Abstract
The phylogenetic distribution of the components comprising the transcriptional machinery in the crenarchaeal and euryarchaeal lineages of the Archaea was analyzed in a systematic manner by genome-wide profiling of transcription complements in fifteen complete archaeal genome sequences. Initially, a reference set of transcription-associated proteins (TAPs) consisting of sequences functioning in all aspects of the transcriptional process, and originating from the three domains of life, was used to query the genomes. TAP-families were detected by sequence clustering of the TAPs and their archaeal homologues, and through extensive database searching, these families were assigned a function. The phylogenetic origins of archaeal genes matching hidden Markov model profiles of protein domains associated with transcription, and those encoding the TAP-homologues, showed there is extensive lineage-specificity of proteins that function as regulators of transcription: most of these sequences are present solely in the Euryarchaeota, with nearly all of them homologous to bacterial DNA-binding proteins. Strikingly, the hidden Markov model profile searches revealed that archaeal chromatin and histone-modifying enzymes also display extensive taxon-restrictedness, both across and within the two phyla.
Keywords: genome profiling, protein families, sequence clustering, transcription-associated proteins
Introduction
Transcription, a core gene expression process, involves different agents participating in initiation, elongation, termination and regulation. The basic principles of transcriptional regulation in the Bacteria and Eukaryota have been outlined (Ptashne and Gann 1997), contrasting with the Archaea, where such mechanisms are less well-understood. The study of the archaeal transcriptional machinery is important to an understanding of both the molecular mechanisms, and the evolutionary history, of transcriptional regulation in all three domains of life. Furthermore, these analyses may reveal how archaea respond to environmental challenges, in particular, given their possible association with various aspects of human disease (Eckburg et al. 2003). Several previous studies have shown that the RNA polymerase core enzyme exhibits structural similarity between the Archaea and the Eukaryota (Puhler et al. 1989). Moreover, the minimal set of factors required for in vitro transcription initiation in archaea consists of TATA-box binding protein (TBP), TFIIB and RNA polymerase II (Werner and Weinzierl 2002). In bacteria, however, the process appears to be fundamentally different (Struhl 1999), with regulation accomplished by an entirely different set of proteins (Gralla 1996).
Evidence from sequence similarity studies between RNA polymerases suggests that archaeal transcription shared certain components with that of the Eukarya (Puhler et al. 1989), a conclusion further supported by the discovery of TFIIB in Pyrococcus furiosus (Ouzounis and Sander 1992) and TBP in P. woesei (Rowlands et al. 1994). The strong similarity of archaeal transcription initiation factors with their eukaryotic counterparts has reinforced the concept that these domains shared an ancestral transcriptional apparatus. However, a rigorous comparison with bacteria was not performed until entire archaeal genomes became available. It was then shown that archaea contain a significant proportion of bacterial transcripional regulators, in addition to their eukaryotic-like transcription initiation factors (Kyrpides and Ouzounis 1999), for example Lrp (Kyrpides and Ouzounis 1995) and sigma-70 (Kyrpides and Ouzounis 1997). Notably, the only transcriptional components common to all three domains of life are the two largest RNA polymerase subunits, SIR2, NifL and TenA (Coulson et al. 2001), the latter found fused with the metabolic enzyme ThiD in eukaryotes (Ouzounis and Kyrpides 1997). With 15 archaeal genomes, encompassing a wider range of genera from the two main archaeal lineages, a systematic analysis of the archaeal transcriptional machinery would test the robustness of the assumption that bacterial-type regulators and eukaryotic initiation factors co-exist in the Archaea (Bell et al. 2001a).
Methods
The reference sequence dataset of transcription-associated proteins (TAPs) was extracted from SwissProt and TrEMBL from entries containing the word “transcription” in their description and keyword records, as previously described (Coulson and Ouzounis 2003, Coulson et al. 2004). Searching and linking database records was facilitated by SRS (Etzold et al. 1996). From a total of 9874 TAP sequences, 4654 are bacterial, 4907 are eukaryotic and 313 are archaeal. Archaeal genomes were obtained from the COGENT database (Janssen et al. 2003).
The following fifteen fully sequenced and published archaeal genome sequences were used (strain names after comma, COGENT identifiers in parentheses): Archaeoglobus fulgidus, DSM4304 (AFUL-DSM-01), Aeropyrum pernix, K1 (APER-XK1-01), Halobacterium sp., NRC-1 (HALO-NRC- 01), Methanosarcina acetivorans, C2A (MACE-C2A-01), Methanoccocus jannaschii, DSM 2661 (MJAN-DSM-01), Methanopyrus kandleri, AV19 (MKAN-AV1-01), Methanosarcina mazei, Go1 (MMAZ-GO1-01), Methanobacterium thermoautotrophicum, deltaH (MTHE-DEL-01), Pyrococcus abyssi, GE5 (PABY-GE5-01), Pyrobaculum aerophilum, IM2 (PAER-IM2-01), Pyrococcus horikoshii, OT3 (PHOR-OT3- 01), Sulfolobus solfataricus, P2 (SSOL-XP2-01), Sulfolobus tokodaii, Str. 7 (STOK-XX7-01), Thermoplasma acidophilum, DSM1728 (TACI-DSM-01) and Thermoplasma volcanium, GSS1 (TVOL-GSS-01). There is only one complete nanoarchaeotal genome available, and its uniqueness precluded it from the intra-lineage analyses performed in these studies.
The TAP reference dataset was compared against the fifteen archaeal genomes (36,194 sequences) using BLASTp with an E-value cut-off of 10–6 (Altschul et al. 1997), after being filtered for composition bias with CAST (Promponas et al. 2000). For each archaeal genome, the numbers of both the TAP-homologues and matching-reference TAPs were recorded (see Table 1). The 1938 archaeal proteins and 2634 TAPs thus obtained (4572 in total) were clustered using the TRIBE-MCL algorithm (Enright et al. 2002), i.e., only the TAPs matching archaeal proteins (2634 in number) and these archaeal homologues (1938 in number) were clustered by sequence similarity. To assess the effect of granularity, two runs were performed with inflation values of 2.0 and 5.0, as previously described (Coulson and Ouzounis 2003). The distribution of both singletons and families across the Archaea and the reference dataset was examined (see Table 2).
Table 1.
BLASTp search results using the reference set to query fifteen archaeal genomes.
| Species name | Size1 | TAP hits2 | % Genome3 |
| Crenarchaeota | |||
| P.aerophilum | 2605 | 91 | 3.49 |
| A. pernix | 2694 | 77 | 2.86 |
| S. tokodaii | 2826 | 121 | 4.28 |
| S. solfataricus | 2995 | 139 | 4.64 |
| Division total | 11120 | 428 | 3.82 ± 0.80 |
| Euryarchaeota | |||
| T.acidophilum | 1478 | 79 | 5.35 |
| T.volcanium | 1526 | 80 | 5.24 |
| M.kandleri | 1687 | 76 | 4.51 |
| P. abyssi | 1765 | 113 | 6.40 |
| M.jannaschii | 1773 | 107 | 6.03 |
| M.thermoautotrophicum | 1871 | 126 | 6.73 |
| P.horikoshii | 2061 | 103 | 5.00 |
| A.fulgidus | 2409 | 176 | 7.31 |
| Halobacterium sp. | 2605 | 159 | 6.10 |
| M. mazei | 3371 | 221 | 6.56 |
| M. acetivorans | 4528 | 270 | 5.96 |
| Division total | 25074 | 1510 | 5.93 ± 0.83 |
| Combined total | 36194 | 1938 | 5.36 ± 1.25 |
1 The number of protein sequence entries. 2 The number of TAP matches identified using the reference set. 3 The percentage of the genome entries identified as TAP hits. Totals are shown, ± the standard deviation.
Table 2.
Distribution of TRIBE-MCL protein families. Shown are the number of identified families at inflation values of 2.0 and 5.0, corresponding to different granularities (see Methods).
| Inflation value | ||
| 2.0 | 5.0 | |
| TAP singletons | 0 | 128 |
| Archaea singletons | 0 | 115 |
| TAPs and Archaea families | 179 | 179 |
| TAP-only families | 42 | 101 |
| Archaea-only families | 60 | 109 |
| Total families | 281 | 632 |
To further establish the validity of this approach, a parallel analysis was conducted with hidden Markov model (HMM) profiles from the Pfam database (version 7.8) (Bateman et al. 2004). The Pfam accession numbers associated with the 9874 reference TAPs were used to link directly to 508 unique Pfam entries. The fifteen archaeal genomes were queried with HMM profiles, using “hmmsearch” from the software package HMMER (Eddy 1998). Hits were considered significant if their score against the corresponding HMM profile was above a trusted cut-off value, equivalent to the lowest-scoring known member of the Pfam alignment. Of the 508 profiles, 173 have detected homologues in archaea, of which 115 are not involved in transcription. The remaining 58 profiles, defined as TA-HMMs, were validated by their InterPro records to confirm that they were transcription-related.
All results are available at: http://www.ebi.ac.uk/research/cgg/projects/transcription/archaea.
Results
The 9874 reference TAP sequences extracted from SWISS-PROT and TrEMBL identified 1938 proteins in the fifteen archaeal genomes (Table 1). On average, about 5% of the genomes matched the reference set, with some slight variation; the TAP matches identified by BLASTp represent about 4% of the gene content of the Crenarchaeota, and about 6% of the gene content of the Euryarchaeota. This observation suggests that the TAP complement of the Crenarchaeota is more divergent from the reference set than that of the Euryarchaeota (Table 1). However, in both archaeal divisions, the abundance of TAP hits appears to increase with genome size.
The extent of sequence divergence between the detected archaeal proteins and the sequences in the reference set was examined by sequence clustering analyses performed on the BLASTp similarity search results. The clustering operations were implemented at inflation values of 2.0 and 5.0, corresponding to different granularities; the lower value produces larger clusters, containing more distantly related sequences. With an inflation value of 2.0, clustering results in 281 families of which 179 (64%) contain at least one TAP and one archaeal sequence (Table 2). With an inflation value of 5.0, clustering results in 632 families of which 179 (28%) also contain at least one TAP and one archaeal sequence. In this second case, 243 singletons were generated (38% of all clusters, Table 2). Hence, attention was focused on clusters generated with an inflation value of 2.0 because these are both more accurate and broader than clusters generated with a larger inflation value (Coulson and Ouzounis 2003). Similar parameter settings have been shown to result in a precision of >90% for InterPro, and >80% for SCOP families (Enright et al. 2002).
Lineage specificity and functional classification
Clusters generated by TRIBE-MCL correspond to protein families whose members are related by common function. Thus, the 179 TAP-containing clusters generated at an inflation value of 2.0 (Table 2), and containing at least one TAP and one archaeal homologue, were annotated manually based on the description records of SwissProt obtained via SRS (Etzold et al. 1996). After this step, 119 clusters were found to be directly related to the transcriptional process, corresponding to 906 archaeal sequences. These transcription-associated (TA-) clusters were examined in terms of the role played in the transcriptional process by their TAP members and the phylogenetic origins of both the TAP and archaeal sequences (Table 3). The term “phylogenetic origin” refers to the phylogenetic domain to which a TAP belongs, and in which lineage the archaeal homologue is observed.
Table 3.
Functional classification and taxonomic distribution of families containing both TAPs and their archaeal homologues. Definitions: TAP domain = phylogenetic domain (Archaea = ARC, Bacteria = BAC, Eukaryota = EUK) of the reference TAP sequences; Code = abbreviation of TAP domains (A = Archaea, B = Bacteria, E = Eukaryota); No. = number of families in each taxonomic grouping; percentage (%) = total number of TRIBE clusters; Functional classification = RNA Polymerase, Basal TFacs (basal transcription factors), Regulators (transcriptional regulators and DNA-binding proteins)—these classifications were derived from the SwissProt functional annotations of the TAPs that are present in the families of a particular TAP domain; Taxonomic distribution = Crenarchaeota (CREN), Euryarchaeota (EURY) or both (CREN & EURY)—lineage origins of the archaeal TAP-homologues within clusters of a particular TAP domain. In both these previous sections, each of the rows in the three columns that comprise the section, adds to 100% per TAP domain grouping—the number of families present in each category is shown in brackets.
| TAP domain | Code | No. | % | Functional classification | Distribution | ||||
| RNA Polymerase | Basal TFacs | Regulators | CREN | EURY | CREN & EURY | ||||
| ARC | A | 43 | 36.1% | 11.6% (5) | 7.0% (3) | 81.4% (35) | 16.3% (7) | 62.8% (27) | 20.9% (9) |
| BAC | B | 36 | 30.3% | - - | - - | 100.0% (36) | 2.8% (1) | 80.6% (29) | 16.7% (6) |
| ARC::BAC | AB | 18 | 15.1% | - - | - - | 100.0% (18) | - - | 44.4% (8) | 55.6% (10) |
| ARC::EUK | AE | 12 | 10.1% | 66.7% (8) | 25.0% (3) | 8.3% (1) | 8.3% (1) | 8.3% (1) | 83.3% (10) |
| ARC::BAC::EUK | ABE | 5 | 4.2% | 20.0% (1) | 40.0% (2) | 40.0% (2) | - - | 40.0% (2) | 60.0% (3) |
| BAC::EUK | BE | 4 | 3.4% | 25.0% (1) | - - | 75.0% (3) | - - | 75.0% (3) | 25.0% (1) |
| EUK | E | 1 | 0.8% | - - | 100.0% (1) | - - | - - | - - | 100.0% (1) |
| Total no. TRIBE clusters | 119 | 100.0% | (15) | (9) | (95) | (9) | (70) | (40) | |
Analysis of the phylogenetic origins of the TAPs in the TA-clusters showed that in just over 80% of the families (97 clusters) the TAPs are found only in prokaryotic genomes (Table 3). In ~44% (i.e., 43 out of 97 families) and ~37% (36 families) of these TA-clusters, the TAP members of an individual cluster originate solely from either the Archaea (A) or the Bacteria (B), respectively. TAP members in the remaining ~19% of these TA-clusters originate from both the archaeal and bacterial domains (AB); so, each of these 18 families contains at least one archaeal and one bacterial TAP. Strikingly, in 89 out of the 97 families (~92%) whose TAPs are present only in prokaryotes (A, B and AB families), the TAPs function as transcriptional regulators (listed as ‘Regulators’ in Table 3): in the B and AB families, the role of every TAP member is to regulate transcription, and in ~82% of the A families, the TAP members perform the same regulatory role. This finding highlights the prevalence of regulators of prokaryotic origin in the Archaea.
The above pattern contrasts with the other 22 TA-clusters, which contain TAPs present in the Eukaryota (codes with E in Table 3). In 10 of these families (~46%) the TAPs comprise RNA polymerase subunits, and in six families (~27%) they function as basal transcription factors, corresponding to a total of 73% of families involved in transcription initiation and synthesis. This compares with just 8% (8 out of 97) of families of prokaryotic origin that contain TAPs of identical function. Hence, few archaeal sequences show homology to eukaryotic regulators of transcription and bacterial proteins associated with RNA synthesis. However, even though a large number of archaeal sequences exhibit homology to bacterial transcriptional regulators, there appears to be a substantial proportion of archaeal-specific regulatory factors represented by the archaeal families (A families in Table 3).
The 119 TA-clusters exhibit a high degree of lineage-specificity; ~63% (27 cases) and ~81% (29 cases) of the A and B families, respectively—the majority of which are transcriptional regulators—detect archaeal homologues that are unique to Euryarchaeotal genomes (Table 3). This contrasts with the TA-clusters containing TAPs originating from the Eukaryota, where the opposite pattern is observed: ~83% and ~60% of the AE and ABE families, respectively, contain archaeal homologues present in both the Crenarchaeota and Euryarchaeota. Furthermore, in over four fifths of these phylogenetic classes of TA-clusters (14 out of 17 families), the TAP members function as either subunits of RNA polymerase or basal transcription factors.
However, among the prokaryotic (A, B and AB) families, two-thirds (64 out of 97) contain archaeal homologues present only in the Euryarchaeota, whereas in 25 (about one-quarter) of such clusters the TAP homologues originate from both archaeal phyla. Very few of these families (~8%, 8 out of 97) have as members archaeal TAP homologues that are present uniquely in the Crenarchaeota. The vast majority of the A, B and AB families (>90%, 89 families) contain TAPs that function as regulators and are mostly present in Euryarchaeota only. In particular, all the TAPs in the B families are DNA-binding proteins, and >80% of their families possess only Euryarchaeota sequences (Table 3). These functional and taxonomic distribution patterns suggest that there are either fewer (known) Crenarchaeota-specific sequences associated with the control of transcription, or that such sequences are more diverged from the bacterial-type regulators in the Euryarchaeota.
Protein domain representation in archaeal phyla
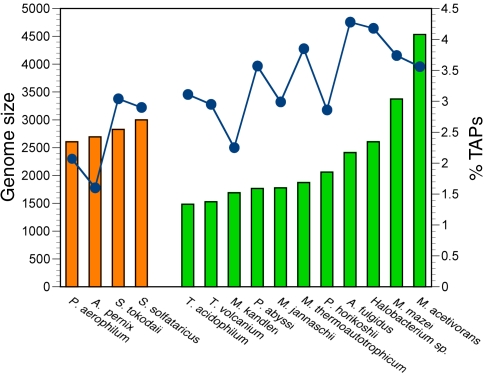
The TRIBE-MCL clustering analyses were complemented by querying the archaeal genomes with 58 HMM profiles that had been validated to be associated with the transcriptional process (TA-HMMs). A large proportion of sequences were identified by both procedures, corresponding to 600 proteins and 81 clusters. (Table 4 displays for each genome the number of TAP-homolgues detected by each method individually, and the number detected by both methods.) Additionally, the two methods of TRIBE-MCL clustering and HMM profile searches uniquely detect 306 and 239 proteins, respectively (Table 4). This degree of overlap indicates a consistent identification of the transcription-associated gene complement in the archaeal genomes under consideration (Table 4). Fewer proteins associated with the transcriptional process were found in the Crenarchaeota than in the Euryarchaeota (~2.40% versus ~3.39%; Table 4). The presence of nearly 1.5-fold more TAP-homologues in the Euryarchaeota is highly significant (P < 2×10–7, Fisher’s exact test). Furthermore, a slight, monotonically-increasing relationship of TAP-homologue abundance with genome size is observed (Tables 1 and 4 and Figure 1). This correlation is significant for the euryarchaeal genomes (P < 0.05, Spearman rank correlation), even though M. acetivorans, containing the highest number of protein-encoding genes, exhibits a similar percentage to M. thermoautotrophicum and P. abyssi, with much smaller genomes (Table 4 and Figure 1).
Table 4.
TAP-homologue abundance in the Crenarchaeota and Euryarchaeota genomes. Definitions: MCL, the number of TAP-homologues detected by the TRIBE-MCL clustering; MCL & HMM, the number of TAP-homologues detected by both the TRIBE-MLC and the profile-HMM searches (the number of families in which these homologues are detected is shown in brackets); HMM, number of sequences that match only the TA-HMMs; TAP homologues, number of genes encoding TAP-homologues; Size, the number of protein sequence entries; % Genome, the percentage of genes identified as TAP-homologues. Totals for % Genome are shown ± the standard deviation.
| Species name | MCL | MCL & HMM | HMM | TAP homologues | Size | % Genome |
| Crenarchaeota | ||||||
| P. aerophilum | 39 (30) | 24 (21) | 15 | 54 | 2605 | 2.07 |
| A. pernix | 36 (32) | 24 (21) | 7 | 43 | 2694 | 1.60 |
| S. tokodaii | 56 (37) | 36 (25) | 30 | 86 | 2826 | 3.04 |
| S. solfataricus | 54 (35) | 39 (28) | 33 | 87 | 2995 | 2.90 |
| Division total | 185 (49) | 123 (34) | 85 | 270 | 11120 | 2.40 ± 0.69 |
| Euryarchaeota | ||||||
| T. acidophilum | 36 (29) | 27 (21) | 10 | 46 | 1478 | 3.11 |
| T. volcanium | 35 (28) | 25 (18) | 10 | 45 | 1526 | 2.95 |
| M. kandleri | 30 (24) | 21 (17) | 8 | 38 | 1687 | 2.25 |
| P. abyssi | 51 (37) | 39 (29) | 12 | 63 | 1765 | 3.57 |
| M. jannaschii | 41 (29) | 27 (22) | 12 | 53 | 1773 | 2.99 |
| M. thermoautotrophicum | 61 (41) | 39 (26) | 11 | 72 | 1871 | 3.85 |
| P. horikoshii | 47 (33) | 34 (24) | 12 | 59 | 2061 | 2.86 |
| A. fulgidus | 83 (51) | 60 (38) | 20 | 103 | 2409 | 4.28 |
| Halobacterium sp. | 75 (40) | 57 (26) | 34 | 109 | 2605 | 4.18 |
| M. mazei | 116 (51) | 64 (31) | 10 | 126 | 3371 | 3.74 |
| M. acetivorans | 146 (54) | 84 (35) | 15 | 161 | 4528 | 3.56 |
| Division total | 721 (110) | 477 (75) | 154 | 875 | 25074 | 3.39 ± 0.62 |
| Combined total | 906 (119) | 600 (81) | 239 | 1145 | 36194 | 3.13 ± 0.76 |
Figure 1.
Transcription-associated protein abundance in archaeal lineages. The genome sizes of the Crenarchaeota (orange) and Euryarchaeota (green) species are displayed as bar graphs, with the percentage of TAPs present in each of the genomes superimposed as blue line plots.
The twenty-one TA-HMMs representing eleven of the twelve polypeptides that constitute RNA polymerase, match approximately equivalent abundances of genes in the Crenarchaeota and Euryarchaeota genomes (~0.44% versus ~0.53% respectively: 49 out of 11,120 and 132 out of 25,074 sequences). Genes encoding subunits A′, A², B′, B², D, E, H and N are detected in all fifteen of the archaeal species by the profile-based method (Supplementary Table 1). Only matches to subunits F, K and L were not observed in all genomes; F is not detected in two of the crenarchaeal (P. aerophilum and S. tokodaii) and just one of the euryarchaeal genomes (M. kandleri), whereas L exhibits the opposite pattern and is undetected in P. horikoshii, T. volcanium (both euryarchaeotes) and P. aerophilum (a crenarchaeote), with a homologue of K absent only from P. horikoshii. This apparent absence of only a small number of RNA polymerase subunits probably arises from their diverged nature as genes encoding these proteins have been observed in these species (Brochier et al. 2004). Furthermore, nearly identical phylogenetic distributions are observed with the RNA polymerase families detected by the TRIBE-MCL clustering (Supplementary Table 2), suggesting little sequence divergence between the RNA polymerase subunits of the euryarchaeota and crenarchaeota.
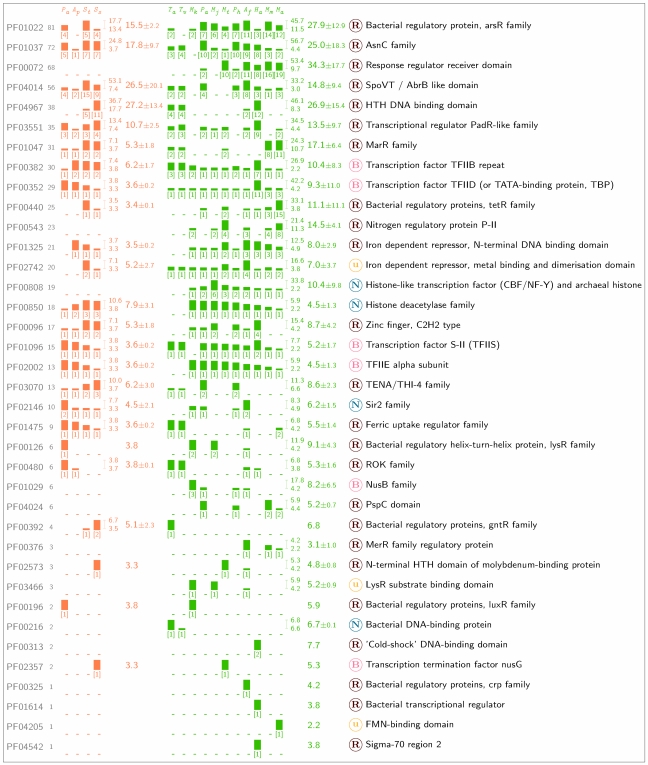
Significantly more sequences in the Euryarchaeota match TA-HMMs of proteins associated with the initiation complex, and transcription elongation and termination, than in the Crenarchaeota (Figure 2): ~0.18% of genes in crenarchaeal genomes (20 genes) and ~0.30% of genes in euryarchaeal genomes (75 genes) encode such transcription factors (P < 0.05, Fisher’s exact test). This difference can mainly be accounted for by the highly amplified TATA-box binding protein (TBP, PF00352) and TFIIB (TFB, PF00382) gene families in Halobacterium sp. (Baliga et al. 2000). The Halobacterium sp. genome contains (when normalized for genome size) ~4.5-fold and ~2.6-fold more genes encoding TBP and TFB, respectively, than the average for this phylum (see Figure 2; namely 42.2 copies per 10,000 genes versus an average of 9.3 for TBP and 26.9 versus 10.4 for TFB). Sequences with homology to the eukaryotic basal transcription factor TFIIE-alpha (TFE, PF02002) are observed in every archaeal species except Thermoplasma, and the transcription elongation TFIIS (TFS, PF01096) is detected in all archaeal genomes except M. kandleri, as previously noted by Brochier et al. (2004). Hence, there appears to be little sequence divergence of these four basal transcription factors across the two archaeal phyla. However, sequences homologous to bacterial transcription termination factors are not widespread in the Archaea: the anti-termination factor NusB (PF01029) is found in only four euryarchaeal genomes including M. kandleri, where there are three homologues, and NusG is found in only one crenarchaeote (S. solfataricus) and one euryarchaeote (M. thermoautotrophicum).
Figure 2.
Phylogenetic distribution of genes matching protein domains associated with transcription. The Pfam accession numbers of the TA-HMMs that match archaeal sequences are shown on the left, with the next column indicating the total number of sequences matched in all the fifteen archaeal genomes. The matches in individual genomes are shown as bar graphs, and grouped into the two lineages (orange representing the Crenarchaeota, and green the Euryarchaeota), with the number of matching sequences normalised by genome size and expressed as number of sequences per 10,000 genes. Beneath each bar graph, in square brackets, is shown the actual number of matching genes. The two numbers above one another and immediately to the right of the two groupings of bar graphs show the maximum and minimum number of matches for the phylum, and the column next to these values shows the mean number of matches with the standard deviation. The letter in the circle indicates which transcriptional functional class the TA-HMM is a member of: basal, elongation and termination factors (B); nucleosomes and HDACs (N); transcriptional regulators (R); unclassified (u). The Pfam description of the TA-HMM is shown at the right. Species codes: P. aerophilum (Pa); A. pernix (Ap); S. tokodaii (St); S. solfataricus (Ss); T. acidophilum (Ta); T. volcanium (Tv); M. kandleri (Mk); P. abyssi (Pa); M. jannaschii (Mj); M. thermoautotrophicum (Mt); P. horikoshii (Ph); A. fulgidus (Af); Halobacterium sp. (Ha); M. mazei (Mm); and M. acetivorans (Ma).
The lack of taxon-restrictedness exhibited by RNA polymerase subunits and basal transcription factors contrasts sharply with proteins that bind DNA nonspecifically and modulate chromatin structure (Figure 2). Sequences encoding archaeal histones (PF00808) are found in all Euryarchaeota genomes except those of T. acidophilum and T. volcanium, where instead the bacterial histone-like HU protein (PF00216) is present. Even though HU protein is ubiquitous and highly conserved across the bacterial kingdom (Sagi et al. 2004), no homologues were observed in the other nine euryarchaeal genomes. None of these proteins associated with compacting prokaryotic chromosomes (archaeal histones and HU protein) are found in the Crenarchaeota. Sequences belonging to the histone deacetylase (HDAC) family (PF00850) are observed in the Crenarchaeota, and all of the euryarchaeal species except Thermoplasma, resulting in the phylogenetic distribution of these HDAC homologues being identical to that of the TFE. None of the HDAC-matching sequences are detected by the TA-HMM representing Sir2 (PF02146), a histone deacetylase present in all three domains of life: the profile identifies Sir2 homologues in all Crenarchaeota species and the four Euryarchaeota genomes that contain homologues of NusB. Hence, the proteins that function to fold DNA, and those that control this process, exhibit considerable diversity across the archaeal domain.
A high degree of phylum-specificity is observed with sequences matching the 24 transcriptional regulator TA-HMMs (Figure 2). There are 127 (~1.14%) and 396 (~1.58%) sequences in Crenarchaeota and Euryarchaeota genomes, respectively, that match these profiles, suggesting that the Euryarchaeota contain nearly 1.5-fold more sequences homologous to well-characterized bacterial proteins associated with controlling gene expression than do the Crenarchaeota (P ≥ 0.0014, Fisher’s exact test). Of the three regulator TA-HMMs that match the highest number of archaeal genes (Figure 2), there are nearly twice the number of euryarchaeaotal as crenarchaeotal sequences (when genome size is accounted for) encoding metalloregulatory repressors (ArsR, PF01022) and specific regulators of amino acid metabolism (AsnC/Lrp, PF01037), with sequences homologous to those involved in the bacterial two-component signal transduction system being absent from the Crenarchaeota (response regulator receiver domain, PF00072). In conjunction with the functional and phylogenetic analyses of the TA-clusters (Table 3), the profile searches imply that the Crenarchaeota contain significantly fewer sequences with homology to well-characterized bacterial repressors than do the Euryarchaeota.
The pattern of elevated abundance of sequences homologous to bacterial regulators in the Euryarchaeota (Figure 2) is also observed with proteins containing the LysR (PF00126), MarR (PF01047), TetR (PF00440) and the iron-dependent repressor (PF01325) DNA-binding domains. The only exception to this trend is a domain (PF04014) present in the Bacillus subtilis abrB and spoVT genes that control developmental gene expression: curiously, twice the number of Crenarchaeotal genes contain this domain than do Euryarchaeotal genes. The TA-HMMs detected no bacterial transcriptional regulators with homologues confined to the Crenarchaeota, contrasting with the Euryarchaeota where nitrogen regulatory protein P-II (PF00543), the transcriptional regulator Phage shock protein C (PspC, PF04024) and bacterial metalloregulatory proteins controlling the transcription of mercury resistance operons (MerR, PF00376) have homologues in two or more of the euryarchaeaotal genomes.
The only eukaryotic DNA-binding domain identified in the archaeal genomes was a variant of the C2H2-zinc finger (PF00096), that detected a total of 17 sequences across all Crenarchaeota and six Euryarchaeota species. However, Halobacterium—the only non-thermophile analyzed—appears most “bacteria-like” in its complement of transcriptional regulators; it is the only species with homologues of cold-shock protein (PF00313), Crp (PF00325) and region 2 of s70 (PF04542). Therefore, although nearly all the archaeal transcriptional regulators identified are related to bacterial repressors, there is considerable heterogeneity across, and within the two phyla in the types of regulator families present within a particular species.
Discussion
The genome-wide profiling of archaeal transcription that was performed indicates that the Euryarchaeota have more proteins associated with the transcriptional process than do the Crenarchaeota. Furthermore, in both phyla, there is a trend showing that TAP-homologue abundance increases with genome size, i.e., smaller genomes appear to contain proportionately fewer transcription factors than larger ones. Evidence of this type of correlation has also been observed in bacteria (Cases et al. 2003). Although a number of functional classes associated with RNA synthesis and transcription initiation are shared both between and within the known archaeal phyla, there are a number of surprising lineage-specific patterns that have not previously been systematically characterized: both the proteins that constitute archaeal chromatin and histone modifying enzymes, as well as transcriptional regulators—which are major contributors to the lineage-specificity—display extensive sequence diversity across the archaeal domain.
Genes encoding eleven out of the twelve RNA polymerase subunits are observed in all fifteen archaeal genomes, with a few exceptions (Slesarev et al. 2002). Subunit F (identified only with the profile-based method) is not observed in three genomes; however, its eukaryotic functional and structural equivalent, Rpb4 (Werner et al. 2000), is not essential for cell viability in yeast (Woychik and Young 1989). The other two subunits that were not found in all of the analysed genomes are K and L, which appear to be absent only from P. horikoshii as homologues of L are detected by the TRIBE-MCL clustering in P. aerophilum and T. volcanium. These data imply that RNA polymerase is very highly conserved in the Archaea.
Transcription initiation in archaea resembles the eukaryotic process in its requirement for TBP, TFB (the archaeal version of eukaryotic TFIIB) (Soppa 1999, Reeve 2003) and TFE (an archaeal protein homologous to the N-terminal region of the a subunit of eukaryotic TFIIE) (Bell et al. 2001b, Hanzelka et al. 2001). Genes encoding these three basal transcription factors are present in all of the genomes, with the only notable absence being that of TFE from T. acidophilum and T. volcanium. However, no archaeal genes encoding homologues of TAFs (Qureshi et al. 1997) or TFIIE (subunit b), TFIIF and TFIIH (Ouhammouch 2004) were identified. TBP is usually encoded by a single copy gene, though Halobacterium sp. contains eleven copies (Ng et al. 2000), with three genes in M. acetivorans and M. mazei coding for this canonical transcription factor. The Halobacterium genome also contains multiple copies of TFB-encoding genes (seven), as do half of the other genomes that contain either two or three copies. Hence, the identification of multiple copies of TBP and TFB genes in a range of archaeal species supports the hypothesis that different types of transcriptional regulation occur in response to environmental challenges (Goo et al. 2004), although the role of multiple TBPs and their relationship with multiple TFBs is not yet clear (Galagan et al. 2002).
The archaeal nucleosome appears to display extensive taxon variablity: with the exception of the Thermoplasma (whose genomes encode bacterial HU-like proteins), the Euryarchaeota use homologues of histones H3 and H4 to assemble chromatin (Geiduschek and Ouhammouch 2005), contrasting with the Crenarchaeota where completely different sets of proteins compact the genome (e.g., Alba in S. solfataricus (Bell et al. 2002)). It is interesting to note, given the apparent lack of histones in Thermoplasma, that no homologues of members of the HDAC family, Sir2 or TFE are present in the genomes of T. acidophilum and T. volcanium. Sir2 homologues are present in all the Crenarchaeotal genomes, but only in the same four Euryarchaeotal genomes as NusB. This may suggest some association between RNA chain elongation and histone deacetylation, as both TFIIE and NusB function in the elongation process, and in eukaryotes many factors can influence transcript elongation on chromatin templates (Sims et al. 2004). This heterogeneity in the constituents of archaeal chromatin is unexpected as trypanosomatids contain the four core histones, and possess a range of chromatin-remodeling activities, also present in mammals even though there is considerable evolutionary distance between the two eukaryotic taxa (Ivens et al. 2005).
The elevated abundance in euryarchaeal genomes of proteins associated with the transcriptional process arises, predominately, from the increased number (when genome size is accounted for) of sequences homologous to bacterial transcriptional regulators within these genomes (Figure 2). Members of the AsnC/Lrp family of regulators have been shown to act as both repressors and activators of archaeal transcription (Geiduschek and Ouhammouch 2005), though whether chromatin remodeling and histone modification play an important role in the control of gene expression, as in eukaryotes, is not known. DNA-compacting proteins and HDACs are observed in all archaeal species except T. acidophilum and T. volcanium, which are only distantly related to the extreme halophilic and methanogenic euryarchaeotes (DeLong 2000). Instead, Thermoplasma contain the bacterial HU-protein, making transcriptional control in this archaeal group reminiscent of Bacteria, i.e., the genomic DNA is readily accessible to DNA-binding proteins, resulting in the default expression state of a gene being “on” (Struhl 1999). This could be in contrast to the other Euryarchaeota and the Crenarchaeota, where the default state is “off” because of genome compacting by chromatin.
Nearly all the Crenarchaeota studied have shown a high degree of niche specialization (most are hyperthermophiles) and, like parasitic protozoa, which occupy only a limited range of environments, appear to contain far fewer proteins that regulate transcription than do organisms adapted to a wider range of environmental conditions (Coulson et al. 2004, Ivens et al. 2005). At present, a lack of experimental data makes it impossible to determine whether the Crenarchaeota possess significant numbers of lineage-specific transcriptional regulators or whether they just contain fewer of the bacterial-type compared with the Euryarchaeota. Nonetheless, these results are consistent with previous genome profiling studies of transcriptional complements, which show that the greater the evolutionary separation between taxa, the greater the taxon-restrictedness of the TAP-families (Coulson and Ouzounis 2003).
Supplementary material
Table S1. Number of archaeal genes matching domains present in RNA polymerase subunits.
Table S2. RNA polymerase subunit gene copy number in archaeal genomes.
Acknowledgments
R.M.R.C. and C.A.O. thank the Medical Research Council for supporting this work through a Special Training Fellowship in Bioinformatics to R.M.R.C.
References
- R1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Baliga N.S., Goo Y.A., Ng W.V., Hood L., Daniels C.J., Dassarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol. Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- R3.Bateman A., Coin L., Durbin R., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32 Database issue:D:138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Bell S.D., Magill C.P., Jackson S.P. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 2001;29:392–395. doi: 10.1042/bst0290392. [DOI] [PubMed] [Google Scholar]
- R5.Bell S.D., Brinkman A.B., Van der oost J., Jackson S.P. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2001;2:133–138. doi: 10.1093/embo-reports/kve021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Bell S.D., Botting C.H., Wardleworth B.N., Jackson S.P., White M.F. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- R7.Brochier C., Forterre P., Gribaldo S. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004;5:R17.01–12. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R8.Cases I., De lorenzo V., Ouzounis C.A. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003;11:248–253. doi: 10.1016/s0966-842x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- R9.Coulson R.M., Ouzounis C.A. The phylogenetic diversity of eukaryotic transcription. Nucleic Acids Res. 2003;31:653–660. doi: 10.1093/nar/gkg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Coulson R.M., Enright A.J., Ouzounis C.A. Transcription-associated protein families are primarily taxon-specific. Bioinformatics. 2001;17:95–97. doi: 10.1093/bioinformatics/17.1.95. [DOI] [PubMed] [Google Scholar]
- R11.Coulson R.M., Hall N., Ouzounis C.A. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum . Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.DeLong E.F. Extreme genomes. Genome Biol. 2000;1:r10291–10293. doi: 10.1186/gb-2000-1-6-reviews1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Eckburg P.B., Lepp P.W., Relman D.A. Archaea and their potential role in human disease. Infect. Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- R15.Enright A.J., Van dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Etzold T., Ulyanov A., Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- R17.Galagan J.E., Nusbaum C., Roy A., et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Geiduschek E.P., Ouhammouch M. Archaeal transcription and its regulators. Mol. Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- R19.Goo Y.A., Roach J., Glusman G., et al. Low-pass sequencing for microbial comparative genomics. BMC Genomics. 2004;5:3. doi: 10.1186/1471-2164-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Gralla J.D. Activation and repression of E. coli promoters. Curr. Opin. Genet. Dev. 1996;6:526–530. doi: 10.1016/s0959-437x(96)80079-7. [DOI] [PubMed] [Google Scholar]
- R21.Hanzelka B.L., Darcy T.J., Reeve J.N. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEalpha. J. Bacteriol. 2001;183:1813–1818. doi: 10.1128/JB.183.5.1813-1818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Ivens A.C., Peacock C.S., Worthey E.A., et al. The genome of the kinetoplastid parasite, Leishmania major . Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.Janssen P., Enright A.J., Audit B., Cases I., Goldovsky L., Harte N., Kunin V., Ouzounis C.A. COmplete GENome Tracking (COGENT): a flexible data environment for computational genomics. Bioinformatics. 2003;19:1451–1452. doi: 10.1093/bioinformatics/btg161. [DOI] [PubMed] [Google Scholar]
- R24.Kyrpides N.C., Ouzounis C.A. The eubacterial transcriptional activator Lrp is present in the archaeon Pyrococcus furiosus . Trends Biochem. Sci. 1995;20:140–141. doi: 10.1016/s0968-0004(00)88988-4. [DOI] [PubMed] [Google Scholar]
- R25.Kyrpides N.C., Ouzounis C.A. Bacterial sigma 70 transcription factor DNA-binding domains in the archaeon Methanococcus jannaschii . J. Mol. Evol. 1997;45:706–707. [PubMed] [Google Scholar]
- R26.Kyrpides N.C., Ouzounis C.A. Transcription in archaea. Proc. Natl. Acad. Sci. USA. 1999;96:8545–8550. doi: 10.1073/pnas.96.15.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Ng W.V., Kennedy S.P., Mahairas G.G., et al. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Ouhammouch M. Transcriptional regulation in Archaea. Curr. Opin. Genet. Dev. 2004;14:133–138. doi: 10.1016/j.gde.2004.01.002. [DOI] [PubMed] [Google Scholar]
- R29.Ouzounis C.A., Kyrpides N.C. ThiD-TenA: a gene pair fusion in eukaryotes. J. Mol. Evol. 1997;45:708–711. doi: 10.1007/pl00013145. [DOI] [PubMed] [Google Scholar]
- R30.Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992;71:189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- R31.Promponas V.J., Enright A.J., Tsoka S., Kreil D.P., Leroy C., Hamodrakas S., Sander C., Ouzounis C.A. CAST: an iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- R32.Ptashne M., Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- R33.Puhler G., Leffers H., Gropp F., Palm P., Klenk H.P., Lottspeich F., Garrett R.A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl. Acad. Sci. USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Qureshi S.A., Bell S.D., Jackson S.P. Factor requirements for transcription in the archaeon Sulfolobus shibatae . EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Reeve J.N. Archaeal chromatin and transcription. Mol. Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- R36.Rowlands T., Baumann P., Jackson S.P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- R37.Sagi D., Friedman N., Vorgias C., Oppenheim A.B., Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU–DNA complexes. J. Mol. Biol. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- R38.Sims R.J., Belotserkovskaya R., Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- R39.Slesarev A.I., Mezhevaya K.V., Makarova K.S., et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R40.Soppa J. Transcription initiation in Archaea: facts, factors and future aspects. Mol. Microbiol. 1999;31:1295–1305. doi: 10.1046/j.1365-2958.1999.01273.x. [DOI] [PubMed] [Google Scholar]
- R41.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- R42.Werner F., Weinzierl R.O. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- R43.Werner F., Eloranta J.J., Weinzierl R.O. Archaeal RNA polymerase subunits F and P are bona fide homologs of eukaryotic RPB4 and RPB12. Nucleic Acids Res. 2000;28:4299–4305. doi: 10.1093/nar/28.21.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R44.Woychik N.A., Young R.A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 1989;9:2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Number of archaeal genes matching domains present in RNA polymerase subunits.
Table S2. RNA polymerase subunit gene copy number in archaeal genomes.