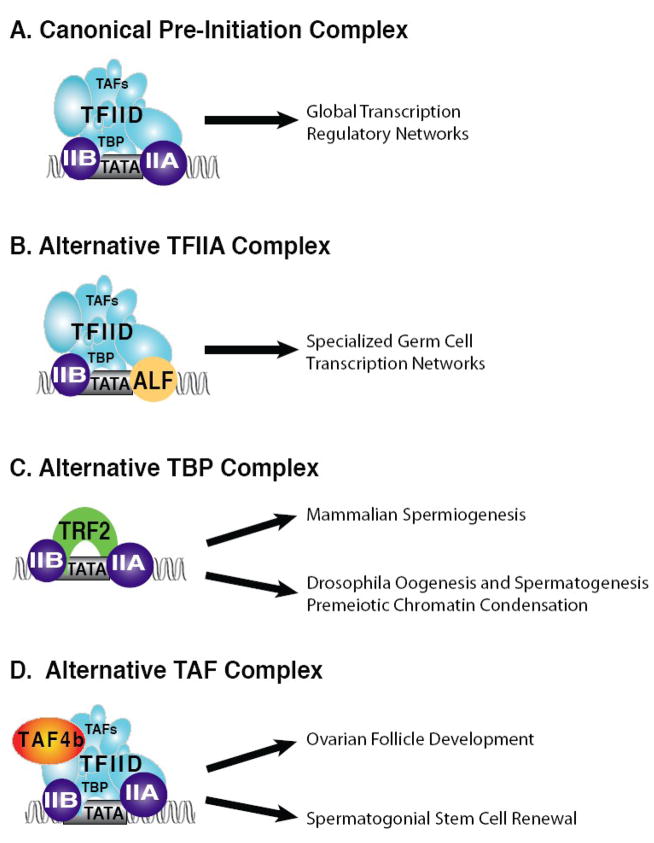
Figure 2.
Alternative core promoter recognition factors in germ cell development. Assembly of the TFII-DAB complex is critical for RNA polymerase II pre-initiation complex formation. A) The canonical RNA polymerase II pre-initiation complex is well conserved from yeast to humans and is involved in regulating global transcription programs. B) ALF is a germ cell-specific TFIIA paralog that regulates germ cell-specific programs of gene expression. C) TRF2 is a TBP paralog that has functions in meiotic chromosome condensation and spermatid differentiation. D) TAF4b is a gonadal-enriched TAF4 paralog that is essential for ovarian follicle development and the maintenance of spermatogenesis in mice.
The TFIID complex is a critical player in the activator-dependent recruitment of Pol II to specific genes in the genome [5–8]. TFIID is a large multi-protein complex containing the TATA-box binding protein (TBP) and approximately 14 TBP-associated factors (TAFs). Biochemical studies have demonstrated roles for TFIID in core promoter recognition and activator-dependent Pol II recruitment [5, 6, 9–12]. The overall architecture of the TFIID complex is largely conserved from yeast to humans, suggesting that it plays a fundamental role in the regulation of eukaryotic gene expression [9, 13, 14]. The broadly expressed transcriptional activator proteins Sp1 and CREB were shown to specifically target the TAF4 (formerly called TAFII110 and TAFII130) subunits of the TFIID complex in Drosophila and mammals, respectively [15–18]. Such selective protein-protein interactions between activators and coactivators are thought to help modulate Pol II recruitment to certain promoters in response to specific activators. TFIID binding to the core promoter is also modulated by a second critical GTF named TFIIA, which together with TFIID nucleates the formation of a stable pre-initiation complex at the start site of transcription [3, 4]. This pre-initiation complex is largely functionally and mechanistically conserved between yeast and mammals highlighting the global role of GTFs in the control of gene transcription across eukaryotes [19, 20].
2. TFIID variation in metazoans
The first example of a GTF paralog was described by Tjian and colleagues in 1993 [21], when they reported an alternative form of Drosophila TBP called TBP-related factor 1 (TRF1). In that study, TRF1 was shown to be an independent TBP-related gene that is expressed in a unique pattern compared to the canonical TBP suggesting that it could play selective rather than general roles in regulating gene-specific programs of transcription [21]. Subsequent biochemical studies provided critical evidence that fly TRF1 was involved for the expression of RNA polymerase III (Pol III) transcripts encoding multiple transfer RNA (tRNA) genes indicative of promoter-selective modes of regulation by TRF1 [22, 23]. In addition to Pol III genes, TRF1 has also been shown to function at selective Pol II promoters, both in vitro and in vivo, suggesting diverse promoter selective functions of TRF1 in the regulation of transcription [24]. Unlike many of the other GTF variants discussed below, TRF1 is apparently restricted to Drosophila genomes and there does not seem to be a vertebrate ortholog to perform similar functions. While these studies do not indicate roles for TRF1 in germ cell development, they illuminated the notion that alternative forms of GTFs could act selectively in regulating transcription and revealed the tip of the iceberg of numerous variant GTFs that evolved to regulate diverse aspects of embryogenesis and germline determination.
3. Expression and Function of TFIIA and TFIID variants in the vertebrate germ line
The connection between variant GTFs and germline regulation involved the discovery of exclusive or enriched expression of GTF paralogs in the gonads of both invertebrate and vertebrate organisms. This gonadal expression pattern was first evident in the testis-specific expression of an alternative form of human TFIIA named TFIIAalpha/beta-like factor (ALF; also called TFIIAtau; Figure 2B) [25–27]. TFIIA is a trimeric protein complex of processed TFIIAalpha and TFIIAbeta subunits encoded within a single polypeptide and an independently encoded TFIIAgamma subunit. Initial characterization of ALF suggested that it could functionally substitute for the canonical TFIIAalpha/beta by binding to TFIIAgamma and TBP and stabilizing TFIIA-TBP interactions on DNA [25–27]. However, what was unique upon ALF’s discovery was that it is exclusively expressed in germ cells and can replace the canonical TFIIAalpha/beta during gametogenesis in mammals and frogs [28, 29]. In fact, a recent comparison of the gonadal expression patterns of multiple GTFs, including TFIIA and TFIID, in frogs and mice has illuminated greater than 10 paralogous GTFs which are exclusively expressed in the gonadal tissues across distant vertebrate organisms [30]. Such an extensive set of gonadal-specific GTF paralogs reveals that in addition to performing their global transcription functions which are well conserved from yeast to mammalian cells, that selective variants of these factors have adopted key roles in executing gonadal-specific gene regulatory events that underlie sexual reproduction.
In addition to TFIIA, a number of paralogous TFIID subunits have been identified with exclusive or enriched expression in mouse gonads and germ cells. The illumination of the developmental functions of a number of these factors has come from reverse genetic studies in the mouse where phenotypic inspection of knockout mice revealed striking defects in gonadal development and fertility [31–33]. For example, disruption of TBP-related factor 2 (also called TLF) revealed a specific block in spermiogenesis, a late stage of sperm maturation that occurs in the semiferous tubules of the mammalian testis [32, 33]. These initial studies implicated TRF2 in the expression of RNA polymerase II-dependent and testis-specific mRNAs required for the completion of spermiogenesis (Figure 2C). Recent evidence suggests TRF2 may play a pivotal role in the chromatin compaction that is required for successful execution of the spermatocyte-spermatid transition where the male germ line becomes progressively more compacted by replacement histones called transition proteins and protamines [34, 35].
While most of the studies on variant GTFs in germ cell development focus on the development of the male germline, the function of the mammalian ovary is equally dependent on similar variation within TFIID. TAF4b (formerly TAFII105) was identified as a sub-stoichiometric TAF4-related paralog subunit of TFIID complexes derived from cultured B lymphocytes [36]. Mouse knockout studies revealed an essential function of TAF4b in ovarian follicle development and female fertility (Figure 2D) [31]. Within the mouse ovary, TAF4b is expressed within both somatic granulosa cells required to nurture the developing egg and the oocyte itself [31]. Recent studies have demonstrated that TAF4b is required for integrating transcriptional activation of multiple ovarian granulosa cell target genes that are required for granulosa cell proliferation in response to diverse hormonal stimulation [37–39]. In addition, TAF4b has also been shown to be required for multiple aspects of oocyte development including the establishment of the primordial follicle pool and oocyte maturation [39, 40]. Interestingly, TAF4b is also required for the proper maintenance of spermtogenesis in the mammalian testis [41]. Again, TAF4b expression is detected in the somatic cells of the testis, including Sertoli and Leydig cells, and in multiple germ cell populations [41]. Gene expression profiling of the juvenile TAF4b-null testis suggests that TAF4b regulates a subset of genes required for spematogonial stem cell (SSC) maintenance and a recent study has revealed the expression of TAF4b in long-term cultured SSCs (Figure 2D) [41, 42]. Strikingly, the paradigm of global TAF4 expression and gonadal TAF4b expression is conserved in Xenopus where TAF4 is broadly expressed and a TAF4-related paralog (xlTAF4b) is exclusively expressed in the frog ovary and testis [30]. The diversification of TAF subunits in reproduction is not limited to the TAF4 family but includes paralogs of many other canonical TAFs that are selectively expressed in the gonads [30, 43]. The extensive array of paralogous TAFs that are conserved between distant vertebrate species suggests that diversification of the transcriptional machinery is a common mode of germ cell regulation across vertebrate evolution.
4. TFIID variation and chromatin structure
One possible mechanism associated with variant TFIID complexes is their unique impact on chromatin structure and function. While molecular genetic studies in diverse vertebrates illuminated the critical roles of variant GTFs in reproduction, are GTF variants conserved amongst invertebrates, are such variants specifically expressed in the gonads and do they inform us about the potential regulation of chromatin structure? The answer to all three questions is yes [44]. TFIID variation in particular is highly extensive in Drosophila and many of these variants are critical for spermatogenesis. Much of this work is largely the result of unbiased forward genetic screens in Drosophila by Fuller and colleagues that identified novel regulators of male meiosis and spermatid differentiation [45]. These screens yielded a number of genes that are expressed in exquisitely specific patterns in the testis and whose protein products are required for meiotic progression and spermatid differentiation. Strikingly, many of these genes encode paralogs of canonical Drosophila TAFs including no hitter (nht; dTAF4 paralog), cannonball (can; dTAF5 paralog), meiosis I arrest (mia; dTAF6 paralog), spermatocyte arrest (sa; dTAF8 paralog) and ryan express (rye; dTAF12 paralog) [46, 47]. These testis-specific TAFs (tTAFs) form a stable alternative TFIID complex that collectively function to regulate transcription of genes required for meiotic progression and spermatid differentiation similar to its vertebrate counterparts TRF2 and TAF4b [45]. Surprisingly however, recent analysis of tTAFs in target gene activation has revealed a novel function of the tTAFs in localization of the polycomb repressive complex 1 (PRC1) to the nucleolus [48]. PRC1 is a sequence-specific multi-protein chromatin regulatory complex that is most often associated with heterochromatin and gene silencing. The nucleolar sequestration of PRC1 by this alternative TAF complex allows for transcriptional activation, rather than repression, of spermatocyte differentiation genes and may be a critical switch in promoting terminal spermatid differentiation (Figure 3). A recent study has also revealed the nucleolar colocalization of a spliced variant of TAF1 and the tTAFs suggesting that alternative TFIID complexes may be localized to the nucleolus in Drosophila. Thus both invertebrates and vertebrates have evolved specialized GTFs subunits that are essential for diverse transcriptional programs associated with gametogenesis that may involve the regulation of chromatin structure and function.
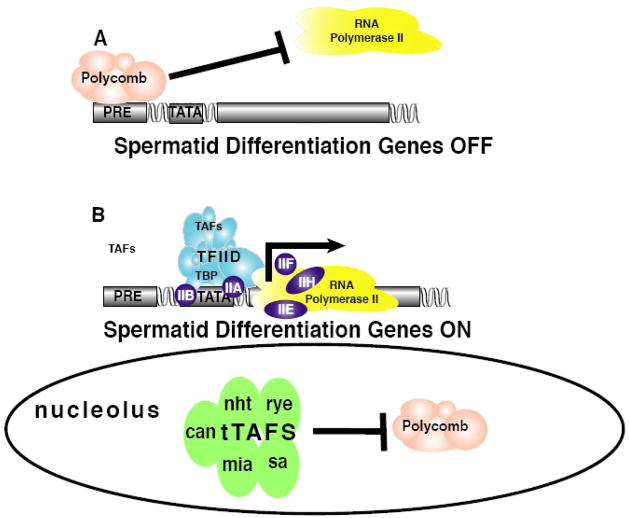
Figure 3.
Proposed mechanism of an alternative testis-specific TAF (tTAF) complex in Drosophila [48]. A) In non-spermatocytes, where tTAFs are not expressed, the polycomb repressive complex 1 (PRC1) is bound to the promoters of spermatid differentiation genes keeping them in a repressed state. B) In spermatocytes, where tTAFs are expressed, PRC1 is sequestered by the tTAFs in the nucleolus whereby it is unable to repress transcription. In the absence of polycomb repression, trithorax-dependent transcriptional activation occurs at the spermatid differentiation genes. Thus, the spermatocyte-to-spermatid transition in Drosophila is regulated by a highly elaborate transcriptional mechanism involving a testis-specific alternative form of TFIID.
The paradigm of increasing regulatory diversity by evolving selective variants is not unique to GTFs, but has been observed in the regulation of chromatin structure and transcription in the form of histone variation. Alternative forms of histones have been identified across evolution and genetic and biochemical studies have demonstrated unique structural and functional properties of specific histone variants [49]. Moreover, germ cells are known to contain specialized chromatin states that arise from a large number of germ cell-specific histone variants and modifying enzymes that selectively function in spermatogenesis and oogenesis [50, 51]. The Drosophila ortholog of TRF2 (dTRF2) has been shown to associate with a large multiprotein complex containing components of the chromatin remodeling complex NURF [52]. A recent study has revealed distinct dTRF2 expression patterns and functions in Drosophila ovaries and testes suggesting that by recruiting specific chromatin remodeling complexes, dTRF2 may help establish specific chromatin structures required for proper germ cell differentiation [53]. Together these studies reveal that in addition to promoter selective initiation, TRF2 may play diverse architectural roles in stabilizing specific chromatin states required for the successful completion of gametogenesis from flies to mammals (Figure 2C).
As TRF2 has been recently implicated in chromatin regulation in flies and in mice, GTF variants may collaborate with other specialized germ cell transcriptional regulators, such as specific histone variants, to coordinate transcriptional activity with specialized chromatin states [34, 53]. In fact, the parallel between histone variants and TAF variants is even more direct given the fact that many TAFs contain histone fold structural domains that are essential for stability of the TFIID complex [54, 55]. Four of the five tTAFs contain histone fold domains [46]. Moreover, nht is a dTAF4 paralog that is more similar in structure to the yeast TAF4 ortholog that lacks the metazoan coactivator domain, than the canonical dTAF4 that contains this activator interaction surface [14, 46]. Perhaps, in addition to functioning as transcriptional coactivator complexes, variant TAF-containing complexes may impart some unique structural identity to chromatin states, like variant histones, that are essential for germ cell differentiation. Future studies elucidating the molecular mechanisms associated with these variant GTFs is thus required to illuminate how such specialized programs of germ cell differentiation are accomplished.
5. Multiple mechanisms for generating general transcription factor diversity
How do we get from the global building blocks of the eukaryotic pre-initiation complex to the specific regulation of germ cell development across diverse multi-cellular organisms? The answer seems to be largely dependent on the duplication, alternative splicing and rapid evolution of paralogous TFIID and TFIIA genes. Such genetic events have yielded related protein products that are used in diverse ways to exquisitely regulate global gene expression patterns as well as those required for sexual reproduction. However, it remains to be determined which of the paralogous genes arose first, those with somatic functions or those with germ line functions. Gene duplication is only one of multiple mechanisms that can give rise to alternative TAFs that function in sexual reproduction. In humans, TAF1 (formerly TAFII250) is the largest subunit of the TFIID complex and is encoded by a gene located on the X chromosome. During male meiosis, the X and Y chromosomes pair and are silenced by post-meiotic sex heterochromatin [56]. As autosomes are transcriptionally active during male meiosis, a testis-expressed intronless version of TAF1 called TAF1L was identified on human chromosome 9 and shown to bind to TBP and rescue a TAF1-deficient cell line [57]. This retrotransposition event occurred recently during evolution as TAF1L is only expressed in a subset of primates including humans. Thus, retrotransposition of a canonical TAF gene has lead to the gonadal-specific expression of paralogous gene in the testis. Given the high degree of conservation in their respective coding regions, TAF1L is predicted to execute similar promoter selective functions as does the broadly expressed TAF1 paralog in spermatocytes.
In addition to gene duplication, alternative splicing of dTAF1 has been shown to provide multiple isoforms of dTAF1 in Drosophila. One specific isoform of dTAF1, dTAF1-2, exhibits enriched expression in the testis. Recent evidence has revealed the localization of dTAF1 in spermatocyte nucleoli, where tTAFs are localized suggesting the testis-expressed dTAF1-2 isoform of dTAF1 might function together with the tTAFs to help elicit testis-specific programs of gene expression [58]. Thus, while multiple mechanisms are utilized to express gonad-specific variants of TFIID, either by gene duplication in the case of the tTAFs or alternative splicing in the case of dTAF1-2, each may ultimately work together to regulate the unique gene expression patterns required for sexual reproduction. These mechanisms, in combination with retrotransposition, present evidence that evolution has taken advantage of multiple mechanisms to diversify aspects of the gloabal transcription machinery to function selectively in the reproductive success of multicellular organisms. Diversification of TAFs is observed in plants where duplicate forms of numerous Arabidopsis TAFs (atTAFs) have been identified that may have arisen by genome duplication [59]. Moreover, where examined functionally for atTAF1 and atTAF6, specific paralogs play non-redundant developmental functions in plants as they do in animals, including integrating light signals and mediating pollen tube growth, a male germline function [60, 61]. Thus, the ability of paralogous TAFs to function both in global transcription programs and specialized reproductive capacities may be a widespread mechanism for increasing the repertoire of gene transcription programs required for sexual reproduction in the context of a finite genome [62].
6. Future considerations
Many questions regarding the evolution and function of diversified basal transcription factors still remain. For example, why is GTF variation so extensive in the germ line and are there somatic tissues that also need to express specific gene programs that use similar mechanisms? In fact, there are now multiple examples of GTF variants that have critical functions in regulating differentiation programs outside of the germline. A prime example is the recent characterization of the mechanism by which the mammalian TBP paralog, TRF3, functions in muscle differentiation. These studies have revealed an eloquent switch from the core TFIID and mediator complexes in proliferating myoblasts, to a very simple but variant core TRF3/TAF3 complex required in differentiated myotubes [63, 64]. Thus, to execute muscle differentiation, mammals are poised to execute wholesale remodeling of the global transcription machinery to specialized forms of this machinery required for the expression of myogenin, a master regulator of the muscle differentiation program [63, 64]. In addition, TRF3 was also shown to be required for mesoderm development and hematopoiesis during zebrafish embryogenesis [65]. These studies suggest that GTF variation may have diverse effects on differentiation programs both within and outside of the gonads. Thus, contrasting the diverse functions associated with multiple GTF variants is critical to dissect transcriptional mechanisms of germ and somatic cell diversification that is fundamental to metazoan development.
Where does the study of gonadal-selective GTF variants lead us in the future? There are at least three equally critical areas of study that will be greatly enhanced by the continued study of developmental and mechanistic functions played by paralogous GTFs in sexual reproduction. First, further detailed studies are required to identify additional players and their precise modes of interactions during the process of transcription initiation. While the biochemical study of transcription factors has been hugely helpful and successful in understanding the fundamental nature of this basal machinery, there is still only limited knowledge of how such factors work in an organ- and cell type-specific manner. These studies thus afford a window of opportunity to understand the fundamental basis of how multi-cellular organisms yield enormous cell type diversity given a limited number of regulatory factors. Second, future studies in this fertile area will uncover novel mechanisms of germ cell development. The process of gametogenesis involves coordinate regulation of mitosis, meiosis and germ cell differentiation. Deciphering regulatory events that underlie these factors will yield new mechanisms associated with stem cell regulation, asymmetric cell division and chromatin regulation of meiotic progression. Third, the understanding of new reproductive developmental players and mechanisms in diverse model systems will identify potential etiologies of reproductive disease states in humans such as infertility and cancer. Insight into how germ cells develop and mature in diverse model systems, from plants to humans, will collectively provide the framework for how the biology of sexual reproduction is normally achieved and illuminate new pathways and targets that may go awry during human reproductive health. While it was initially a surprise that gonadal-selective GTFs could contribute to germ line determination, numerous examples have now been identified across many distantly related organisms. Understanding precisely how these factors work collectively is a challenging but exciting area of current research in transcriptional regulatory mechanisms of cell differentiation and development.
Acknowledgments
I thank Lindsay Lovasco, Kimberly Seymour, Diana Donovan and Mark Johnson for critical comments on the manuscript and the reviewers for their helpful critiques and suggestions. My lab is supported by an NIH/NCRR COBRE Award #P20RR015578, an Ellison Medical Foundation Junior Scholar Award in Aging and the National Center of Excellence in Women’s Health at Brown University. This review is dedicated to the fond memory of a dear friend and colleague Ruby Bell.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 4.Reinberg D, Horikoshi M, Roeder RG. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987;262:3322–3330. [PubMed] [Google Scholar]
- 5.Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 6.Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 7.Hoey T, Weinzierl RO, Gill G, Chen JL, Dynlacht BD, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 8.Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 9.Moqtaderi Z, Yale JD, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci U S A. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 11.Tanese N, Saluja D, Vassallo MF, Chen JL, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci U S A. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 13.Sanders SL, Garbett KA, Weil PA. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol. 2002;22:6000–6013. doi: 10.1128/MCB.22.16.6000-6013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders SL, Weil PA. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 15.Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felinski EA, Quinn PG. The CREB constitutive activation domain interacts with TATA-binding protein-associated factor 110 (TAF110) through specific hydrophobic residues in one of the three subdomains required for both activation and TAF110 binding. J Biol Chem. 1999;274:11672–11678. doi: 10.1074/jbc.274.17.11672. [DOI] [PubMed] [Google Scholar]
- 18.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci U S A. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komarnitsky PB, Michel B, Buratowski S. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 1999;13:2484–2489. doi: 10.1101/gad.13.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auty R, Steen H, Myers LC, Persinger J, Bartholomew B, Gygi SP, Buratowski S. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J Biol Chem. 2004;279:49973–49981. doi: 10.1074/jbc.M409849200. [DOI] [PubMed] [Google Scholar]
- 21.Crowley TE, Hoey T, Liu JK, Jan YN, Jan LY, Tjian R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993;361:557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- 22.Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 23.Isogai Y, Takada S, Tjian R, Keles S. Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. Embo J. 2007;26:79–89. doi: 10.1038/sj.emboj.7601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes MC, Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- 25.Ozer J, Moore PA, Lieberman PM. A testis-specific transcription factor IIA (TFIIAtau) stimulates TATA-binding protein-DNA binding and transcription activation. J Biol Chem. 2000;275:122–128. doi: 10.1074/jbc.275.1.122. [DOI] [PubMed] [Google Scholar]
- 26.Upadhyaya AB, Khan M, Mou TC, Junker M, Gray DM, DeJong J. The germ cell-specific transcription factor ALF. Structural properties and stabilization of the TATA-binding protein (TBP)-DNA complex. J Biol Chem. 2002;277:34208–34216. doi: 10.1074/jbc.M204808200. [DOI] [PubMed] [Google Scholar]
- 27.Upadhyaya AB, Lee SH, DeJong J. Identification of a general transcription factor TFIIAalpha/beta homolog selectively expressed in testis. J Biol Chem. 1999;274:18040–18048. doi: 10.1074/jbc.274.25.18040. [DOI] [PubMed] [Google Scholar]
- 28.Han S, Xie W, Hammes SR, DeJong J. Expression of the germ cell-specific transcription factor ALF in Xenopus oocytes compensates for translational inactivation of the somatic factor TFIIA. J Biol Chem. 2003;278:45586–45593. doi: 10.1074/jbc.M302884200. [DOI] [PubMed] [Google Scholar]
- 29.Han SY, Zhou L, Upadhyaya A, Lee SH, Parker KL, DeJong J. TFIIAalpha/beta-like factor is encoded by a germ cell-specific gene whose expression is up-regulated with other general transcription factors during spermatogenesis in the mouse. Biol Reprod. 2001;64:507–517. doi: 10.1095/biolreprod64.2.507. [DOI] [PubMed] [Google Scholar]
- 30.Xiao L, Kim M, DeJong J. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns. 2006;6:409–419. doi: 10.1016/j.modgep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 32.Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 34.Brancorsini S, Davidson I, Sassone-Corsi P. TIPT, a male germ cell-specific partner of TRF2, is chromatin-associated and interacts with HP1. Cell Cycle. 2008;7:1415–1422. doi: 10.4161/cc.7.10.5835. [DOI] [PubMed] [Google Scholar]
- 35.Martianov I, Brancorsini S, Gansmuller A, Parvinen M, Davidson I, Sassone-Corsi P. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002;129:945–955. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- 36.Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 37.Geles KG, Freiman RN, Liu WL, Zheng S, Voronina E, Tjian R. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc Natl Acad Sci U S A. 2006;103:2594–2599. doi: 10.1073/pnas.0510764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E, Tjian R. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voronina E, Lovasco LA, Gyuris A, Baumgartner RA, Parlow AF, Freiman RN. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 2007;303:715–726. doi: 10.1016/j.ydbio.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falender AE, Shimada M, Lo YK, Richards JS. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol. 2005;288:405–419. doi: 10.1016/j.ydbio.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 41.Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, Shinkai Y, Oshimura M, Ishino F, Ogura A, Shinohara T. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyagi N, Wassarman DA. Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J Cell Biol. 2000;150:F45–50. doi: 10.1083/jcb.150.2.f45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- 46.Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- 47.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 49.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 50.Bromfield J, Messamore W, Albertini DF. Epigenetic regulation during mammalian oogenesis. Reprod Fertil Dev. 2008;20:74–80. doi: 10.1071/rd07181. [DOI] [PubMed] [Google Scholar]
- 51.Khalil AM, Wahlestedt C. Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics. 2008;3:21–28. doi: 10.4161/epi.3.1.5555. [DOI] [PubMed] [Google Scholar]
- 52.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 53.Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, Korochkin LI, Tora L, Georgiev PG, Georgieva SG. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann A, Chiang CM, Oelgeschlager T, Xie X, Burley SK, Nakatani Y, Roeder RG. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 55.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 57.Wang PJ, Page DC. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet. 2002;11:2341–2346. doi: 10.1093/hmg/11.19.2341. [DOI] [PubMed] [Google Scholar]
- 58.Metcalf CE, Wassarman DA. Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev Dyn. 2007;236:2836–2843. doi: 10.1002/dvdy.21294. [DOI] [PubMed] [Google Scholar]
- 59.Lago C, Clerici E, Mizzi L, Colombo L, Kater MM. TBP-associated factors in Arabidopsis. Gene. 2004;342:231–241. doi: 10.1016/j.gene.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 60.Bertrand C, Benhamed M, Li YF, Ayadi M, Lemonnier G, Renou JP, Delarue M, Zhou DX. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J Biol Chem. 2005;280:1465–1473. doi: 10.1074/jbc.M409000200. [DOI] [PubMed] [Google Scholar]
- 61.Lago C, Clerici E, Dreni L, Horlow C, Caporali E, Colombo L, Kater MM. The Arabidopsis TFIID factor AtTAF6 controls pollen tube growth. Dev Biol. 2005;285:91–100. doi: 10.1016/j.ydbio.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 63.Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]